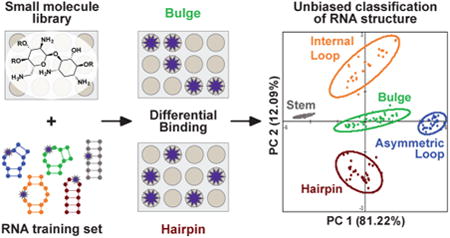
Abstract
Three-dimensional RNA structures are notoriously difficult to determine, and the link between secondary structure and RNA conformation is only beginning to be understood. These challenges have hindered the identification of guiding principles for small molecule:RNA recognition. We herein demonstrate that the strong and differential binding ability of aminoglycosides to RNA structures can be used to classify five canonical RNA secondary structure motifs through principal component analysis (PCA). In these analyses, the aminoglycosides act as receptors, while RNA structures labeled with a benzofuranyluridine fluorophore act as analytes. Complete (100%) predictive ability for this RNA training set was achieved by incorporating two exhaustively guanidinylated aminoglycosides into the receptor library. The PCA was then externally validated using biologically relevant RNA constructs. In bulge-stem-loop constructs of HIV-1 transactivation response element (TAR) RNA, we achieved nucleotide-specific classification of two independent secondary structure motifs. Furthermore, examination of cheminformatic parameters and PCA loading factors revealed trends in aminoglycoside:RNA recognition, including the importance of shape-based discrimination, and suggested the potential for size and sequence discrimination within RNA structural motifs. These studies present a new approach to classifying RNA structure and provide direct evidence that RNA topology, in addition to sequence, is critical for the molecular recognition of RNA.
Graphical abstract
Introduction
RNA molecules have recently been revealed to play regulatory roles in a wide array of biological processes, ranging from the initial steps of embryonic development to the progression of metastatic cancer.1,2 At the same time, myriad questions remain regarding their fundamental biochemistry and the principles behind RNA molecular recognition.3,4 Experimentally, the recognition of RNA by proteins has largely been probed from a sequence-dependent point of view to date,5,6 although several cases of structure-based recognition have been reported7 and a lack of sequence conservation in several long noncoding RNA protein recognition elements suggests that RNA topology can play an equally important role in their regulatory activities.8,9 Elucidation of the regulatory and/or disease-related roles of these RNA:protein interactions thus requires a fundamental understanding of how shape impacts RNA recognition. At the same time, RNA structural characterization remains a difficult problem. For 2D structures, that is, base-pairing, computational predictions can be vastly improved using chemical probing techniques such as selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) and related techniques.10–12 These techniques are widely used and have led to the elucidation of an impressive range of RNA structures,13–15 although in some cases these techniques can yield multiple possible structures and require input from other experimental and phylogenetic analyses.16,17 For 3D RNA structures, computational prediction is currently limited to short sequences,18 while experimental characterization can be difficult, time-consuming, and often impossible using traditional methods such as NMR and X-ray diffraction. Ongoing work in the combination of 2D probing and 3D predictions19 as well as NMR-observable connections between junction topology and RNA dynamics20,21 demonstrate the extant need for a wide range of tools to understand RNA three-dimensional structure and its connection to molecular recognition. Indeed, recent NMR and computational studies have suggested that the shape or topology of internal bulge and loop motifs, that is, two-way junctions, is critical to determining global RNA conformations.20,22 A relationship between such topologies and the molecular recognition of RNA has yet to be defined.
Small molecule probes offer a promising solution for RNA studies and have proven highly effective tools for investigating proteins. While the difficulties in the characterization of RNA 3D structure often hinder the rational design of RNA ligands, interest in the targeting of RNA with small molecules has surged.23–25 Indeed, recent successes of in vivo RNA targeting suggest that RNA structures other than the ribosome may constitute plausible drug targets.26–29 In contrast to synthetic oligonucleotides, which are limited to targeting single-stranded sequences, small molecules offer the opportunity for three-dimensional structure-based targeting.30–32 Many unanswered questions remain, however, regarding both the small molecule properties and the RNA structures that allow selective interactions, and insights into this relationship would facilitate both RNA-targeted drug discovery and the development of small molecule probes for RNA structure and function.
Molecular-scale pattern-based sensing offers an opportunity to elucidate complex recognition properties through the use of receptors that interact differentially with the analyte of interest without the need for highly specific receptor:analyte pairing. This method mimics the olfactory sense, where hundreds of cross-reactive receptors are thought to distinguish up to 1 trillion stimuli.33 Small molecule receptors have been employed to detect and classify analytes ranging from inorganic ions34 to protein functional classes35,36 to whole cells,37 allowing for single-step optical assays that can answer questions ranging from the differentiation of red wines38 to the disease state of a human cell.39 To our knowledge, however, this technique has not been used to differentiate structural motifs of biomacromolecules. We sought to determine if RNA-binding small molecules could be used as receptors to differentially sense RNA structures as analytes. This method has the potential to reveal both the required small molecule properties for selective RNA binding and the components of RNA topology that define the molecular recognition of RNA.
To test this hypothesis, we selected a library of known RNA-binding ligands and designed a training set of small RNA constructs that contained five canonical RNA secondary structure motifs: bulge, asymmetrical internal loop, symmetric internal loop, hairpin, and stem. The RNA analyte was fluorescently labeled at the structural motif of interest, and changes in the emission intensity at different concentrations of ligand were recorded. These fluorescence data were used as the input for principal component analysis (PCA), which revealed unbiased clustering of the RNA training set according to the canonical secondary structure definitions. In addition, comparison of the chemical and physical properties of aminoglycosides with their differentiation ability explained some but not all of the recognition trends. A deeper look within each RNA structural class revealed a potential to further distinguish motif size and sequence. These results reveal that three-dimensional shape and topology are key determinants for RNA recognition at the molecular level, even outside of differences in the sequence and size of a motif, and suggest that small molecules may be able to differentiate more complex RNA topologies in the future.
Results and Discussion
Initial Aminoglycoside Receptor Library
Aminoglycosides are arguably the best-known commercially available RNA ligands and generally feature a central 2-deoxystreptamine (2-DOS) core decorated with one to four amine-rich pyranose or furanose ring structures (Figure 1). While first recognized as antibiotics that target the ribosome, aminoglycosides are now known to bind a wide range of RNA structures with dissociation constants as low as 40 nM.40–46 At the same time, aminoglycoside:RNA selectivity is often limited by the largely electrostatic nature of these interactions.32,47,48 One of the advantages of pattern recognition, however, is that it does not require highly selective interactions but rather differential binding affinities between the analytes and receptors. For our initial experiments, we purchased 12 inexpensive aminoglycosides with varying substitution patterns of the 2-DOS core and with established literature precedent to bind multiple RNA structures with varying affinity (Figure 1).32,49–52
Figure 1.
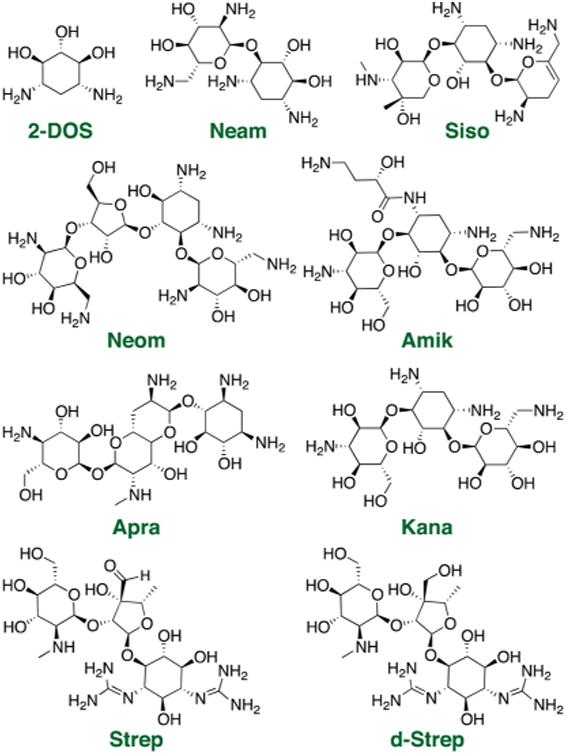
Example of commercially available aminoglycoside receptors used in the pattern recognition assay: 2-deoxystreptamine (2-DOS), neamine (Neam), sisomicin (Siso), neomycin (Neom), amikacin (Amik), apramycin (Apra), kanamycin (Kana), streptomycin (Strep), and dihydrostreptomycin (d-Strep). The entire receptor library is shown in Figure S1-1.
RNA Training Set Design and Synthesis
We next designed an RNA training set that would sample the five most common secondary structure motifs: bulge, symmetric internal loop, asymmetrical internal loop, stem, and hairpin. For each secondary structure motif, the RNA constructs were designed to vary the size and/or sequence of the motif while keeping the flanking regions constant (Figure 2, Table S2-1). To ensure structural stability and facilitate solid-phase synthesis, each RNA construct also maintained ∼50% G–C content and a length less than 40 n.t. (Table S2-2). Initially, potential RNA sequences were analyzed with RNAStructure,53 an online secondary structure prediction software, to ensure the designed fold was predicted at greater than 95% probability. Similar results were observed using the MC-FOLD software54 (see S2). A representative set of 16 RNA sequences was chosen, taking at least 3 sequences for each secondary structure motif while ensuring diversity in size and sequence. Importantly, a lack of sequence conservation in the variable regions was confirmed via motif mapping (Figure S2-1-5). Next, the three-dimensional structures were computationally modeled using Rosetta's FARFAR algorithm, which allows for de novo structural modeling of small RNAs.18,55 An ensemble of conformations was generated for each RNA construct and then visually examined to identify flexible positions within each motif at which to insert a solvatochromic fluorophore (Figure S2-6). Flexible sites were expected to be the most sensitive to small molecule-induced conformational changes.
Figure 2.
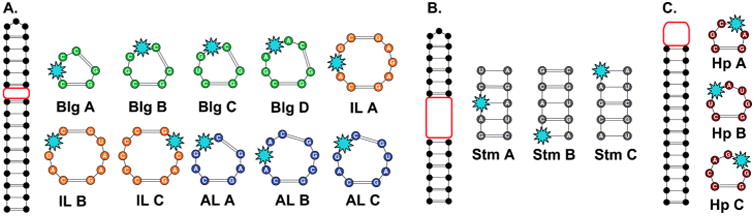
Training set of 16 RNA constructs synthesized by solid-phase synthesis. The fluorophore position is indicated by the light blue star. Red boxes indicate variable regions. (A) The bulge (Blg), symmetric internal loop (IL), and asymmetric internal loop (AL) motif library. (B) Stem (Stm) library. (C) Hairpin (Hp) library. For full sequences, see Table S2-2.
We chose benzofuranyluridine (BFU) as the most practical solvatochromic fluorophore to incorporate into the RNA motifs of interest. First reported by Srivatsan and co-workers, BFU offers native Watson–Crick hydrogen bonding to adenine, while commercially available and commonly used 2-amino-purine (2-AP) can pair with either uracil or cytosine, the latter through protonation or wobble base pairing.56 In addition, BFU offers a higher quantum yield and longer wavelength emission than 2-AP.57 Indeed, the higher sensitivity of BFU allowed for increased reproducibility in our multiwell plate experiments. To efficiently and site-specifically incorporate BFU, we utilized solid-phase oligonucleotide synthesis,58 which required the nucleoside monomer to be synthesized with regiospecific protecting groups at the 2′- and 5′-positions as well as a 3′-O-phosphoramidite. Using a modified synthetic procedure, we started with 5-iodouridine and installed the benzofuran via a microwave-assisted Suzuki–Miyaura coupling reaction in water (2) (Scheme 1).59 Using methods adapted from Burrows, Beal, and co-workers,60 the nucleoside was selectively 3′,5′-O-bissilyated and successively 2′-O-TBDMS protected in a two-step, one-pot reaction to obtain intermediate 3. The silyl acetal was selectively removed using hydrogen fluoride (4), which was then 5′-O-4,4′-dimethoxytrityl (DMT) protected (5).60 The 3′-O-phosphoramidite was installed via a microwave-assisted synthesis protocol developed by Krishnamurthy and co-workers to afford BFU-phosphoramidite 6.61,62 The overall yield from commercial starting materials was 35–45%, as compared to the ∼28% overall yield reported for an alternate method reported after the start of this work.63 Solid-phase synthesis and purification are described in the Supporting Information, S3.
Scheme 1. Microwave-Assisted Suzuki-Miyaura Coupling Reactiona.
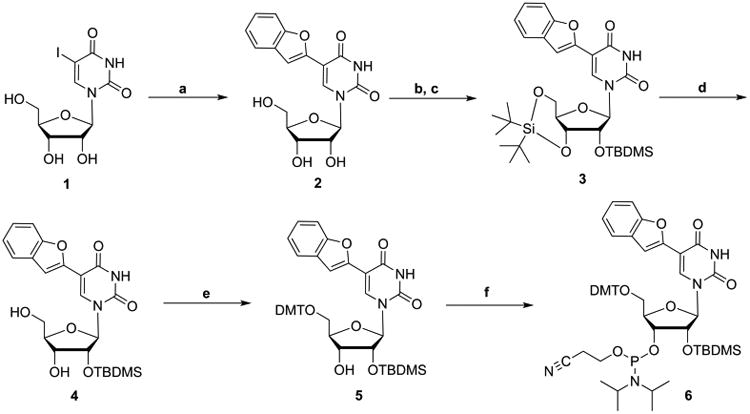
aReagents and conditions: (a) 2-benzofuranylboronic acid, KOH, Na2PdCl4, H2O, 100 °C, μW, 1 h; (b) (t-Bu)2Si(OTf)2, imidazole, DMF, 0 °C, 1.3 h; (c) TBDMS-Cl, imidazole, DMF, 60 °C, 1 h; (d) HF-pyridine, pyridine, CH2Cl2, −10 °C, 2 h; (e) DMT-Cl, pyridine, 0 °C, 6 h; (f) 2-cyanoethyl N,N,N′,N′-tetraisopropyl phosphorodiamidite, 5-(ethylthio)-1H-tetrazole, CH2Cl2, 65 °C, μW, 1 h.
Small Molecule:RNA Interaction Assays and Principal Component Analysis
Each BFU-labeled RNA construct was incubated with each of the 12 initial aminoglycoside receptors at multiple concentrations in a 384-well plate (Figure 3A, S4). The fluorescence intensity of each well was used as the input for PCA, a method that reduces the dimensionality of a given data set by determining the maximum variance within the data without taking into account the identity of the analyte.64,65 PCA thus offers the opportunity to visually examine analyte clustering based only on the measured properties. Gratifyingly, the five canonical secondary structure motifs were found to be clustered into five distinct groups. The successful unbiased clustering of these constructs supports our computational predictions that the label itself does not significantly disrupt the predicted RNA structure or related topology. The predictive power of the PCA was calculated using leave-one-out cross validation analysis (LOOCV).66 With the 12 commercially available aminoglycosides, the PCA plot was 78% predictive. On the basis of loading plot analysis, hygromycin B (Hygro), tobramycin (Tobra), and paromomycin (Paro) were found to bind similarly to all 16 RNA constructs (Figure S5-1). After data were removed for these three receptors, PCA with data from the nine remaining aminoglycosides increased the predictive power to 87% (Figure 3B).
Figure 3.

(A) Relative fluorescence measurements of RNA training set constructs in the presence of kanamycin. All titrations were performed in 10 mM NaH2PO4, 25 mM NaCl, 4 mM MgCl2, 0.5 mM EDTA, pH 7.3 buffer, at 25 °C from 0 to 4 μM aminoglycoside concentration. The titrations were performed in triplicate and averaged for input to PCA. Error bars have been removed for clarity, and error analysis can be found in Table S4-1. Remaining curves can be found in Figure S4-2. (B) PCA plot with nine commercially available aminoglycosides. The predictive power of the PCA plot is 87%. Ovals indicate 95% confidence intervals for each cluster.
Expansion of the Small Molecule Receptor Library
To increase the differentiation and thus predictive power of the RNA training set, we next sought to expand the chemical diversity of our small molecule receptor library. Specifically, we exhaustively guanidinylated two aminoglycosides on the basis of reported procedures from Tor and co-workers.67 Importantly, guanidinylated aminoglycosides are known to bind RNA with higher affinity than their unguanidinylated counterparts.67 To begin, Paro and Kana were functionalized at each amine position using 1,3-di-Boc-2-(trifluoromethylsulfonyl)guanidine followed by deprotection with HCl to yield guanidinoparomomycin (G-Paro) and guanidinokanamycin (G-Kana) (Figure 4A). The RNA training set was assayed against the guanidinylated aminoglycosides under conditions identical to those described previously. Using LOOCV, 100% predictive power was achieved with the addition of the guanidinylated aminoglycosides (Figure 4B).66 On the basis of the successful naive clustering of all five secondary structure motifs, we decided to further test the predictive power of this analysis using a biologically relevant RNA sequence containing multiple secondary structure motifs.
Figure 4.
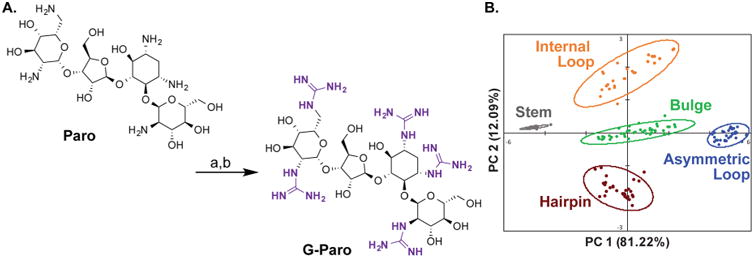
(A) Synthesis of G-Paro (G-Kana followed analogous procedures): (a) 1,3-di-Boc-2-(trifluoromethylsulfonyl)guanidine, triethylamine, 5:1 dioxane:water, rt, 3 days; (b) HCl, ethyl acetate, rt, 4 h. (B) PCA plot of RNA training set with expanded aminoglycoside receptor library. A total of nine commercially available aminoglycosides and 2 guanidino-aminoglycosides were used as receptors. Ovals indicate 95% confidence intervals for each cluster.
Accurate Distinction of Two Secondary Structure Motifs in TAR RNA
The HIV-1 trans-activation response element (TAR) is a well-characterized RNA and a therapeutic target due to its role in promoting transcription of the HIV genome.68–70 Particularly relevant to these studies, TAR contains two distinct secondary structure motifs that can bind small molecules: a 3 n.t. bulge and a 6 n.t. hairpin.70,71 In two separate RNA constructs, BFU was incorporated at either the U25 or the G33 position within the bulge or hairpin motif, respectively, using the same site-selection criteria for labeling as was used for the training set (Figure 5A, S2). The two TAR RNA constructs, Blg-TAR and Hp-TAR, were then assayed against the expanded aminoglycoside receptor library and used as external validation versus the PCA plot of the RNA training set (Figure 5B). Even though both motifs were present in each structure, we were able to independently discriminate the respective labeled motif within each construct with 100% accuracy. These analyses demonstrate that the BFU fluorophore is a location-specific reporter, and we thus propose our pattern recognition technology as an orthogonal approach to rapidly gather site-specific information on RNA structure.
Figure 5.
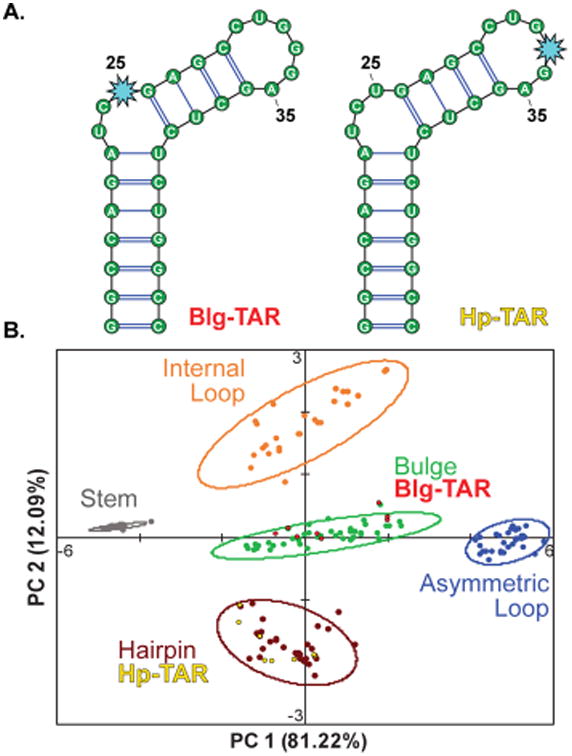
(A) Secondary structures of TAR RNA constructs. The BFU nucleotide (light blue star) was inserted at the U25 and G33 positions, respectively. (B) Training set PCA plot along with the external validation of TAR (red and yellow).
Binding Trends and Cheminformatics Analysis of Aminoglycosides
To gain insight into the small molecule properties important for differentiation, we analyzed the PCA factor loadings and cheminformatic parameters of each aminoglycoside. Factor loadings in a PCA plot reveal which receptors (i.e., aminoglycosides) contribute the most to the variance explained by each principal component (PC) (Figure 6A, Table S5-1). PC1, for example, relied on the strongest contributions from 2-DOS, Neom, Neam, and Kana, although all of the aminoglycosides contributed strongly. PC2 relied on the strongest contribution from Strep and d-Strep followed by Siso, while PC3 demonstrated positive correlations with G-Paro and G-Kana, followed by Strep and d-Strep, which all contain guanidine groups. The trend in guanidine number may reflect the differences in molecular recognition of guanidines versus amines, as guanidines are more basic, planar, and offer more directionality in hydrogen bonding.67 The loading plots (Figure S5-2) visually confirmed these trends and revealed that similar contributions are consistently observed from (a) 2-DOS and Apra; (b) d-Strep, Strep, and Siso; and (c) Neom, Neam, and Kana. We re-evaluated the PCA with the removal of each single receptor and found that, while clusters were visually closer, the LOOCV value remained at 100% (Table S7-2). Removal of two from a redundant group of three, however, did reduce the LOOCV value (91–97%). These analyses supported the robustness of our system as well as the value of modest redundancy.
Figure 6.
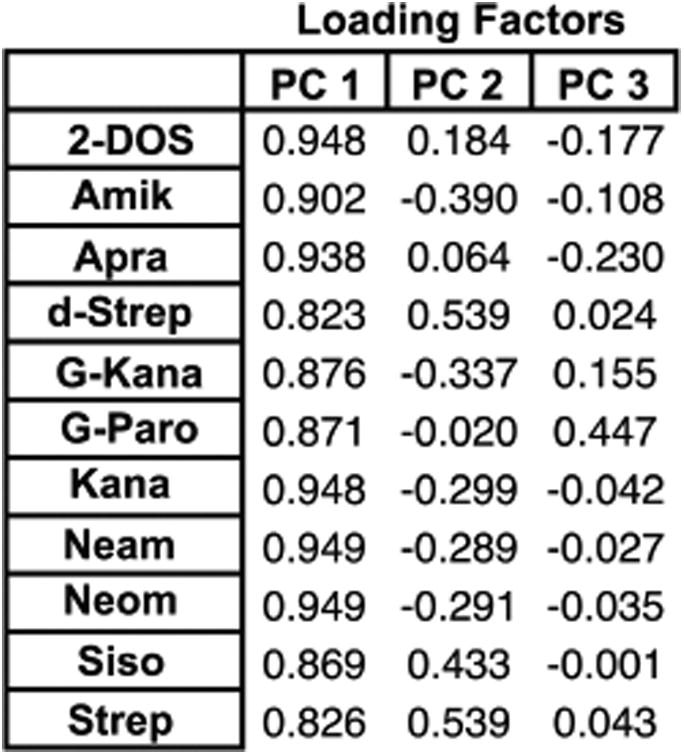
Loading factors of the aminoglycoside receptors for the first three principal components (PCs) of the training set PCA (all PCs available in Table S5-1).
Toward a more detailed analysis, we explored this apparent redundancy through the calculation of two-dimensional path-based fingerprints via the Tanimoto method, where a Tanimoto coefficient (Tc) between two molecules with a score greater than 0.85 general indicates similarity (Figure S7-1).72,73 The observation that Neam, Kana, and Neom, which, respectively, contain 2, 3, and 4 total rings, have similar path-based fingerprints reveals a chemical redundancy in these structures that coincides with the similarity of their loading factors for PC1–3. The comparable contribution of these three aminoglycosides to PC1–3 implies that the calculated redundancies are reflected experimentally as similarities in RNA binding preferences. While Apra was somewhat similar in fingerprint to these aminoglycosides (Tc = 0.76–0.84), it shows very distinct contributions to PC1–3, presumably due to a difference in binding mode. As Apra is the only aminoglycoside with a bicyclic ring, this may highlight the limitations of two-dimensional chemical analyses to represent three-dimensional interactions.74 Furthermore, calculation of standard cheminformatic parameters75 as well as predicted charge did not generally demonstrate correlations with factor loadings. One exception was found in guanidinylated aminoglycosides and PC3 loading factors, presumably due to increased nitrogen number and increased molecular weight but similar total charge relative to standard aminoglycosides (Table S7). The general lack of dependence on molecular weight or total charge is consistent with the more complex view of structure-electrostatic complementarity that has been reported for structured RNAs such as rRNA76,77 and hammerhead ribozymes78 and supports the hypothesis that three-dimensional properties of amino-glycosides must be taken into account in the recognition of a wide range of secondary structure motifs.
Analysis of Differentiation within the RNA Structural Motifs
We also examined the PCA plot of the RNA training set for evidence of structural discrimination within each set of secondary structure motifs differentiated in Figure 2. As previously mentioned, single nucleotide additions and deletions have been shown to strongly influence RNA conformations and are also expected to impact topology.20,22 To begin, all RNA constructs were separately labeled on the PCA plot, and both the 95% confidence intervals and centroid (average) positions were calculated (Figure 7). The bulge, asymmetric internal loop, symmetric internal loop, and hairpin libraries showed clear separation in the centroid values of each RNA construct, although some overlap in the respective 95% confidence intervals was also observed. Along PC1, the position of the centroid values of 3 bulge constructs and 3 asymmetric loop constructs correlated to the size of the secondary structure motif. The less defined clustering of the smaller 2-nucleotide bulge (Bulge A), seen in its wide 95% confidence interval, precluded analysis and may have been due to weaker overall interactions with the aminoglycoside library as compared to the 3- and 4-nucleotide bulge structures. At the same time, the two smallest receptors, 2-DOS and Neam, demonstrated dramatically stronger changes in signal with Bulge A relative to other aminoglycosides (Figure S4-2). When the unusually specific 2-DOS and Neam were removed from the PCA, the 95% confidence interval of Bulge A became more condensed, and the centroid aligned with the general trend in size (Figure S5-3). Dramatic differences in binding affinities among receptors are indeed known to cause skewing effects in PCA,65 although we did not see improvement in our global PCA analysis (Figure S5-4), likely due to the competing effect of reducing the overall number of receptors. While the internal loop constructs were all the same size (3 × 3 nucleotides), separation of the centroid values on PC2 followed differences in purine/pyrimidine ratio; that is, loops with more purine nucleobases had rightward centroids. Finally, the hairpin constructs were separated on PC2 by both loop size and purine/pyrimidine ratio and will require a larger training set to separately evaluate these parameters. Minimal separation was observed among stem constructs, which by definition varied only by sequence and would be expected to differ least in three-dimensional structure among the motifs. Interestingly, it was found that the TAR bulge and hairpin correlated to the 95% confidence intervals of the respective RNA motifs closest in size within the RNA training set (Figure S6-3–5). Taken together, these data suggest that further differentiation may be possible with expanded libraries and training sets, including size and sequence variations within motif classes.
Figure 7.
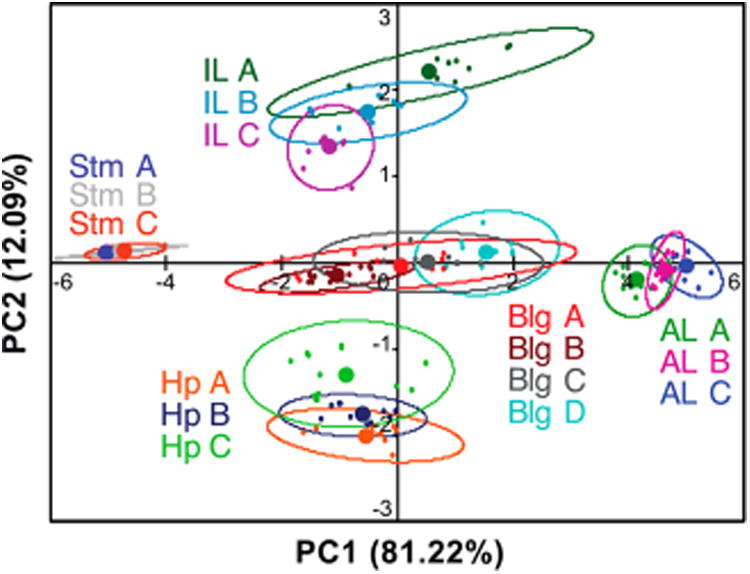
PCA plot with individual RNA constructs separately labeled. Open ovals indicate 95% confidence intervals, while solid circles represent the centroid (average) positions. PC1 is generally proportional to size of secondary structure. The IL motifs are separated by purine/pyrimidine ratio on PC1 and PC2, and the Hp constructs are separated by both the size and the purine/pyrimidine ratio along PC2. See Figure 2 for sequences.
Conclusions
We have provided the first evidence that RNA-binding small molecules can be used as receptors to differentiate and predict canonical RNA secondary structure motifs. Using principal component analysis to reduce the dimensionality of the data, we have created and validated an experimental method that provides insight into both the RNA topological elements and the small molecule ligand properties critical to RNA recognition. Specifically, the clustering of basic secondary structure motifs implies that the general class definitions generated by the RNA Ontology Consortium79 have common topologies that dictate their molecular recognition, independent of size and sequence differences. This classification is particularly notable between bulges and asymmetric internal loops, which impart similar constraints on RNA conformation, presumably due to the availability of noncanonical base pairing between the two sides of the asymmetric loop that can effectively mimic a bulge structure.20,22 The distinct classification of these two motifs in our studies implies that these similarities are less influential in small molecule recognition. Our studies further lend insight into aminoglycoside:RNA interactions, including the observation that differences in total charge and size alone cannot explain the differences in binding interactions over a range of RNA secondary structures. Furthermore, the preliminary separations within RNA secondary structure classes based on size and sequence suggest a secondary layer of complexity in RNA recognition that may be differentiated in future work. In summary, these results support shape complementarity as a critical component of general small molecule:RNA recognition despite the dynamic nature of RNA structure. Expansion of this technology to include a wider range of RNA structure sizes and sequences as well as a more diverse receptor library is expected to reveal further links between RNA topology and molecular recognition as well as elements crucial to the development of small molecule RNA probes. Additional directions include exploring patterns in the combination of secondary structural elements, ranging from simple stem-loop positions to bona fide tertiary structures.
Supplementary Material
All experimental and computational methods and procedures, as well as additional data and analysis (PDF)
Acknowledgments
We thank the members of the Hargrove lab as well as Prof. Hashim Al-Hashimi and co-workers for stimulating discussion and input. A.E.H. wishes to acknowledge financial support for this work from Duke University and the U.S. National Institute of Health (P50GM103297). C.S.E. was supported in part through the U.S. Department of Education GAANN Fellowship (P200A150114). We also wish to thank Dr. George R. Dubay, Director of Analytical Services, Duke University, for performing the high-resolution mass spectroscopy analyses and the Duke University NMR Center for use of the 500 and 400 MHz spectrometers.
Footnotes
Supporting Information: The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b11087.
Notes: The authors declare no competing financial interest.
References
- 1.Cech TR, Steitz JA. Cell. 2014;157:77. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Morris KV, Mattick JS. Nat Rev. Genet. 2014;15:423. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFadden EJ, Hargrove AE. Biochemistry. 2016;55:1615. doi: 10.1021/acs.biochem.5b01141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu Z, Chang HY. Curr Opin Struct. Biol. 2016;36:142. doi: 10.1016/j.sbi.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenan DJ, Query CC, Keene JD. Trends Biochem Sci. 1991;16:214. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 6.Re A, Joshi T, Kulberkyte E, Morris Q, Workman CT. Methods Mol Biol (N Y, NY, U S) 2014;1097:491. doi: 10.1007/978-1-62703-709-9_23. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Kazan H, Lipshitz HD, Morris QD. Wiley Interdiscip Rev: RNA. 2014;5:111. doi: 10.1002/wrna.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR. Mol Cell. 2015;57:552. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkes EJ, Hennelly SP, Novikova IV, Irwin JA, Dean C, Sanbonmatsu KY. Cell Rep. 2016;16:3087. doi: 10.1016/j.celrep.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seetin MG, Mathews DH. Methods Mol Biol (N Y, NY, U S) 2012;905:99. doi: 10.1007/978-1-61779-949-5_8. [DOI] [PubMed] [Google Scholar]
- 11.Siegfried NA, Busan S, Rice GM, Nelson JAE, Weeks KM. Nat Methods. 2014;11:959. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weeks KM. Biopolymers. 2015;103:438. doi: 10.1002/bip.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. Proc Natl Acad Sci U S A. 2013;110:5498. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavender CA, Gorelick RJ, Weeks KM. PLoS Comput. Biol. 2015;11:1. doi: 10.1371/journal.pcbi.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. Proc Natl Acad Sci U S A. 2016;113:10322. doi: 10.1073/pnas.1600008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kladwang W, VanLang CC, Cordero P, Das R. Biochemistry. 2011:50, 8049. doi: 10.1021/bi200524n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, Bolden NC, McCowin S, Case DA, Johnson BA, Salemi M, Telesnitsky A, Summers MF. Science. 2015;348:917. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das R, Karanicolas J, Baker D. Nat Methods. 2010;7:291. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng CY, Chou FC, Kladwang W, Tian S, Cordero P, Das R. eLife. 2015;4:e07600. doi: 10.7554/eLife.07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mustoe AM, Bailor MH, Teixeira RM, Brooks CL, Al-Hashimi HM. Nucleic Acids Res. 2012;40:892. doi: 10.1093/nar/gkr751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mustoe AM, Brooks CL, 3rd, Al-Hashimid HM. Nucleic Acids Res. 2014;42:11792. doi: 10.1093/nar/gku807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailor MH, Sun X, Al-Hashimi HM. Science. 2010;327:202. doi: 10.1126/science.1181085. [DOI] [PubMed] [Google Scholar]
- 23.Guan L, Disney MD. ACS Chem Biol. 2012;7:73. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 24.Morgan BS, Hargrove AEIn. In: Synthetic Receptors for Biomolecules: Design Principles and Applications. BD S, editor. The Royal Society of Chemistry; Cambridge: 2015. p. 253. [Google Scholar]
- 25.Connelly CM, Moon MH, Schneekloth JS. Cell Chem Biol. 2016;23:1077. doi: 10.1016/j.chembiol.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, Ling KKY, Karp GM, Qi H, Woll MG, Chen G, Zhang N, Gabbeta V, Vazirani P, Bhattacharyya A, Furia B, Risher N, Sheedy J, Kong R, Ma J, Turpoff A, Lee CS, Zhang X, Moon YC, Trifillis P, Welch EM, Colacino JM, Babiak J, Almstead NG, Peltz SW, Eng LA, Chen KS, Mull JL, Lynes MS, Rubin LL, Fontoura P, Santarelli L, Haehnke D, McCarthy KD, Schmucki R, Ebeling M, Sivaramakrishnan M, Ko CP, Paushkin SV, Ratni H, Gerlach I, Ghosh A, Metzger F. Science. 2014;345:688. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen L, Luu LM, Peng S, Serrano JF, Chan HYE, Zimmerman SC. J Am Chem Soc. 2015;137:14180. doi: 10.1021/jacs.5b09266. [DOI] [PubMed] [Google Scholar]
- 28.Palacino J, Swalley SE, Song C, Cheung AK, Shu L, Zhang X, Van Hoosear M, Shin Y, Chin DN, Keller CG, Beibel M, Renaud NA, Smith TM, Salcius M, Shi X, Hild M, Servais R, Jain M, Deng L, Bullock C, McLellan M, Schuierer S, Murphy L, Blommers MJJ, Blaustein C, Berenshteyn F, Lacoste A, Thomas JR, Roma G, Michaud GA, Tseng BS, Porter JA, Myer VE, Tallarico JA, Hamann LG, Curtis D, Fishman MC, Dietrich WF, Dales NA, Sivasankaran R. Nat Chem Biol. 2015;11:511. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- 29.Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, Murgolo N, Barbieri CM, Mann PA, Carr D, Xia E, Zuck P, Riley D, Painter RE, Walker SS, Sherborne B, de Jesus R, Pan W, Plotkin MA, Wu J, Rindgen D, Cummings J, Garlisi CG, Zhang R, Sheth PR, Gill CJ, Tang H, Roemer T. Nature. 2015;526:672. doi: 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- 30.Disney MD, Yildirim I, Childs-Disney JL. Org Biomol Chem. 2014;12:1029. doi: 10.1039/c3ob42023j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shortridge MD, Varani G. Curr Opin Struct Biol. 2015;30:79. doi: 10.1016/j.sbi.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermann T. Wiley Interdiscip Rev: RNA. 2016;7:726. doi: 10.1002/wrna.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bushdid C, Magnasco MO, Vosshall LB, Keller A. Science. 2014;343:1370. doi: 10.1126/science.1249168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palacios MA, Nishiyabu R, Marquez M, Anzenbacher P. J Am Chem Soc. 2007;129:7538. doi: 10.1021/ja0704784. [DOI] [PubMed] [Google Scholar]
- 35.Wright AT, Griffin MJ, Zhong Z, McCleskey SC, Anslyn EV, McDevitt JT. Angew Chem Int Ed. 2005;44:6375. doi: 10.1002/anie.200501137. [DOI] [PubMed] [Google Scholar]
- 36.Zamora-Olivares D, Kaoud TS, Dalby KN, Anslyn EV. J Am Chem Soc. 2013;135:14814. doi: 10.1021/ja407397z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Khaldi SF, Mossoba MM, Burke TL, Fry FS. Foodborne Pathog Dis. 2009;6:1001. doi: 10.1089/fpd.2009.0276. [DOI] [PubMed] [Google Scholar]
- 38.Umali AP, LeBoeuf SE, Newberry RW, Kim S, Tran L, Rome WA, Tian T, Taing D, Hong J, Kwan M, Heymann H, Anslyn EV. Chemical Sci. 2011;2:439. [Google Scholar]
- 39.Bajaj A, Miranda OR, Kim IB, Phillips RL, Jerry DJ, Bunz UH, Rotello VM. Proc Natl Acad Sci U S A. 2009;106:10912. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendrix M, Priestley ES, Joyce GF, Wong CH. J Am Chem Soc. 1997;119:3641. doi: 10.1021/ja964290o. [DOI] [PubMed] [Google Scholar]
- 41.Sannes-Lowery KA, Griffey RH, Hofstadler SA. Anal Biochem. 2000;280:264. doi: 10.1006/abio.2000.4550. [DOI] [PubMed] [Google Scholar]
- 42.Blount KF, Tor Y. Nucleic Acids Res. 2003;31:5490. doi: 10.1093/nar/gkg755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Means JA, Hines JV. Bioorg Med Chem Lett. 2005;15:2169. doi: 10.1016/j.bmcl.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Scheunemann AE, Graham WD, Vendeix FA, Agris PF. Nucleic Acids Res. 2010;38:3094. doi: 10.1093/nar/gkp1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tran T, Disney MD. Biochemistry. 2010;49:1833. doi: 10.1021/bi901998m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran T, Disney MD. Biochemistry. 2011;50:962. doi: 10.1021/bi101724h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroeder R, Waldsich C, Wank H. EMBO J. 2000;19:1. doi: 10.1093/emboj/19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulik M, Goral AM, Jasinski M, Dominiak PM, Trylska J. Biophys J. 2015;108:655. doi: 10.1016/j.bpj.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tor Y. ChemBioChem. 2003;4:998. doi: 10.1002/cbic.200300680. [DOI] [PubMed] [Google Scholar]
- 50.Hermann T, Tor Y. Expert Opin Ther Pat. 2005;15:49. [Google Scholar]
- 51.Thomas JR, Hergenrother PJ. Chem Rev. 2008;108:1171. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 52.Aboul-ela F. Future Med Chem. 2010;2:93. doi: 10.4155/fmc.09.149. [DOI] [PubMed] [Google Scholar]
- 53.Reuter JS, Mathews DH. BMC Bioinf. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parisien M, Major F. Nature. 2008;452:51. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 55.Lyskov S, Chou FC, Conchir S, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, Borgo B, Havranek JJ, Kuhlman B, Kortemme T, Bonneau R, Gray JJ, Das R. PLoS One. 2013;8:e63906. doi: 10.1371/journal.pone.0063906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sowers LC, Boulard Y, Fazakerley GV. Biochemistry. 2000;39:7613. doi: 10.1021/bi992388k. [DOI] [PubMed] [Google Scholar]
- 57.Tanpure AA, Srivatsan SG. ChemBioChem. 2012;13:2392. doi: 10.1002/cbic.201200408. [DOI] [PubMed] [Google Scholar]
- 58.Beaucage SL, Iyer RP. Tetrahedron. 1992;48:2223. [Google Scholar]
- 59.Gallagher-Duval S, Herve G, Sartori G, Enderlin G, Len C. New J Chem. 2013;37:1989. [Google Scholar]
- 60.Ghanty U, Fostvedt E, Valenzuela R, Beal PA, Burrows CJ. J Am Chem Soc. 2012;134:17643. doi: 10.1021/ja307102g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Efthymiou T, Krishnamurthy R. Current Protocols in Nucleic Acid Chemistry. John Wiley & Sons, Inc.; New York: 2001. [Google Scholar]
- 62.Meher G, Efthymiou T, Stoop M, Krishnamurthy R. Chem Commun (Cambridge, U K) 2014;50:7463. doi: 10.1039/c4cc03092c. [DOI] [PubMed] [Google Scholar]
- 63.Tanpure AA, Srivatsan SG. Nucleic Acids Res. 2015;43:e149. doi: 10.1093/nar/gkv743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ringner M. Nat. Biotechnol. 2008;26:303. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 65.Stewart S, Adams Ivy M, Anslyn E. Chem Soc Rev. 2014;43:70. doi: 10.1039/c3cs60183h. [DOI] [PubMed] [Google Scholar]
- 66.Baumann K. TrAC, Trends Anal Chem. 2003;22:395. [Google Scholar]
- 67.Luedtke NW, Baker TJ, Goodman M, Tor Y. J Am Chem Soc. 2000;122:12035. doi: 10.1021/jo001142e. [DOI] [PubMed] [Google Scholar]
- 68.Aboul-ela F, Karn J, Varani G. J Mol Biol. 1995;253:313. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 69.Stevens M, De Clercq E, Balzarini J. Med Res Rev. 2006;26:595. doi: 10.1002/med.20081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stelzer A, Frank A, Kratz J, Swanson M, Gonzalez-Hernandez M, Lee J, Andricioaei I, Markovitz D, Al-Hashimi H. Nat Chem Biol. 2011;7:553. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar S, Ranjan N, Kellish P, Gong C, Watkins D, Arya DP. Org Biomol Chem. 2016;14:2052. doi: 10.1039/c5ob02016f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patterson DE, Cramer RD, Ferguson AM, Clark RD, Weinberger LE. J Med Chem. 1996;39:3049. doi: 10.1021/jm960290n. [DOI] [PubMed] [Google Scholar]
- 73.O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. J Chem inf. 2011;3:1. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koutsoukas A, Paricharak S, Galloway WRJD, Spring DR, Ijzerman AP, Glen RC, Marcus D, Bender A. J Chem Inf Model. 2014;54:230. doi: 10.1021/ci400469u. [DOI] [PubMed] [Google Scholar]
- 75.Wenderski TA, Stratton CF, Bauer RA, Kopp F, Tan DS. Methods Mol Biol (N Y, NY, U S) 2015;1263:225. doi: 10.1007/978-1-4939-2269-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kondo J, Hainrichson M, Nudelman I, Shallom-Shezifi D, Barbieri CM, Pilch DS, Westhof E, Baasov T. ChemBioChem. 2007;8:1700. doi: 10.1002/cbic.200700271. [DOI] [PubMed] [Google Scholar]
- 77.Vicens Q, Westhof E. ChemBioChem. 2003;4:1018. doi: 10.1002/cbic.200300684. [DOI] [PubMed] [Google Scholar]
- 78.Hermann T, Westhof E. Biopolymers. 1998;48:155. doi: 10.1002/(SICI)1097-0282(1998)48:2<155::AID-BIP5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 79.Batchelor C, Bittner T, Eilbeck K, Mungall C, Richardson J, Knight R, Stombaugh J, Zirbel C, Westof E, Leontis N. Proc Natl Acad Sci USA. 2009;6 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All experimental and computational methods and procedures, as well as additional data and analysis (PDF)


