Significance
Eps15 (epidermal growth factor receptor pathway substrate 15)-homology domain containing proteins (EHDs) are molecular machines that use the energy of ATP binding and ATP hydrolysis to remodel shallow membranes into highly curved membrane tubules. This activity is required in many cellular membrane trafficking pathways. In this work, we have determined a high-resolution structure of an EHD machine in the active state. The structure indicates how EHDs assemble at the membrane surface into ring-like scaffolds that deform the underlying membrane. By comparing this active state with a previously determined autoinhibited conformation, we can deduce the mechanistic details how recruitment of EHDs to membranes is regulated. A comparison with other membrane-associated molecular machines reveals commonalities and differences in the activation mechanism.
Keywords: dynamin superfamily, endocytic pathways, protein structure, membrane remodeling, autoinhibition
Abstract
Eps15 (epidermal growth factor receptor pathway substrate 15)-homology domain containing proteins (EHDs) comprise a family of dynamin-related mechano-chemical ATPases involved in cellular membrane trafficking. Previous studies have revealed the structure of the EHD2 dimer, but the molecular mechanisms of membrane recruitment and assembly have remained obscure. Here, we determined the crystal structure of an amino-terminally truncated EHD4 dimer. Compared with the EHD2 structure, the helical domains are 50° rotated relative to the GTPase domain. Using electron paramagnetic spin resonance (EPR), we show that this rotation aligns the two membrane-binding regions in the helical domain toward the lipid bilayer, allowing membrane interaction. A loop rearrangement in GTPase domain creates a new interface for oligomer formation. Our results suggest that the EHD4 structure represents the active EHD conformation, whereas the EHD2 structure is autoinhibited, and reveal a complex series of domain rearrangements accompanying activation. A comparison with other peripheral membrane proteins elucidates common and specific features of this activation mechanism.
Eps15 (epidermal growth factor receptor pathway substrate 15)-homology domain containing proteins (EHDs) comprise a highly conserved eukaryotic family of dynamin-related ATPases, which regulate diverse membrane trafficking pathways (1). The single Caenorhabditis elegans homolog receptor-mediated endocytosis 1 (Rme-1) localizes to the endocytic recycling compartment and mediates the exit of cargo proteins to the cell surface (2, 3). Similar functions were subsequently shown for mammalian EHD1 and EHD3, which in addition control early endosome to Golgi transport (4–6). EHD4/Pincher facilitates the macroendocytic uptake of tropomyosin receptor kinase (Trk) receptors (7, 8). In contrast, EHD2 localizes to caveolae and, together with the Bin Amphiphysin Rvs (BAR)-domain containing binding partner PACSIN2 (9), stabilizes caveolae at the cell surface (10–12). Despite possessing a dynamin-related GTPase domain, EHDs bind to adenine rather than guanine nucleotides (13, 14). Similar to other dynamin superfamily members, EHD2 tubulates negatively charged liposomes and forms ring-like oligomers around the tubulated membranes (14, 15). Furthermore, the slow ATPase rate of EHD2 is stimulated by the assembly on membrane surfaces.
The crystal structure of the EHD2 dimer in the presence of the nonhydrolyzable ATP analog adenylyl imidodiphosphate (AMPPNP) revealed a dynamin-related extended GTPase domain that mediates stable dimerization via an EHD family-specific dimerization interface (14). The helical domain is composed of amino acid sequences lining the GTPase domain at its N and C terminus. We previously demonstrated by mutagenesis and electron paramagnetic spin resonance (EPR) that helix α9 at the tip of the helical domain participates in membrane binding (14, 16). The Eps15-homology (EH) domains at the C terminus interact with linear peptide sequences containing Asn-Pro-Phe (NPF) motifs (17, 18). In the EHD2 dimer, they bind to an internal Gly-Pro-Phe (GPF) motif in the linker between the helical domain and the EH domain. This interaction locks the EH domain to the opposing GTPase domain, from where it delivers its C-terminal tail into the active site (14). In this way, the EH domains were suggested to block a highly conserved second assembly site in the GTPase domain, which extends across the nucleotide binding site. GTPase domain assembly via this “G interface” is a conserved feature in the dynamin superfamily and is often accompanied by activation of the GTPase activity (19).
Our recent X-ray and EPR structural analysis demonstrated that an N-terminal stretch of 8 aa folds back into a highly conserved hydrophobic pocket of the GTPase domain in the EHD2 dimer (16). Upon its recruitment to membranes, the N-terminal residues are released and may insert into the lipid bilayer. This switch mechanism appears to negatively regulate membrane interaction of EHDs, because an EHD2 variant without the N-terminal residues showed enhanced membrane binding when overexpressed in mammalian cells. We suggested that the switch may also influence the partly disordered “KPF loop” (containing a Lys-Pro-Phe amino acid stretch) at the distal side of the GTPase domain that is required for caveolar targeting of EHD2 (10).
In this study, we elucidated the structural basis for the switch mechanism by determining the crystal structure of an N-terminally truncated EHD4 dimer in the oligomerized state. We reveal a complex series of domain rearrangements during activation and uncover common and specific features of this activation mechanism compared with other membrane-activated systems.
Results
Crystal Structure of the EHD4 Dimer.
To obtain mechanistic insights into the activation mechanism of EHD proteins, we expressed and purified an N-terminally truncated EHD4 variant (amino acids 22–541, EHD4ΔN; Fig. 1A). We reasoned that this variant may mimic the situation when EHDs are activated by membrane recruitment and release their N terminus from the GTPase domain (16). An EHD4 full-length construct was insoluble when expressed in Escherichia coli.
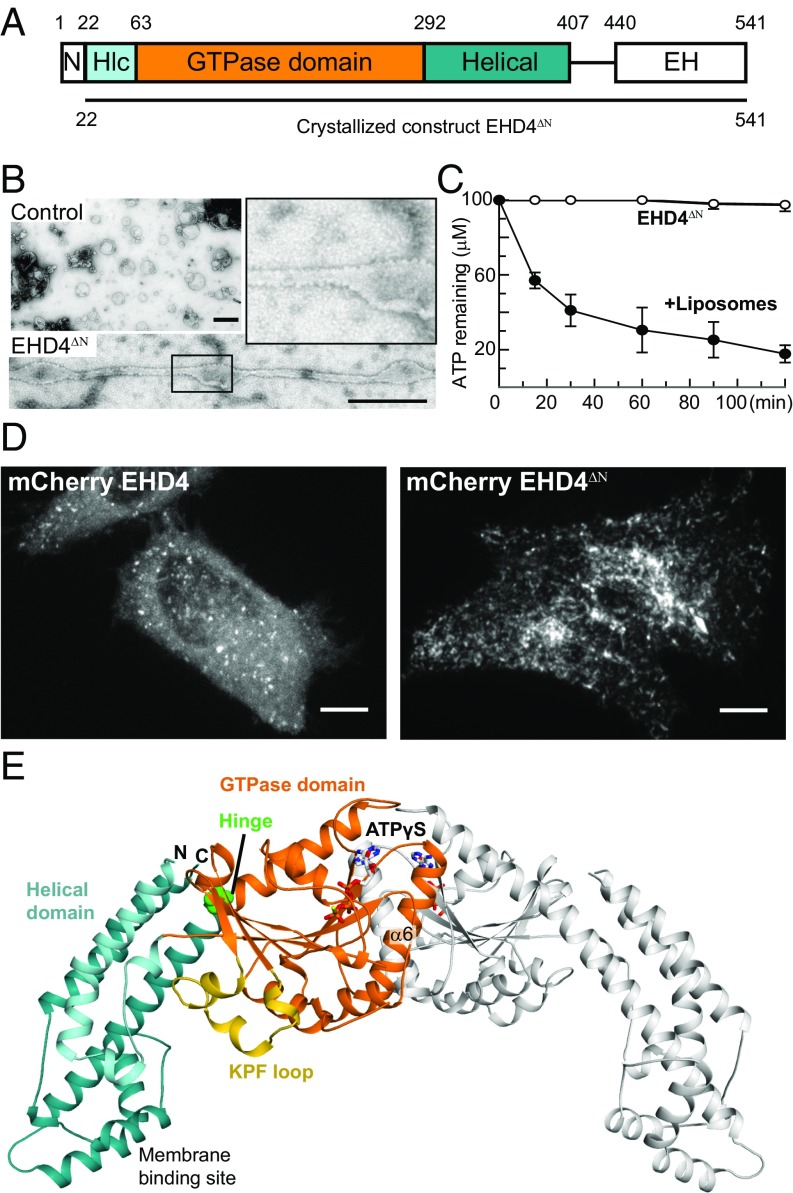
Fig. 1.
Structure of the activated EHD4 dimer. (A) Domain architecture of EHD proteins. Numbers refer to the amino acid sequence of mouse EHD4. (B) EM micrographs of negatively stained EHD4ΔN in the presence of liposomes. As a control, liposomes were stained without the addition of EHD4ΔN. (Scale bars: B, 500 nm; D, 10 μm.) (C) ATPase assays of EHD4ΔN in the absence and presence of Folch liposomes were carried out at 30 °C. Error bars represent the range of two independent measurements. (D) EHD4 and EHD4ΔN were expressed in HeLa cells with a C-terminal mCherry tag. (E) Structure of the ATPγS-bound EHD4ΔN. In the left molecule, domains are colored according to the domain architecture, the conserved hinge around Pro289 is indicated in green. The right molecule is shown in gray. Note that the EH domains were not resolved in the electron density.
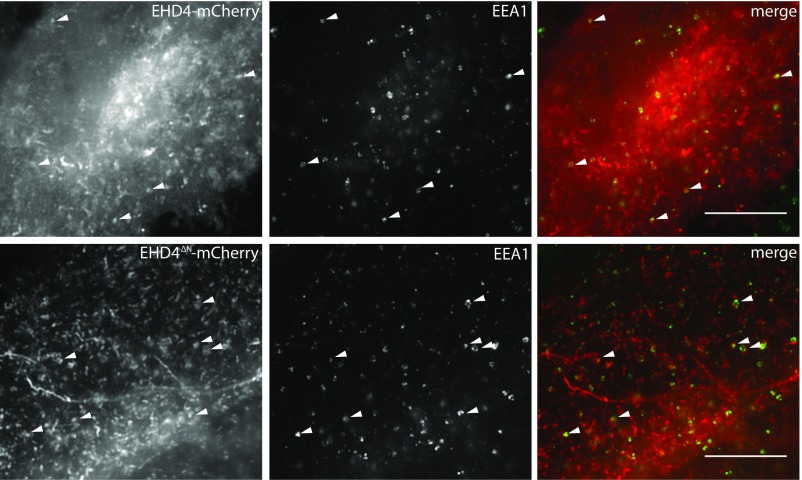
Similar to EHD2, EHD4ΔN bound to the nonhydrolyzable ATP analog adenosine 5′-(γ-thio)-triphosphate (ATPγS) with a micromolar affinity in isothermal titration calorimetry (ITC) experiments (Fig. S1A). Furthermore, EHD4ΔN cosedimented with liposomes derived from bovine brain (Folch) lipids (Fig. S1B). Membrane binding resulted in liposome tubulation, and regular EHD4ΔN oligomers were formed at the surface of the lipid tubules (Fig. 1B and Fig. S1C). In addition, the slow ATPase activity of EHD4ΔN was 200-fold stimulated by the addition of Folch liposomes (Fig. 1C). The stimulated ATPase rate of EHD4ΔN is sevenfold higher compared with EHD2 under identical conditions (14). When C-terminally GFP-tagged EHD4 was overexpressed in HeLa cells, it localized to endosomal structures that partially overlapped with early endosomal antigen1 (EEA1) (Fig. 1D, Left and Fig. S2, see also ref. 20). The N-terminally truncated EHD4 variant showed increased membrane association (Fig. 1D, Right, quantified in Fig. S1 D and E). Many membranous structures appeared tubulated in these cells, pointing to an increased membrane remodelling activity of EHD4ΔN. Taken together, these results indicate that EHD2 and EHD4 use related mechanisms of membrane recruitment and oligomerization. In particular, the N-terminal residues appear to negatively control membrane recruitment in both proteins.
Fig. S1.
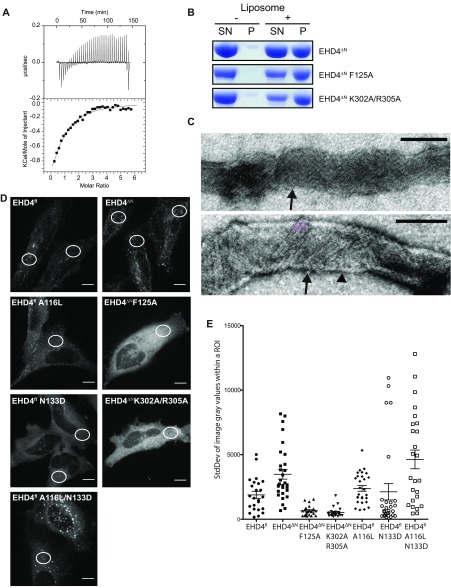
Functional characterization of EHD4ΔN. (A) In ITC experiments, a 2 mM ATPγS solution was titrated into a 70 µM EHD4ΔN solution. The resulting heat changes were integrated and fitted to a quadratic binding model. A KD of 77 µM, with a binding number n of 0.67 was obtained. Note that this experiment was carried out at 500 mM NaCl, which was required to prevent protein precipitation. The high salt concentration may negatively influence the binding affinity. (B) Folch liposome cosedimentation assays of EHD4ΔN and the indicated mutants. P, Pellet fraction; SN, supernatant. (C) Liposome tubulation experiments as in Fig. 1B. Some tubulated liposomes (arrowhead) contained a regular EHD4 coat with striations of 9 nm width (black arrows), which corresponds to the width of our EHD4 oligomeric model (Fig. 4A). (D) Representative maximum intensity projections of live HeLa cells expressing mCherry-tagged protein as indicated. White circles illustrate typical placings of the region of interest (ROI) within the cells. (E) SD of gray values derived from one ROI per cell from at least 20 cells for each indicated construct. Error bars show SEM. Two-tailed t tests was performed on EHD4ΔN to each set of data to determine significance *P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0001. (Scale bars: 10 μm.)
Fig. S2.
Deletion of the N terminus increases EHD4 localization at early endosomes. Endogenous staining of EEA1 in HeLa cells expressing mCherry-tagged protein as indicated. Arrowheads exemplify colocalization of mCherry-tagged protein and EEA1. (Scale bars: 10 μm.)
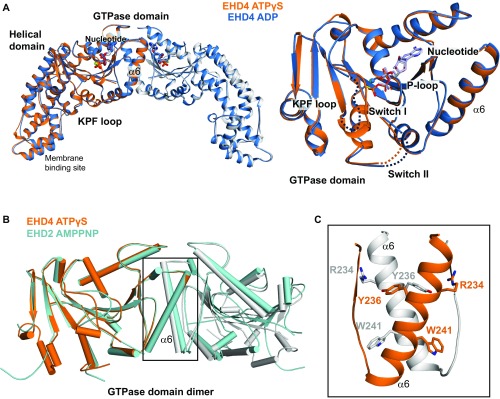
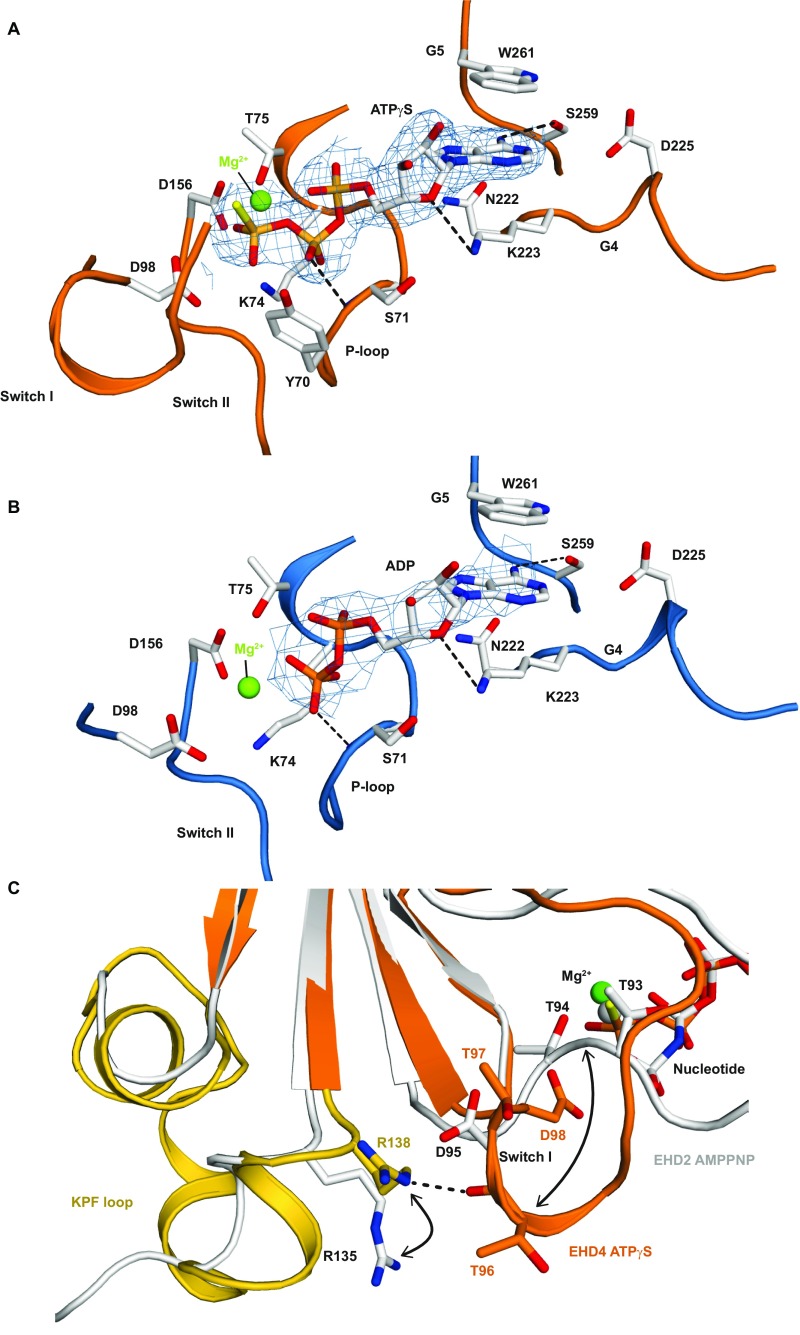
For EHD4ΔN, crystals of the same space group were obtained in the presence of ATPγS and ADP and diffracted to a maximal resolution of 2.8 Å and 3.3 Å, respectively (Table S1). The structures were solved by molecular replacement and refined to Rwork/Rfree of 22.7%/24.3% and 20.8%/25.0%, respectively. Besides minor changes in the nucleotide binding site, both structures were essentially identical, with a rms deviation (rmsd) of 0.30 Å for all built Cα atoms (Fig. S3A). If not otherwise mentioned, we refer in the following to the higher resolution ATPγS-bound structure.
Table S1.
Data collection and refinement statistics
| Crystals | EHD4ΔN ATPγS | EHD4ΔN ADP |
| Data collection | ||
| Space group | P42212 | P42212 |
| Cell dimensions | ||
| a, b, c, Å | 199.97, 199.97, 41.54 | 199.47 199.47,41.8 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 |
| Resolution, Å | 48.5–2.79 (2.96–2.79)* | 47.02–3.27 (3.4–3.27) |
| Rmerge, % | 9.5 (91.9) | 28.8 (211.5) |
| I/σI | 20.0 (1.9) | 9.53 (1.02) |
| Completeness, % | 99.8 (98.6) | 99.54 (96.51) |
| Redundancy | 7.2 | 10.5 |
| Refinement | ||
| Resolution, Å | 48.5–2.79 (2.89–2.79) | 48.4–3.27 (3.4–3.27) |
| No. of reflections | 21,733 (2,091) | 13,642 (1,273) |
| Rwork/Rfree, % | 22.7/24.3 (32.8/34.2) | 20.8/25.0 (35.8/39.1) |
| No. of atoms | ||
| Protein | 3,072 | 3,026 |
| Ligand/ion | 32 | 28 |
| Water | 21 | 1 |
| B factors, Å2 | ||
| Protein | 76 | 105 |
| Ligand/ion | 100 | 146 |
| Water | 50 | 87 |
| rms deviations | ||
| Bond lengths, Å | 0.002 | 0.003 |
| Bond angles, ° | 0.54 | 0.59 |
n.a., not applicable.
Values in parentheses are for highest-resolution shell.
Fig. S3.
Structural comparisons within the EHD family. (A) A superposition of the EHD4ΔN ATPγS and ADP-bound structures. Only minor changes in the GTPase domains were apparent (Right). Note that in the ADP-bound EHD4 structure, switch I is disordered. (B) A superposition of EHD2 and EHD4 GTPase domain dimers indicates an almost identical architecture. (C) Interaction details in the GTPase domain dimerization interface of EHD4ΔN.
The overall architecture of EHD4ΔN is similar to that of AMPPNP-bound EHD2 (Fig. 1E). For example, the GTPase domains of EHD4 and EHD2 show a highly related fold (rmsd of 0.92 Å for 194 aligned Cα atoms; Fig. S3B). They form a dimer via an interface involving helix α6, including Arg234, Tyr236, and Trp241 (Fig. 1E and Fig. S3C). The interface is highly conserved in EHD proteins (Fig. S4) (14), but not in other dynamin-related proteins.
Fig. S4.
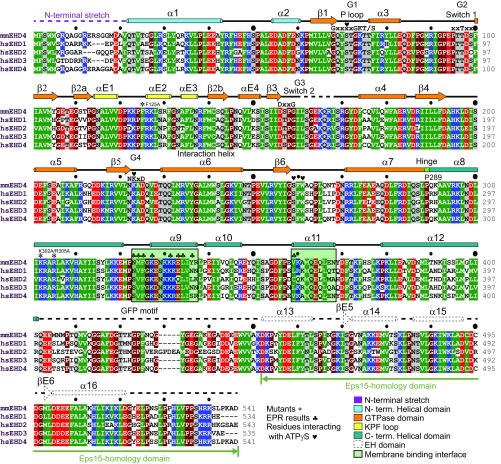
Sequence alignment of the crystallized mmEHD4 and human EHDs. Amino acid sequences of Mus musculus (mm) EHD4 (Q9EQP2), Homo sapiens (hs) EHD1 (Q9H4M9), hsEHD2 (Q9NZN4), hsEHD3 (Q9NZN3), and hsEHD4 (Q9H223) were aligned by using CLUSTAL W and manually adjusted. Residues with a conservation greater than 70% are color-coded (D, E in red; R, K, H in blue N, Q, S, T in gray; A, L, I, V, F, Y, W, M, C in green and P, G in brown). α-Helices are shown as cylinders and β-strands as arrows. The secondary structure of the EH domain is from PDB ID code 4CID. The domains are colored as in Fig. 1E. Mutated residues in this study are indicated with an asterisk (*), residues interacting with the membrane are indicated with a clover (♣), and residues interacting with ATPγS are indicated with a heart (♥).
Electron density for ATPγS and ADP was clearly discernible in the nucleotide-binding pocket (Fig. S5 A and B). Switch I in the ATPγS-bound EHD4ΔN structure adopts a different conformation compared with EHD2 (Fig. S5C). For example, the completely invariant Thr97 (Thr94 in EHD2), which coordinates the Mg2+ ion in EHD2 (14) and, in analogy to dynamin (21), may position a catalytic water molecule for nucleotide hydrolysis, points away from the nucleotide in the EHD4ΔN structure. The reorientation of switch I may be caused by a newly formed contact of Arg138 from the rearranged KPF loop (see below) with the main chain of Thr96 in switch I. In the ADP-bound structure, switch I is disordered. These observations suggest an ATP-dependent conformational cross-talk between the KPF loop and the nucleotide-binding site in EHD proteins.
Fig. S5.
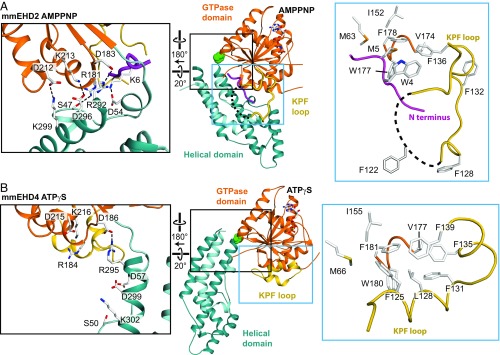
Structural details of the GTPase domain. (A and B) Detailed view into the catalytic site of the ATPγS and ADP-bound structures, with selected residues from the five conserved nucleotide binding motifs (G1–G5) shown in stick representation. Hydrogen bonds are indicated by dashed lines. An aspartate in the G4 motif is responsible for guanine specificity in dynamin superfamily members (19). In EHD4ΔN, the corresponding Asp225 does not participate in binding to the adenine base. The final 2Fo-Fc density was contoured at 1 σ around the nucleotides. (C) Superposition of the GTPase domains of AMPPNP-bound EHD2 and ATPγS-bound EHD4ΔN. In the EHD4 dimers (colored), the KPF loop (gold) folds back into a hydrophobic pocket of the GTPase domain. Inset shows the interaction of the R138 with the backbone of the switch I in EHD4, which is not present in EHD2 (white). Note in particular that the highly conserved catalytic threonine residue (T97 in EHD4) points away from the nucleotide in EHD4ΔN.
The EH domains were not resolved in the electron densities, despite being present in the crystallized construct. SDS/PAGE analysis of dissolved crystals showed no hints for proteolytic degradation. This observation indicates that the EH domains are disordered in the crystals and do not bind to their autoinhibitory site on top of the GTPase domains.
A Large-Scale Rotation of the Helical Domain Promotes Activation.
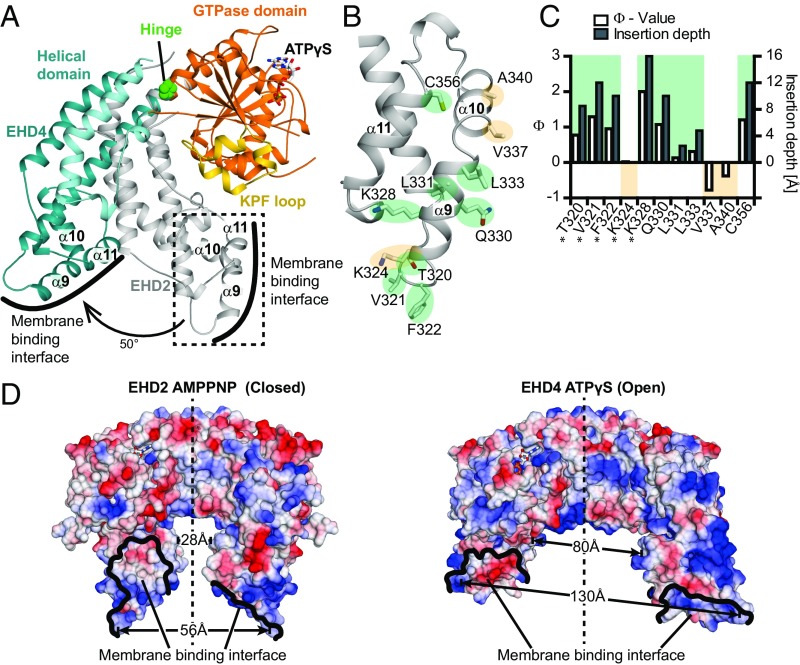
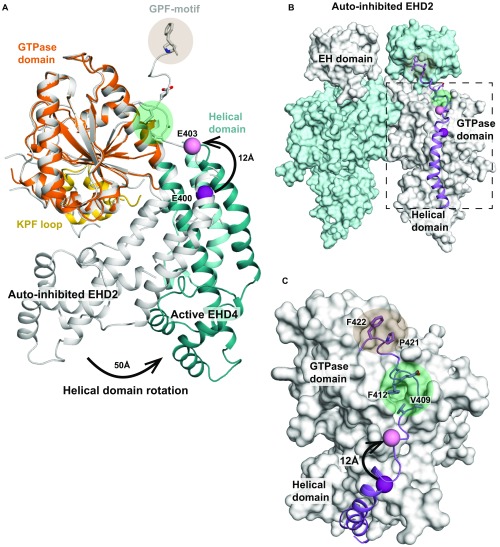
The helical domain of EHD4ΔN has an almost identical architecture as its counterpart in EHD2 (rmsd of 0.67 Å for 142 Cα atoms). However, compared with EHD2, it is rotated by ∼50° relative to the GTPase domain around a hinge featuring the invariant Pro289 (Fig. 2A and Movie S1). This rotation pushes the N-terminal residues of the linker to the EH domain 12 Å away (Fig. S6 A–C). In the EHD2 structure, the linker makes prominent contacts to the GTPase domain and binds via a GPF motif to the EH domain of the opposing monomer. The rotation may thus displace the linker, therefore releasing the EH domain from its autoinhibitory site. It also reorients the membrane binding site in α9 including Thr320, Val321, Phe322, and Lys328, which, in EHD2, were shown to directly interact with the membrane (16) (Fig. 2 A–C).
Fig. 2.
A rotation of the helical domains allows membrane binding. (A) Superposition of the GTPase domains of ATPγS-bound EHD4ΔN and AMPNP-bound EHD2 (PDB ID code 4CID, only the helical of EHD2 is shown). Pro289 acts as the hinge for a 50° rotation of the helical domain. (B) Magnification of the boxed area in A showing details of the membrane binding interface of EHD2. Membrane inserting (green)/noninserting (yellow) residues are highlighted. (C) The logarithmic ratio Φ of the accessibilities of spin labels to the paramagnetic colliders O2 and NiEDDA was calculated for EHD2 labeled at the indicated positions in the presence of Folch-SUVs (open bars referring to the left y axis). Results from residues 277, 320–324, and 328 (*) are from ref. 16. Positive Φ values indicate membrane insertion based on prior calibration with spin-labeled lipids. This calibration was used to convert Φ values into membrane insertion depth of each residue (filled bars to the right y axis). (D) Electrostatic potentials (±10 kcal/mol × e, where e is the charge of an electron) were plotted on the surfaces of the EHD2 (Left) and EHD4ΔN (Right) dimers, with the GTPase domains of both dimers in the same orientation. The distances between Phe322-Phe322 in EHD2 (56 Å), Phe325-Phe325 in EHD4 (130 Å), Pro350-Pro350 in EHD2 (28 Å), and Ala353-Ala353 in EHD4 (80 Å) are indicated. The membrane binding surface of EHD2 is more positively charged compared with EHD4, possibly reflecting different lipid binding specificities (Fig. S4).
Fig. S6.
Reorientation of the linker in the active conformation. (A) The GTPase domains of EHD2 and EHD4ΔN were superimposed. Highlighted is the position of Glu403 (represented as pink ball), the last resolved residue in the EHD4ΔN structure, corresponding to Glu400 in EHD2 (represented as purple ball). This residue is displaced by 12 Å through the rotation of the helical domain. (B) Autoinhibited EHD2 dimer (PDB ID code 4CID), in which the final helix α12 and the adjacent linker is shown in purple. (C) Detailed view showing the interaction of the helical domain-EH domain linker with the GTPase domain. The displacement of Glu400/Glu403 is indicated as in A. We suggest that the rotation pushes the linker away from its position in the autoinhibited state EHD2 dimer. In particular, Val409 and Phe412 will be shifted away from their hydrophobic binding pocket (marked as green circle). Consequently, also the GPF motif (residues 420–422, marked as orange circle), which binds to the opposing EH domain, may be displaced.
To understand the consequences for membrane binding, we used the previously established EPR assay for EHD2 as a model for the EHD family. In these experiments, a single paramagnetic spin probe is attached to a single cysteine introduced at a specific site in an otherwise cysteine-free EHD2 variant. The accessibility of the spin label toward paramagnetic spin colliders, such as oxygen and nickel ethylenediamine diacetic acid (NiEDDA), can provide information on the membrane immersion of the spin label (22). Using this assay, we now demonstrate that not only the N-terminal part of α9 at the tip of the stalk, but also Gln330, Leu331, and Leu333 at the C-terminal end of α9, contribute to membrane binding (Fig. 2 B and C). Furthermore, we show that Cys356 in the adjacent helix α11 directly interacts with the membrane. In contrast, Val337 and A340 in α10, which is bent away from α9 and α11, did not penetrate into the membrane (Fig. 2 B and C). These results suggest that the membrane interaction site in EHD proteins extends along the parallel helices α9 and α11, which, together with the entire helical domain, move en bloc during activation. In the autoinhibited EHD2 structure, the membrane binding sites from two opposing monomers point away from each other (Fig. 2D). In contrast, in the EHD4ΔN structure, the lipid binding regions reorient in a parallel fashion toward the membrane surface for binding. These results suggest that the EHD4ΔN structure represents a membrane-binding competent, active conformation.
A Switch in the GTPase Domains Allows Oligomerization.
In the EHD2 structure, the helical domains form several salt bridges to the GTPase domain of the same monomer (Fig. 3A, black box). The corresponding salt bridges are broken in the EHD4ΔN structure (Fig. 3B, black box). Furthermore, we previously demonstrated that the N-terminal 8 aa in the autoinhibited EHD2 dimer fold back into a hydrophobic pocket of the GTPase domain (Fig. 3A) (16). In EHD4ΔN, the hydrophobic pocket in the GTPase domain is occupied by the adjacent KPF loop, which undergoes a large-scale reorientation (Fig. 3B, blue box, Movie S1). Conserved residues in this loop, such as Phe125, Leu128 and Phe131, anchor the helix into this pocket, whereas in the auto-inhibited EHD2 structure, the highly conserved Trp4 and Met5 of the N terminus (Fig. S4) occupy the equivalent space. These observations suggest that during the activation process, the KPF loop switches into the hydrophobic binding pocket of the GTPase domain.
Fig. 3.
The KPF loop undergoes a large-scale reorientation upon activation. (A) Structure of EHD2 (PDB ID code 4CID). The black box highlights the contacts of the helical domain with the GTPase domain and the N terminus in the autoinhibited state. The blue box shows the localization of the N terminus in a hydrophobic groove of the GTPase domain. (B) Structure of EHD4ΔN, with the same orientation of the GTPase domain as in A. The black box features the broken contacts between the helical domain and the GTPase domain in the active state. The blue box shows that the KPF loop occupies the hydrophobic groove in the GTPase domain, with Phe125, Leu128, and Phe131 (corresponding to Phe122, Leu125, and Phe128 in EHD2) acting as anchor points. Because of three extra amino acids in the N-terminal region of EHD4, the numbering of residues on EHD4 (residues 13–541) corresponds to the EHD2 residue number plus three (Fig. S4).
When analyzing the crystal packing, we noticed that EHD4ΔN dimers assembled in a linear fashion in the crystals (Fig. 4A and Fig. S7 A and B). In these oligomers, the membrane binding sites of the helical domain were oriented in the same direction, suggesting a physiologically plausible assembly. The oligomer had the same width (90 Å) as single EHD4 filaments sometimes observed on tubulated liposomes (Fig. S1C). Furthermore, the GTPase domains of adjacent EHD4ΔN dimers directly opposed each other via the highly conserved G interface, although they were separated by a small gap (Fig. 4A, Right). We previously proposed a similar oligomerization model for EHD2, but without the rotation of the helical domains (14).
Fig. 4.
Oligomerization via a new interface between the KPF loop and the helical domain. (A) Two views on the EHD4ΔN oligomers in the crystals represented by two dimers. One dimer is colored as in Fig. 1E, whereas the other is colored in light blue. Note the gap between the G interfaces of assembling dimers (Right). (B) Inset shows details of the oligomerization surface involving the KPF loop and the helical domain of the opposing dimer. (C) Liposome tubulation of EHD4ΔN, EHD4ΔN F125A and EHD4ΔN K302A/R305A, as in Fig. 1B. (D) Basic and stimulated ATP hydrolysis reactions of EHD4ΔN F125A and EHD4ΔN K302A/R305A were carried out as in Fig. 1C. (E) Expression of mCherry-tagged EHD4ΔN F125A and EHD4ΔN K302A/R305A in HeLa cells, as in Fig. 1D. (Scale bars: C, 500 nm; E, 10 μm.)
Fig. S7.
Assembly of EHD4. (A) Crystal packing of ATPγS-bound EHD4ΔN, shown in two different views. The crystals are built of four EHD4 filaments, which are rotated 90° to each other (Left). In Right, alternate dimers are colored in blue and red (light/dark for the monomers). (B) Schematic arrangement of EHD4 dimers in the oligomer. At Right, the linear oligomer is shown from a side view. The distance between the first amino acid of EHD4ΔN (Gln22) and the membrane interacting Val359 (corresponding to Cys356 in EHD2; Fig. 2) is 42 Å. To allow the N-terminal residues to insert into the same membrane leaflet as the primary membrane binding site in the helical domain, additional structural rearrangements, such as a partial unfolding of helix α1 may be required. Our previously published EPR data (16) showed that spin labels attached to the N terminus of EHD2 experience highly increased accessibility to oxygen in the presence of membranes, indicating that the N terminus is immersing into the membrane in the presence of liposomes (hence its denotation as “secondary membrane binding site”). Furthermore, continuous wave measurements revealed that N-terminal residues display increased mobility upon membrane binding, consistent with a release of the N terminus in the presence of membranes. In general, the N-terminal residues in EHD proteins are hydrophobic and highly conserved in the EHD family, suggesting a conserved function. However, in a cellular context (e.g., at caveolar or endosomal membranes), the exact position of the N terminus relative to the primary membrane binding site in the helical domain is not known. (C) Surface conservation plot of EHD4, using 39 EHD sequences from 20 different species. Conserved residues are shown in purple and nonconserved residues in cyan. Note that the oligomerization interface between the GTPase domain and helical domain is highly conserved.
When analyzing the oligomerization determinants in these linear EHD4ΔN assemblies, we found that the rearranged KPF loop interacted via a highly conserved interface with the helical domain of the adjacent dimer (Fig. 4B and Fig. S7 B and C). To probe the involvement of this new interface for oligomerization, we aimed to disrupt it by introducing the F125A and K302A/R305A mutations in EHD4ΔN (Figs. 3 and 4B). Both mutants still bound to liposomes (Fig. S1B). Whereas the F125A completely lost its ability to remodel membranes, the K302A/R305A mutant showed reduced remodeling of membranes, as indicated by the formation of less and irregular membrane tubules (Fig. 4C). Both mutants showed slightly increased basal ATPase rates, which, however, were not or only to a minor extent stimulated by the addition of liposomes (Fig. 4D). When these N-terminal deletion variants were overexpressed in HeLa cells, they showed a severe loss of membrane recruitment (Fig. 4E and Fig. S1 D and E), and similar results were previously obtained for the F122A mutant of EHD2 (corresponding to F125A in EHD4; Fig. S4) (10). We also attempted to stabilize the oligomerization interface in full-length EHD4 by introducing the A116L and N133D mutations to enhance hydrophobic interactions and create an additional salt bridge to Lys302, respectively. When expressed in HeLa cells, the single mutants showed similar membrane recruitment compared with EHD4 (Fig. S1 D and E). Strikingly, the double mutant showed greatly enhanced membrane recruitment, consistent with an increased oligomerization activity. Taken together, these data suggest that upon membrane recruitment and release of the N terminus, the KPF loop switches into the hydrophobic pocket of the GTPase domains and promotes oligomerization. A contact of the rearranged KPF loop with the open helical domain of the adjacent EHD dimer contributes to the regular assembly of EHDs on membranes.
Discussion
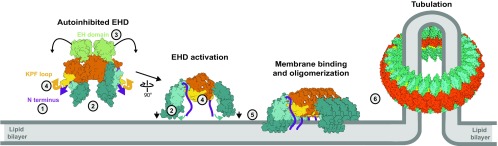
Membrane-associated assembly processes of peripheral membrane proteins are challenging to study at the structural level, and only in very few cases, high-resolution structures of such proteins in the autoinhibited and the activated, oligomerized form have been described. By removing the autoinhibitory N-terminal sequence, we obtained a membrane-binding and oligomerization-competent conformation of EHD4, which we consider as the active conformation [see also related manuscript by Hoernke et al. (23)]. In contrast, the membrane binding sites in the previous EHD2 structure were oriented away from the membrane. We therefore refer to the EHD2 structure as an autoinhibited conformation. A comparison between the active and autoinhibited form reveals a complex domain rearrangement in EHD proteins accompanying activation, resulting in an activation model for EHDs (Fig. 5). In the autoinhibited cytosolic form, the N terminus is locked in the GTPase domain and the EH domains block the assembly. Upon membrane recruitment, a series of conformational changes is triggered: (i) The N terminus of EHDs is released from the GTPase domain and may switch into the membrane (Fig. S7B for further discussion). This switch appears to activate EHDs because the EHD4ΔN construct is in the active conformation even in the absence of membranes and in both the ATP- and ADP-bound states (Fig. S3A). (ii) The helical domains rotate around the conserved Pro289. A similar rotation mechanism was observed for dynamin and suggested to act as a power stroke (19). In contrast, the domain rotation of the helical domains in EHDs appears to regulate membrane binding by adjusting the position of the membrane-binding site in α9 and α11. (iii) Concomitant with the rotation of the helical domain, the linker to the EH domain is dislodged from the GTPase domain. Consequently, the EH domains are displaced from their autoinhibitory site on the GTPase domain, as observed in the EHD4ΔN structure. In cells, this release may be further promoted by interactions of the EH domain with NPF motif-containing binding partners, such as syndapins/PACSINs (9, 12) and MICAL-L1 (24). (iv) Our structural analysis indicates that the KPF loop moves into the hydrophobic pocket of the GTPase domain. This loop creates a new assembly interface with the helical domain of the adjacent EHD dimer, therefore stabilizing the active conformation and promoting oligomerization of EHDs at the membrane. (v) In addition, the nucleotide-loading state of EHDs may affect the activation by nucleotide-dependent stabilization of the N-terminal loop or the KPF loop in the GTPase domain. Supporting this hypothesis, the KPF loop and switch I interact in an ATP-dependent manner. Furthermore, the removal of the EH domain tail from the active site may allow the ATP-dependent oligomerization of EHDs in ring-like structures via the conserved G-interface, which could explain the strict ATP dependency of assembly (Movie S2) (16). Such assembly would facilitate (vi) a direct coupling of EHD oligomerization with the creation of membrane curvature.
Fig. 5.
Activation model of EHDs. For details see Discussion; the numbers refer to release of the N terminus into the membrane (1), rotation of the helical domain (2), release of the EH domains from the autoinhibitory site (3), insertion of the KPF-loop into the hydrophobic pocket of the GTPase domain (4), membrane binding and oligomerization (5), and membrane tubulation (6).
Whereas the overall architecture of EHDs is related to dynamin, the oligomerization modes of these proteins differ fundamentally: Oligomerization of dynamin/MxA/DNM1L in helical filaments is mediated by three assembly interfaces in the helical stalk (19). The GTPase domains contribute to assembly by mediating GTPase-dependent contacts between adjacent filaments. In this way, nucleotide binding and hydrolysis can induce the rearrangement of adjacent filaments assembled via the stalk, leading to constriction of the underlying membrane. In contrast, EHDs use a unique interface in the GTPase domain for dimerization and use the ATP-dependent G interface for further oligomerization. Contacts between the GTPase domain and the helical domain of the next dimer contribute to oligomer formation. The architecture of the EHD oligomers excludes a nucleotide-driven sliding mechanism because nucleotide-dependent contacts are formed within and not between adjacent filaments. However, the assembly mode is well suited for the stabilization of membrane curvature by oligomerizing into ring-like tubular oligomers.
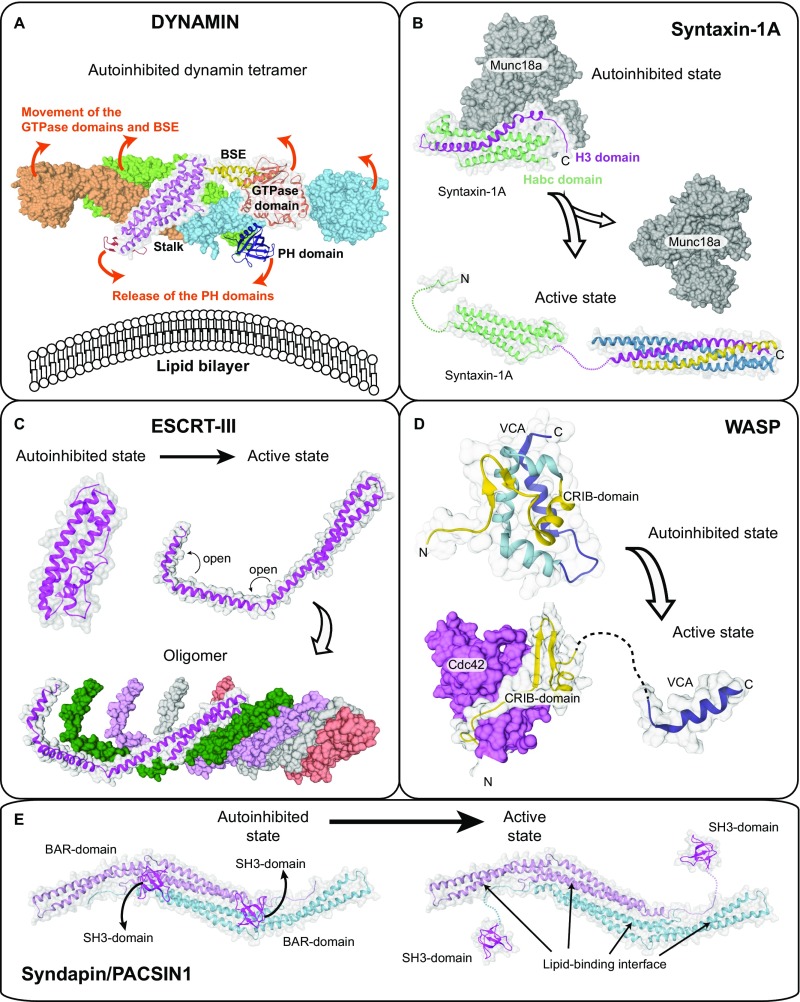
Despite the involvement of different domains, there are striking parallels in the activation mechanisms of EHDs and dynamin (Fig. S8). In the autoinhibited dynamin tetramer, an intramolecular contact of the pleckstrin homology (PH) domain with the oligomerization surface of the stalk prevents the assembly in the cytosol (25), whereas an autoinhibitory contact of the C-terminal EH domain with the GTPase domains takes over this function in EHDs. Such intramolecular inhibitory contacts are also observed in other peripheral or integral membrane proteins, such as BAR-domain containing proteins (26, 27), SNAREs (28), ESCRT-III (29), or the WASP protein (30) (Fig. S8 B–E). Binding of specific elements to membranes or interaction partners shifts the equilibrium toward the active state. The formation of new interactions in the membrane-bound oligomer is accompanied by the loss of intramolecular interactions in the autoinhibited dimer. Such mechanism may allow reversibility of membrane recruitment and oligomerization.
Fig. S8.
Activation mechanism of other membrane-associated proteins. (A) Crystal structure of the autoinhibited dynamin-3 tetramer (PDB ID code 5A3F). Based on a low-resolution EM reconstruction of oligomerized dynamin, it was suggested that the PH domains are released from the autoinhibitory stalk interface during activation and bind to the membrane, whereas the GTPase domains dimerize via the G-interface across helical turns (suggested movements are indicated by arrows). The PH domain release is thought to expose oligomerization interfaces in the stalk, therefore permitting oligomerization. Because there is no high-resolution structure of dynamin in the oligomerized state available, the exact mechanistic details of activation are still elusive. (B) The SNARE subunit Syntaxin-1A forms a closed autoinhibited four helix-bundle conformation (PDB ID code 3C98) and an open three helix-bundle conformation (comprising the “Habc domain,” PDB ID code 1S94). In the open state, the released H3 helix (magenta) associates with other SNARE partners to form the assembled SNARE complex. Binding of the regulatory Munc18a/Sec1-subunit stabilizes the autoinhibited conformation of syntaxin1A. (C) The ESCRTIII-component CHMP1B in the closed (modeled based on PDB ID code 3GGY) and open state (based on an electron microscopy reconstruction of the IST1-CHMP1B ESCRT-III copolymer, PDB ID code 3JC1). Domain opening exposes oligomerization interfaces in CHMP1B and allows assembly of a heteromeric scaffold. (D) Autoinhibited (PDB ID code 1EJ5) and active conformation (PDB ID code 1CEE) of the WASP protein. In the autoinhibited state, the CRIB domain tightly interacts with the VCA peptide. The small GTPase Cdc42 binds to the CRIB domain, therefore releasing the VCA peptide to induce actin polymerization. Domain opening is further promoted by PIP2 binding. (E) Crystal structure of the autoinhibited Syndapin/PACSIN-1 dimer comprising BAR and SH3 domains (PDB ID code 2X3W). Only one SH3 domain was observed in the crystal structure; the localization of the second SH3 domains was modeled. Upon binding to proline-rich sequences of target proteins such as dynamin, the SH3 domains are released, therefore exposing the membrane binding site in the BAR domain to allow formation of a membrane-associated BAR domain scaffold. Similar to EHDs, these peripheral membrane proteins show a closed, autoinhibited conformation and an active open conformation. In the open conformation, self-assembly (dynamin, ESCRT-III), membrane binding (dynamin, PACSIN-1, ESCRT-III, WASP) or protein–protein interaction motifs (all examples) become exposed. However, the complex coupling of membrane binding to domain releases, rotations, and loop switches is specific for EHDs and possibly other dynamin proteins.
Taken together, our study elucidates a complex activation mechanism of EHD family proteins, revealing common and specific features how peripheral membrane proteins scaffold cellular membranes.
Materials and Methods
Protein expression and purification were carried out as described before (14). A detailed description of the purification, crystallization, structure solution, refinement, biochemical experiments, electron microscopy, cell biology and microscopy analysis can be found in SI Materials and Methods.
SI Materials and Methods
Protein Expression and Purification.
Mouse EHD4 (residues 22–541, EHD4ΔN) and the indicated mutants were expressed from a modified pET28 vector as N-terminal His6-tag fusions followed by a PreScission protease cleavage site, according to ref. 14. Expression plasmids were transformed in Escherichia coli host strain BL21(DE3)-Rosetta2 (Novagen). Cells were grown at 37 °C in TB medium, and protein expression was induced at an optical density of 0.5 by the addition of 40 µM isopropyl-β-d-thiogalactopyranoside (IPTG), followed by overnight incubation at 18 °C. Upon centrifugation, cells were resuspended in 50 mM Hepes/NaOH (pH 7.5), 500 mM NaCl, 25 mM imidazole, 2 mM MgCl2, 2.5 mM β-mercaptoethanol (β-ME) including 1 mM Pefabloc protease inhibitor (Carl Roth) and 1 µM DNase I (Roche) and lysed in a microfluidizer (Microfluidics). Following centrifugation (30,000 × g, 1 h, 4 °C), cleared lysates were applied to a NiNTA column (GE Healthcare). The column was then extensively washed with 50 mM Hepes/NaOH (pH 7.5), 700 mM NaCl, 10 mM CaCl2, 1 mM ATP, 10 mM MgCl2, 10 mM KCl, and afterward with 50 mM Hepes/NaOH (pH 7.5), 500 mM NaCl, 25 mM imidazole, 2 mM MgCl2, 2.5 mM β-ME. The protein was eluted with 50 mM Hepes/NaOH (pH 7.5), 500 mM NaCl, 2 mM MgCl2, 2.5 mM β-ME, and 300 mM imidazole. One hundred fifty micrograms of PreScission protease per 5 mg of the EHD constructs was added and the protein was dialyzed overnight against 50 mM Hepes/NaOH (pH 7.5), 500 mM NaCl, 1 mM MgCl2 and 2.5 mM β-ME. Following reapplication of the protein to a NiNTA column to remove the His tag and uncleaved protein, EHD4ΔN was concentrated by using a 50-kDa molecular mass cutoff concentrators (Amicon) and applied to a Superdex200 gel filtration column (GE Healthcare) equilibrated with 50 mM Hepes/NaOH (pH 7.5), 500 mM NaCl, 1 mM MgCl2, and 2.5 mM β-ME. Fractions containing the EHD constructs were pooled, concentrated, and flash-frozen in liquid nitrogen. The purified protein was nucleotide-free, as judged by HPLC analysis.
Crystallization and Structure Determination.
Initial crystallization trials by the sitting-drop vapor-diffusion method were performed at 20 °C, using a Gryphon LCP pipetting robot (Art Robbins Instrument) and Rock Imager storage system (Formulatrix). Subsequently, conditions were refined in the 24-well format by using hanging drops. One microliter of mouse EHD4ΔN at a concentration of 10 mg/mL was mixed with 2 µL of the reservoir solution containing 26% (wt/vol) sodium polyacrylate 5100, 200 mM MgCl2, 100 mM Hepes/NaOH (pH 7.5) in the presence of 2 mM ATPγS, or 900 mM sodium malonate (pH 7) in the presence of 2 mM ADP. In both cases, rod-shaped crystals appeared after 1 d at 20 °C. Crystals were cryoprotected by transfer into a solution containing 50% (vol/vol) of the buffer and reservoir components and 20% (vol/vol) glycerol and flash-cooled in liquid nitrogen. Data were recorded at beamline BL-14.1 at BESSY-II (Berlin) at a temperature of 100 K and a wavelength of 0.9184 Å. Data from single crystals were processed and scaled by using the program package XDS (31) and XDSAPP (32). The structure was solved by molecular replacement with Phaser (33) using the individual G domain and helical domain of EHD2 (PDB ID code 4CID) as search models. The model was built by using Coot (34) and iteratively refined by using Phenix (35) with three translation, libration, and screw-rotation (TLS) parameters per molecule. For the ATPγS-bound structure, 96% of all residues are in the most favored regions of the Ramachandran plot and 0.5% in the disallowed regions; for the ADP-bound structure, 93.5% of all residues are in the most favored regions of the Ramachandran plot and 1.1% in the disallowed regions. Figures were prepared with the PyMOL Molecular Graphics System Version 1.8, Schrödinger, LLC and Chimera (36). The molecular morph was created in Chimera. Domain superpositions were performed with Coot. The surface conservation plot was calculated and visualized by using Chimera.
Liposome Cosedimentation Assays.
Liposomes were prepared as described (www.endocytosis.org). Folch liposomes (total bovine brain lipids fraction I from Sigma) in 20 mM Hepes/NaOH (pH 7.5), 300 mM NaCl, and 1 mM DTT were sonicated in a water bath for 30 s. Liposomes (0.2 mg/mL) were incubated at room temperature with 10 µM of the indicated EHD4 construct for 10 min in 40 µL of reaction volume, followed by a 213,000 × g spin for 10 min at 20 °C. The final reaction buffer contained 25 mM Hepes/NaOH (pH 7.5), 300 mM NaCl, and 0.5 mM MgCl2.
ATP Hydrolysis Assay.
ATPase activities of 10 µM of the indicated EHD4 constructs were determined at 30 °C in 25 mM Hepes/NaOH (pH 7.5), 300 mM NaCl, and 0.5 mM MgCl2 in the absence and presence of 1 mg/mL nonextruded Folch liposomes, using 100 µM ATP as the substrate. Reactions were initiated by addition of protein to the reaction. At different time points, reaction aliquots were fivefold diluted in reaction buffer and quickly transferred to liquid nitrogen. Nucleotides in the samples were separated via a reversed-phase Hypersil ODS-2 C18 column (250 × 4 mm) with 100 mM potassium phosphate buffer pH 6.5, 10 mM tetrabutylammonium bromide, 7.5% (vol/vol) acetonitrile as running buffer. Denatured proteins were adsorbed on a C18 guard column. Nucleotides were detected by absorption at 254 nm and quantified by integration of the corresponding peaks. Rates were derived from a linear fit to the initial reaction (<40% ATP hydrolyzed).
Electron Microscopy.
For membrane tubulation assays, 10 µM EHD4ΔN in 25 mM Hepes/NaOH (pH 7.5), 300 mM NaCl, 1 mM MgCl2, and 1 mM ATP were incubated at room temperature for 20 min with 1 mg/mL liposomes. Samples were spotted on carbon-coated copper grids (Plano) and negatively stained with 2% uranyl acetate. Electron grids were imaged with a transmission electron microscope at 80 kV (EM 910; Zeiss) and acquisition was done with a CCD camera (Quemesa; Olympus Viewing System).
EPR Power Saturation Experiments.
These experiments were carried out as described in ref. 16 at a concentration of ∼2 g/L. Accessibilities to O2 (from air, ΠOx) and 10 mM NiEDDA (ΠNiEDDA) were obtained from power saturation experiments by using a Bruker EMX X-Band ESR spectrometer fitted with ER4123D dielectric resonator. The depth parameter Φ was calculated from Φ = ln[ΠOx/ΠNiEDDA] (16) and the membrane insertion depth was obtained as described in ref. 16.
Cell Biology and Microscopy.
HeLa cells (ATCC-CRM-CCL-2) were grown in DMEM (Gibco) supplemented with 10% (vol/vol) FBS (Invitrogen). For confocal imaging, 80,000 cells were seeded on 1.5 high tolerance glass coverslips 25 mm (Warner Instruments) 24 h before transfection. EHD4-mCherry constructs were transiently transfected by using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions 16–24 h before the experiment. The microscope stage and objective lenses were maintained at 37 °C within an environmental chamber supplemented with 5% (vol/vol) CO2. Images were acquired with a Zeiss Cell Observer Spinning Disk Confocal controlled by ZEN interface with an Axio Observer.Z1 inverted microscope, equipped with a CSU-X1A 5000 Spinning Disk Unit and a EMCCD camera iXon Ultra from ANDOR. Cells were imaged in DMEM lacking Phenol Red (Gibco) supplemented with 10% (vol/vol) FBS. For epifluorescence analysis, cells were fixed in 3% (vol/vol) paraformaldehyde in PBS for 20 min at room temperature, then washed and blocked in 5% (vol/vol) goat serum with 0.05% saponin in PBS before staining with mouse anti-EEA1 (clone 14, 610456, BD Biosciences) in 1% goat serum, 0.05% saponin in PBS using standard protocols. Images were acquired by using a Zeiss Axio Imager Z1 system with Zen software. The representative microscopic images were cropped by using ImageJ (NIH).
Image Analysis and Statistics.
The SDs of the image histogram from maximum intensity projections of confocal stacks were used to measure the textures of the different mutants. A ROI of fixed size was applied within an area with representative texture and the SDs of the gray values were measured by using ImageJ. Statistical analysis was performed byusing Prism5 (GraphPad Software).
Supplementary Material
Acknowledgments
We thank Vivian Schulz and Chris van Hoorn for technical support, Irene Martinez from the Biochemical Imaging Center Umeå for help with image analysis, Bettina Purfürst for help with electron microscopy, and the staff at BESSY beamline 14.1 for help during data collection. This project was supported by Deutsche Forschungsgemeinschaft Grant SFB958/A12 (to O.D.); European Research Council Consolidator Grant ERC-2013-CoG-616024 (to O.D.); NIH Grant GM115736 (to R. Langen); and the Swedish Research Council, Swedish Foundation for Strategic Research (to R. Lundmark).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors of ATPγS- and ADP-bound mouse EHD4 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5MTV and 5MVF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614075114/-/DCSupplemental.
References
- 1.Naslavsky N, Caplan S. EHD proteins: Key conductors of endocytic transport. Trends Cell Biol. 2011;21(2):122–131. doi: 10.1016/j.tcb.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin SX, Grant B, Hirsh D, Maxfield FR. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol. 2001;3(6):567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- 3.Grant B, et al. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol. 2001;3(6):573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- 4.Caplan S, et al. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21(11):2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galperin E, et al. EHD3: A protein that resides in recycling tubular and vesicular membrane structures and interacts with EHD1. Traffic. 2002;3(8):575–589. doi: 10.1034/j.1600-0854.2002.30807.x. [DOI] [PubMed] [Google Scholar]
- 6.Naslavsky N, McKenzie J, Altan-Bonnet N, Sheff D, Caplan S. EHD3 regulates early-endosome-to-Golgi transport and preserves Golgi morphology. J Cell Sci. 2009;122(Pt 3):389–400. doi: 10.1242/jcs.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao Y, et al. Pincher, a pinocytic chaperone for nerve growth factor/TrkA signaling endosomes. J Cell Biol. 2002;157(4):679–691. doi: 10.1083/jcb.200201063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Philippidou P, et al. Trk retrograde signaling requires persistent, Pincher-directed endosomes. Proc Natl Acad Sci USA. 2011;108(2):852–857. doi: 10.1073/pnas.1015981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun A, et al. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell. 2005;16(8):3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morén B, et al. EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Mol Biol Cell. 2012;23(7):1316–1329. doi: 10.1091/mbc.E11-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoeber M, et al. Oligomers of the ATPase EHD2 confine caveolae to the plasma membrane through association with actin. EMBO J. 2012;31(10):2350–2364. doi: 10.1038/emboj.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senju Y, et al. Phosphorylation of PACSIN2 by protein kinase C triggers the removal of caveolae from the plasma membrane. J Cell Sci. 2015;128(15):2766–2780. doi: 10.1242/jcs.167775. [DOI] [PubMed] [Google Scholar]
- 13.Lee DW, et al. ATP binding regulates oligomerization and endosome association of RME-1 family proteins. J Biol Chem. 2005;280(17):17213–17220. doi: 10.1074/jbc.M412751200. [DOI] [PubMed] [Google Scholar]
- 14.Daumke O, et al. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449(7164):923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- 15.Pant S, et al. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11(12):1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah C, et al. Structural insights into membrane interaction and caveolar targeting of dynamin-like EHD2. Structure. 2014;22(3):409–420. doi: 10.1016/j.str.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salcini AE, et al. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11(17):2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieken F, et al. Structural insight into the interaction of proteins containing NPF, DPF, and GPF motifs with the C-terminal EH-domain of EHD1. Protein Sci. 2009;18(12):2471–2479. doi: 10.1002/pro.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daumke O, Praefcke GJ. Invited review: Mechanisms of GTP hydrolysis and conformational transitions in the dynamin superfamily. Biopolymers. 2016;105(8):580–593. doi: 10.1002/bip.22855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma M, Naslavsky N, Caplan S. A role for EHD4 in the regulation of early endosomal transport. Traffic. 2008;9(6):995–1018. doi: 10.1111/j.1600-0854.2008.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature. 2010;465(7297):435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margittai M, Langen R. Spin labeling analysis of amyloids and other protein aggregates. Methods Enzymol. 2006;413:122–139. doi: 10.1016/S0076-6879(06)13007-4. [DOI] [PubMed] [Google Scholar]
- 23.Hoernke M, et al. EHD2 restrains dynamics of caveolae by an ATP-dependent, membrane-bound, open conformation. Proc Natl Acad Sci USA. 2017;114:E4360–E4369. doi: 10.1073/pnas.1614066114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma M, Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol Biol Cell. 2009;20(24):5181–5194. doi: 10.1091/mbc.E09-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reubold TF, et al. Crystal structure of the dynamin tetramer. Nature. 2015;525(7569):404–408. doi: 10.1038/nature14880. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, et al. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci USA. 2009;106(31):12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao Y, et al. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci USA. 2010;107(18):8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404(6776):355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 29.McCullough J, et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 2015;350(6267):1548–1551. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404(6774):151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krug M, Weiss MS, Heinemann U, Müller U. XDSAPP: A graphical user interface for the convenient processing of diffraction data using XDS. J Appl Cryst. 2012;45:568–572. [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.