During DNA replication, the major DNA replicative polymerases Polδ and Polε introduce noncomplementary nucleotides approximately once in every 100,000 polymerization events. Proofreading properties of these polymerases improve their fidelity by 10- to 100-fold, and postreplicative DNA mismatch repair (MMR), which acts as a spell checker to remove mismatches that escape polymerase proofreading, improves this fidelity even further, resulting in mutation rates as low as 2 × 10−10 substitutions per base per cell division (1). Mutations in genes encoding MMR proteins and DNA polymerase proofreading activities are linked to elevated mutation rates and diseases such as cancer, and these rates are synergistically increased in double mutants (2, 3). In PNAS, Schmidt et al. (4) describe work in which they screened for defects in genome stability in genetic backgrounds where processes that limit mutagenesis were compromised. Their work provides new insights for how cellular metabolism and nucleotide pool homeostasis interact to avoid mutation and maintain genome stability. They also provide evidence that DNA polymerase fidelity and MMR functions are sufficiently robust to compensate at least partially for defects in cellular metabolism (Fig. 1).
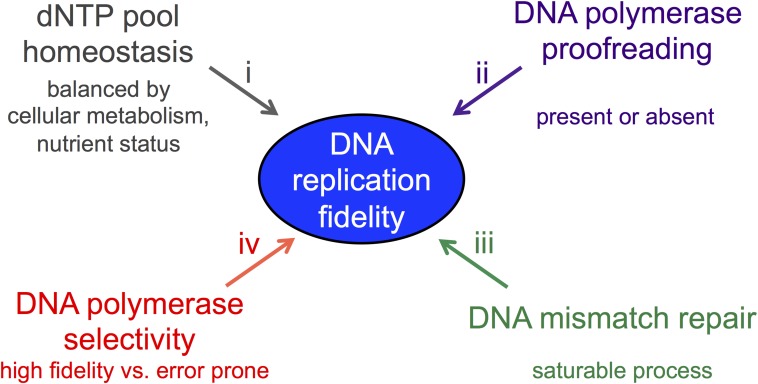
Fig. 1.
Removing the buffering capabilities of DNA replication and DNA MMR uncovers new factors that contribute to nucleotide pool homeostasis. Multiple mechanisms contribute to the overall fidelity of DNA replication. For example, each of the four mechanisms outlined in this figure can compensate for defects in the others. (i) A homeostasis mechanism responds to cellular metabolism to regulate the biosynthesis of dNTP and dNTP precursors to maintain a balanced concentration of dNTPs. (ii) DNA polymerase proofreading activities: During eukaryotic DNA replication, the major replicative polymerases Polε and Polδ both contain proofreading activities that enable them to excise misincorporation errors. However, Polα, which initiates DNA synthesis at origins and at Okazaki fragments, does not contain such a function. (iii) DNA MMR: MMR identifies mismatches resulting primarily from DNA replication errors. Nascent DNA is excised by the MMR machinery and is subsequently repaired by DNA synthesis. The MMR protein machinery can become saturated when there is an abundance of replication errors, allowing mismatches to escape repair. (iv) DNA polymerase selectivity: Polε and Polδ are high-fidelity replicative polymerases but can differ by ∼10-fold in their ability to incorporate the correct base (14). In contrast, translesion synthesis DNA polymerases are highly error-prone. When multiple mechanisms are defective, buffering fails and mutation rates increase significantly.
Schmidt et al. (4) performed a genome-wide screen in the baker’s yeast Saccharomyces cerevisiae. This screen involved analyzing strains bearing knockout mutations in nonessential genes in a genetic background containing DNA polymerase active-site mutations (pol1-L868M, pol2-M644G, and pol3-L612M). They then assayed for increases in mutation rate using a reporter in which loss-of-function mutations in the CAN1 gene confer resistance to the arginine analog canavanine and a lys2-A10 insertion construct that primarily detects frameshift mutations to yield Lys+ revertants. Previous screens of this type, performed in functional DNA polymerase backgrounds, identified genome stability factors that are likely to be important targets in cancer (5–7). Schmidt et al. (4) used this approach to identify new targets whose effects on mutation rate are normally buffered by the fidelity of DNA polymerases. Their approach identified knockout mutations in the EXO1, RRM3, GLN3, URA7, and SHM2 genes. Interestingly, gln3, ura7, and shm2 knockout alleles were not previously linked to mutation avoidance and did not confer an increase in mutation rate independent of the polymerase mutations. Furthermore, for four of the five knockouts (exo1 knockout was the exception), the mutator phenotype seen in the DNA polymerase active-site mutant backgrounds was detected only in the canavanine resistance assay. Because elevated mutation rates were not observed in the Lys+ reversion assay, Schmidt et al. (4) surmised that the mutator phenotypes seen in polymerase mutants lacking RRM3, GLN3, URA7, and SHM2 were likely the result of base substitutions; they confirmed this hypothesis by analyzing mutation rates in MMR-deficient backgrounds that have strong defects in the repair of base–base mismatches (msh6 knockout). This work also encouraged them to understand the roles of the GLN3, URA7, and SHM2 gene products better. Here, we will focus on the roles of the Gln3 and Ura7 proteins.
DNA replication and repair require the presence of precisely balanced cellular nucleotide (NTP) and deoxynucleotide (dNTP) pools; in fact, changes in the concentration and balance of dNTPs affect mutation rates in vivo (e.g., ref. 8). Factors that act upstream of dNTP biosynthesis, particularly in cellular metabolism, have been linked to maintaining genome integrity (9). Gln3 protein is a transcription factor that activates genes dependent on nitrogen catabolite repression in glutamine-limiting conditions. Interestingly, glutamine is a precursor for nucleotide base biosynthesis. Ura7, the major CTP synthase isozyme in baker’s yeast, converts UTP to CTP, which is primarily a precursor for the synthesis of dCTP. This information led Schmidt et al. (4) to hypothesize that the Gln3 and Ura7 proteins contribute to mutational avoidance in early steps of nucleotide metabolism.
Cells subjected to genotoxic stress activate a DNA damage response that results in the phosphorylation of the cell cycle checkpoint protein Dun1, and subsequent activation of ribonucleotide reductase, an enzyme that catalyzes the formation of dNTPs from NTPs. The dun1 knockout mutants have reduced dNTP pools (10). Schmidt et al. (4) observed that the dun1 knockout mutation suppressed the elevated mutation rate exhibited by gln3 and ura7 knockout mutants in a replication fidelity-defective background (pol3-L612M), providing an important clue that the gln3 and ura7 knockout mutations altered cellular dNTP pools. One of their most exciting observations was that supplementing yeast growth media with glutamine partially suppressed the mutator phenotype of gln3 mutants in replication fidelity-defective backgrounds (pol3-L612M, pol2-04, or exo1 knockout), suggesting that metabolic defects seen in gln3 mutants can be partially corrected. This observation is also interesting because it provides a potential avenue for future therapeutic research. Schmidt et al. (4) then measured nucleotide concentrations in gln3 and ura7 knockout strains to test their hypothesis that the genome instability defects seen in these strains were the result of imbalances in nucleotide pools. They observed reduced CTP and dCTP levels and elevated dATP, dGTP, and dTTP levels in gln3 and ura7 knockout strains. These data were further supported by their finding that primarily G:C-to-A:T transitions were observed at the CAN1 locus in gln3 and ura7 mutants analyzed in a genetic background defective in the repair of base–base mismatches (msh6 knockout).
Glutamine is used as a cellular source of carbon and nitrogen, which are used for biosynthetic and energetic processes. It is also an activator of regulatory proteins involved in the uptake of other amino acids and factors involved in cell growth. Cancer cells can take advantage of increased glutamine concentrations to synthesize nitrogenous compounds to support tumor growth. In fact, some cancer lines show increased uptake of glutamine and exhibit glutamine addiction, a behavior that could serve as an Achilles’ heel for therapeutic intervention (11). In baker’s yeast, the Gln3 protein is activated at low glutamine concentrations to mediate glutamine synthesis, and ultimately stimulates the production of nucleotides, particularly CTP and dCTP. The finding by Schmidt et al. (4) of a link between nutrient deprivation and high-fidelity DNA synthesis is reminiscent of work showing that meiotic gene conversion, a DNA repair event initiated by programmed double-strand breaks in DNA, can be regulated by metabolic states (12). Abdullah and Borts (12) showed that meiotic gene conversion frequencies at a specific locus (HIS4) could be modulated by altering nutritional states through the disruption of genes involved in leucine, lysine, and adenine biosynthesis (LEU2, LYS2, and ADE1) or by a gene encoding a transcription factor (GCN4) that regulates HIS4 expression. Their data suggest that “both direct (gene disruption, overexpression) and indirect (metabolic state of the cell) modulation of Gcn4p levels affect frequencies of gene conversion…at susceptible sites” (12). Together, these observations suggest that additional links will be discovered between nutritional states and DNA metabolism.
Interestingly, Schmidt et al. (4) found that null mutations in GLN3, SHM2, URA7, and EXO1 conferred effects on mutation rate primarily in conjunction with defects in DNA polymerases thought to act primarily in lagging strand synthesis (Polα and Polδ). They provide many possible explanations for why this finding could be, including that DNA replication on the leading strand may be more faithful; active site mutations could affect the three polymerases differently with respect to their fidelities; and the possibility that the canonical interpretation of leading strand synthesis being performed by Polε and lagging strand synthesis being performed by Polδ may not be so strict, which has been suggested previously (13). These data add to the growing body of evidence in the field that there are differences in the error rate and repair of the leading and lagging strands (1), and provide a motivation for future studies.
By taking advantage of synergistic defects that occur when polymerase fidelity or MMR is compromised in conjunction with other factors, Schmidt et al. (4) identify new factors that contribute to nucleotide homeostasis, and advance our understanding of how polymerases and MMR shield against the formation of mutations. Their work also suggests a new approach to identify factors that impact mutation rates, and how the results obtained from these efforts could possibly be used to manipulate nucleotide pools for therapeutic purposes.
Acknowledgments
C.M.M. and E.A. are supported by the National Institute of General Medical Sciences of the NIH (Grant GM53085 to E.A. and Grant F32 GM112435 to C.M.M.). The content of this review is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no conflict of interest.
See companion article on page E4442.
References
- 1.Kunkel TA, Erie DA. Eukaryotic mismatch repair in relation to DNA replication. Annu Rev Genet. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loeb LA, Monnat RJ., Jr DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt TT, et al. Alterations in cellular metabolism triggered by URA7 or GLN3 inactivation cause imbalanced dNTP pools and increased mutagenesis. Proc Natl Acad Sci USA. 2017;114:E4442–E4451. doi: 10.1073/pnas.1618714114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang M-E, Rio A-G, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci USA. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith S, et al. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 8.Chabes A, et al. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 9.MacFarlane AJ, et al. Nuclear localization of de novo thymidylate biosynthesis pathway is required to prevent uracil accumulation in DNA. J Biol Chem. 2011;286:44015–44022. doi: 10.1074/jbc.M111.307629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasullo M, Tsaponina O, Sun M, Chabes A. Elevated dNTP levels suppress hyper-recombination in Saccharomyces cerevisiae S-phase checkpoint mutants. Nucleic Acids Res. 2010;38:1195–1203. doi: 10.1093/nar/gkp1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wise DR, Thompson CB. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdullah MF, Borts RH. Meiotic recombination frequencies are affected by nutritional states in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2001;98:14524–14529. doi: 10.1073/pnas.201529598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson RE, Klassen R, Prakash L, Prakash S. A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St Charles JA, Liberti SE, Williams JS, Lujan SA, Kunkel TA. Quantifying the contributions of base selectivity, proofreading and mismatch repair to nuclear DNA replication in Saccharomyces cerevisiae. DNA Repair (Amst) 2015;31:41–51. doi: 10.1016/j.dnarep.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]