Abstract
Numerous deaths of koi and common carp (Cyprinus carpio) were observed on many farms throughout Israel, resulting in severe financial losses. The lethal viral disease observed is highly contagious and extremely virulent, but morbidity and mortality are restricted to koi and common carp populations. Diseased fish exhibit fatigue and gasping movements in shallow water. Infected fish had interstitial nephritis and gill necrosis as well as petechial hemorrhages in the liver and other symptoms that were not consistent with viral disease, suggesting a secondary infection. Here we report the isolation of carp nephritis and gill necrosis virus (CNGV), which is the etiologic agent of this disease. The virus propagates and induces severe cytopathic effects by 5 days postinfection in fresh koi or carp fin cell cultures (KFC and CFC, respectively), but not in epithelioma papillosum cyprini cells. The virus harvested from KFC cultures induced the same clinical signs, with a mortality of 75 to 95%, upon inoculation into naive koi and common carp. Using PCR, we provide final proof that the isolated virus is indeed the etiologic agent of food and ornamental carp mortalities in fish husbandry. Electron microscopy revealed viral cores with icosahedral morphology of 100 to 110 nm that resembled herpesviruses. Electron micrographs of purified pelleted CNGV sections, together with viral sensitivities to ether and Triton X-100, suggested that it is an enveloped virus. However, the genome of the isolated virus is a double-stranded DNA (dsDNA) molecule of 270 to 290 kbp, which is larger than known herpesviruses. The viral DNA seems highly divergent and bears only small fragments (16 to 45 bp) that are similar to the genomes of several DNA viruses. Nevertheless, amino acid sequences encoded by CNGV DNA fragments bear similarities primarily to members of the Poxviridae and Herpesviridae and to other large dsDNA viruses. We suggest, therefore, that the etiologic agent of this disease may represent an as yet unclassified virus species that is endemic in C. carpio (carp).
During the spring of 1998, numerous deaths of Cyprinus carpio (carp) fish were observed in several food and ornamental-trade fish farms along Israel's coastal region (3). Since then, there has been a continual threat of this disease in most Israeli fisheries. The disease has now spread to numerous farms throughout the country, causing severe financial losses. Mortality rates have consistently been above 80% for all ponds (23, 26). Although the disease is highly contagious and extremely virulent, morbidity and mortality are restricted to koi and common carp populations. Several closely related species, including other cyprinids such as goldfish (Carassius auratus), were found to be completely asymptomatic, even after long-term cohabitation with diseased fish sharing the same tank (23). The disease frequently appears during the transient stages of the spring and fall. Thus, it seems that the agent causing the disease has quite a narrow host range and that its effect is restricted to specific temperature conditions of 18 to 28°C (11, 23).
The intensive farming of koi and common carp results in frequent distributions of viral diseases in these populations. A corona-like virus (19), rhabdovirus (17), iridovirus (30), and herpesviruses (5, 15, 16, 28) have been described as the causes of severe diseases in cyprinids. A herpesvirus was detected in papillomatous skin growths on koi carp (C. carpio) in North America (5, 15). This carp herpesvirus is consistent with the herpesvirus cyprini, which is present in koi carp populations in Japan (28, 29). A newly emerging disease that is responsible for the mass mortality of koi and common carp has been observed since 1998 in Israel, the United States, and several European and Asian countries and has resulted in severe financial losses (3, 10, 11, 13, 16, 22, 23). The virus causes nephritis as early as 2 days postinfection which increases up to 10 days postinfection. Infected fish have a severe gill disease that is evidenced by a loss of villi and inflammation of the gill rakers (24). Previously, it was suggested that this disease is caused by a koi herpesvirus (KHV), but so far the virus has only been partly characterized (3, 10, 11, 13, 16, 24, 26).
In this report, we describe the isolation of a carp nephritis and gill necrosis virus (CNGV), the causative agent of the lethal disease in carp in Israel. Cohabitation experiments demonstrated that the disease is contagious under controlled conditions and that mortality occurs at 8 to 18 days postchallenge of carp. The virus was propagated in koi and carp fin cells (KFC and CFC, respectively), inducing a cytopathic effect at 4 to 6 days postinfection of cultures, and virus harvested in culture induced an identical disease in carp following intraperitoneal injection or immersion. The virus was released into the culture medium during the appearance of a cytopathic effect, but a significant amount remained associated with the cell. Electron microscopy revealed enveloped particles bearing cores with electron-dense regions. The viral core has an icosahedral morphology of 100 to 110 nm that resembles a herpesvirus. However, the genome of the isolated virus is a double-stranded DNA (dsDNA) of ca. 277 kbp. So far, we have found that the viral DNA bears only small fragments (16 to 45 bp) that are similar to the genomes of several DNA viruses. However, amino acid sequences encoded by CNGV DNA fragments bear similarities primarily to members of the Poxviridae and Herpesviridae and to other large dsDNA viruses. Thus, the genome of the etiologic agent of the carp disease is larger than those of the other members of the Herpesviridae and contains highly divergent DNA sequences which encode polypeptides resembling those of other large dsDNA viruses. Although CNGV is similar to the previously described KHV (13, 16) and even though the morphology of CNGV is similar in some aspects to that of Herpesviridae, for the time being we prefer to designate it CNGV, according to its pathological manifestation (24) rather than its phylogenetic classification, which has yet to be determined.
MATERIALS AND METHODS
Fish.
Koi or common carp with an average weight of 42 g were grown in 500-liter tanks with the water temperature kept at 22 to 24°C, and they received fresh well water at 0.9 liter/min.
Cell cultures.
Koi fin cell (KFC) cultures were prepared as previously described by Hasegawa et al. (14) and Neukirch et al. (21). Briefly, the caudal fins of 50-g anesthetized koi or common carp were removed, bathed in a 1% sodium hypochloride solution for 1 min, and then rinsed in 70% ethanol for a few seconds. The fins were then washed three times for 0.5 min each in phosphate-buffered saline (PBS) containing penicillin and streptomycin. The fins were transferred to petri dishes and extensively minced with scissors, and semidry small tissue pieces of approximately 1 mm3 were placed in dry 50-ml culture flasks (Nunc, Roskilde, Denmark). After 60 min of incubation at room temperature, the clumps adhering to the flasks were covered with culture medium containing 60% Dulbecco's modified Eagle's medium, 20% Leibovitz (L-15) medium, 10% fetal calf serum (Biological Industries, Kibbutz Beit Haemek, Israel), and 10% tryptose phosphate (Difco) and supplemented with 1% HEPES and antibiotics in sealed flasks. During incubation for 10 to 14 days at 22°C, cells grew out from the tissue to form a monolayer around each clump. The monolayer cultures were trypsinized and transferred into new flasks with fresh medium. The fin clumps could have been transferred to new flasks to form a new monolayer culture of primary cells.
EPC cultures.
Epithelioma papulosum cyprini (EPC) cells (9) were kindly supplied by A. Eldar and H. Bercovier (The Hebrew University-Hadassah Medical School) and grown in the medium mixture described above.
Viruses.
Vaccinia virus (WR strain) and herpes simplex virus type 1 (HSV-1) (kindly supplied by E. Katz and Y. Becker, The Hebrew University-Hadassah Medical School) were propagated in CV-1 cells.
Purification of virus from culture medium.
The medium harvested from CNGV-infected KFC or CFC or from HSV-1- or vaccinia virus-infected CV-1 cells was cleared of cells and cell debris by centrifugation for 10 min at 7,000 × g. The virus was then pelleted by centrifugation in a Beckman Ti-60 rotor for 50 min at 100,000 × g. Pellets were suspended in PBS, loaded on a 15 to 65% (wt/vol) sucrose gradient prepared in PBS, and centrifuged for 60 min at 110,000 × g in a Beckman SW28 rotor. Bands were visualized, aspirated from tubes, diluted 10-fold in PBS, and repelleted. The pellets were suspended in PBS and frozen at −70°C until further investigation.
Electron microscopy analysis.
Purified virus preparations were negatively stained with 2% phosphotungstate. Grids were examined with a Philips 120 electron microscope operating at 80 kV.
For examinations of CNGV by transmission electron microscopy, purified virus pellets were fixed with 3% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.2) for 2 h and then rinsed five times in phosphate buffer (pH 7.2). The virus pellets were postfixed in 1% OsO4 in phosphate buffer and dehydrated with increasing concentrations of ethanol. The pellets were then washed twice with 100% propylene oxide and treated with propylene oxide-Epon (3:1) for 30 min, followed by propylene oxide-Epon (1:1) for 15 min. Finally, the pellets were embedded in 100% Epon and left overnight. Thin sections (70 to 90 nm) were placed on Formvar-coated copper grids and stained with uranyl acetate followed by lead citrate. The sections were examined with a calibrated Philips 420 electron microscope operating at 80 kV.
Viral DNA purification, plasmid construction, and DNA sequencing.
Purified viral pellets were suspended in TNE buffer (10 mM Tris [pH 7.8], 100 mM NaCl, and 1 mM EDTA) containing 0.5% sodium dodecyl sulfate (SDS). The virus preparations were treated with 50 μg of proteinase K (Boehringer)/ml for 2 to 3 h in the presence of 0.5% SDS, 0.01 M Tris-Cl (pH 7.8), and 0.005 M EDTA at 50°C. Viral DNAs were extracted with phenol and precipitated with ethanol, and DNA pellets were suspended in TNE (27).
CNGV DNA was cleaved with BamHI and EcoRI, and the fragments were cloned into the Bluescript II SK(−) plasmid (Stratagene) (1). Sequencing of the inserted viral DNA fragments was performed for both strands by use of the plasmid-derived T3 and T7 primers. Sequencing was performed by the dideoxynucleotide terminator cycle sequencing method by use of a Prism BigDye Ready Reaction Terminator cycle sequencing kit (PE Applied Biosystems). Reaction and cycling conditions were chosen according to the manufacturer's protocol. Sequencing reactions were run on an ABI 3700 DNA analyzer (PE Applied Biosystems) and subsequently analyzed with the standard BLAST program (2, 31) (sequencing was performed at the Center for Genomic Technologies, Hebrew University, Jerusalem, Israel). Internal primers from each sequenced clone and purified viral DNA templates were used to verify that the inserted fragments were derived from the CNGV genome.
PCR analysis.
Cellular DNA preparations were extracted from cells with phenol and were precipitated with ethanol. DNA pellets were suspended in Tris-EDTA buffer, pH 7.4, as described above (27).
The following primers, derived from viral DNAs, were used for the amplification of viral DNA fragments by PCR: AP1 (forward), 5′-CCCATGAGGCTGAGGAACGCCG-3′; AP2 (reverse), 5′-GCACCCCCGTGATGGTCTTGC-3′; AP3 (reverse), 5′-GGAACATGAGGGCCAGTATGTGG-3′; NH1 (forward), 5′-GGATCCAGACGGTGACGGTCACCC-3′; NH2 (reverse), 5′-GCCCAGAGTCACTTCCAGCTTCG-3′; S-1 (forward), 5′-TTCGTTTTGCACATTGTGGC-3′; and S-2 (reverse), 5′-AGACACTGAGAGCGTCATCGG-3′. AP and NH primers were derived from clone A and clone E, respectively (see Fig. 5). S primers were generated from KHV sequences submitted to GenBank by Kurita et al. (accession no. AB178324) (17a).
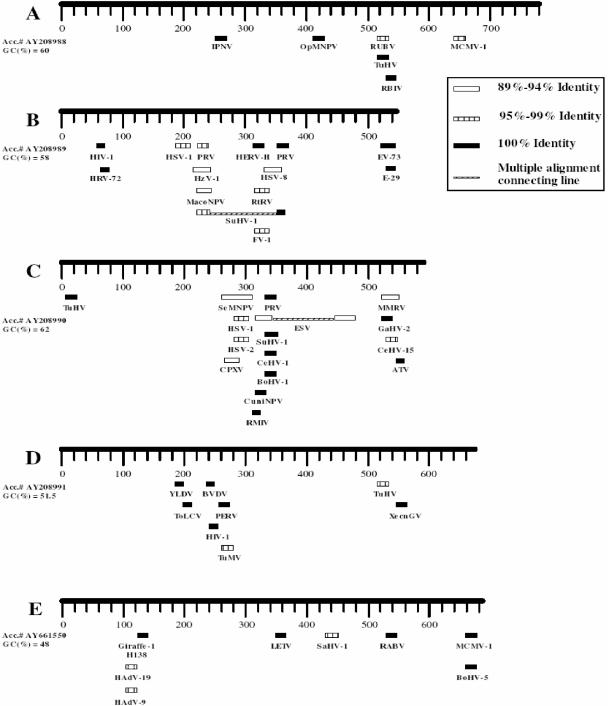
FIG. 5.
Sequences in CNGV genome with similarities to other viral sequences. The viral DNA clones (A through E) were sequenced and analyzed with the BLAST program of PubMed (National Institutes of Health). Herpes simplex virus types 1, 2, and 8 (HSV-1, HSV-2 and HSV-8, respectively); pseudorabies virus (PRV); gallid herpesvirus (GaHV); Macaca mulata rhadinovirus (MMRV); Tupaia herpesvirus (TuHV); mouse cytomegalovirus (MCMV-1); suidherpesvirus 1 (SuHV-1); bovine herpesvirus (BoHV); squirrel monkey herpesvirus (SaHV); lung-eye-trachea disease-associated herpesvirus (LETV); and cercopithecine herpesvirus 1 (CeHV-1) and cercopithicine herpesvirus 15 (CeHV-15) all belong to the herpesvirus family. Spodoptera exigua nucleopolyhedrovirus (SeMNPV), Mamestra configurata nucleopolyhedrovirus (MacoNPV), Xestia c-nigrum granulovirus (XecnGV), Orgyia pseudosugata nucleopolyhedrovirus (OpMNPV), and Culex nigripapus baculovirus (CuniNPV) are members of the baculovirus family. Ectocarpus siliculosus virus (ESV) is an algal virus. Yaba-like disease virus (YLDV) and cowpox virus (CPXV) are poxviruses. Rock bream iridovirus (RBIV), Ambystoma tigrinum stebbensi virus (ATV), frog virus 1 (FV-1), iridovirus RMIV, and Rana Tigrina Rana virus (RtRV) belong to the iridovirus family. Human immunodeficiency virus type 1 (HIV-1), human endogenous virus (HERV-H), and porcine endogenous retrovirus (PERV) are retroviruses. Human adenovirus (HAdV) is an adenovirus. Human rhinovirus 72 (HRV-72), human enterovirus (EV-73), and human echovirus 29 (E-29) are picornaviruses. Bovine viral diarrhea virus (BVDV) and pestivirus giraffe (Giraffe-1 H138) are flaviviruses. Infectious pancreatic necrosis virus (IPNV) is a birnavirus. Rubella virus (RUBV) is a togavirus. Rabies virus (RABV) is a rhabdovirus. Tomato leaf curl virus (ToLCV) is a geminivirus. Turnip mosaic virus (TuMV) is a potyvirus, and Heliothis zea virus 1 (HzV-1) is an unassigned virus.
DNA preparations were used as templates in reaction mixtures containing a 0.5 μM concentration (each) of forward and reverse primers, 4 mM MgCl2, a 0.1 mM concentration of each deoxynucleoside triphosphate, 1 U of FastStart Taq DNA polymerase (Roche), and 10× reaction buffer (Roche). Cycling was performed as follows: 30 cycles at 94°C for 45 s, 55°C for 30 s, and 72°C for 45 s. PCR products were resolved in 1% (wt/vol) agarose gels and 0.5× TAE (40 mM Tris-acetate and 1 mM EDTA).
Analysis of DNA by PFGE.
Cleaved and uncleaved viral DNA preparations were resolved in a 1% agarose gel by pulsed-field gel electrophoresis (PFGE) via counter-clamped homogenous electric field gels (CHEF-DRII; Bio-Rad) (6). The separation conditions were as follows: 6 V/cm at 14°C for 14 h. The switch time ramped from 5 to 35 s. After electrophoresis, the gels were immersed in 1 μg of ethidium bromide/ml in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA) for 30 min at room temperature or stained with SYBR gold (Molecular Probes) in 0.5× TBE according to the manufacturer's instructions. The gels were then washed with distilled water and photographed under UV light.
Detection of viral DNA by Southern blotting.
DNAs were extracted from purified CNGV and CNGV-infected and -uninfected KFC by standard techniques (27). Twenty micrograms of cellular purified DNA and 5 μg of viral DNA were cleaved with EcoRI and separated by gel electrophoresis in a 0.8% agarose gel with a 23-cm length (0.5 V/cm for 18 h). The gel was subsequently denatured prior to nucleic acid transfer onto a nylon membrane (Hybond-N). A radiolabeled probe was generated by using a pBluescript II SK(−) plasmid containing viral fragment E (see Fig. 5) as a template and the NH1 and NH2 as primers for PCR, as described above, in the presence of 20 μM [α-32P]dCTP (125 Ci/mmol) (NEN). Hybridization was performed overnight at 65°C according to the method of Sambrook and Russell (27). The blot was washed and dried at room temperature, and viral DNAs were detected with a phosphorimager (Bas1000; Fujix, Tokyo, Japan).
Nucleotide sequence accession numbers.
DNA sequences were submitted to GenBank and were assigned the following accession numbers: AY208988, AY208989, AY208990, AY208991, and AY661550.
RESULTS
Transmission and spread of CNGV.
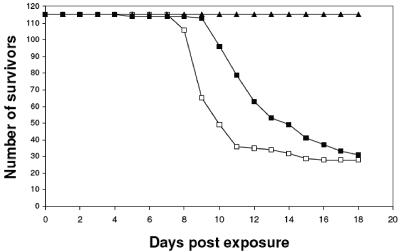
Adult fish were divided into three groups of 114 or 115 fish each which were hatched at 22 to 24°C in separate tanks. The exposure of two groups to infected specimens taken from a contaminated hatchery caused mortality rates of 75.6 and 73.7% (87 and 84 fish, respectively) at 8 to 13 days postinfection (Fig. 1). All fish in the third control group survived during the 21-day period of the experiments.
FIG. 1.
Kinetics of mortality in koi carp after cohabitation challenge. Adult fish were divided into three groups of 114 or 115 fish. Groups 1 (□) and 2 (▪) were exposed to a contaminated specimen, and group 3 (▴) was used as an uninfected control. The numbers of survivors were monitored daily.
Three to four days before death, the fish ceased eating and displayed abnormal behaviors, such as lassitude and gasping movements in shallow water. In addition, fish exhibited uncoordinated movements and erratic swimming, which are characteristic of neurological disorders. Neurological signs, such as a decline in the frequency of tail movements and a loss of equilibrium, were observed for several fish. Similar signs in koi and common carp in the United States were previously described (11, 16). These signs were followed by the appearance of severe interstitial nephritis and gill necrosis, with minimal focal inflammation in the liver and brain (24). In similar experiments conducted in tanks, where fish were hatched at a temperature of 29°C, all exposed fish survived during the 22-day period (data not shown). These results clearly show that the virus causing the disease is easily transmitted through water and is highly contagious.
Isolation of virus from infected specimens.
The cocultivation of cells taken from kidneys (11 specimens) and livers (5 specimens) of infected fish with KFC resulted in the appearance of cytopathic effects (CPE) at 5 to 6 days postinoculation, while the cocultivation of blood cells or sera taken from diseased carp did not result in CPE. At 10 days postcocultivation, cultured cells lost their attachment to the flask bottom and died. Medium harvested from KFC that had been cocultivated with kidney or liver cells (5 to 7 days postinfection) was first clarified of cells and cell debris by centrifugation and then was used to titrate the virus in fresh KFC cultures. The initial titer of the virus in cocultivated cultures was 103 PFU/ml. The virus that was released into the medium was serially transferred from culture to culture in a similar manner, and the titer in each transfer was determined by a plaque assay and/or end-point dilution. The viral titer increased by 2 orders of magnitude during five serial transfers in cell culture. Isolation of the virus by this method was repeated independently three times, while control cells taken from normal fish never caused a CPE or plaques in KFC cultures. The cocultivation of liver, spleen, or kidney cells taken from infected fish with EPC did not show any effect in these cultures. Unlike the KHV described by Hedrick et al. (16), which propagates in EPC, all of our efforts to isolate a virus from sick fish by cocultivation with EPC have failed so far.
Isolation of the etiologic agent of the disease.
Table 1 shows that naive fish inoculated with clarified culture medium from uninfected KFC remained asymptomatic. However, 75% of fish that were inoculated with infected cell extracts and 82% of those that were inoculated with clarified medium harvested from infected cultures died within 15 days post-intraperitoneal injection. These fish developed typical pathological signs and lesions identical to those observed in infected fish grown in ponds. Kidney cells taken from inoculated specimens exhibiting disease symptoms were cocultivated with KFC. The titers of virus harvested from cocultivated cultures were 1.5 × 102 to 1.8 × 102 PFU/ml on KFC, as determined by a plaque assay. The medium harvested from these infected KFC was used for reinfection of naive juvenile fish. Four of 10 fish died from the disease by 9 to 14 days postinfection. This “ping-pong” type of experiment, which was serially repeated three times (Table 1), clearly verifies that the virus isolated from infected fish and propagated in KFC is indeed the etiologic agent of the disease. Final proof that the isolated virus is the cause of the disease was obtained by injecting fish with a single plaque-isolated virus (results not shown).
TABLE 1.
Isolation of the virus responsible for mortality in carpa
| Subjects for expt | Result of indicated expt (no. of dead fish/no. of inoculated fish [%] or no. of plaques/culture [for expts with KFC])
|
||
|---|---|---|---|
| Inoculation with infected cell extracts | Inoculation with medium from uninfected KFC | Inoculation with medium from infected KFC | |
| Fish | 15/20 (75) | 0/15 (0) | 14/17 (82) |
| KFC cocultivated with 104 fish cells | 1.8 × 102 | 0 | 1.5 × 102 |
| Fish | Not done | 0/10 (0) | 4/10 (40) |
Medium and cells extracts harvested from the fifth passage of the virus in cell culture were used for intraperitoneal injection of juvenile naive carp (top row). Kidney cells taken from an inoculated fish from each group were cocultivated with KFC (middle row). The medium harvested from KFC that were cocultivated with carp cells (middle row, last column) was transferred three times in KFC and then used to reinfect juvenile carp (bottom row).
Electron microscopy.
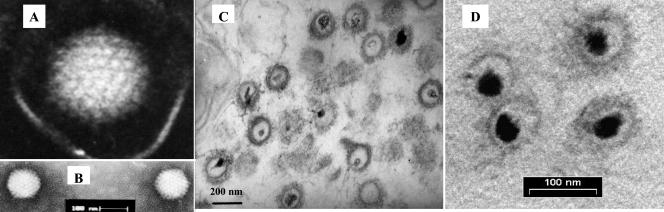
The medium harvested from infected KFC cultures was purified on sucrose gradients. A distinct band at 37 to 39% sucrose was removed from the gradients, diluted 10-fold in PBS, and pelleted by centrifugation. The pellets were suspended in 500 μl of PBS, and samples were taken for titration on KFC and for electron microscopy analysis. The titers of the purified virus preparations in the medium were 106 to 107 PFU/ml. The negatively stained particles had a symmetric icosahedral morphology (Fig. 2A and B), with an average core diameter of 103 nm, resembling the cores of herpesviruses (20, 25). Several negatively stained particles were surrounded by envelope-like structures (Fig. 2A). However, to determine whether CNGV is an enveloped virus, we prepared thin sections through a pellet of purified virus (Fig. 2C and D). The micrographs show that our CNGV preparations were purified, had abundant viral particles, and contained small amounts of cell membranes. The thin sections showed the presence of thread-like structures on the surface of the core. The finding that the virus is enveloped was supported by treatments of the virus preparation with 25% ether or 0.1% Triton X-100 (Baker) (8). These treatments reduced the titer of the virus from 104 PFU/ml to 2.5 × 102 and 4 × 100 PFU/ml, respectively. CNGV contains a relatively small asymmetrical electron-dense region within the viral core, which is probably the genomic DNA and nucleoprotein complex. Previously, we detected similar viral particles near the nuclear membrane in fixed intestinal tissues of sick fish (23).
FIG. 2.
Electron micrographs of CNGV harvested from infected KFC. The purified virus was negatively stained with 2% phosphotungstate (A and B). The particle size was in the range of 96 to 105 nm, with an average of 103 nm. Bar, 0.1 μm. Thin sections (C and D) were made for ultrastructural analysis by transmission electron microscopy. Purified virus pellets were fixed with 3% glutaraldehyde in 0.1 M sodium cacodylate and then stained with uranyl acetate and lead citrate.
Viral DNA.
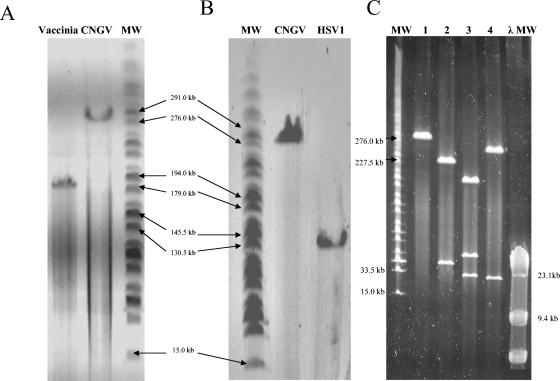
To obtain a close estimate of the molecular weight of the CNGV genome, we compared it to the genomic DNAs of HSV-1 and vaccinia virus by PFGE analysis. Viral DNA genomes were extracted from purified viruses by phenol extraction after incubation with proteinase K and SDS. Figure 3 clearly shows that the molecular size of the genomic CNGV DNA is ca. 277 kbp, while HSV-1 and vaccinia virus migrate as ca. 150-kbp (Fig. 3B) and 185-kbp (Fig. 3A) DNA molecules, respectively, as previously described (12, 18).
FIG. 3.
Analysis of CNGV DNA by PFGE. Viral DNAs were obtained from purified viruses after incubation with proteinase K and SDS and phenol extraction. (A) Comparison of CNGV DNA and vaccinia virus DNA. (B) Comparison of CNGV DNA and HSV-1 DNA. (C) CNGV DNA cleaved with PmeI, PacI, and SwaI restriction enzymes. The molecular weight markers (MW) used were HindIII-cleaved λ phage DNA and a MidRange I PFGE marker (New England Biolabs).
This estimation of the molecular size of the CNGV genome was further confirmed by cleaving the CNGV DNA with the PmeI, PacI, or SwaI restriction enzyme. These eight-cutter restriction enzymes cleaved the viral DNA into fragments. The sum of the molecular weights of the fragments equals the size of the uncleaved CNGV DNA molecule (Fig. 4C). These results strongly suggest that the genome of CNGV is a large linear dsDNA molecule of ca. 277 kbp.
FIG. 4.
Identification of CNGV genome fragments by PCR (A and B) and Southern blotting (C). The AP1-AP2 (A) and AP1-AP3 (B) primers derived from clone A (Fig. 5) were used for PCR amplification. The templates for the PCR were as follows: total DNAs extracted from infected (TCin) and uninfected (TCun) KFC, CNGV DNA (CNGV), and DNAs from livers of sick and naïve fish (LI and LU, respectively) and from kidneys of sick and naïve fish (KI and KU, respectively). MW, HindIII-cleaved λ phage DNA. (C) A [α-32P]dCTP-labeled probe derived from clone E (Fig. 5) was hybridized to a pBluescript II SK(−) plasmid containing viral fragment E (pCNGV), vaccine virus (VAC), HSV-1, CNGV, TCin, and TCun DNA samples. Labeled viral DNAs were detected with a phosphorimager.
Sequence of CNGV DNA fragments.
BamHI-EcoRI CNGV DNA fragments were cloned into pBluescript plasmids (1), and the viral DNA clones were sequenced, analyzed with the BLAST program (2, 31), and submitted to GenBank. Figure 4 shows that the internal primers AP1-AP2 and AP1-AP3 (panels A and B, respectively) were efficient at amplifying appropriate fragments of DNA preparations extracted from purified CNGV (A), infected cultured cells (A and B), and kidneys and livers removed from sick fish (A and B) but not from uninfected KFC (B) and naive fish organs (A and B). Similar results were also achieved by using primers from clones B to E (results not shown).
To exclude any suspicion that the sequenced fragments were derived from a contaminant, whether viral or cellular DNA, we performed Southern blotting. Figure 4C shows that a probe generated from clone E (accession no. AY661550) (Fig. 5) hybridized with the original plasmid, CNGV, and CNGV-infected KFC DNAs but not with HSV-1 or vaccinia DNAs or DNAs extracted from uninfected KFC. By using PCR with internal primers derived from the sequences of clones A to E, as shown in Fig. 5, and by Southern blotting, we ascertained that all of these clones represent CNGV DNA.
An analysis of >4,500 bp derived from the CNGV genome did not reveal any fragments of longer than 45 bp that were similar to viral genomes described in GenBank (Fig. 5 and data not shown). Although the CNGV clones bear only small fragments similar to other viruses, many of these sequences are homologous to members of the Herpesviridae, Adenoviridae, Baculoviridae, and Poxviridae families. The G+C content in each one of the viral DNA fragments analyzed so far varies from 48 to 62%, with an overall average of 55.9%.
An analysis of the CNGV DNA fragments (Fig. 5) by Blastx (2) suggested that clones B, C, D, and E are primarily similar to the poxviruses and that clone A bears sequences similar to those of the channel catfish virus. Clones B and C encode ribonucleotide reductase (RR). Clone C is mostly similar to the cowpox virus RR gene, while fragment B bears genes encoding amino acid sequences similar to those of baculoviruses and poxviruses (Table 2). Fragment D may be a hybrid encoding poxvirus and herpesvirus proteins, since the N-terminal section of this fragment is 32% identical and 48% similar to Yaba-like disease virus, followed by a section that is similar to human herpesvirus 8 (25% identical and 51% similar) (Table 2). Fragment A contains a segment encoding amino acid sequences similar to ictalurid herpesvirus 1 (channel catfish herpesvirus), and fragment E bears small sequences similar to those of Herpesviridae members and adenoviruses.
TABLE 2.
Comparison of putative CNGV-derived peptides by using Blastx
| Clone (accession no.) | Family | Genus | Protein | Accession no. | Identity (no. of amino acids with identity/total no. of amino acids [%]) | Conservation (no. of conserved amino acids/total no. of amino acids [%]) | Gaps (no. of gaps/total no. of amino acids [%]) |
|---|---|---|---|---|---|---|---|
| A (AY208988) | Herpesviridae | Ictalurid herpesvirus 1 | Hypothetical protein ORF56 | NP041147 | 37/138 (26) | 57/138 (41) | |
| B (AY208989) | Baculoviridae | Spodoptera littoralis nucleopolyhedrovirus | Ribonucleotide reductase | CAA67423 | 74/163 (45) | 102/163 (62) | |
| Poxviridae | Monkeypox virus | 14L | NP536492 | 74/162 (45) | 101/162 (62) | ||
| Poxviridae | Variola virus | Ribonucleoside-diphosphate reductase large chain | NP042102 | 74/162 (45) | 99/162 (61) | ||
| Poxviridae | Ectromelia virus | EVM057 | NP671575 | 73/162 (45) | 99/162 (61) | ||
| Baculoviridae | Ecotropis obliqua nucleopolyhedrovirus | Ribonucleotide reductase | AAQ88175 | 64/123 (52) | 90/123 (73) | ||
| Poxviridae | Vaccinia virus (strain WR) | Ribonucleoside-diphosphate reductase large chain | WZVZH4 | 73/162 (45) | 98/162 (60) | ||
| Poxviridae | Camelpox virus | Ribonucleotide reductase large subunit | NP570461 | 73/162 (45) | 98/162 (60) | ||
| Nimaviridae | Shrimp white spot syndrome virus | Ribonucleotide reductase large subunit | AAK69359 | 64/181 (35) | 98/181 (54) | 18/181 (9) | |
| Asfarviridae | African swine fever virus (strain Malawi) | Ribonucleoside-diphosphate reductase large chain | P26685 | 45/109 (41) | 69/109 (63) | ||
| Iridoviridae | Invertebrate iridescent virus 6 | Ribonucleoside-diphosphate reductase large chain | NP149548 | 48/113 (42) | 75/113 (66) | 4/113 (3) | |
| Herpesviridae | Equine herpesvirus 1a | Ribonucleoside-diphosphate reductase large chain | NP041030 | 38/70 (54) | 51/70 (72) | 1/70 (1) | |
| C (AY208990) | Poxviridae | Cowpox virus | CPXV083 | NP619870 | 124/190 (65) | 151/190 (79) | |
| Poxviridae | Camelpox virus | Ribonucleotide reductase large subunit | NP570461 | 126/190 (66) | 151/190 (79) | ||
| Poxviridae | Variola virus | Ribonucleoside diphosphate reductase large chain | NP042102 | 123/190 (64) | 150/190 (78) | ||
| Poxviridae | Vaccinia virus (strain WR) | Ribonucleosite reductase large subunit | P12848 | 124/190 (65) | 149/190 (78) | ||
| Nimaviridae | Shrimp white spot syndrome virus | Large subunit of ribonucleotide reductase | NP477694 | 104/193 (53) | 143/193 (74) | 1/193 (0) | |
| Asfarviridae | African swine fever virus (strain Malawi) | Ribonucleoside-diphosphate reductase large chain | P26685 | 78/203 (38) | 107/203 (52) | 19/203 (9) | |
| Herpesviridae | Meleagrid herpesvirus 1a | UL39 ribonucleotide reductase large subunit | NP073333 | 62/168 (36) | 90/168 (53) | 1/168 (0) | |
| D (AY208991) | Poxviridae | Yaba-like disease virus | 135R protein | NP073520 | 24/74 (32) | 36/74 (48) | 2/74 (2) |
| Herpesviridae | Human herpesvirus 8a | K1 glycoprotein | AAD26403 | 17/66 (25) | 34/66 (51) | ||
| E (AY661550) | Poxviridae | Yaba monkey tumor virus | 135R protein serine protease inhibitor | NP938338 | 21/55 (38) | 30/55 (54) | |
| Poxviridae | Myxoma virus | M134R protein | NP051848 | 19/56 (33) | 29/56 (51) | ||
| Poxviridae | Swinepox virus | SPV131 VAR B22R homologue | NP57029.1 | 19/56 (33) | 29/56 (51) | ||
| Poxviridae | Lumpy skin disease virus | LD131 | AAN02720.1 | 18/56 (32) | 32/56 (57) | ||
| Poxviridae | Rabbit fibroma virus | gp134R | NP052023.1 | 19/56 (33) | 29/56 (51) | ||
| Poxviridae | Sheeppox virus | VARV B22R homologue | NP659705.1 | 18/56 (32) | 35/56 (57) | ||
| Poxviridae | Molluscom contagiosum virus | MCO35R | NP043986.1 | 15/35 (42) | 23/35 (65) | ||
| Poxviridae | Cowpox virus | B7.5R | AAB57929.1 | 17/55 (30) | 31/55 (56) | ||
| Herpesviridae | Human herpesvirus 8 | ORF17 capsid assembly protein homologue | AAB62670 | 12/27 (44) | 16/27 (59) |
In general, similarity to Herpesviridae was not significant, but Herpesviridae is represented in the table as the most similar member of the family.
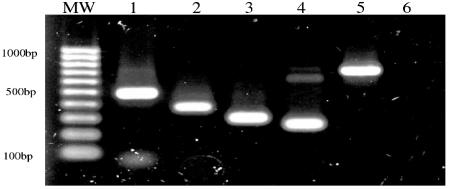
We next asked whether CNGV resembles KHV, which was previously described by Hedrick and coworkers (10, 16). To this end, we amplified DNA fragments from the CNGV genome by PCR, using primers derived from KHV as described by Gilad et al. (10), Gray et al. (13), and Sano and his coworkers (28) (accession no. AB178537). Figure 6 clearly shows that all of the primers used for PCR were appropriate to amplify DNA fragments from the CNGV DNA. The resulting fragments had the molecular weights that were expected according to the sequenced fragments derived from KHV. These results suggest that CNGV and KHV refer to the same virus.
FIG. 6.
Amplification of CNGV DNA fragments by PCR using KHV derived primers. Sets of primers were used to amplify purified CNGV DNA in a PCR. Primers are as follows: Lane 1, primers derived by Gilad et al. 2002. Lanes 2 and 3, primers derived by Gray et al. 2002. Lane 4, primers S1-2 (see Materials and Methods). Lane 5, primers AP1-2 (see Materials and Methods), and lane 6, negative control (NC), no primers were added to the reaction. Lane MW, EZ load 100bp molecular ruler (Bio-Rad).
DISCUSSION
A massive mortality of carp (C. carpio) was observed at many fish farms throughout Israel. A similar disease was also observed in North America, Europe, Indonesia, Korea, and Japan, causing severe economic losses to the fish farming industry (3, 4, 10, 11, 13, 16, 22, 23). It was therefore especially important to determine the disease-causing agent in order to develop protection against this agent. In this report, we verified that the causative agent of the disease in koi and common carp is a large DNA virus. By establishing a new cell line from common carp and koi fish species, which allows virus propagation, we were able to isolate the etiologic agent. Final evidence that the virus that was isolated and propagated in KFC is indeed the cause of the disease was based on the following data. (i) The virus was successfully isolated from infected fish, but not from naive specimens. (ii) Inoculation of the virus that was propagated in KFC culture into naive fish induced a similar disease. (iii) The cocultivation of kidney cells taken from specimens with the induced disease, but not from mock-infected fish, yielded a similar virus, which was propagated in KFC cultures. (iv) Three cycles of this ping-pong technique were successfully applied. (v) The cloned virus isolated in tissue culture induced the same disease in fish. (vi) Rabbit sera prepared against the purified virus interacted specifically with tissues from infected fish as well as with sick fish from ponds (24; data not shown). (vii) Viral DNA was identified in infected KFC and in sick, but not naive, fish. Thus, the results of our work will facilitate diagnosis of this disease by infections of KFC, PCR, and immunological methods.
The following characteristics of this virus are noteworthy. (i) The virus is very contagious. (ii) It is transmitted through water. (iii) Induction of the disease is restricted to 18 to 28°C (11), as observed in open-air ponds and under laboratory conditions (3, 23). (iv) It has a narrow host range, as even closely related cyprinid fish are resistant to this virus (23) and as it was unable to propagate in EPC cultures. (v) The virus morphology and diameter are consistent with herpesviruses (16), although the virus contains a relatively small nonsymmetrical electron-dense region in the viral core. (vi) However, the DNA fragment sequences analyzed so far do not bear significant similarities to any known viral genomes. (vii) Finally, the virus contains a very large dsDNA molecule of ca. 277 kbp, which is larger than the known herpesvirus genomes.
At present, we have sequenced only a small percentage of the CNGV genome (about 4,100 bp). In agreement with the work of Gray et al. (13), no significant similarity between CNGV and fish viruses, such as channel catfish virus (7) or other fish herpesviruses, nor any other viruses has come to light so far. However, the CNGV genome bears small fragments of 16 to 45 bp which are similar mainly to members of the Herpesviridae, Adenoviridae, Poxviridae, and Baculoviridae families. Moreover, the DNA sequence of the koi herpesvirus thymidine kinase gene (TK) (accession no. AJ535112) described by Bercovier and coworkers bears only a 21-bp DNA fragment similar to the Shope fibroma virus TK gene. In addition, the koi herpesvirus amplicon sequence (accession no. AF411803) described by Hedrick and coworkers bears only slight similarities to other dsDNA viruses. Recently, Sano and his coworkers sequenced about 7,000 bp of the KHV genome (accession no. AB178324) and showed some similarities to ictalurid herpesvirus (channel catfish virus). These results suggest that the CNGV DNA genome is highly divergent from those of the known dsDNA viruses.
By using the Blastx program, which allows comparisons of translation products of the DNA sequences to viral protein databases, we revealed several similarities to several DNA viruses (e.g., clones B and C [accession no. AY208989 and AY208990, respectively], encode amino acid sequences similar to a ribonucleotide reductase of poxviruses and other viruses). The TK gene (accession no. AJ535112) is similar to that of cowpox virus. It is interesting that some similarity was also noted for African swine fever virus and shrimp white spot syndrome virus, which are the sole representatives of their phylogenetic groups. However, the high molecular weight of the CNGV DNA and the divergence of its DNA and protein sequences do not allow a phylogenetic classification of the virus. Additional data are required to solve this enigma.
It is conceivable that fish viruses, which evolved in separate and distinct ecological environments, may be genetically highly divergent from each other. It can be expected, therefore, that CNGV, which thrives in captivity and is restricted to limited species of Cyprinus, is probably diverse from other viruses. Intensive fish farming with overcrowding facilitates the invasion of new unclassified viruses, which may pose a serious economic threat.
In conclusion, the results of this study verified that the CNGV morphology and diameter are similar but not identical to those of Herpesviridae. However, the results demonstrate that the genome of the virus isolated in Israel is composed of a ca. 277-kbp linear DNA molecule, which is larger than those of the other Herpesviridae members. Its high molecular weight and analyses of the DNA and amino acid sequences revealed so far suggest that CNGV, although it is yet unclassified, may be a novel virus.
Acknowledgments
We are indebted to H. Bercovier, Department of Clinical Microbiology, The Hebrew University of Jerusalem, for encouraging us to initiate this project and for supplying the EPC line. We thank Neomi Bahat from the Electron Microscopy Unit of the Faculty of Agriculture, The Hebrew University, Rehovot, Israel, for preparing the electron microscopy micrographs. We thank Y. Becker and Y. Asher for their thoughtful comments and suggestions during this study and S. Amir for editing the manuscript.
This work was supported by grant 02-0082 from the Chief Scientist's Office, Ministry of Agriculture and Rural Development, Israel.
REFERENCES
- 1.Alting-Mees, M. A., and J. M. Short. 1989. pBluescript II: gene mapping vectors. Nucleic Acids Res. 17:9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariav, R., S. Timan, I. Paperna, and I. Bejerano. 1998. First report of newly emerging viral disease of Cyprinus carpio species in Israel. Presented at the EAFP 9th International Conference, Rhodes, Greece.
- 4.Bretzinger, A., T. Fischer-Scherl, M. Oumouna, R. Hoffman, and U. Truyen. 1999. Mass mortalities in koi, Cyprinus carpio, associated with gill and skin disease. Bull. Eur. Assoc. Fish Pathol. 19:182-185. [Google Scholar]
- 5.Calle, P. P., T. McNamara, and Y. Kress. 1999. Herpesvirus-associated papillomas in koi carp (Cyprinus carpio). J. Zoo. Wildl. Med. 30:165-169. [PubMed] [Google Scholar]
- 6.Chu, G., D. Vollrath, and R. W. Davis. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582-1585. [DOI] [PubMed] [Google Scholar]
- 7.Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Dougherty, E. M., S. L. Vaughn, and C. F. Reichelderfer. 1975. Characteristics of the non-occluded form of a nuclear polyhedrosis virus. Intervirology 5:109-121. [DOI] [PubMed] [Google Scholar]
- 9.Fijan, N., M. Sulimanovic, M. Bearzotti, D. Muzinic, O. Zwillenberg, S. Chilmonczyk, F. Vautherot, and P. de Kinkelin. 1983. Some properties of the epithelioma papulosum cyprini (EPC) cell line from Cyprinus carpio. Ann. Virol. 134:207-220. [Google Scholar]
- 10.Gilad, O., S. Yun, K. B. Andree, M. A. Adkison, A. Zlotkin, H. Bercovier, A. Eldar, and R. P. Hedrick. 2002. Initial characteristics of koi herpesvirus and development of a polymerase chain reaction assay to detect the virus in koi, Cyprinus carpio koi. Dis. Aquat. Organ. 48:101-108. [DOI] [PubMed] [Google Scholar]
- 11.Gilad, O., S. Yun, M. A. Adkinson, K. Way, N. H. Willits, H. Bercovier, and R. P. Hedrick. 2003. Molecular comparison of isolates of an emerging fish pathogen, koi herpesvirus, and the effect of water temperature on mortality of experimentally infected koi. J. Gen. Virol. 84:2661-2668. [DOI] [PubMed] [Google Scholar]
- 12.Goebel, S. F., G. P. Johnson, M. E. Perkus, S. W. Davis, J. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-266, 517-563. [DOI] [PubMed] [Google Scholar]
- 13.Gray, W. L., L. Mullis, S. E. LaPatra, J. M. Groff, and A. Goodwin. 2002. Detection of koi herpesvirus DNA in tissues of infected fish. J. Fish Dis. 25:171-178. [Google Scholar]
- 14.Hasegawa, S., T. Somamoto, C. Nakayasu, T. Nakanishi, and N. Okamoto. 1997. A cell line (CFK) from fin of isogeneic ginbuna crusian carp. Fish Pathol. 32:127-128. [Google Scholar]
- 15.Hedrick, R. P., J. M. Groff, M. S. Okihiro, and T. S. McDowell. 1990. Herpesviruses detected in papillomatous skin growths of koi carp (Cyprinus carpio). J. Wildl. Dis. 26:578-581. [DOI] [PubMed] [Google Scholar]
- 16.Hedrick, R. P., O. Gilad, S. Yun, J. Spangenberg, G. Marty, R. Nordhausen, M. Kebus, H. Bercovier, and A. Eldar. 2000. A herpesvirus associated with mass mortality of juvenile and adult koi, a strain of common carp. J. Aquat. Anim. Health 12:44-55. [DOI] [PubMed] [Google Scholar]
- 17.Kim, C. H., D. M. Dummer, P. P. Chiou, and J. A. Leong. 1999. Truncated particles produced in fish surviving infectious hematopoietic necrosis virus infection: mediators of persistence? J. Virol. 73:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 19.Miyazaki, T., H. Okamoto, T. Kageyama, and T. Kobayashi. 2000. Viremia-associated ana-aki-byo, a new viral disease in color carp Cyprinus carpio in Japan. Dis. Aquat. Organ. 39:183-192. [DOI] [PubMed] [Google Scholar]
- 20.Murphy, F., and N. Nathanson. 1997. An atlas of viral disease pathogenesis, p. 433-463. In N. Nathanson, R. Ahmed, K. Holmes, D. Griffin, F. Gonzale, and H. Robinson (ed.), Viral pathogenesis. Lippincott-Raven, Philadelphia, Pa.
- 21.Neukirch, M., K. Bottcher, and S. Bunnajirakul. 1999. Isolation of a virus from koi with altered gills. Bull. Eur. Assoc. Fish Pathol. 19:221-224. [Google Scholar]
- 22.Pearson, H. 2004. Carp virus crisis prompts moves to avert global spread. Nature 427:577. [DOI] [PubMed] [Google Scholar]
- 23.Perelberg, A., M. Smirnov, M. Hutoran, A. Diamant, Y. Bejerano, and M. Kotler. 2003. Epidemiological description of a new viral disease afflicting cultured Cyprinus carpio in Israel. Isr. J. Aquacult. (Bamidge) 55:5-12. [Google Scholar]
- 24.Pikarsky, E., A. Ronen, J. Abramowitz, B. Levavi-Sivan, M. Hutoran, Y. Shapira, M. Steinitz, A. Perelberg, D. Soffer, and M. Kotler. 2004. The pathogenesis of the acute viral disease in fish induced by the carp interstitial nephritis and gill necrosis virus. J. Virol. 78:9544-9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roizman, B. 1990. Whither herpesviruses? Adv. Exp. Med. Biol. 278:285-291. [DOI] [PubMed] [Google Scholar]
- 26.Ronen, A., A. Perelberg, J. Abramowitz, M. Hutoran, S. Tinman, I. Bejerano, M. Steinitz, and M. Kotler. 2003. Efficient vaccine against the virus causing a lethal disease in cultured Cyprinus carpio. Vaccine 21:4677-4684. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, p. 6.7-6.10. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sano, T., H. Fukuda, and M. Furukawa. 1985. A herpesvirus isolated from carp papilloma in Japan. Fish Shellfish Pathol. 32:307-311. [Google Scholar]
- 29.Sano, T., N. Morita, N. Shima, and M. Akimoto. 1991. Herpesvirus cyprini: lethality and oncogenicity. J. Fish Dis. 14:533-543. [Google Scholar]
- 30.Shchelkunov, I., and T. Shchelkunova. 1990. Infectivity experiments with Cyprinus carpio iridovirus (CCIV), a virus unassociated with carp gill necrosis. J. Fish Dis. 13:475-484. [Google Scholar]
- 31.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]