Significance
Carnivorous plants capture and digest animal prey for nutrition. In addition to being carnivorous, the humped bladderwort plant, Utricularia gibba, has the smallest reliably assembled flowering plant genome. We generated an updated genome assembly based on single-molecule sequencing to address questions regarding the bladderwort’s genome adaptive landscape. Among encoded genes, we segregated those that could be confidently distinguished as having derived from small-scale versus whole-genome duplication processes and showed that conspicuous expansions of gene families useful for prey trapping and processing derived mainly from localized duplication events. Such small-scale, tandem duplicates are therefore revealed as essential elements in the bladderwort’s carnivorous adaptation.
Keywords: plant genomics, evolution, polyploidy, carnivorous plant, Utricularia
Abstract
Utricularia gibba, the humped bladderwort, is a carnivorous plant that retains a tiny nuclear genome despite at least two rounds of whole genome duplication (WGD) since common ancestry with grapevine and other species. We used a third-generation genome assembly with several complete chromosomes to reconstruct the two most recent lineage-specific ancestral genomes that led to the modern U. gibba genome structure. Patterns of subgenome dominance in the most recent WGD, both architectural and transcriptional, are suggestive of allopolyploidization, which may have generated genomic novelty and led to instantaneous speciation. Syntenic duplicates retained in polyploid blocks are enriched for transcription factor functions, whereas gene copies derived from ongoing tandem duplication events are enriched in metabolic functions potentially important for a carnivorous plant. Among these are tandem arrays of cysteine protease genes with trap-specific expression that evolved within a protein family known to be useful in the digestion of animal prey. Further enriched functions among tandem duplicates (also with trap-enhanced expression) include peptide transport (intercellular movement of broken-down prey proteins), ATPase activities (bladder-trap acidification and transmembrane nutrient transport), hydrolase and chitinase activities (breakdown of prey polysaccharides), and cell-wall dynamic components possibly associated with active bladder movements. Whereas independently polyploid Arabidopsis syntenic gene duplicates are similarly enriched for transcriptional regulatory activities, Arabidopsis tandems are distinct from those of U. gibba, while still metabolic and likely reflecting unique adaptations of that species. Taken together, these findings highlight the special importance of tandem duplications in the adaptive landscapes of a carnivorous plant genome.
The architectural evolution of flowering plant genomes includes a long history of gene duplication and diversification. Tandem gene duplication is an ongoing but nonglobal process that generates coding sequence diversity in eukaryotic genomes through subfunctionalization or neofunctionalization of gene copies on an individual basis (1). On the other hand, polyploidy events provide scores of genomically balanced duplicate genes all at once, on which divergent selection pressures can act to generate phenotypic diversity (2, 3). Evidence from available plant genomes supports the theory that modular, dosage-sensitive functions such as transcriptional regulation are enriched among duplicates surviving polyploidy events, whereas single-gene survivors of local duplication events have the opportunity to be enriched for dosage responsive functions, such as secondary metabolite production (e.g., refs. 4–7). Although it has been repeatedly noted that polyploidy events correlate with some major plant radiations (2, 8, 9), the specific roles that tandem duplicates play in species- or lineage-specific plant adaptation remain more poorly explored.
Utricularia gibba is an aquatic carnivorous plant with an unusually small but highly dynamic nuclear genome that experienced at least two whole-genome duplication (WGD) events during its evolutionary history since divergence from grapevine, tomato, and other species (10). Carnivorous plants are interesting model systems not only for understanding the molecular mechanisms underlying nutrient acquisition strategies, but also for discovering the regulatory underpinnings of their unique trapping morphologies. U. gibba is of particular interest given the previous publication of an ∼82-Mb short-read assembly (10), which revealed that its genome gained and deleted gene duplicates significantly faster than those of other genomes (11). Given that the U. gibba genome likely descended via considerable shrinkage from an ancestral genome up to 1.5 Gb in size (12), duplicates that survived deletion during its evolutionary history arguably evolved under greater purifying selection pressure compared with the more expansive genomes of most angiosperms. Therefore, we hypothesized that the deletion-prone genome of U. gibba could be particularly illustrative regarding the adaptive legacy of differential duplicate survival following their two modes of generation, with tandems highlighting aspects of the carnivorous lifestyle and syntenic duplicates highlighting transcriptional functions.
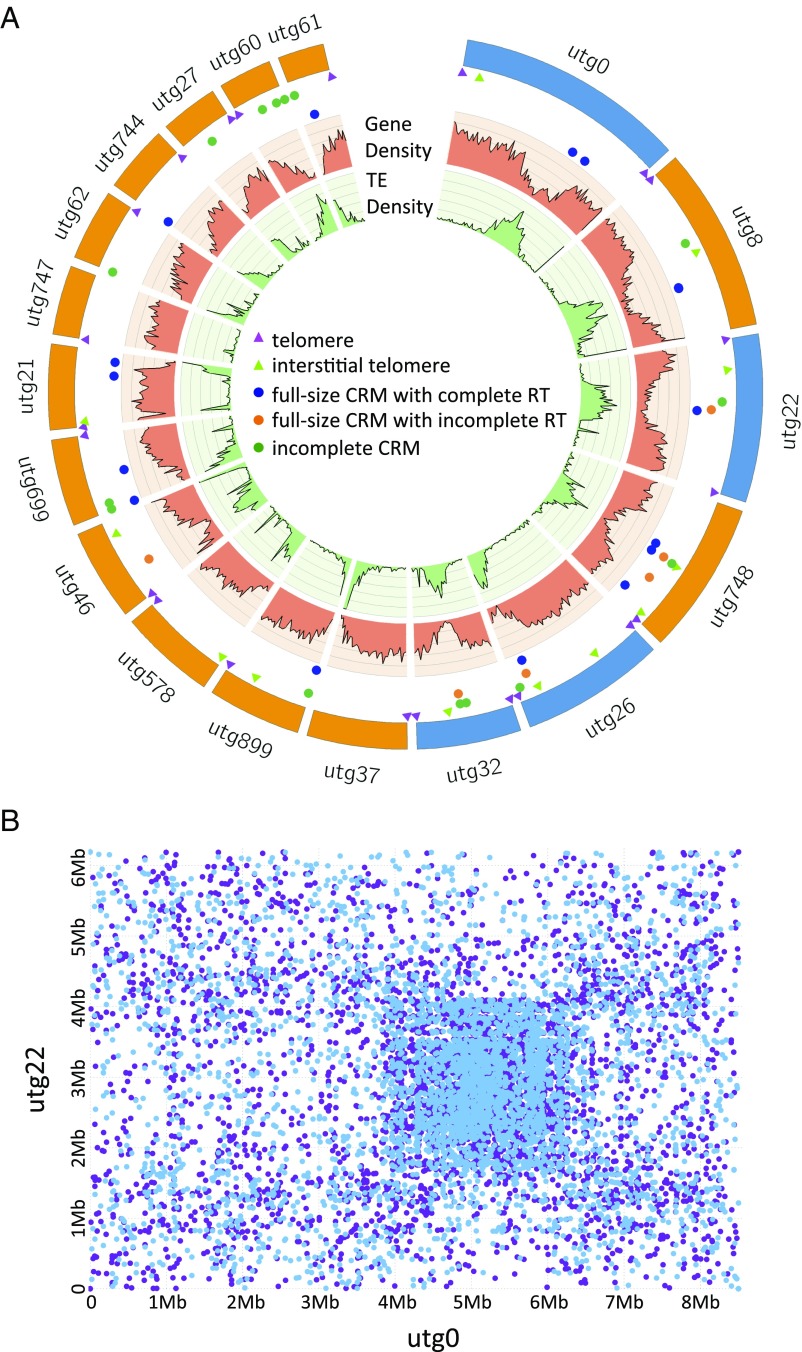
To explore this possibility, we generated a highly contiguous nuclear genome assembly for U. gibba based on Pacific Biosciences (PacBio) Single Molecule, Real-Time (SMRT) technology. We used 10 SMRT cells and P6-C4 PacBio chemistry to produce 521,937 raw and 702,640 filtered subreads with N50 values of 21,825 and 15,244 bp, respectively. After assembly with HGAP.3 (13), we produced a genome of 581 contigs with an N50 of 3,424,836 bp and 101,949,210 total bases (SI Appendix, Fig. S2). Remarkably, base pair correction using either the PacBio data or Illumina MiSeq reads from our previous assembly (10) led to extremely minor improvements, only 0.071% and 0.01% of total bases, respectively (SI Appendix, section 1.5). Four contigs represented complete chromosomes marked on either end by telomeres, including the longest contig of the assembly at 8,502,017 bp (Fig. 1). Twenty additional contigs had telomere repeats on one end, the 14 largest being ≥1 Mb in size (Fig. 1). Arabidopsis-type telomeric repeats (TTTAGGG) were identified in these 24 contigs. Two variants, the Chlamydomonas type (14) (TTTTTAGGG) and TTCAGGG (similar to the variants TTCAGG and TTTCAGG known from the close carnivorous plant relative Genlisea) (15), were also found sporadically intermingled with the Arabidopsis-type telomeric repeats. Ten contigs were observed to have interstitial telomeric repeats, which were identified by searching for (CCCTAAA)3 and (TTTAGGG)3 within chromosomal arms (Fig. 1A). After filtration for bacterial and other contamination (SI Appendix, section 1.6), the assembled genome amounted to 100,688,548 bp (on 518 contigs), including a complete 172,489-bp plastid genome on a single contig and a 283,823-bp partial mitochondrial genome (SI Appendix, section 1.6.2). Therefore, our newly assembled nuclear genome gained 18,356,750 bp from the former assembly size of 81,875,486 bp.
Fig. 1.
A chromosome-scale view of the architecture of the U. gibba genome. (A) Gene density, TE density tracks, telomeres, and the locations of CRM centromeric retrotransposon sequences are shown for all U. gibba contigs >1 Mb in size. Four complete chromosomal contigs are shown in blue, and partial chromosomes that have at least one end with telomere sequence are shown in orange. Putative centromeric regions are visible as peaks of increased TE density and decreased gene density. Most CRMs are localized at putative centromeric regions. (B) MUMmer (82) pairwise dot-plot alignment of contigs 0 and 22, which represent complete chromosomes. Blue and purple dots indicate hits on each DNA strand, respectively. Putative centromeric regions of strong sequence similarity are apparent as a densely hit square.
Calculation of the genome space occupied by transposable elements (TEs) uncovered almost 9 Mb (∼8.9%) complete TEs, with up to 59 Mb (∼59%) of the nuclear genome possibly TE-derived (SI Appendix, Dataset S1); the latter amounted to ∼16.6 Mb more TE-related genome space than was found in the previously published short-read assembly (SI Appendix, section 2.1). We found that ∼2.9 Mb of the genome (on 115 contigs) was composed of ribosomal DNA repeats (SI Appendix, section 2.2). Indeed, a syntenic path alignment with the short-read assembly demonstrated that most of the DNA gained by PacBio sequencing contained repeated elements, particularly surrounding putative centromeres (Fig. 1B and SI Appendix, Figs. S4–S8).
To identify signature centromeric repeats in U. gibba, we selected tandem repeat clusters with average period size of 50–500 bp for identification as putative centromere repeats (SI Appendix, Fig. S5B), as described previously (16). The top 10 most abundant tandem repeat clusters were considered prime candidates for centromeric repeats, but these were not even preferentially located in our chromosome-sized contigs. We then manually checked the locations of the next 10 most abundant tandem repeat clusters in the genome, and found that none of these clusters showed unique localization in putative centromeric regions. Therefore, we conclude that U. gibba centromeres are devoid of high-copy tandem repeat arrays such as those known from Arabidopsis and maize (16). Similar findings also have been reported for the centromeres of several plant and animal species (17–19), including two closely related carnivorous plants, Genlisea hispidula and Genlisea subglabra (15).
Although plant retrotransposon families generally are randomly dispersed, there are families distinctly concentrated in centromeric regions, such as the CRM centromeric chromoviruses. CRMs, a lineage of Ty3/gypsy retrotransposons, have been well characterized as centromeric retrotransposons in many species (20–25), including G. hispidula and G. subglabra (15). Using phylogenetic analysis, we found that 55 U. gibba sequences are grouped within the subgroup A CRMs, which include the centromere-specific CRMs (SI Appendix, section 3.3.3). All but one of the U. gibba sequences form a single, monophyletic CRM subfamily. To investigate the chromosomal localization of the 55 U. gibba CRMs, we plotted them on the complete and near-complete chromosomes together with the TE and gene model tracks. As depicted in Fig. 1A, most U. gibba CRMs are located in the putative centromeric regions; however, not all putative centromeres have CRM elements. It has been proposed that CRMs may play an important role in stabilizing centromere structure and maintaining centromere function (26, 27), whereas an opposing hypothesis holds that they are merely parasitic and tend to accumulate in recombination-poor centromeric regions to escape negative selection against insertions in distal regions (28). Our finding that some putative centromeric regions in U. gibba lack CRMs or other high-copy centromeric tandem repeats suggests that neither CRMs nor tandem repeats are crucial for maintaining functional centromeres in the species.
Our highly contiguous genome assembly also permitted a much finer account of protein-coding gene number than previously available, which amounted to 30,689, 7.7% more than reported for our short-read assembly (10). Unlike the far shorter scaffolds from that assembly (10), our largely chromosome-sized contigs permitted us to conservatively distinguish the WGD-derived and tandem duplicate portions of U. gibba’s genome adaptive landscape. In both cases, we were concerned with duplicates that could still be discerned within their formative genome structural contexts, not with duplicates that might have migrated to other chromosomal positions after their generation via small-scale or WGD events, because such genes could be only indirectly assigned to one duplicative process versus the other.
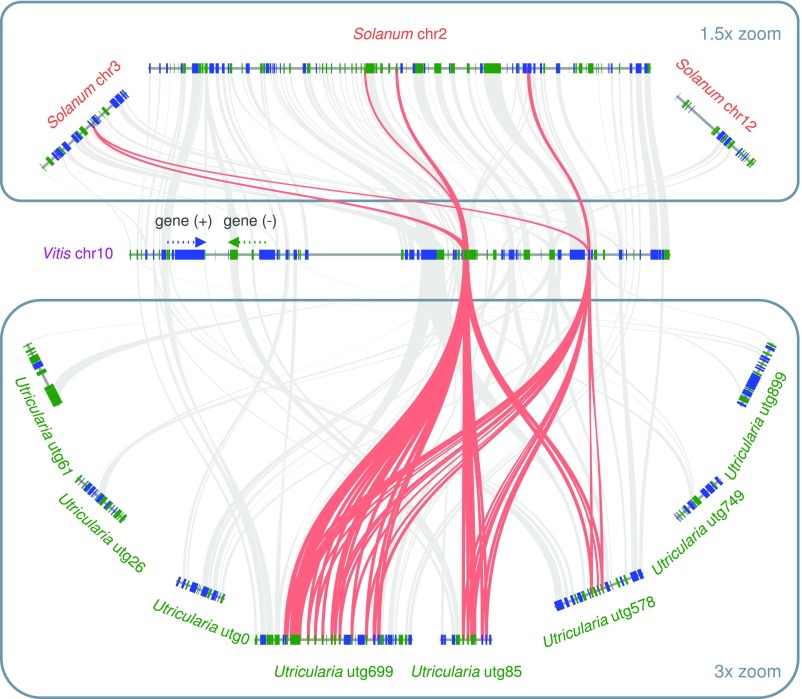
Through syntenic analysis using CoGe (29, 30), we were able to identify 54 syntenic block pairs descending from the most recent U. gibba WGD event (SI Appendix, Fig. S11). We were then able to reconstruct the immediate, nine-chromosome prepolyploid ancestor of the modern genome, following which numerous large-scale inversion events were required to account for modern gene order (SI Appendix, section 4.1). Further analysis permitted deconstruction of this ancestral genome into an earlier, six-chromosome pre-WGD ancestor that existed immediately before U. gibba’s second most recent polyploidy event (SI Appendix, Fig. S12); however, we could not reconstruct the third WGD event that was previously described based on visual inspection of syntenic dot plots and syntenic depth calculations (10). Nonetheless, microsynteny analyses did reveal many examples of eight (or more)-to-one syntenic block relationships with the Vitis vinifera genome (Fig. 2 and SI Appendix, section 4.5), some of which may include blocks dating to the gamma hexaploidy event at the base of all core eudicots (31).
Fig. 2.
Syntenic relationships among V. vinifera, S. lycopersicum, and U. gibba regions containing tandemly duplicated cysteine protease genes. Some parts of these tandem arrays clearly preexisted in U. gibba’s prepolyploid ancestral genomes, with further tandem duplications having occurred since those events, together increasing functional potential for U. gibba’s carnivory. A typical ancestral region in Vitis can be traced to up to three regions in Solanum (through the latter's genome triplication) and up to eight regions in U. gibba (where as many as three WGDs are possible). Red connecting lines highlight matching cysteine proteases in the selected regions; genes otherwise syntenic are shown in gray.
We analyzed the duplicate block pairs from the most recent WGD event to assess the degree of fractionation (gene loss) experienced by each subgenome following polyploidization (SI Appendix, Fig. S13). This analysis yielded a clear pattern of deletion bias characteristic of subgenome dominance inherited through a polyploidy event (32, 33). Fractionation bias was matched by both subgenome expression dominance (34) and fewer single nucleotide polymorphisms on dominant blocks (35, 36) (SI Appendix, section 4.4, Figs. S13 and S14, and Datasets S3 and S4), indicating the influence of stronger purifying selection. Taken together, these data suggest that the most recent WGD in U. gibba’s past was an allopolyploidization event resulting from a broad cross (37), because autopolyploidies are not expected to show such strong biases; for example, unbiased fractionation has been discovered in the genomes of poplar, banana, and soybean (37, 38). Hybridization of two species accompanied by genome doubling can instantly generate a third species with novel and transcendent phenotypic traits (39). Moreover, the modern U. gibba genome displays highly heterogeneous patterns of heterozygosity (SI Appendix, Dataset S4) that do not correlate with the structural limits of syntenic blocks, suggesting that outcrossing events subsequent to the most recent WGD were broad, but were not followed by ploidy changes. Given the highly clonal nature of aquatic Utricularia species (e.g., refs. 40, 41), this state could represent “frozen” heterozygosity in a particularly adaptive genotype, such as seen in unisexual hybrid vertebrates (42).
To examine polyploid adaptive genetic features in U. gibba, we evaluated gene ontology (GO) functional enrichments among syntenically retained gene duplicates descending from U. gibba’s lineage-specific WGDs. Duplicates retained following WGD were mostly enriched for transcriptional regulatory functions (SI Appendix, Dataset S5). As expected based on earlier studies, very similar results were obtained for Arabidopsis WGD duplicates analyzed in the same manner (SI Appendix, Dataset S6) (4, 43, 44); however, comparing the 522 U. gibba WGD duplicates annotated with the GO “regulation of transcription, DNA-templated” with all U. gibba genes with this GO revealed no significant enrichment of any biological process category (SI Appendix, Dataset S14). Similar analysis of Arabidopsis WGD duplicates yielded only one significant biological process category, “response to jasmonic acid” (SI Appendix, Dataset S15), suggesting that in both species, transcriptional regulatory enrichment is functionally generic.
In contrast to functional enrichments of WGD duplicates, U. gibba genes filtered out by the blast_to_raw script in the QUOTA-ALIGN package [https://github.com/tanghaibao/quota-alignment (45), included in CoGe SynMap (29, 30)] as tandem duplicates in the modern genome (and thus ignored in syntenic dot plot comparison) were enriched for many secondary metabolic functions, including specific functions that could be anticipated for a carnivorous plant (SI Appendix, Datasets S7 and S8). Arabidopsis tandems discovered in the same manner were similarly enriched for secondary metabolic activities, as anticipated based on earlier results (5). However, in many cases the Arabidopsis activities were entirely different (SI Appendix, Dataset S9). Among the most significantly enriched categories in U. gibba was the category “oligopeptide transporter activity,” assigned to 23 members of the OPT gene family (46). Importantly, oligopeptide transport was also among the most significantly enriched functional categories of genes specifically and strongly expressed in the bladder traps (47), with 13 genes showing 4- to 400-fold trap-enhanced transcription (SI Appendix, Dataset S8). Peptide transporters, which are involved in the plant nitrogen budget, have been identified as expressed in the trap fluid of the carnivorous pitcher plant Nepenthes (48, 49). The Nepenthes gene identified in that study is, however, a member of the PTR family, a group itself highlighted among U. gibba tandems by the significantly enriched term “dipeptide transporter activity,” wherein there are 22 family members, including three homologs of the Arabidopsis nitrate transporter gene NPF5.5 (50); unitig_52.g17408.t1 and unitig_26.g9035.t1 had >65-fold trap-enhanced expression (SI Appendix, Dataset S8). Carnivorous plants, bladderworts included, typically grow in nitrogen-poor habitats, where they compensate for deficiencies via prey capture and uptake of released nitrogen.
Another highly enriched functional category among tandem duplicates was “ATPase activity, coupled to transmembrane movement of substances,” comprising 58 genes, mostly ABC transporters. Proteins encoded by such genes are known from Nepenthes traps, where they are hypothesized to be responsible for maintaining trap acidity and various molecular transport functions (51). Several of these genes show greater than ninefold trap-specific expression, including unitig_85.g27344.t1, unitig_85.g27345.t1, unitig_750.g28500.t1, and unitig_750.g28501.t1 (SI Appendix, Dataset S8). Another enriched category was “transmembrane transport,” which highlighted all of the foregoing genes and also included eight phosphate transporter genes homologous to PHT1 (52). PHT1 family genes are induced during nutritional phosphate deficiency, a condition characteristic of the carnivorous plant lifestyle (53). Of these, unitig_747.g21685.t1 and unitig_747.g21690.t1 showed 2- to 24-fold trap-enhanced expression (SI Appendix, Dataset S8).
Another significantly enriched tandem duplicate functional category was “hydrolase activity, hydrolyzing O-glycosyl compounds.” This GO category included a gene encoding a class III chitinase (unitig_60.g25630.t1, showing >20-fold trap-enhanced expression) (SI Appendix, Dataset S8), representing one of the chitinase families [glycoside hydrolase (GH) family 18] active within the digestive fluid of both open and closed traps of various carnivorous plant species. In Nepenthes, the GH family 18 enzyme is encoded by a single-copy gene that is up-regulated in response to prey in both the pitted glands and surrounding tissues (54). Galactosidases and xylosidases (55) are also among the genes with the hydrolase annotation, and enzymes encoding both have been identified in the Nepenthes trap fluid proteome (56, 57). Nepenthes and Drosera (carnivorous sundew plant) digestive mucilage contains galactose and xylose (58), which may require breakdown for peptide and other nutrient absorption in U. gibba traps as well (59). Three xylosidase genes—unitig_62.g23624.t1, unitig_62.g23625.t1, and unitig_748.g7352.t1—show 4- to 35-fold trap-enhanced expression (SI Appendix, Dataset S8).
The traps of Utricularia operate through an intricate triggering mechanism (60). High-speed snap-buckling movements (61, 62) occur following triggered release of negative internal trap pressure achieved by active pumping out of water (63). Prey is engulfed with the influx of liquid, after which the trap may reset itself with a new negative pressure potential. This repeating process likely demands highly dynamic cell-wall changes. Indeed, the tandems-enriched GO category “cell wall” annotated 17 genes encoding expansins (64) (none of which, however, showed uniformly trap-enriched expression) and 8 genes encoding xyloglucan endotransglycosylases (65) (of which unitig_749.g14196.t1 and unitig_26.g9135.t1 showed greater than sixfold trap-enhanced expression) (SI Appendix, Dataset S8). Seventeen encoded peroxidases homologous to PRX52, which cross-link cell-wall strengthening extensins (unitig_26.g8978.t1 and unitig_22.g6605.t1 were >14-fold trap-enhanced), and 21 encoded polygalacturonases, which degrade cell-wall pectin (66) (unitig_8.g3155.t1 and unitig_8.g3156.t1 were >fourfold trap-enhanced) (SI Appendix, Dataset S8). Indeed, members of these protein families have been identified as candidates for involvement in plant mechanical stimulation or movements (62, 67, 68). Another cell-wall modification-related gene family under this GO term encoded a group of 19 pectin methylesterases and their inhibitors (69) (unitig_899.g15179.t1 and unitig_22.g5384.t1 were 2- to 32-fold trap-enhanced) (SI Appendix, Dataset S8). Interestingly, a second class of chitinases, the class IV enzymes, was also highlighted as an expanded gene family under the GO category “cell wall,” but none of these five genes showed trap-enhanced expression. Class IV chitinases are defense response proteins that represent a second family of chitinase (GH family 19) involved in plant carnivory (70, 71). Finally, four genes encoding β-galactosidases (known from Nepenthes pitcher fluid) (57) appeared under the same GO category but did not have trap-enhanced expression in U. gibba. Another expanded GO category, “lipid catabolic process,” comprised members of various lipase gene families, among them genes encoding patatin-like and GDSL lipases (unitig_736.g22657.t1, unitig_37.g12702.t1, unitig_736.g22658.t1, and unitig_37.g12699.t1 showed 35- to 180-fold trap-enhanced expression) (SI Appendix, Dataset S8). A GDSL lipase likely related to carnivory was identified in the trap fluid of Nepenthes pitchers (57).
Strikingly, the most significantly enriched GO category among all tandemly duplicated genes, “senescence-associated vacuole,” pointed to a specific expansion in one gene family encoding cysteine proteases that had nearly trap-specific expression patterns (SI Appendix, Datasets S2 and S8). Several other significantly enriched GOs are associated with this gene family. Cysteine proteases have been identified as major functional components of Venus flytrap (Dionaea muscipula) digestive fluid (72), reported in three D. muscipula transcriptomes (70, 73, 74), and structurally annotated for both Cape sundew (Drosera capensis) draft genome sequences (75, 76) and D. muscipula (77). We found tandem clusters of homologous protease-encoding genes in the U. gibba genome that had demonstrably undergone tandem duplication both before and after the most recent WGD event in U. gibba’s evolutionary history (Fig. 2). These tandem cysteine protease arrays are assignable to both dominant and recessive subgenomic blocks and are more preserved on the dominant block, where enhanced purifying selection on gene space is expected (SI Appendix, Fig. S13). Genome-wide BLAST search revealed that in general, U. gibba cysteine proteases have become nearly totally restricted to this single, specific subfamily, clearly indicating that diverse, related cysteine proteases known from various other species have become expendable during U. gibba’s genome evolution.
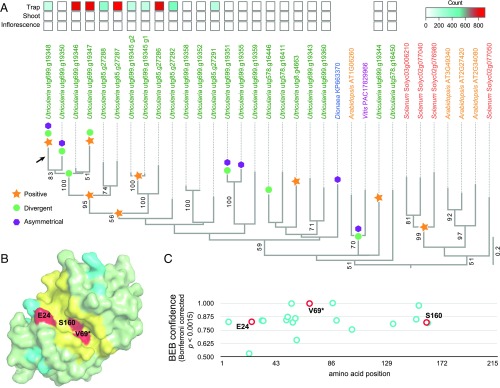
We further examined the cysteine proteases for molecular evolutionary features (SI Appendix, section 6.1), given that gene family members would have diversified in sequence and function to be retained by selection in the dynamically shrinking U. gibba genome. The alternative would be that the observed duplicates were extremely recent and functionally redundant; however, analyses of protein evolution showed this to not be the case, although tandem duplications did continue following the most recent WGD event that yielded arrays on contigs 85 and 699 (Fig. 3A). Instead, we detected evidence for positive selection acting on specific amino acid residues in a lineage leading to several of the U. gibba cysteine protease duplicates (Fig. 3A). When homology modeling these changes onto the D. muscipula cysteine protease structure (77) (Protein Data Bank ID code 5a24), we found some of these amino acids located within the substrate-binding cleft, near residues with known functions in protease activity (Fig. 3 B and C). These substitutions could affect polarity and charge within the cleft, as well as hydrogen bonding between residues essential for catalytic activity and the ligand.
Fig. 3.
Molecular and structural evolutionary analysis of U. gibba cysteine proteases suggests adaptive protein evolution accompanying WGD and tandem duplication events. (A) Best-scoring tree from maximum-likelihood based searches, with bootstrap support (BS) values ≥50 indicated at branches. Symbols on branches indicate significant evidence for positive selection (orange stars), divergent selection (green circles), or asymmetrical sequence evolution (purple hexagons) as determined using PAML (83) (SI Appendix, Dataset S10). The heatmap above the phylogeny shows trap-dominant expression of particular homologs in U. gibba, based on trap, shoot, and inflorescence transcriptome data (47) (SI Appendix, Dataset S2). Note that two tandem duplicates (g1 and g2) were repredicted at locus utg699.g19345. (B) The protein homology surface model for the catalytic domain of utg699.g19348 (encoded by the gene annotated by an arrow in A; based on the Venus flytrap [D. muscipula] enzyme structure (77)) shows that some residues under positive selection lie within or near the substrate-binding cleft. The cleft is depicted in yellow, and amino acid sites identified as under positive selection are indicated in red or cyan. Three (E24, V69, and S160) amino acid sites under positive selection (BEB confidence >0.82, Bonferroni-corrected P < 0.0015) are within five amino acids of known D. muscipula functional residues, where they line the substrate-binding cleft (red). (C) Plot of utg699.g19348 amino acid sites under positive selection, with colors corresponding to specific sites in the surface model (SI Appendix, Fig. S4B).
SHORT VEGETATIVE PHASE (SVP) MADS box gene homologs and homologs of the cuticle biosynthesis gene 3-KETOACYL-COA SYNTHASE 6 (KCS6; highlighted by the significantly enriched GO category among tandems, “wax biosynthetic process”) (SI Appendix, Dataset S8) are two additional cases of tandem duplicate arrays for which some members exhibit trap-enhanced gene expression. Both of these examples have been described previously, based on simple orthogroup clustering methods, as generic gene family expansions derived from unknown duplication mechanisms (11). However, only our highly contiguous PacBio genome provides the structural context necessary to discern that these duplicates are tandems. The SVP-like gene cluster may be involved with flowering phenology, and the KCS6-like genes may be involved in cuticle buttressing of the thin, two-celled trap wall (78–80). The SVP-like genes appear to have diversified anciently, whereas the KCS6-like array occurs in a region of the genome without internal synteny, so it is likely more recent than the last U. gibba WGD. Similar to the cysteine protease clusters, we discovered likely evidence of protein functional divergence in both of these array types (SI Appendix, Dataset S10). Also of note, both the cysteine protease and KCS6-like gene clusters occur within islands of mobile elements (SI Appendix, section 2.5) annotated as large retrotransposon derivatives (LARDs) (81). Serving as a good illustration of the repeat discovery power of PacBio sequencing, ∼47% of the total TE assembly space comprised LARDs, whereas these elements amounted to only ∼14.6% of TEs in the previous short-read assembly (SI Appendix, Dataset S1). We hypothesize that LARDs and other DNA repeats may have facilitated the tandem duplications that gave rise to metabolic gene arrays, as illustrated in the foregoing examples. Finally, we hypothesize that such tandem gene clusters could be coregulated to act in concert, perhaps at particular plant developmental stages or under particular environmental stimuli.
Taken together, our findings regarding the size-limited U. gibba genome highlight the important role that tandemly duplicated genes, under sufficiently substantial purifying selection to survive continual deletion pressure, may play in the individualized adaptive genomic architecture of a plant uniquely adapted for carnivorous morphology and physiology. Although WGD duplicates are not enriched for such niche-specific functions, polyploidy events clearly potentiated the evolutionary influence of preexisting tandem arrays.
Materials and Methods
U. gibba material was sourced from Umécuaro municipality, Michoacán, México, and grown in sterile tissue culture before nuclear DNA extraction. DNA was sequenced using PacBio SMRT technology and assembled using HGAP.3. Genome features were then annotated and analyzed using various bioinformatic tools. GO enrichments were analyzed within different gene pools. For selected gene families, molecular evolutionary pressures were evaluated using codon models and likelihood ratio tests. Detailed information is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Thomas J. Givnish for an insightful additional review. Funding for this work was provided by National Science Foundation Grants 0922742 and 1442190 (to V.A.A.).
Footnotes
The authors declare no conflict of interest.
Data deposition: This Whole Genome Shotgun project has been deposited at the DNA Data Bank of Japan/European Nucleotide Archive/GenBank (accession no. NEEC00000000). The version described in this paper is version NEEC01000000. The assembly and gene models are also available at https://genomevolution.org/coge/GenomeInfo.pl?gid=29027.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702072114/-/DCSupplemental.
References
- 1.Lynch M. The Origins of Genome Architecture. Sinauer Associates; Sunderland, MA: 2007. [Google Scholar]
- 2.Soltis DE, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 3.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 4.Freeling M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- 5.Chae L, Kim T, Nilo-Poyanco R, Rhee SY. Genomic signatures of specialized metabolism in plants. Science. 2014;344:510–513. doi: 10.1126/science.1252076. [DOI] [PubMed] [Google Scholar]
- 6.Myburg AA, et al. The genome of Eucalyptus grandis. Nature. 2014;510:356–362. doi: 10.1038/nature13308. [DOI] [PubMed] [Google Scholar]
- 7.Sollars ES, et al. Genome sequence and genetic diversity of European ash trees. Nature. 2017;541:212–216. doi: 10.1038/nature20786. [DOI] [PubMed] [Google Scholar]
- 8.Albert VA, et al. Amborella Genome Project The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 9.Soltis PS, Soltis DE. Ancient WGD events as drivers of key innovations in angiosperms. Curr Opin Plant Biol. 2016;30:159–165. doi: 10.1016/j.pbi.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Ibarra-Laclette E, et al. Architecture and evolution of a minute plant genome. Nature. 2013;498:94–98. doi: 10.1038/nature12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carretero-Paulet L, et al. High gene family turnover rates and gene space adaptation in the compact genome of the carnivorous plant Utricularia gibba. Mol Biol Evol. 2015;32:1284–1295. doi: 10.1093/molbev/msv020. [DOI] [PubMed] [Google Scholar]
- 12.Veleba A, et al. Genome size and genomic GC content evolution in the miniature genome-sized family Lentibulariaceae. New Phytol. 2014;203:22–28. doi: 10.1111/nph.12790. [DOI] [PubMed] [Google Scholar]
- 13.Chin C-S, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 14.Fulnecková J, et al. A broad phylogenetic survey unveils the diversity and evolution of telomeres in eukaryotes. Genome Biol Evol. 2013;5:468–483. doi: 10.1093/gbe/evt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran TD, et al. Centromere and telomere sequence alterations reflect the rapid genome evolution within the carnivorous plant genus Genlisea. Plant J. 2015;84:1087–1099. doi: 10.1111/tpj.13058. [DOI] [PubMed] [Google Scholar]
- 16.Melters DP, et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013;14:R10. doi: 10.1186/gb-2013-14-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wade CM, et al. Broad Institute Genome Sequencing Platform Broad Institute Whole Genome Assembly Team Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR. Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci USA. 2005;102:9842–9847. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, et al. Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma. 2008;117:445–456. doi: 10.1007/s00412-008-0161-9. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Z, et al. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell. 2002;14:1691–1704. doi: 10.1105/tpc.003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagaki K, et al. Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol Biol Evol. 2005;22:845–855. doi: 10.1093/molbev/msi069. [DOI] [PubMed] [Google Scholar]
- 23.Zhong CX, et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell. 2002;14:2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudakova S, et al. Sequence organization of barley centromeres. Nucleic Acids Res. 2001;29:5029–5035. doi: 10.1093/nar/29.24.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorinšek B, Gubenšek F, Kordiš D. Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol. 2004;21:781–798. doi: 10.1093/molbev/msh057. [DOI] [PubMed] [Google Scholar]
- 26.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 27.Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008;18:359–369. doi: 10.1101/gr.7146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyons E, et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008;148:1772–1781. doi: 10.1104/pp.108.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang H, et al. SynFind: Compiling syntenic regions across any set of genomes on demand. Genome Biol Evol. 2015;7:3286–3298. doi: 10.1093/gbe/evv219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang H, et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sankoff D, Zheng C, Zhu Q. The collapse of gene complement following whole genome duplication. BMC Genomics. 2010;11:313. doi: 10.1186/1471-2164-11-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas BC, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA. 2011;108:4069–4074. doi: 10.1073/pnas.1101368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnable JC, Wang X, Pires JC, Freeling M. Escape from preferential retention following repeated whole genome duplications in plants. Front Plant Sci. 2012;3:94. doi: 10.3389/fpls.2012.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng F, et al. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One. 2012;7:e36442. doi: 10.1371/journal.pone.0036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garsmeur O, et al. Two evolutionarily distinct classes of paleopolyploidy. Mol Biol Evol. 2014;31:448–454. doi: 10.1093/molbev/mst230. [DOI] [PubMed] [Google Scholar]
- 38.Joyce BL, et al. FractBias: A graphical tool for assessing fractionation bias following polyploidy. Bioinformatics. 2017;33:552–554. doi: 10.1093/bioinformatics/btw666. [DOI] [PubMed] [Google Scholar]
- 39.Soltis PS. Hybridization, speciation and novelty. J Evol Biol. 2013;26:291–293. doi: 10.1111/jeb.12095. [DOI] [PubMed] [Google Scholar]
- 40.Kameyama Y, Toyama M, Ohara M. Hybrid origins and F1 dominance in the free-floating, sterile bladderwort, Utricularia australis f. australis (Lentibulariaceae) Am J Bot. 2005;92:469–476. doi: 10.3732/ajb.92.3.469. [DOI] [PubMed] [Google Scholar]
- 41.Chormanski TA, Richards JH. An architectural model for the bladderwort Utricularia gibba (Lentibulariaceae) J Torrey Bot Soc. 2012;139:137–148. [Google Scholar]
- 42.Lampert KP, Schartl M. The origin and evolution of a unisexual hybrid: Poecilia formosa. Philos Trans R Soc Lond B Biol Sci. 2008;363:2901–2909. doi: 10.1098/rstb.2008.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang H, et al. Screening synteny blocks in pairwise genome comparisons through integer programming. BMC Bioinformatics. 2011;12:102. doi: 10.1186/1471-2105-12-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lubkowitz M. The OPT family functions in long-distance peptide and metal transport in plants. In: Setlow JK, editor. Genetic Engineering: Principles and Methods. Vol 27. Springer Science and Business Media; Berlin: 2006. pp. 35–55. [DOI] [PubMed] [Google Scholar]
- 47.Ibarra-Laclette E, et al. Transcriptomics and molecular evolutionary rate analysis of the bladderwort (Utricularia), a carnivorous plant with a minimal genome. BMC Plant Biol. 2011;11:101. doi: 10.1186/1471-2229-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulze W, Frommer WB, Ward JM. Transporters for ammonium, amino acids and peptides are expressed in pitchers of the carnivorous plant Nepenthes. Plant J. 1999;17:637–646. doi: 10.1046/j.1365-313x.1999.00414.x. [DOI] [PubMed] [Google Scholar]
- 49.Adlassnig W, et al. Endocytotic uptake of nutrients in carnivorous plants. Plant J. 2012;71:303–313. doi: 10.1111/j.1365-313X.2012.04997.x. [DOI] [PubMed] [Google Scholar]
- 50.Léran S, et al. AtNPF5.5, a nitrate transporter affecting nitrogen accumulation in Arabidopsis embryo. Sci Rep. 2015;5:7962. doi: 10.1038/srep07962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brownlee C. Carnivorous plants: Trapping, digesting and absorbing all in one. Curr Biol. 2013;23:R714–R716. doi: 10.1016/j.cub.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 52.Stigter KA, Plaxton WC. Molecular mechanisms of phosphorus metabolism and transport during leaf senescence. Plants (Basel) 2015;4:773–798. doi: 10.3390/plants4040773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nussaume L, et al. Phosphate import in plants: Focus on the PHT1 transporters. Front Plant Sci. 2011;2:83. doi: 10.3389/fpls.2011.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rottloff S, et al. Functional characterization of a class III acid endochitinase from the traps of the carnivorous pitcher plant genus, Nepenthes. J Exp Bot. 2011;62:4639–4647. doi: 10.1093/jxb/err173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goujon T, et al. AtBXL1, a novel higher plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development. Plant J. 2003;33:677–690. doi: 10.1046/j.1365-313x.2003.01654.x. [DOI] [PubMed] [Google Scholar]
- 56.Hatano N, Hamada T. Proteome analysis of pitcher fluid of the carnivorous plant Nepenthes alata. J Proteome Res. 2008;7:809–816. doi: 10.1021/pr700566d. [DOI] [PubMed] [Google Scholar]
- 57.Rottloff S, et al. Proteome analysis of digestive fluids in Nepenthes pitchers. Ann Bot (Lond) 2016;117:479–495. doi: 10.1093/aob/mcw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erni P, Varagnat M, Clasen C, Crest J, McKinley GH. Microrheometry of sub-nanolitre biopolymer samples: Non-Newtonian flow phenomena of carnivorous plant mucilage. Soft Matter. 2011;7(22):10889–10898. [Google Scholar]
- 59.Vintéjoux C, Shoar-Ghafari A. Sécrétion de mucilages par une plante aquatique. Acta Bot Gallica. 1997;144(3):347–351. [Google Scholar]
- 60.Poppinga S, Weisskopf C, Westermeier AS, Masselter T, Speck T. Fastest predators in the plant kingdom: Functional morphology and biomechanics of suction traps found in the largest genus of carnivorous plants. AoB Plants. 2015;8:plv140. doi: 10.1093/aobpla/plv140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skotheim JM, Mahadevan L. Physical limits and design principles for plant and fungal movements. Science. 2005;308:1308–1310. doi: 10.1126/science.1107976. [DOI] [PubMed] [Google Scholar]
- 62.Forterre Y. Slow, fast and furious: Understanding the physics of plant movements. J Exp Bot. 2013;64:4745–4760. doi: 10.1093/jxb/ert230. [DOI] [PubMed] [Google Scholar]
- 63.Llorens C, Argentina M, Bouret Y, Marmottant P, Vincent O. A dynamical model for the Utricularia trap. J R Soc Interface. 2012;9:3129–3139. doi: 10.1098/rsif.2012.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Jones L, McQueen-Mason S. Expansins and cell growth. Curr Opin Plant Biol. 2003;6:603–610. doi: 10.1016/j.pbi.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 65.Campbell P, Braam J. Xyloglucan endotransglycosylases: Diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 1999;4:361–366. doi: 10.1016/s1360-1385(99)01468-5. [DOI] [PubMed] [Google Scholar]
- 66.Yadav S, Yadav PK, Yadav D, Yadav KDS. Pectin lyase: A review. Process Biochem. 2009;44:1–10. [Google Scholar]
- 67.Humphrey TV, Bonetta DT, Goring DR. Sentinels at the wall: Cell wall receptors and sensors. New Phytol. 2007;176:7–21. doi: 10.1111/j.1469-8137.2007.02192.x. [DOI] [PubMed] [Google Scholar]
- 68.Zonia L, Munnik T. Life under pressure: Hydrostatic pressure in cell growth and function. Trends Plant Sci. 2007;12:90–97. doi: 10.1016/j.tplants.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 69.Micheli F. Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001;6:414–419. doi: 10.1016/s1360-1385(01)02045-3. [DOI] [PubMed] [Google Scholar]
- 70.Schulze WX, et al. The protein composition of the digestive fluid from the Venus flytrap sheds light on prey digestion mechanisms. Mol Cell Proteomics. 2012;11:1306–1319. doi: 10.1074/mcp.M112.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Renner T, Specht CD. Inside the trap: Gland morphologies, digestive enzymes, and the evolution of plant carnivory in the Caryophyllales. Curr Opin Plant Biol. 2013;16:436–442. doi: 10.1016/j.pbi.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Libiaková M, Floková K, Novák O, Slováková L, Pavlovič A. Abundance of cysteine endopeptidase dionain in digestive fluid of Venus flytrap (Dionaea muscipula Ellis) is regulated by different stimuli from prey through jasmonates. PLoS One. 2014;9:e104424–e104424. doi: 10.1371/journal.pone.0104424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jensen MK, et al. Transcriptome and genome size analysis of the Venus flytrap. PLoS One. 2015;10:e0123887. doi: 10.1371/journal.pone.0123887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bemm F, et al. Venus flytrap carnivorous lifestyle builds on herbivore defense strategies. Genome Res. 2016;26:812–825. doi: 10.1101/gr.202200.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Butts CT, Bierma JC, Martin RW. Novel proteases from the genome of the carnivorous plant Drosera capensis: Structural prediction and comparative analysis. Proteins. 2016;84:1517–1533. doi: 10.1002/prot.25095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Butts CT, et al. Sequence comparison, molecular modeling, and network analysis predict structural diversity in cysteine proteases from the Cape sundew, Drosera capensis. Comput Struct Biotechnol J. 2016;14:271–282. doi: 10.1016/j.csbj.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Risør MW, et al. Enzymatic and structural characterization of the major endopeptidase in the Venus flytrap digestion fluid. J Biol Chem. 2015;291:2271–87. doi: 10.1074/jbc.M115.672550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mateos JL, et al. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015;16:31. doi: 10.1186/s13059-015-0597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gregis V, et al. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 2013;14:R56. doi: 10.1186/gb-2013-14-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Todd J, Post-Beittenmiller D, Jaworski JG. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999;17:119–130. doi: 10.1046/j.1365-313x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- 81.Havecker ER, Gao X, Voytas DF. The diversity of LTR retrotransposons. Genome Biol. 2004;5:225. doi: 10.1186/gb-2004-5-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics. 2003 doi: 10.1002/0471250953.bi1003s00. Chapter 10: Unit 10.13. [DOI] [PubMed] [Google Scholar]
- 83.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.