Abstract
Mutations in the isocitrate dehydrogenase 1 (IDH1) and IDH2 genes are reported recently in AML. Here we investigate the frequency and the clinicopathologic features of IDH1 and IDH2 mutations in AML. Mutations in IDH1 (IDH1R132) and IDH2 (IDH2R172) were assessed by Sanger sequencing in 199 AML cases. Point mutations in IDH1R132 were detected in 12/199 (6%) cases, and in IDH2R172 in 4/196 (2%) cases. Fifteen out of the 16 (94%) mutated cases were cytogenetically normal, for an overall frequency in this group of 12%. IDH1R132 and IDH2R172 mutations were mutually exclusive. Concurrent mutations in NPM1, FLT3, CEBPA and NRAS were detected only in AML with IDH1R132 mutation. The clinical and laboratory variables of AML patients with IDH mutations showed no significant differences compared with patients with wild-type IDH. We conclude that IDH1R132 and IDH2R172 mutations occur most often in cytogenetically normal AML with an overall frequency of approximately 10%.
Keywords: Acute Myeloid Leukemia, Isocitrate Dehydrogenase, IDH1, IDH2, mutation
Introduction
Identification of somatically acquired gene mutations has provided critical insights into the pathogenesis of acute myeloid leukemia (AML).1 Gene mutations in AML provide useful markers for diagnosis and for monitoring response to therapy, and also provide information useful in assessing prognosis and making therapeutic decisions.2–6 The most recent WHO classification of myeloid neoplasms acknowledges the clinical significance of gene mutations in AML and has proposed separate entities for AML with mutations in NPM1 and CEBPA.7 Interestingly, mutations AML are detected commonly in cytogenetically normal (CN) cases which account for 40–50% of all AML.1, 8 Currently, no known mutations are identified in about 20–30% of CN-AML cases suggesting the possibility that more mutations likely exist.
Recently, the entire genome of a patient with CN-AML was sequenced and a total of 64 somatic mutations, 12 within coding sequences of genes and 52 in conserved or regulatory regions, were identified.9 In particular, a novel mutation was detected in isocitrate dehydrogenase 1 (IDH1), a metabolic gene frequently mutated in gliomas.10–12 The mutation occurred consistently at an evolutionary conserved arginine residue at codon 132 (R132) within the substrate binding site of the enzyme and was strongly associated with normal cytogenetic status. A limited number of studies examining the frequency of IDH1 mutation in AML were performed subsequently.13–16 In addition to IDH1R132, mutations in codon 172 of IDH2 (IDH2R172), a mitochondrial isoform of IDH1, have been documented in AML.14, 17 Published studies have focused on examining the frequency and correlation with mutational status and clinical outcome. A very limited amount of information is available on the histomorphologic features and immunophenotypic profiles associated with IDH1R132 and IDH2R172 mutations in AML. Only one study, involving a Chinese population, reported clinical and biologic features of AML with IDH1R132 mutation.16 There are no such reports available for the western population and for AML with IDH2R172 mutation.
We report the frequency of IDH1R132 and IDH2R172 mutations in 199 AML cases with clinical, histologic and immunologic characterization of the mutated cases. We also performed a meta-analysis of available studies that have assessed AML cases for IDH1R132 mutations.
Materials and Methods
Study Group
DNA was extracted from diagnostic bone marrow aspirate samples of AML with 20% or greater blasts using methods described previously.18 All samples had been sent to the clinical Molecular Diagnostics Laboratory at The University of Texas M.D. Anderson Cancer Center at time of diagnosis. Residual DNA was used under an approved Institutional Review Board protocol. Cases of AML with favorable-risk cytogenetics were excluded from analysis based on data from an earlier study that showed absence of IDH1 mutations in this group.9 Pertinent laboratory information was obtained from the laboratory information system. The patient characteristics are listed in Tables 1 and 2.
Table 1.
Clinical features of AML with IDH1R132 mutation
| Total Cases N=199 |
IDH1 mutant (N=12) N (% of IDH1R132) |
IDH1 wild type (N=187) N (% of IDH1wt) |
p value† | |
|---|---|---|---|---|
| No. of Cases (N=199) | 199 | 12/199 (6) | 187/199 (94) | |
| Age in years (Mean, Range) | 55 (17–89) | 55 (37–77) | 54 (17–89) | 0.917†† |
| M:F | 95:104 (1:1.1) | 3:9 (1:3) | 92:95 (1:1.03) | 0.134 |
| WHO Classification | ||||
| AML, NOS | 163 | 10 (83) | 153 (82) | 1.00 |
| AML, MDS | 30 | 2 (17) | 28 (15) | 1.00 |
| t-AML | 4 | – | 4 (2) | |
| AML, Biphenotypic | 2 | – | 2 (1) | |
| FAB Classification | ||||
| M0 | 11 | – | 11 (6) | – |
| M1 | 32 | 4 (33) | 28 (15) | 0.106 |
| M2 | 46 | 4 (33) | 42 (22) | 0.482 |
| M4 | 42 | – | 42 (22) | – |
| M5 | 20 | 3 (25) | 17 (9) | 0.106 |
| M6 | 5 | – | 5 (3) | – |
| M7 | 1 | – | 1 (0.5) | – |
| RAEB-T | 14 | 1 (9) | 13 (7) | 0.596 |
| NA | 28 | – | 28 (15) | – |
| WBC Count (K/ml) | 19 (0.3–204) | 15 (0.3–83.1) | 20 (0.3–204) | 0.515†† |
| % Blast (Mean, Range) | 53 (20–98) | 55 (22–88) | 53 (20–98) | 0.732†† |
| Karyotype | ||||
| Normal | 127 | 11 (92) | 116 (62) | 0.059 |
| Abnormal | 72 | 1 (8) | 71 (38) | |
| Cytogenetic Risk Group | ||||
| Intermediate | 166 | 12 (100) | 154 (82) | 0.223 |
| Poor | 33 | – | 33 (18) | |
| Additional Mutations* | ||||
| NPM1 | 5/11 (45.5) | 13/54 (24.1) | 0.161 | |
| FLT3-ITD | 3/12 (25) | 43/179 (24) | 1.000 | |
| FLT3-D835 | 1/12 (8.5) | 9/179 (5) | 0.486 | |
| CEBPA | 2/10 (20) | 0/4 | ||
| NRAS | 2/12 (16.7) | 12/154 (7.8) | 1.000 | |
| KRAS | 0/12 | 7/154 (4.5) | 0.267 | |
| KIT | 1/9 (11.1) | 0/78 | 0.103 | |
| IDH2 | 0/11 | 4/185 (2.2) | 1.000 | |
| Molecular Risk Group‡ | 0.114 | |||
| Low | 11 | 3/11 (27) | 8/84 (10) | |
| High | 84 | 8/11 (73) | 76/84 (90) |
data presented as number of positive/number of tested;
Fisher exact test unless specified;
two-tailed t-test;
Molecular low risk: NPM1 mutated and FLT3-ITD wild type; high risk: NPM1 wild-type and/or FLT3-ITD positive; AML, NOS, AML not otherwise specified; AML, NOS, AML not otherwise specified; AML, MLD; AML with myelodysplasia-related changes; t-AML, therapy-related AML; RAEB-T, refractory anemia with excess blast in transformation; NA, not available
Table 2.
Clinical features of AML with IDH2R172 mutation
| Total Cases N=196 |
IDH2 mutant (N=4) N (% of IDH2R172) |
IDH2 wild type (N=192) N (% of IDH1wt) |
p value† | |
|---|---|---|---|---|
| No. of Cases (N=196) | 196 | 4/196 (2) | 192/196 (98) | |
| Age in years (Mean, Range) | 55 (17–89) | 51 (22–76) | 55 (17–89) | 0.778†† |
| M:F | 93:103 (1:1.1) | 1:3 | 92:99 (1:1.1) | 0.623 |
| WHO Classification | ||||
| AML, NOS | 160 | 4 (100) | 156 (81) | 1.000 |
| AML, MLD | 30 | – | 30 (16) | – |
| t-AML | 4 | – | 4 (2) | – |
| AML, Biphenotypic | 2 | – | 2 (1) | – |
| FAB Classification | ||||
| M0 | 11 | – | 11 (6) | – |
| M1 | 30 | 3 (75) | 27 (14) | 0.010 |
| M2 | 45 | 1 (25) | 44 (23) | 1.000 |
| M4 | 42 | – | 42 (22) | – |
| M5 | 19 | – | 19 (10) | – |
| M6 | 5 | – | 5 (3) | – |
| M7 | 1 | – | 1 (0.5) | – |
| RAEB-T | 15 | – | 15 (8) | – |
| NA | 28 | – | 28 (15) | – |
| WBC Count (K/ml) | 20 (0.3–204.2) | 4 (1.7–11.8) | 20 (0.3–204.2) | 0.001†† |
| % Blast (Mean, Range) | 53 (20–98) | 59 (42–73) | 53 (20–98) | 0.390†† |
| Karyotype | ||||
| Normal | 124 | 4 (100) | 120 (63) | 0.299 |
| Abnormal | 72 | – | 72 (37) | |
| Cytogenetic Risk Group | ||||
| Intermediate | 166 | 4 (100) | 159 (83) | 1.000 |
| Poor | 33 | – | 33 (17) | |
| Additional Mutations* | ||||
| NPM1 | 0/4 | 17/60 (28.3) | 0.566 | |
| FLT3-ITD | 0/4 | 46/185 (24.9) | 0.574 | |
| FLT3-D835 | 0/4 | 10/185 (5.4) | 1.000 | |
| CEBPA | 0/4 | 2/10 (20) | 1.000 | |
| NRAS | 0/4 | 13/159 (8.2) | 1.000 | |
| KRAS | 0/4 | 7/159 (4.4) | 1.000 | |
| KIT | 0/4 | 1/81 (1.2) | 1.000 | |
| IDH1 | 0/4 | 11/192 (5.7) | 1.000 | |
| Molecular Risk Group‡ | 1.000 | |||
| Low | 11 | 0/4 | 11/91 (12) | |
| High | 84 | 4/4 (100) | 80/91 (88) |
data presented as number of positive/number of tested;
Fisher exact test unless specified;
two-tailed t-test;
Molecular low risk: NPM1 mutated and FLT3-ITD wild type; high risk: NPM1 wild-type and/or FLT3-ITD positive; AML, NOS, AML not otherwise specified; AML, MLD; AML with myelodysplasia-related changes; t-AML, therapy-related AML; RAEB-T, refractory anemia with excess blast in transformation; NA, not available
IDH1 and IDH2 Mutation Detection
Exon 4 mutations in codon R132 of IDH1 and codon R172 of IDH2 were detected using PCR amplification followed by Sanger sequencing. Previously described PCR primers were modified with addition of M13 sequence.19 PCR primers utilized were forward-5′- TGTAAAACGACGGCCAGTCGGTCTTCAGAGAAGCCATT-3′ and reverse-5′- CAGGAAACAGCTATGACCGCAAAATCACATTATTGCCAAC-3′ for IDH1R132 and, forward-5′-TGTAAAACGACGGCCAGTAGCCCATCATCTGCAAAAAC-3′ and reverse-5′-CAGGAAACAGCTATGACCCTAGGCGAGGAGCTCCAGT-3′ for IDH2R172. All primers were purchased from Integrated DNA Technologies (Coralville, IA). For both IDH1R132 and IDH2R172, 2 μl patient DNA (100 ng/μl) was added to 48 μl PCR master mix that consisted of 31.7 μl molecular grade water, 5 ml 10× PCR buffer II (Applied Biosystems, Carlsbad, CA, USA), 4 μl 25 mM MgCl2, 10 mM dNTP mix, 1 μl each of M13-tagged forward and reverse primers (10 μM), 0.3 μl amplitaq gold (5U/μl). PCR conditions for both IDH1R132 and IDH2R172 included initial denaturing at 95°C for 10 min, 40 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec and, final extension at 72°C for 7 min. Post-PCR purification of the products was performed using QIAquick kit (Valencia, CA). PCR products were analyzed on a 2% agarose gel (115V for 30–40 minutes) using 8μl of PCR product and 2μl of 1× gel loading dye. Sanger sequencing of IDH1R132 and IDH2R172 amplicons was performed using M13-tagged primers in 20 μl final volume that contained 4 μl PCR product, 6.8 μl H2O, 2 μl 5× sequencing buffer, 3.2 μl M13 forward or reverse primer (1μM) and 4 μl Big Dye v1.1. Sanger sequencing was performed using M13-forward: 5′-TGTAAAACGACGGCCAGT-3′ and M13-reverse: 5′- CAGGAAACAGCTATGACC-3′ primers.
Sequencing conditions included initial denaturing at 96°C for 1 min, 25 cycles of 96°C for 10 sec, 50°C for 5 sec and 60°C for 4 sec. Sequencing products were purified using Qiagen DyeEx 96 Spin Kit (Valencia, CA) and analyzed by capillary gel electrophoresis on 3130 genetic analyzer (Applied Biosystems, Foster City, CA).
NPM1 Mutation Detection
Mutations in coding regions of exon 12 of NPM1 were detected using PCR amplification of a 168-bp segment followed by capillary gel electrophoresis. PCR primers included, forward: 5′-FAM-GATGTCTATGAAGTGTTGTGGTTCC -3′ and reverse: 5′- GGACAGCCAGATCAACTG-3′. PCR was performed in 50 μl reaction volume that contained 2 μl patient DNA (100 ng/μl), 5 μl 10× Thermol Pol Buffer with MgSO4, 5 μl 10 mM dNTPs, 1 μl NPM1-forward primer (10 μM), 1 μl NPM1-reverse primer (10 μM), 35.2 μl H2O and 0.75 μl Vent DNA polymerase (2U/μl). PCR conditions included initial denaturing at 95°C for 10 min, 40 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec and, final extension at 72°C for 7 min. PCR products were analyzed by capillary electrophoresis on 3100 or 3130 Genetic analyzer (Applied Biosystems).
RAS Mutation Detection
Mutations in codons 12, 13 and 61 of KRAS were detected using pyrosequencing as described earlier.18 Mutations in codons 12, 13 and 61 of NRAS were detected with the same protocol, but using the following primers: codons 12, 13: forward: 5′-GTTCTTGCTGGTGTGAAATGA-3′, reverse: 5′-BIOTIN-CTCTATGGTGGGATCATATTC-3′, sequencing: 5′-CAAACTGGTGGTGGTTGGAGCA-3′; codon 61: forward: 5′-GGACATACTGGATACAGCT-3′, reverse: 5′-BIOTIN-CTGTAGAGGTTAATATCCGCA-3′, sequencing: 5′-GGACATACTGGATACAGCT-3′
Detection of KIT, FLT3-ITD, FLT3-D835, CEBPA, and TP53 Mutations
Mutation analysis for KIT, FLT3-ITD, FLT3-D835, CEBPA and TP53 mutations were performed using Sanger sequencing as described previously.20–23 For CEBPA and TP53, a M13 sequence was added to the previously described PCR primers for Sanger sequencing. M13 tag primers were utilized for Sanger sequencing.
Morphologic, Immunophenotypic and Cytogenetic analysis
Bone marrow aspirate smears were stained with Wright-Giemsa and aspirate clot and biopsy specimens were stained with hematoxylin-eosin. Flow cytometric immunophenotypic analysis was performed using 4-color staining and conventional G-banded karyotyping was performed as described previously.24
Results
IDH1R132 and IDH2R172 mutations in AML
A total of 199 AML cases were tested for IDH1R132 mutation and 196 cases were tested for IDH2R172 mutation (Tables 1 and 2). IDH1R132 mutation was detected in 12 (6%) cases and IDH2R172 mutation was detected in 4 of 196 (2%) cases (Figure 1). No mutated cases had both IDH1 and IDH2 mutations suggesting that these mutations are mutually exclusive.
Figure 1. Detection of IDH1 R132 mutation by Sanger sequencing.
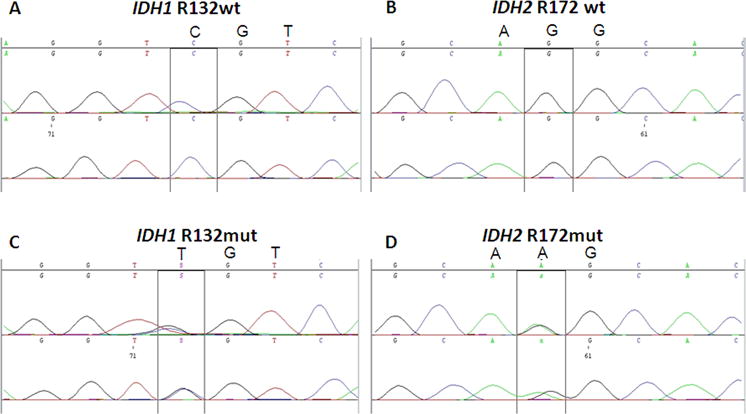
Sanger-sequencing showing (A) wild-type IDH1 codon 132: CGT, R132; (B) wild-type IDH2 codon 172: AGG, R172; (C) mutant IDH1 codon 132: TGT, R132C and (D) mutant IDH2 codon 172: AAG, R172K. Note that both IDH1R132 and IDH2R172 mutations are heterozygous missense point mutations.
Cytogenetic Features of IDH Mutated AML Cases
The AMLs tested included 127 (64%) with normal and 72 (36%) with abnormal cytogenetics. Subdivided by cytogenetic risk group, 166 (83%) cases were intermediate-risk and 33 (17%) were poor-risk. Eleven of 12 (92%) AML cases with IDH1 mutation had normal cytogenetics and all 12 (100%) were in the intermediate-risk cytogenetic group. The association between IDH1 mutation and normal cytogenetics approached statistical significance (Fisher’s exact test, p=0.059). All 4 cases with IDH2 mutation had normal cytogenetics and belonged to the intermediate-risk group.
Molecular Features of IDH Mutated AML Cases
The 12 AML with IDH1R132 mutations were equally distributed between R132H and R132C substitutions (Table 3). All IDH2R172 mutations resulted in R172K substitutions (Table 3). IDH1R132 mutated cases showed higher frequency of concurrent NPM1 mutation compared with wild-type cases (46% vs. 21%), this did not achieve statistical significance (Fisher’s exact test; p=0.161). Additional gene mutations were identified in IDH1 mutated cases including NPM1 in 5 of 11 (45.5%, FLT3-ITD in 3 of 12 (25%), CEBPA in 2 of 10 (20%), NRAS in 2 of 12 (16.7%), KIT in 1 of 9 (11.1%), and FLT3-D835 in 1 of 12 (8.5%), Eight of 11 (73%) of IDH1 mutated cases met the criteria for the high-risk molecular group. There was no significant association of IDH1R132 mutation with FLT3-ITD, FLT3-D835, CEBPA, NRAS, KRAS, or KIT mutations or a high-risk molecular profile (Table 1, 3).1, 25 None of the 4 IDH2R172 mutated cases showed a concurrent mutation in NPM1, FLT3-ITD, FLT3-D835, CEBPA, NRAS, KRAS, KIT or IDH1 (Table 2, 3). All 4 IDH2R172 mutated AML met the criteria for the high-risk molecular group based on the absence of NPM1 mutation.
Table 3.
Cytogenetic and molecular features of AML with IDH1R132 and IDH2R172 mutations
| Patient No. | Age/Sex | WHO | FAB | Karyotype | Cytogenetic Risk-Group | Mutation | Nucleotide Change | Amino Acid Change | Other Mutations |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 37/F | AML, NOS | M1 | N | I | IDH1 | CGT-CAT | R132H | NPM1, FLT3-ITD, CEBPA |
| 2 | 37/F | AML, NOS | M1 | N | I | IDH1 | CGT-TGT | R132C | N-RAS, KIT |
| 3 | 51/F | AML, MLD | M1 | N | I | IDH1 | CGT-CAT | R132H | NPM1, FLT3-ITD |
| 4 | 73/F | AML, NOS | M1 | N | I | IDH1 | CGT-TGT | R132C | FLT3-D835 |
| 5 | 52/F | AML, NOS | M2 | N | I | IDH1 | CGT-TGT | R132C | FLT3-ITD |
| 6 | 52/M | AML, NOS | M2 | N | I | IDH1 | CGT-CAT | R132H | NPM1 |
| 7 | 53/M | AML, NOS | M2 | N | I | IDH1 | CGT-TGT | R132C | – |
| 8 | 55/F | AML, NOS | M2 | N | I | IDH1 | CGT-CAT | R132H | CEBPA |
| 9 | 52/F | AML, NOS | M5 | N | I | IDH1 | CGT-TGT | R132C | – |
| 10 | 58/F | AML, NOS | M5 | N | I | IDH1 | CGT-CAT | R132H | NPM1, RAS |
| 11 | 77/F | AML, NOS | M5 | N | I | IDH1 | CGT-CAT | R132H | NPM1 |
| 12 | 62/M | AML, MLD | RAEB-T | A | I | IDH1 | CGT-TGT | R132C | – |
| 13 | 48/F | AML, NOS | M1 | N | I | IDH2 | AGG-AAG | R172K | – |
| 14 | 59/M | AML, NOS | M1 | N | I | IDH2 | AGG-AAG | R172K | – |
| 15 | 76/F | AML, NOS | M1 | N | I | IDH2 | AGG-AAG | R172K | – |
| 16 | 22/F | AML, NOS | M2 | N | I | IDH2 | AGG-AAG | R172K | – |
AML, NOS, AML not otherwise specified; AML, MLD; AML with myelodysplasia related changes; RAEB-T, refractory anemia with excess blast in transformation; N, normal; I, intermediate-risk; R, arginine; H, histidine; C, Cysteine; K, Lysine; A, abnormal 46, XY, del(5)(q13q33), inv(12)(p11.2q24.1)[6]/46,XY[14]
TP53 mutations in AML cases with IDH1R132 mutation
In glioma, non-R132H mutations of IDH1 are associated with higher frequency of TP53 mutations and a distinct gene expression profile as compared to R132H mutation. We therefore tested 11 available IDH1R132 mutant AML cases for TP53 mutations. We detected a P72R polymorphism, known to predispose to a variety of human cancers, in 4 of 5 R132C cases and in 5 of 6 R132H cases. We did not find any significant difference in frequency of TP53 mutations in R132C and R132H subgroups, as each subgroup had only 1 case with TP53 mutation.
Morphologic and Immunophenotypic Features of AML with IDH mutations
For AMLs with IDH1R132 mutation, the mean blast count was 55% (range, 22–88%). Using the WHO classification, the mutated cases were classified as AML NOS (n=10) or AML with myelodysplasia-related changes (n=2). The AML NOS group was further classified as AML without maturation (FAB M1), AML with maturation (FAB M2), and acute monocytic leukemia (FAB M5). Auer rods were detected in 5 of 10 (50%) cases. Ten of 12 (83%) AML cases with IDH1R132 mutation and 3/4 (75%) AML with IDH2 mutation showed dysplastic features in one of more hematopoietic lineages (Table 4). Bone marrow cellularity ranged from 25% to 100% (mean, 71%).
Table 4.
Hematologic and morphologic features of AML with IDH1R132 and IDH2R172 mutations
| Patient No. | Mutation | Age/Sex | WHO | FAB | WBC (K/mL) | Hb (g/dL) | Platelets (K/ml) | Marrow Cellularity (%) | Blast (%) | Auer Rods | Dysplastic Features |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IDH1 | 37/F | AML, NOS | M1 | 45.9 | 7.9 | 180 | 100 | 84 | +++ | Absent |
| 2 | IDH1 | 37/F | AML, NOS | M1 | 4.9 | 9.6 | 42 | 100 | 84 | + | Mega |
| 3 | IDH1 | 51/F | AML, MLD | M1 | 4.9 | 9.6 | 31 | 80 | 53 | NA | E, Mega |
| 4 | IDH1 | 73/F | AML, NOS | M1 | 16.9 | 10 | 92 | 95 | 67 | ++ | E |
| 5 | IDH1 | 52/F | AML, NOS | M2 | 6.7 | 11.2 | 254 | 25 | 37 | − | E (slight), Mega |
| 6 | IDH1 | 52/M | AML, NOS | M2 | 0.5 | 13.9 | 127 | 30 | 22 | + | M, E (slight) |
| 7 | IDH1 | 53/M | AML, NOS | M2 | 0.3 | 12.5 | 45 | 90 | 27 | − | M, E, Mega |
| 8 | IDH1 | 55/F | AML, NOS | M2 | 2.6 | 13.5 | 213 | 30 | 56 | + | M |
| 9 | IDH1 | 52/F | AML, NOS | M5 | 1.4 | 13.6 | 119 | 80 | 24 | − | M, E (Slight), Mega |
| 10 | IDH1 | 58/F | AML, NOS | M5 | 83.1 | 9.3 | 69 | 90 | 86 | − | M, Mega |
| 11 | IDH1 | 77/F | AML, NOS | M5 | 8.6 | 12.8 | 225 | 90 | 80 | NA | Mega |
| 12 | IDH1 | 62/M | AML, MLD | RAEB-T | 0.7 | 10 | 291 | 40 | 44 | − | Absent |
| 13 | IDH2 | 48/F | AML, NOS | M1 | 1.7 | 8.2 | 99 | 80 | 61 | + | M, E |
| 14 | IDH2 | 59/M | AML, NOS | M1 | 1.8 | 9.7 | 238 | 60 | 61 | + | E |
| 15 | IDH2 | 76/F | AML, NOS | M1 | 11.8 | 9.4 | 18 | 70 | 73 | − | Absent |
| 16 | IDH2 | 22/F | AML, NOS | M2 | 1.7 | 8.4 | 79 | 70 | 42 | +++ | M, E |
AML, NOS, AML not otherwise specified; AML, MLD; AML with myelodysplasia-related changes; RAEB-T, refractory anemia with excess blast in transformation; NA, not available; M, myeloid; E, erythroid; Mega, megakaryocytes
For AMLs with IDH2R172 mutations, the mean blast count was 53% (range, 20–98%). These 4 mutated cases were classified as AML without maturation (n=3) and AML with maturation (n=1). IDH2 mutated cases were significantly more often classified as AML without maturation compared with IDH2 wild-type AML cases 3/4 (75%) versus 27/192 (14%); (Fisher’s exact test; p<0.01). Auer rods were detected in 3 of 4 (75%) cases. Three of four (75%) AML cases with IDH2 mutation showed dysplastic features in one of more hematopoietic lineages (Table 4). Bone marrow cellularity ranged from 60% to 80% (mean:70%).
Flow cytometric Immunophenotypes of AML with IDH1R132 and IDH2R172 mutations are similar and are listed in Table 5. Overall AMLs with IDH1R132 or IDH2R172 mutation showed a myeloid immunophenotype: CD117+, HLA-DR+/−, CD34 −/+, CD38+, CD13+, CD33+ and myeloperoxidase +/− CD64 was present in AML-M5 consistent with monocytic differentiation.
Table 5.
Flow-cyteometric immunophenotype of AML with IDH1R132 and IDH2R172 mutations
| Patient No. | Mutation | Age/Sex | WHO | FAB | CD34 | CD117 | HLA-DR | CD38 | CD13 | CD33 | CD14 | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IDH1 | 37/F | AML, NOS | M1 | − | + | + | + | + | + | − | CD9 + |
| 2 | IDH1 | 37/F | AML, NOS | M1 | + | + | + | + | + | + | − | MPO+ |
| 3 | IDH1 | 51/F | AML, MLD | M1 | + | + | + | + | + | + | − | NA |
| 4 | IDH1 | 73/F | AML, NOS | M1 | − | + | − | + | + | + | − | NA |
| 5 | IDH1 | 52/F | AML, NOS | M2 | + | + | + | + | + | + | NA | MPO + |
| 6 | IDH1 | 52/M | AML, NOS | M2 | − | NA | NA | + | NA | + | − | MPO+, CD16(dim) |
| 7 | IDH1 | 53/M | AML, NOS | M2 | − | + | − | + | + | NA | NA | MPO + |
| 8 | IDH1 | 55/F | AML, NOS | M2 | + | + | + | + | + | + | − | CD4(partial), CD7(partial) |
| 9 | IDH1 | 52/F | AML, NOS | M5 | NA | NA | + | NA | + | + | Dim | CD64+ |
| 10 | IDH1 | 58/F | AML, NOS | M5 | − | + | + | + | + | + | NA | CD9+, CD64(dim), MPO+ |
| 11 | IDH1 | 77/F | AML, NOS | M5 | − | NA | + | + | − | + | − | CD56 +, CD64+ |
| 12 | IDH1 | 62/M | AML, MLD | RAEB-T | + | + | + | − | + | + | − | CD56+, CD64−, MPO− |
| 13 | IDH2 | 48/F | AML, NOS | M1 | + | + | + | + | + | + | NA | MPO+ |
| 14 | IDH2 | 59/M | AML, NOS | M1 | + | + | + | + | + | + | NA | MPO+, TdT− |
| 15 | IDH2 | 76/F | AML, NOS | M1 | + | + | + | + | + | + | − | MPO+ |
| 16 | IDH2 | 22/F | AML, NOS | M2 | + | + | + | + | + | + | − | MPO+ |
AML, NOS, AML not otherwise specified; AML, MLD; AML with myelodysplasia-related changes; RAEB-T, refractory anemia with excess blast in transformation; NA, not available; MPO, myeloperoxidase; TdT, terminal deoxyribonucleotide transferase
Clinical Features of IDH Mutated AML cases
For the 12 IDH1 mutated AMLs, the mean age was 55 years (range, 37–77) and the male to female ratio was 1 to 3. The mean WBC count was 15 × 109/L. For the 4 IDH2 mutated cases, the mean age was 51 years (range, 22–76) and the male to female ratio was 1 to 3. The mean WBC was 4 × 109/L. There was no significant difference in age, sex, WBC count, platelet count, hemoglobin level, bone marrow blast count and cellularity, dysplastic features, and morphologic classification between the IDH1 or IDH2 mutated AML (Table 1).
These clinical variables in IDH1 or IDH2 mutated AML cases were also compared with wild type AMLs. IDH2R172 mutant AMLs were also significantly associated with a lower WBC count compared with wild-type AMLs (4.3 vs. 20 × 109/L; student’s t test; p<0.001). There was no significant difference in age, sex, and blast count between either IDH1 mutated versus unmutated AML cases, or between IDH2R172 mutated versus unmutated cases (Table 2).
Meta-Analysis of AML with IDH1R132 mutation
We performed a meta-analysis of all available studies that assessed for IDH mutations in AML cases. To date, virtually all studies have focused on IDH1 mutations in AML, with a total of 1512 AMLs reported; 146 (9.7%) cases being mutated (Table 6). Since IDH1R132 mutation is predominantly restricted to CN-AML, inclusion of AML cases with abnormal karyotypes in earlier studies most likely under-represents the true frequency of IDH1 mutations in CN-AML. We extracted the fraction of CN-AML with IDH1R132 mutation from the studies reported. Out of 1152 CN-AML tested, 130 (11%) showed IDH1R132 mutation compared with 16 of 360 (4%) AML with an abnormal karyotype. Review of cytogenetic studies showed that all 66 IDH1 mutated AML cases were limited to the intermediate-risk category. IDH1R132 mutations resulted in a higher R132H substitution (73/146, 50%) than R132C substitution (45/143, 31%).
Table 6.
Meta-Analysis of AML with IDH1R132 mutation
| Our Study (N=12) N (%) |
Mardis et al11, 2009 (N=16) N (%) |
Chou et al18, 2010 (N=27) N (%) |
Marcucci et al16, 2010 (N=49)‡ N (%) |
Wagner et al15, 2010 N=30 N (%) |
Ho et al17, 2010 N=12 N (%) |
Total Cases N=146 N (%) |
|
|---|---|---|---|---|---|---|---|
| % of all cases in the study | 12/199 (6) | 16/188 (9) | 27/493 (6) | 49/358 (14) | 30/275 (11) | 12/274 (4) | 146/1512(10) |
| Age in years | 55 | 48.9 ± | 52.5 | 62* | 50* | 61* | 52** |
| (Mean, Range) | (37–77) | 15.4 | (25–75) | (21–82) | (33–80) | (34–81) | |
| M:F | 1:3 | 1.3:1 | 1.1:1 | 1:1.1 | 1:1.7 | 2:1 | 1:1.1 |
| FAB Classification | NA | NA | |||||
| M1 | 4 (33) | 10 (62) | 14 (52) | 6 (20) | 28/55 (51) | ||
| M2 | 4 (33) | 3 (19) | 9 (33) | 9 (30) | 16/55 (29) | ||
| M4 | 0 | 3 (19) | 2 (7) | 8 (27) | 5/55 (9) | ||
| M5 | 2 (17) | 0 | 2 (7) | 4 (13) | 4/55 (7) | ||
| RAEB-T | 2 (17) | 0 | 0 | – | 2/55 (4) | ||
| % Blast (Mean, Range) | 55 (22–88) | 76.7 ± 16.4 | NA | 73* (33–99) | 80* (20–99) | 80* (38–99) | 67** |
| Cytogenetic Findings | NA† | NA† | |||||
| Normal | 11 (92) | 13 (81) | 20/26 (77) | 6/10(60) | 50/64 (78) | ||
| Abnormal | 1 (8) | 3 (19) | 6/26 (23) | 4/10(40) | 14/64 (12) | ||
| Cytogenetic Risk-Group | – | 0 | 0 | NA† | NA† | 0 | 0 |
| Favorable | 12 (100) | 16 (100) | 26/26 (100) | 12 (100) | 66/66(100) | ||
| Intermediate | 0 | 0 | 0 | 0 | 0 | ||
| Poor | |||||||
| Frequency in CN- AML | 11/127 (9) | 13/80(16) | 20/227 (8) | 49/358 (14) | 30/275 (11) | 6/85 (7) | 130/1152 (11) |
| Types of IDH1 Mutation | |||||||
| R132H | 6 (50) | 7 (44) | 7 (26) | 24 (49) | 21 (70) | 8 (67) | 73/146(50) |
| R132C | 6 (50) | 8 (50) | 10 (37) | 15 (31) | 5 (17) | 1 (9) | 45/146(31) |
| R132S | – | 1 (6) | 5 (19) | 5 (10) | 3 (10) | – | 13/146(9) |
| R132L | – | – | 1 (4) | – | – | 2 (18) | 3/146 (2) |
| R132G | – | – | 4 (15) | – | 1 (3) | 1 (9) | 6/146 (4) |
| Coexisting Mutations | |||||||
| NPM1 | 5/11 (46) | 7 (44) | 15/27 (56) | 34/48(71) | 17/30(57) | 9/12(75) | 87/144(60) |
| FLT3-ITD | 3/12 (25) | 4 (25) | 10/27 (37) | 10/49(20) | 4/30 (13) | 6/12(50) | 29/146(20) |
| FLT3-D835 | 1/12 (9) | 1 (6) | 3/27 (11) | 3/48 (6) | NA | NA | 8/103 (8) |
| CEBPA | 2/10 (20) | NA | 1/27 (4) | 2/36 (6) | 8/30 (27) | 1/12(9) | 14/115(12) |
| NPM1+FLT3-ITD | 2/12 (17) | NA | 9/27 (33) | NA | NA | NA | 11/39 (28) |
| NRAS | 2/12 (17) | 1 (6) | 4/27 (15) | NA | NA | 2/12(18) | 9/67 (13) |
| KRAS | 0/12 | NA | 0/27 | NA | NA | NA | 0/39 |
| KIT | 1/9 (11) | NA | 0/27 | NA | NA | NA | 1/36 (3) |
| IDH2 | 0/11 | NA | NA | 0/49 | NA | NA | 0/60 |
median age;
average of mean values only;
only cytogenetically normal cases tested in this study;
includes two non-R132 mutations; RAEB-T, refractory anemia with excess blast in transformation; NA, not available; R, arginine; H, histidine; C, cysteine; L, leucine; G, glycine
Analysis of available test-results for co-existing mutations shows NPM1 to be the most common concurrent mutation, in 87 of 144 (60%), followed by FLT3-ITD (29/146, 20%) and CEBPA (14/155,12%). None of the 60 AML with IDH1R132 mutation showed a concurrent IDH2 mutation. The AML cases assessed for IDH2 mutation is too few to perform similar meta-analysis.
Discussion
IDH1 and IDH2, NADP+-dependent isocitrate dehydrogenases, catalyze the oxidative carboxylation of isocitrate to α-ketoglutarate. IDH1 plays a key role in cytosolic NADPH production necessary for the regeneration of reduced glutathione, a main antioxidant in mammalian cells.26 Recurring mutations in IDH1 and IDH2 occur at a very high frequency in many different types of glioma, especially in secondary glioblastoma.12, 27 IDH mutations in gliomas are always reported at the same arginine residues, R132 in IDH1 and R172 in IDH2, responsible for hydrophilic interactions with isocitrate.28 Initial analyses showed that IDH mutations were restricted primarily to gliomas, with rare cases of prostate cancer and B-lymphoblastic leukemia showing IDH mutations.12, 29, 30 More recently, Masrdis and colleagues sequenced the entire genome of a cytogenetically normal case of AML (CN-AML) and detected a novel IDH1R132 mutation.9 They subsequently screened 187 AML cases and showed a heterozygous IDH1R132 mutation in 15 (8%) cases. Sixteen (original case plus 15 additional cases) IDH1R132 mutated cases had available cytogenetic data and 13 (81%) were CN-AML. In addition, all 16 IDH1 mutated AMLs had intermediate-risk cytogenetics. These findings were in contrast to previous studies that did not detect IDH1R132 mutation in 145 AML cases.12, 30 Differences in the cytogenetic findings in the study groups and sensitivities of the detection assays could underlie these discrepancies. For this reason, we chose to undertake this study reviewing our experience in approximately 200 cases of AML at our institution.
Based on the available data in the literature, we specifically studied AML cases with intermediate-risk or poor risk cytogenetic findings.9 Our results show that the frequency of IDH1 mutations is lower in CN-AML cases. We detected IDH1R132 mutations in 11 of 127 (9%) CN-AML, compared to the initial report of 16%. We show that all AML cases with IDH1 mutation have intermediate-risk cytogenetics. In addition, we identified IDH2R172 mutations in 4 of 196 (2%) AML cases, all of which were CN-AML. In no case did we find both IDH1 and IDH2 mutations, strongly suggesting that IDH1R132 and IDH2R172 are mutually exclusive. Only one other study has assessed for both IDH1 and IDH2 mutations in a large group of CN-AML cases. Marcucci and colleagues have shown that IDH mutations occur in 33% of cases, with IDH1 in 14% and IDH2 in 19%.14 The explanation for the higher rate of IDH1 and particularly IDH2 mutations in CN-AML cases in this study, compared with our own data, is explained in large part because Marcucci and colleagues assessed for IDH2 mutations involving the arginine residue at codon 140 (R140). Over 80% of the IDH2 mutations they detected were R140 mutations. We did not test for IDH2R140 mutations in our study as the significance of mutations at this codon is unknown. These mutations have not been shown to have prognostic significance in AML and are not seen in other human cancers. Other authors have suggested that further studies are needed to determine whether R140 substitutions represent a true pathogenic mutation or a polymorphism.14
Since IDH1R132 is mainly limited to CN-AML, the frequency of IDH1R132 mutation in AMLs is likely to vary form one study to the next, depending on the percentage of AML cases with abnormal karyotypes. For example, IDH1R132 mutation was detected in 7% of CN-AML, but in only 4% of all AML cases in a study by Ho and colleagues.15 It is therefore important to account for the cytogenetic composition of the study group when interpreting the frequency of IDH1R132 in AML. We performed meta-analysis of our study and other available studies to obtain a more global view of IDH1 mutations in AML. A total of 146 AML with IDH1R132 mutation have been reported. Most cases with IDH1 mutation have been CN-AML and restricted to the intermediate-risk cytogenetic group. The overall frequency of IDH1R132 mutation in CN-AML is 11% (141 of 1279). The male to female ratio is 1 to 1.1.
Despite the recent interest in IDH mutations in AML, limited amount of information is available on morphologic and immunophenotypic features. Morphologic classification using the FAB system is available in 55 cases. The most common morphologic types were M1 in 28 (51%) and M2 in 16 (29%) cases. Very few cases have been classified using the 2008 WHO system. The classification of cases in our study closely matches what has been reported. The IDH1 mutated cases were classified as AML NOS (n=10) or AML (n=2) with myelodysplasia related changes. Interestingly, the IDH2 mutated cases were all AML NOS. In this category, 7 cases were AML without maturation (FAB M1), 5 cases were AML with maturation (FAB M2) and 3 cases were acute monocytic leukemia (FAB M5). The immunophenotype of most cases in our study was typical myeloid, with expression of CD13, CD33, and CD117 in most cases, and expression of CD34 in 9 of 15 assessed. These findings are generally consistent with a recent report in a Chinese population by Chou and colleagues.16 However, Chou et al report that monocytic differentiation (FAB M4) and expression of CD13, CD14, and HLA-DR are unusual in AQML with IDH mutation. Our experience differs as 3 cases showed monocytic differentiation (FAB M5) and most cases expressed CD13 and HLA-DR.
Our study is the first to report bone marrow aspirate and biopsy findings of AML with IDH mutations in some detail. It is interesting to note that over 80% AML cases with IDH1 or IDH2 mutations showed varying degrees of dysplastic findings in erythroid, myeloid and megakaryocytic lineages. IDH1 mutations are shown to be early events in development of astrocytomas and oligodendrogliomas. It seems plausible that IDH1 mutations are also an early event in myeloid neoplasia. A recent publication has reported IDH1 mutations in early myelodysplastic syndrome (MDS) and in secondary AML (sAML) arising from MDS or MDS/myeloproliferative neoplasms (MPN).31 In our study, one AML patient with IDH1R132 mutation had a history of myelodysplastic syndrome (patient 12, Tables 3, 4 and 5). In two available studies on IDH1 mutations in MPN, one showed IDH1 mutation with coexisting JAK2 mutation, while the other failed to detect IDH1 mutation in MPN that transformed to leukemia.32, 33 The involvement of IDH1 in myeloid neoplasms could be explained by a high requirement of glutamate, an essential amino acid that is converted into α-ketoglutarate and act as a substrate for the mutant IDH1 as described below, in myeloid cells.17, 26, 34 Indeed, Acivicin, a glutamine antagonist, decreased the growth and viability of treatment of a variety of leukemia cell lines.35
We detected equal distribution of R132H and R132C mutations in 12 AML with IDH1R132 mutation. In the meta-analysis, R132H (73/146, 50%) and R132C (45/146, 31%) were the most frequent mutations detected. These findings suggest a different pattern of distribution of IDH1R132 mutations in AMLs than in than gliomas, where R132H mutation constitutes ~90% of IDH1R132 mutations.12 In addition, we did not detect differences in TP53 mutation profiles of R132H and R132C AML cases, unlike those shown in gliomas.36 These findings suggest possible differences in the role of IDH1 mutations between glioma and AML. In fact, IDH1 mutations are associated with good prognosis in glioma, whereas the limited amount of literature in AML suggests lower disease-free survival in young patients with IDH1mutant/NPM1mutant/FLT3-ITDwild-type group.14, 37
IDH1R132 mutations are frequently accompanied by NPM1, FLT3, CEBPA, RAS and KIT mutations. We found NPM1 (5/11, 46%) and FLT3-ITD (2/12, 25%) mutations to be the two most frequent. Comparable frequencies of NPM1 (87/144, 60%) and FLT3-ITD (29/146, 20%) were detected in the meta-analysis. Unlike a recent study, we did not find statistically significant correlation of IDH1R132 and IDH2R172 mutations with NPM1-mutated and FLT3-ITD-negative low-risk molecular profile.14 Frequent presence of coexisting mutations suggest that IDH1 mutations may act cooperatively in leukomogenesis. In contrast, no additional mutations were detected with IDH2R172 mutations in 4 cases. This finding is consistent with 13 cases in the only other available study.14
The details of a possible pathogenic role of IDH mutations are just beginning to emerge. Traditionally, up regulation of a cancer associated transcription factor, hypoxia induced factor (HIF) has been considered to be a major pathogenic mechanisms.26, 38 More recently, accumulation of 2-hydeoxyglutamate (2-HG) in the cells and the serum of glioma and AML patients with IDH1 mutation has been shown.17, 28, 39, 40 This could be used as a potential diagnostic test in the management of patients with IDH mutations.
All reported studies have used Sanger sequencing based assays, which have sensitivity of about 20%.9, 13–17 Since both IDH1 and IDH2 are heterozygous mutations, a minimum blast count of 40% and/or enrichment of myeloblasts will be needed for the Sanger sequencing-based detection of IDH mutations. In practice, however, we have detected IDH mutations in samples with minimum blast count of 22%. Since there is an indication that the mutation is retained at relapse, highly sensitive laboratory assays will be needed for monitoring therapy response and early relapses.16, 39 The involvement of specific codons allows the use of sensitive approaches such as high resolution melt curve analysis and pyrosequencing-based assays, which are currently under development in our laboratory.
In summary, IDH1R132 and IDH2R172 mutations represent a novel class of point mutations in CN-AML. Both mutations occur predominantly in CN-AML leading to overproduction of an oncometabolite, 2-HG. These mutations are heterozygous in nature and mutually exclusive. Despite many similarities, it is possible that molecularly and clinically, they represent distinct subgroups. IDH1R132 is frequently accompanied by other mutations, whereas IDH2R172 is commonly the only mutation detected. Most AML cases with IDH mutation, either IDH1 or IDH2, are morphologically classified as AML with or without myeloid maturation (FAB M1 or M2), have morphologic evidence of dysplasia, and have a non-distinctive myeloid immunophenotype.
Acknowledgments
The authors wish to acknowledge excellent technical services of Neelima Reddy and Xue Ao in performing the IDH1 and IDH2 assays.
Source of support: Departmental funds
Footnotes
Disclosure/Conflict of Interest
The authors do not have any conflicts of interests to disclose.
References
- 1.Gaidzik V, Dohner K. Prognostic implications of gene mutations in acute myeloid leukemia with normal cytogenetics. Semin Oncol. 2008;35:346–355. doi: 10.1053/j.seminoncol.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara F, Palmieri S, Leoni F. Clinically useful prognostic factors in acute myeloid leukemia. Crit Rev Oncol Hematol. 2008;66:181–193. doi: 10.1016/j.critrevonc.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Marcucci G, Mrozek K, Bloomfield CD. Molecular heterogeneity and prognostic biomarkers in adults with acute myeloid leukemia and normal cytogenetics. Curr Opin Hematol. 2005;12:68–75. doi: 10.1097/01.moh.0000149608.29685.d1. [DOI] [PubMed] [Google Scholar]
- 4.Schlenk RF, Dohner K. Impact of new prognostic markers in treatment decisions in acute myeloid leukemia. Curr Opin Hematol. 2009;16:98–104. doi: 10.1097/MOH.0b013e3283257adb. [DOI] [PubMed] [Google Scholar]
- 5.Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, Habdank M, Spath D, Morgan M, Benner A, Schlegelberger B, Heil G, Ganser A, Dohner H. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 6.Scholl S, Fricke HJ, Sayer HG, Hoffken K. Clinical implications of molecular genetic aberrations in acute myeloid leukemia. J Cancer Res Clin Oncol. 2009;135:491–505. doi: 10.1007/s00432-008-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swerdlow CESH, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. Who classification of tumors of haematopoietic and lymphoid tissue. IARC; Lyon: 2008. [Google Scholar]
- 8.Bienz M, Ludwig M, Leibundgut EO, Mueller BU, Ratschiller D, Solenthaler M, Fey MF, Pabst T. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005;11:1416–1424. doi: 10.1158/1078-0432.CCR-04-1552. [DOI] [PubMed] [Google Scholar]
- 9.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the idh1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 11.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. Idh1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 12.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. Idh1 and idh2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner K, Damm F, Gohring G, Gorlich K, Heuser M, Schafer I, Ottmann O, Lubbert M, Heit W, Kanz L, Schlimok G, Raghavachar AA, Fiedler W, Kirchner HH, Brugger W, Zucknick M, Schlegelberger B, Heil G, Ganser A, Krauter J. Impact of idh1 r132 mutations and an idh1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: Snp rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010 doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- 14.Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S, Metzeler KH, Powell BL, Carter TH, Kolitz JE, Wetzler M, Carroll AJ, Baer MR, Caligiuri MA, Larson RA, Bloomfield CD. Idh1 and idh2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A cancer and leukemia group b study. J Clin Oncol. 2010 doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho PA, Alonzo TA, Kopecky KJ, Miller KL, Kuhn J, Zeng R, Gerbing RB, Raimondi SC, Hirsch BA, Oehler V, Hurwitz CA, Franklin JL, Gamis AS, Petersdorf SH, Anderson JE, Reaman GH, Baker LH, Willman CL, Bernstein ID, Radich JP, Appelbaum FR, Stirewalt DL, Meshinchi S. Molecular alterations of the idh1 gene in aml: A children’s oncology group and southwest oncology group study. Leukemia. 2010 doi: 10.1038/leu.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou WC, Hou HA, Chen CY, Tang JL, Yao M, Tsay W, Ko BS, Wu SJ, Huang SY, Hsu SC, Chen YC, Huang YN, Chang YC, Lee FY, Liu MC, Liu CW, Tseng MH, Huang CF, Tien HF. Distinct clinical and biologic characteristics in adult acute myeloid leukemia bearing the isocitrate dehydrogenase 1 mutation. Blood. 2010;115:2749–2754. doi: 10.1182/blood-2009-11-253070. [DOI] [PubMed] [Google Scholar]
- 17.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated idh1 and idh2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo Z, Chen SS, Chandra PK, Galbincea JM, Soape M, Doan S, Barkoh BA, Koeppen H, Medeiros LJ, Luthra R. Application of cold-pcr for improved detection of kras mutations in clinical samples. Mod Pathol. 2009;22:1023–1031. doi: 10.1038/modpathol.2009.59. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, Weller M, Herold-Mende C, Unterberg A, Jeuken JW, Wesseling P, Reifenberger G, von Deimling A. Type and frequency of idh1 and idh2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 20.Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, Behre G, Hiddemann W, Tenen DG. Dominant-negative mutations of cebpa, encoding ccaat/enhancer binding protein-alpha (c/ebpalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 21.Lasa A, Carricondo MT, Carnicer MJ, Perea G, Aventin A, Nomdedeu JF. A new d816 c-kit gene mutation in refractory aml1-eto leukemia. Haematologica. 2006;91:1283–1284. [PubMed] [Google Scholar]
- 22.Lin P, Jones D, Medeiros LJ, Chen W, Vega-Vazquez F, Luthra R. Activating flt3 mutations are detectable in chronic and blast phase of chronic myeloproliferative disorders other than chronic myeloid leukemia. Am J Clin Pathol. 2006;126:530–533. doi: 10.1309/JT5BE2L1FGG8P8Y6. [DOI] [PubMed] [Google Scholar]
- 23.Agell L, Hernandez S, de Muga S, Lorente JA, Juanpere N, Esgueva R, Serrano S, Gelabert A, Lloreta J. Klf6 and tp53 mutations are a rare event in prostate cancer: Distinguishing between taq polymerase artifacts and true mutations. Mod Pathol. 2008;21:1470–1478. doi: 10.1038/modpathol.2008.145. [DOI] [PubMed] [Google Scholar]
- 24.Yin CC, Medeiros LJ, Glassman AB, Lin P. T(8;21)(q22;q22) in blast phase of chronic myelogenous leukemia. Am J Clin Pathol. 2004;121:836–842. doi: 10.1309/H8JH-6L09-4B9U-3HGT. [DOI] [PubMed] [Google Scholar]
- 25.Mrozek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: Are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. N Engl J Med. 2009;360:813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. Idh1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frezza C, Tennant DA, Gottlieb E. Idh1 mutations in gliomas: When an enzyme loses its grip. Cancer Cell. 2010;17:7–9. doi: 10.1016/j.ccr.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 29.Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, Frattini M, Molinari F, Knowles M, Cerrato A, Rodolfo M, Scarpa A, Felicioni L, Buttitta F, Malatesta S, Marchetti A, Bardelli A. Idh1 mutations at residue p.R132 (idh1(r132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 30.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, Lee JY, Yoo NJ, Lee SH. Mutational analysis of idh1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 31.Kosmider O, Gelsi-Boyer V, Slama L, Dreyfus F, Beyne-Rauzy O, Quesnel B, Hunault-Berger M, Slama B, Vey N, Lacombe C, Solary E, Birnbaum D, Bernard OA, Fontenay M. Mutations of idh1 and idh2 genes in early and accelerated phases of myelodysplastic syndromes and mds/myeloproliferative neoplasms. Leukemia. 2010 doi: 10.1038/leu.2010.52. [DOI] [PubMed] [Google Scholar]
- 32.Green A, Beer P. Somatic mutations of idh1 and idh2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med. 2010;362:369–370. doi: 10.1056/NEJMc0910063. [DOI] [PubMed] [Google Scholar]
- 33.Abdel-Wahab O, Manshouri T, Patel J, Harris K, Yao J, Hedvat C, Heguy A, Bueso-Ramos C, Kantarjian H, Levine RL, Verstovsek S. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 2010;70:447–452. doi: 10.1158/0008-5472.CAN-09-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitoh T, Kubota M, Takimoto T, Hashimoto H, Shimizu T, Sano H, Akiyama Y, Mikawa H. Metabolic basis for differential glutamine requirements of human leukemia cell lines. J Cell Physiol. 1990;143:150–153. doi: 10.1002/jcp.1041430120. [DOI] [PubMed] [Google Scholar]
- 35.Nichols KE, Chitneni SR, Moore JO, Weinberg JB. Monocytoid differentiation of freshly isolated human myeloid leukemia cells and hl-60 cells induced by the glutamine antagonist acivicin. Blood. 1989;74:1728–1737. [PubMed] [Google Scholar]
- 36.Gravendeel LA, Kloosterhof NK, Bralten LB, van Marion R, Dubbink HJ, Dinjens W, Bleeker FE, Hoogenraad CC, Michiels E, Kros JM, van den Bent M, Smitt PA, French PJ. Segregation of non-R132h mutations in idh1 in distinct molecular subtypes of glioma. Hum Mutat. 2010 doi: 10.1002/humu.21201. [DOI] [PubMed] [Google Scholar]
- 37.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan H, Bigner DD, Velculescu V, Parsons DW. Mutant metabolic enzymes are at the origin of gliomas. Cancer Res. 2009;69:9157–9159. doi: 10.1158/0008-5472.CAN-09-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, Dang L, Fantin VR, Mak TW. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated idh1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]


