Abstract
Mitochondrial homeostasis is regulated by a balance between mitochondrial biogenesis and degradation. Emerging evidence suggests that mitophagy, a selective form of autophagy that degrades mitochondria, plays a key role in the physiology and pathophysiology of mitochondria-enriched cells, such as brown and beige adipocytes. This review discusses findings regarding the roles of autophagy and mitophagy in cellular development, maintenance, and functions of metabolic organs, including adipose tissue, liver, and pancreas. A better understanding of the molecular links between mitophagy and energy metabolism will help to identify promising targets for the treatment of obesity and obesity-associated disorders.
Introduction: Autophagy and Mitophagy
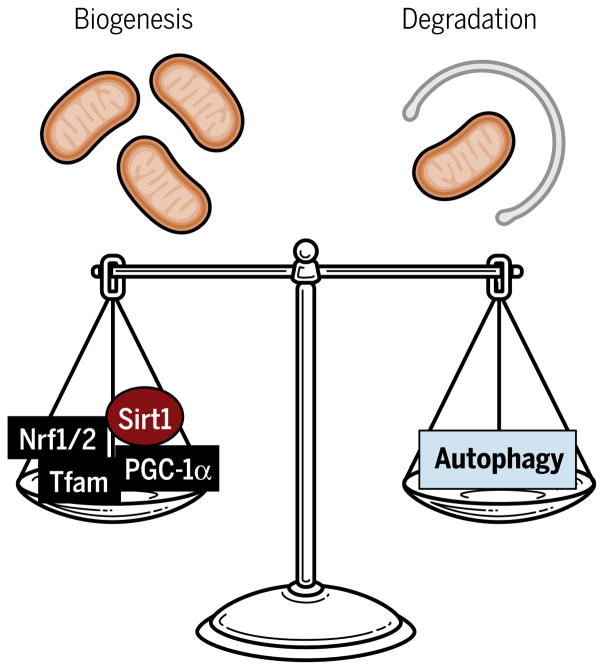
Mitochondria are double-membrane organelles that serve as the central source of ATP (adenosine 5′-triphosphate) with which cells carry out various functions. The cell must undergo both mitochondrial biogenesis and mitochondrial degradation to maintain “healthy” mitochondria in response to varying energetic demands of the cell (Fig. 1). On one end, mitochondrial biogenesis is tightly regulated by various transcriptional regulators encoded by nuclear genes, including peroxisome proliferator–activated receptor γ (PPARγ) coactivator 1α (PGC-1α), nuclear respiratory factor 1 (Nrf1) and Nrf2, and mitochondrial transcription factor A (Tfam). Transcriptional control of mitochondrial biogenesis has been previously discussed in detail (1).
Fig. 1. Regulation of mitochondrial dynamics.
Mitochondrial content is regulated by abalancebetweenmitochondrialbiogenesisanddegradation. Nuclear-codedtranscriptional regulators, such as PGC-1α, Nrf1 and Nrf2 (Nrf1/2), and Tfam, control mitochondrial biogenesis, whereas autophagy removes damaged or unwanted mitochondria. Sirt1, sirtuin 1.
On the other end, mitochondrial degradation is carried out through autophagy, a process of intracellular degradation to break down unwanted or damaged cellular components. The main hallmark of autophagy that distinguishes it from other degradation processes is the formation of a double-membrane vesicle, the autophagosome, to deliver large cytoplasmic components to the lysosome for degradation. The detailed processes of autophagosome formation are described elsewhere (2, 3). In short, the molecular signal from the mechanistic target of rapamycin complex 1 (mTORC1) triggers the activation of unc-51–like autophagy activating kinase 1 (ULK1) complex, consisting of ULK1, autophagy-related protein 13 (ATG13), and focal adhesion kinase family interacting protein of 200 kDa (FIP200), to initiate the formation of the isolation membrane from existing membrane sources such as the endoplasmic reticulum (ER) or Golgi (Fig. 2A). The membrane further expands to produce a completely enclosed, double-membraned vesicle known as the autophagosome. Autophagosome formation is orchestrated by a number of core autophagy-related proteins. A key step for autophagosome formation is the conjugation of phosphatidylethanolamine (PE) to microtubule-associated protein 1 light chain 3 (LC3), an ATG8 homolog, to generate a lipidated form of LC3, LC3-PE. This conjugation is mediated in part by ATG7 and the ATG5-ATG12-ATG16L1 complex (3). LC3 is retained inside the autophagosome and, when expressed as a green fluorescent protein (GFP) fusion protein, serves as a common marker of autophagy (4). Once the autophagosome is developed, it fuses with the lysosome, forming an autolysosome, a single-membraned acidic vesicle where lysosomal hydrolytic enzymes, such as cathepsins, degrade the enclosed contents. Lysosome biogenesis is an important component of autophagy machinery and regulated by the microphthalmia/transcription factor E (MiT/TFE) family of transcription factors, which includes microphthalmia-associated transcription factor (MITF), transcription factor EB (TFEB), and transcription factor binding to IGHM enhancer 3 (TFE3) (5–8). Although the later elements of the autophagy machinery are pivotal in the regulation of degradation, the initial selective degradation of cytoplasmic components through autophagy is worth exploring in depth.
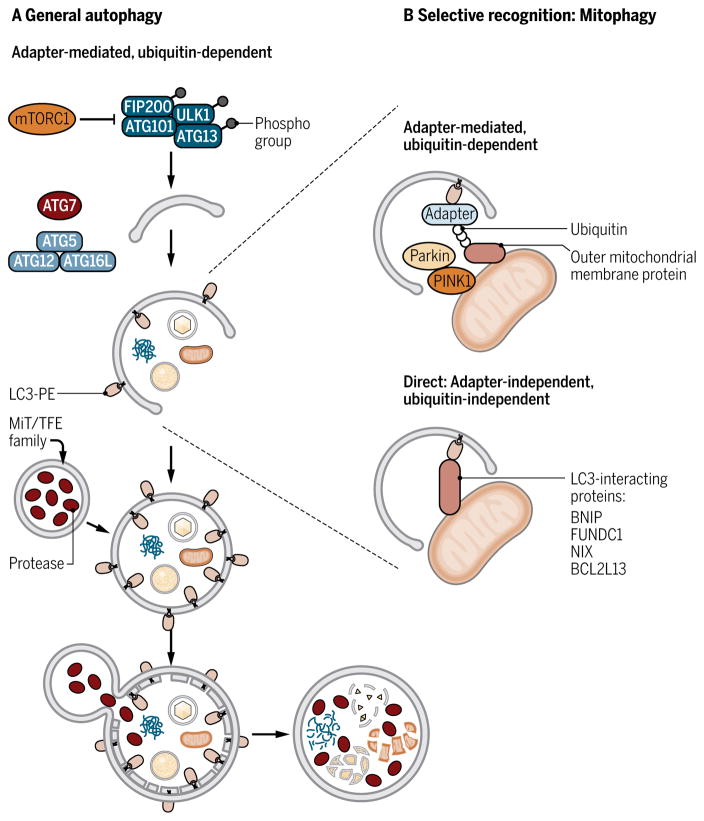
Fig. 2. Overview of the autophagy and mitophagy pathways.
(A) Autophagy begins with the formation of the isolation membrane. Initiation of the isolation membrane requires the ULK1 complex, which is regulated by mTORC1. The isolation membrane then encloses cytosolic components and elongates to completely enclose and form the autophagosome. The elongation and closure of the autophagosome involve two ubiquitin-like conjugation systems: One forms the ATG5-ATG12-ATG16L complex, and the other one forms the PE-conjugated LC3 (LC3-PE). LC3-PE is required for autophagosome formation and serves as a marker of autophagy. Subsequently, the autophagosome fuses with the lysosome, and the enclosed components are degraded by the lysosomal enzymes. The MiT/TFE family of transcription factors regulates transcription of lysosomal autophagy genes. (B) Selective mitochondrial degradation, or mitophagy, relies on autophagy receptors that can interact with LC3-PE proteins (green). In adapter-mediated, ubiquitin-dependent mitophagy (top), PINK1 stabilization recruits Parkin and promotes ubiquitination of proteins in the outer mitochondrial membrane. Ubiquitin chains are recognized by adapter proteins that also contain the LIR and promote encapsulation of the mitochondria by the autophagosome. In adapter-independent, ubiquitin-independent mitophagy, specific mitochondrial proteins, several of which have been identified, directly interact with LC3.
A particularly interesting example is mitophagy, the selective clearance of mitochondria through autophagy. Selectivity is driven by specific proteins that physically connect the intended target (such as mitochondria) with the autophagosomal protein LC3. These receptors interact with the autophagosome through the LC3-interacting region (LIR) (Fig. 2B). Mitochondrial damage is a major physiological trigger for selective mitochondrial clearance. Damage-induced mitophagy can occur through two different mechanisms: (i) adapter-mediated, ubiquitin-dependent mitophagy and (ii) direct, ubiquitin-independent mitophagy (Fig. 2B). Adapter-mediated mitophagy, which is mediated by phosphatase and tensin homolog (PTEN)–induced putative kinase 1 (PINK1) and the E3 ubiquitin ligase Parkin, requires the ubiquitination of the target. Damage to the mitochondria leads to reduced mitochondrial membrane potential, stabilization of PINK1 on the outer mitochondrial membrane, and subsequent recruitment of Parkin, which ubiquitinates outer mitochondrial proteins (9). The ubiquitinated substrates are then recognized by autophagy adapter proteins including p62, optineurin, NDP52 (nuclear dot protein 52 kDa), and NBR1 (neighbor of Brca1 gene 1), which link the ubiquitinated targets to LC3 (10–13). Adapter proteins contain two defining domains: a ubiquitin-binding domain for cargo recognition and an LIR domain that interacts with LC3 to promote encapsulation by the autophagosome. Whether adapter proteins have tissue- or cell type–specific functions have yet to be revealed. Damage-induced mitophagy can also occur through the direct interaction of mitochondria-localized proteins with LC3 independent of ubiquitination. For example, BCL2/E1B 19 kDa–interacting protein 3 (BNIP3) and FUN14 domain-containing protein 1 (FUNDC1) directly interact with LC3 to promote mitophagy in response to hypoxia-triggered mitochondrial damage (14, 15). The mitophagy receptor BCL2-like 13 (BCL2L13), which is a mammalian homolog of Atg32, directly interacts with LC3 through the LIR domain, but the mechanism that activates BCL2L13 remains to be determined (16).
Mitophagy can take place independently of mitochondrial damage during developmental processes, although fewer models of this process have been established. For instance, the BNIP3 homology NIP3-like protein X (NIX; also known as BNIP3L) is required for mitochondrial clearance during erythrocyte maturation (17, 18). NIX mediates mitophagy in a ubiquitin-independent manner, and blocking the direct interaction between NIX and LC3 leads to accumulation of mitochondria in maturing erythrocytes (19). In addition, mitochondrial degradation occurs when sperm mitochondria are removed during fertilization. The mechanism of paternal mitochondrial degradation is not conserved between species. In Caenorhabditis elegans, this process requires autophagosome formation and is independent of ubiquitination (20, 21). In Drosophila, paternal mitophagy also requires autophagosome formation that is ubiquitin- and p62-dependent but does not require Parkin (22). Examples in C. elegans and Drosophila suggest that there are uncharacterized proteins that target mitochondria for degradation. In mammals, including the mouse, pig, and rhesus monkey, mitochondrial degradation appears to occur through the ubiquitin-proteosome system independently of LC3-mediated autophagy (23–25). In a slightly different vein, we have identified a developmentally important process of mitochondrial clearance: selective mitochondrial degradation during beige adipocyte conversion to white adipocytes after the withdrawal of cold exposure or β3-adrenergic receptor (β3-AR) stimulation. The mechanism underlying the recognition of mitochondria for selective degradation awaits future investigation.
The Role of Autophagy in Adipose Biology
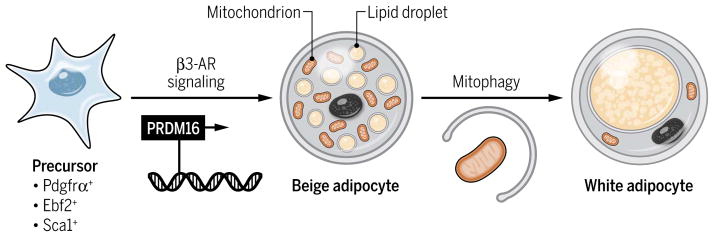
Mammals have two functionally distinct types of adipocytes: white adipocytes, which store excess energy as triglycerides, and brown adipocytes, which dissipate energy in the form of heat and thus can counteract obesity and obesity-associated diseases such as type 2 diabetes (26). Adult humans and rodents have a “recruitable” form of brown adipocytes, termed “beige adipocytes,” the development of which can be induced by certain environmental stimuli such as chronic cold exposure (27–29). Although brown and beige adipocytes have similar biochemical and morphological characteristics, including the brown and beige fat-specific protein uncoupling protein 1 (UCP1), high mitochondrial content, and multilocular lipid droplets, their developmental origins are distinct (29–34). Brown adipocytes arise early during development from a subset of dermomyotome precursors, and their development is stalled during postnatal stages. On the other hand, beige adipocytes arise postnatally in part through the action of the transcription factor progesterone receptor (PR) (PRD1-BF1-RIZ1 homologous) domain–containing 16 (PRDM16) in response to environmental cues from precursors that are positive for early B cell factor 2 (Ebf2), platelet-derived growth factor receptor α (Pdgfrα), and stem cell antigen–1 (Sca1) (Fig. 3) (35–37). Cold exposure, which stimulates the β3-AR signaling pathway, is a dominant activator of brown and beige adipocyte development. Notably, beige adipocytes lose their morphological and thermogenic characteristics and acquire “white-like” characteristics shortly after the requisite stimuli (cold exposure or β3-AR stimulation) are removed (38, 39). We have reported that the conversion from beige adipocyte to white adipocyte is direct, circumventing an intermediate precursor state, and involves active mitochondrial clearance (Fig. 3) (39). Given the crucial role of the mitochondria in the thermogenic function of beige and brown adipocytes, it is important to understand the mechanisms underlying the regulation of mitochondrial homeostasis in brown and beige adipocytes.
Fig. 3. Beige adipocyte development.
Activation of the β3-AR signaling by cold exposure or agonists induces differentiation of precursors into UCP1-positive beige adipocytes. Activation of autophagy after the withdrawal of the stimulus triggers loss of mitochondrial content and conversion from beige to white adipocytes.
Autophagy has been implicated in remodeling mitochondrial contents and thus regulating adipocyte differentiation as well as the maintenance of differentiated adipocytes. Accordingly, it is important to use an appropriate Cre mouse line that can target specific adipocytes at different differentiation stages to dissect the roles of autophagy in defined cell types (for example, preadipocytes or differentiated adipocytes). Several genetic autophagy-deficient animal models have been used to study the function of autophagy in adipose tissues but exhibit inconsistent phenotypes (Table 1). For instance, a total knockout of Atg5 results in a differentiation defect of white adipose tissue (WAT), whereas deletion of Atg7 through a muscle- and brown adipocyte–specific Cre, Myf5-Cre, promotes beige adipocyte development and impairs brown adipocyte differentiation (40, 41). These models assess the role of autophagy during adipogenesis and preclude insight into autophagy function in terminally differentiated adipocytes. Similarly, aP2-Cre–mediated deletion of Atg7 or p62 affects not only mature brown and white adipocytes but also some nonadipose tissues (42–44). Nonspecific expression of aP2-Cre in skeletal muscle, liver, brain, and macrophages can cause indirect effects on adipocyte differentiation and/or function (45, 46). In addition, inhibiting autophagy in proopiomelanocortin (POMC) neurons and skeletal muscle causes browning of WAT (41, 47). We have used Ucp1-Cre to selectively target autophagy in differentiated brown and beige adipocytes to show that autophagy is required specifically for beige-to-white adipocyte conversion after cold or β3-AR withdrawal (39).
Table 1.
Overview of adipose tissue phenotypes in animals that lack autophagy-related genes.
| Molecule | System | Adipocyte phenotype | References |
|---|---|---|---|
| Atg5 | Total knockout | WAT development is impaired. | (40) |
| Atg5 | MEFs in vitro | MEFs from total knockout mice have defects in white adipocyte differentiation in culture. | (40) |
| Atg5 | Ucp1-Cre | Beige adipocytes are maintained in the absence of browning stimuli and retain mitochondria and UCP1. | (39) |
| Atg7 | aP2-Cre | WAT is reduced and adipocytes have increased mitochondrial content with small lipid droplets. WAT in knockout mice contains increased numbers of beige adipocytes. Mice are protected from diet-induced obesity and insulin resistance. | (42, 43) |
| Atg7 | Myf5-Cre | Deletion in Myf5+ progenitors impairs BAT development and promotes beige fat development. | (41) |
| Atg7 | Mlclf-Cre | Deletion in skeletal muscle induces browning of WAT. | (47) |
| Atg7 | Pomc-Cre | Deletion in POMC neurons induces browning of WAT. | (71) |
| Atg12 | Ucp1-Cre | Beige adipocytes are maintained in the absence of browning stimuli and retain mitochondria and UCP1. Mice are protected from diet-induced obesity and insulin resistance. | (39) |
| p62 | Total knockout | Enlarged adipocytes due to lipid accumulation. Mice are obese, glucose intolerant, and have decreased insulin sensitivity. | (106) |
| p62 | aP2-Cre | Brown adipocytes accumulate defective mitochondria and have more lipid droplets. Mice are obese, glucose intolerant, and have decreased insulin sensitivity. | (44) |
| Parkin | Total knockout | Mice are resistant to high-fat diet–induced obesity. EpiWAT and BAT of these mice accumulate less lipid in response to a high-fat diet. | (107) |
| Raptor | Adiponectin-Cre | Browning of WAT is disrupted in response to chronic cold exposure. | (56) |
Another potential cause for various phenotypes observed in autophagy-deficient models may reside in the promiscuity of autophagy machinery components, many of which have broad cellular functions beyond autophagy (48). Deletion of either Atg5 or Atg12 using Ucp1-Cre results in a consistent phenotype: retention of mitochondrial content and high UCP1 abundance in inguinal WAT containing beige adipocytes even after withdrawal of β3-AR stimulation (39). In contrast, deletion of p62, a cargo receptor that mediates selective autophagy, results in impaired mitochondrial function including reduced UCP1 abundance in both brown adipose tissue (BAT) and inguinal WAT (44). These apparently conflicting phenotypes can be explained by the role that p62 plays in various signaling pathways, including those of nuclear factor κB (NF-κB), extra-cellular signal–regulated kinase 1 (ERK1), and nuclear factor erythroid 2–related factor 2 (Nrf2) (49, 50). Assessing multiple components of the autophagy process using the mature adipocyte-specific Cre lines, such as Adiponectin- or Ucp1-Cre, would clarify the specific biological processes that are specifically regulated by each autophagy regulator.
Physiological Regulation of Autophagy in Adipose Tissues
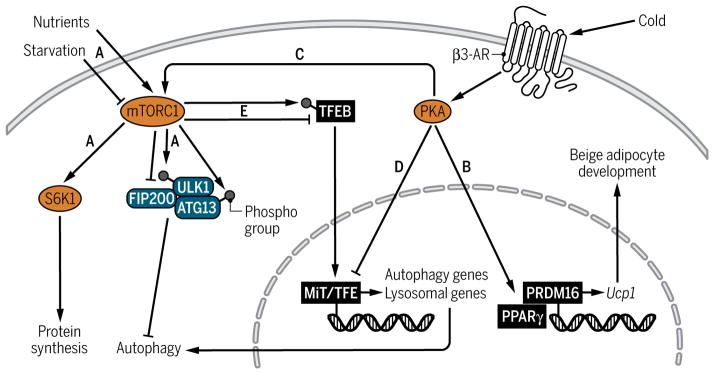
Autophagy is initiated in concordance with responses to nutrient availability and is thus tightly regulated by the mTORC1 complex of the mTOR signaling pathway, which acts as a nutrient sensor to coordinate cellular responses (Fig. 4). High nutrient abundance leads to the activation of mTORC1 and its downstream targets, including ribosomal S6 kinase 1 (S6K1) to promote protein synthesis for anabolic functions. In the absence of nutrients, mTORC1 is inhibited, leading to autophagy activation through the regulation of the ULK1, FIP200, and ATG13 complex (Fig. 4A) (51).
Fig. 4. Cross-talk between the mTOR and β3-AR signaling pathways in beige adipocytes.
(A) Under nutrient-rich conditions, mTORC1 is activated and phosphorylates ULK1 and ATG13 to repress the ULK1 complex and block autophagy. In response to starvation, mTORC1 is inhibited, inducing autophagy. (B and C) Activation of PKA in response to β3-AR stimulation induces transcription of brown/beige adipocyte program and promotes mTORC1 activity to inhibit autophagy partly through regulation of MiT/TFE family of transcription factors. (D) PKA activation suppresses the expression of genes encoding MiT/TFE transcription factors, and its lysosomal and autophagy targets. (E) mTORC1 alters lysosomal and autophagy gene expression through regulating the nuclear-cytoplasmic shuttling of TFEB (an MiT/TFE family member). Active mTORC1 phosphorylates TFEB and blocks its translocation to the nucleus, preventing transcription of lysosomal and autophagy targets.
Another form of physiological regulation of autophagy is β3-AR signaling, which triggers protein kinase A (PKA) signaling. PKA signaling functions to inhibit autophagy in yeast and mammals (52), and feedback loops between autophagy and PKA have been uncovered (53, 54). Notably, the β3-AR signaling pathway is a key mediator of beige adipocyte biogenesis in the face of cold exposure. Beige adipogenesis is promoted when environmental cues, such as cold exposure, trigger the release of norepinephrine from the sympathetic nervous system, which, in turn, acts on the β3-AR, increases intracellular cyclic adenosine 3′,5′-monophosphate (cAMP) concentrations, and triggers the PKA signaling pathway, leading to transcriptional activation of the thermogenic program in brown and beige fat (Fig. 4B) (55). In response to cold or β3-AR stimulation, PKA directly phosphorylates mTOR and its binding partner, regulatory-associated protein of mTOR (RAPTOR), to stimulate the activity of mTORC1, a major regulator of autophagy (Fig. 4C) (56). Our group has further shown that PKA, in turn, represses autophagy in beige adipocytes (Fig. 4D), which is partially mediated through repression of Mitf, a member of the MiT/TFE family of transcription factors that regulates lysosome biogenesis. Pharmacological inhibition of PKA relieves repression of Mitf and genes encoding autophagy and lysosome components, confirming that PKA is responsible for inhibiting autophagy in beige adipocytes. In addition, regulation of Mitf and autophagy-related genes by PKA occurs even under starvation conditions, suggesting that PKA represses autophagy independently of nutritional signals (39). Notably, mTORC1 inhibits transcription of autophagy and lysosome target genes by phosphorylating TFEB, another member of the MiT/TFE family, which blocks its translocation to the nucleus (Fig. 4E) (57–59). It is conceivable that the PKA-mTORC1 signaling axis simultaneously regulates beige adipocyte development and autophagy through MiT/TFE transcription factors.
How is mitochondrial clearance by mitophagy initiated during the beige-to-white adipocyte conversion? One possibility is that the mitophagy activation may be mediated through the PINK1-Parkin pathway. To support the notion, PKA inhibits PINK1-Parkin–mediated mitophagy through phosphorylation of a mitochondrial membrane protein, MIC60 (also known as mitofilin) (60). Another compatible possibility is that the mitochondrial degradation is regulated by the BAT-specific mitochondrial protein UCP1, which reduces mitochondrial membrane potential through proton “leak.” During the beige-to-white adipocyte conversion, the loss of UCP1 protein correlates with the loss of mitochondrial respiratory chain complexes. Inhibition of autophagy by genetic ablation of Atg5 or Atg12 maintains expression of mitochondrial DNA transcripts and abundance of mitochondrial proteins, including UCP1 (39). Hence, it is possible that mitochondrial removal through autophagy during the beige-to-white adipocyte conversion requires the transient induction of UCP1 protein, which acts as an upstream regulator of mitophagy by uncoupling the proton gradient in the mitochondria. To support the hypothesis, UCP1 abundance is tightly associated with mitochondrial content and cristae density in adipose tissues (61, 62).
In addition, the cAMP-PKA signaling pathway has been implicated in regulating adipocyte lipophagy. Lipophagy, which is reviewed by Evans et al. (63) in the same issue, is a selective autophagic degradation of lipids. Although it was first demonstrated in hepatocytes, where pharmacological and genetic inhibition of autophagy in cultured hepatocytes lead to increased triglyceride accumulation (64), lipophagy has also been detected in hepatic stellate cells, neurons, and brown adipocytes (65–69). Lipophagy in brown adipocytes is regulated by cold exposure, although the effect appears to be dependent on the length of exposure. Whereas chronic cold exposure inhibits lipophagy and associated autophagy components through the cAMP-PKA signaling pathway (70), acute cold exposure activates autophagy-mediated lipid degradation (71). An aspect of concern is the shortage of tools to monitor lipophagy, making it difficult to assess selective lipophagy. The biological importance of lipophagy over lipolysis for lipid breakdown awaits future studies.
Pathological Regulation of Autophagy in Metabolic Disease
Autophagy plays a central role in the function and maintenance of metabolic tissues such as liver, pancreas, and adipose tissues. Emerging evidence suggests that dysregulation of autophagy contributes to the initiation or progression of metabolic disorders in the following organs.
Adipose tissue
Obesity (increased adiposity or body mass index) is inversely correlated with the thermogenic activity of BAT in response to cold exposure (72). Beige adipocyte biogenesis is impaired in obese mice partly because of the activation of negative regulators such as the transforming growth factor–β (TGF-β) and Notch signaling pathways (26). In turn, blockade of the TGF-β or Notch signaling pathways by genetic or pharmacological approaches promotes beige adipocyte biogenesis and protects mice from diet-induced obesity (73–75). Furthermore, autophagy blocks beige adipocyte development. Inhibiting autophagy through deletion of Atg7 in adipocytes leads to increased beige adipocyte differentiation, resistance to diet-induced obesity, and improved insulin sensitivity (42, 43).
Dysregulation of beige adipocyte maintenance is likely to contribute to the development of obesity. We have found that diet-induced obesity accelerates the conversion of beige adipocytes to white adipocytes after β3-AR agonist withdrawal, which correlates with increased autophagy in subcutaneous WAT. Genetic deletion of Atg12 or Atg5 specifically in UCP1-positive adipocytes substantially prolongs the retention of beige adipocytes in vivo. The maintained beige fat is thermogenically active and suppresses diet-induced obesity and obesity-induced insulin resistance (39). Notably, increased adipose tissue autophagy has been observed in human obesity and type 2 diabetes (76–79). In addition, activation of autophagy in human subjects with type 2 diabetes and obesity is partly attributed to repression of mTORC1 activity (76). These studies suggest that dysregulation of autophagy in the adipose tissues of obese subjects may contribute to the accelerated beige-to-white adipocyte conversion.
Pancreas
Defects in pancreatic islet β cell function are the fundamental cause for type 1 and type 2 diabetes. Pancreatic β cells rely heavily on mitochondria and the ER to maintain glucose-stimulated insulin production and secretion, and autophagy maintains β cell homeostasis by removing damaged mitochondria and/or ER. Mice with a β cell–specific deletion of Atg7 accumulate defective mitochondria and distended ER in their β cells, leading to impaired glucose tolerance and reduced insulin secretion (80, 81). Autophagy-deficient β cells fail to proliferate as an adaptation to increased insulin demand in obesity (81, 82). The mechanism by which autophagy controls β cell proliferation remains unclear.
A limited number of studies have investigated the metabolic consequences of dysfunctional mitophagy in β cells. Total Parkin-null mice display reduced insulin secretion after glucose challenge under pathological stress conditions such as streptozotocin exposure, whereas Parkin overexpression in pancreatic β cell lines maintains insulin secretion under diabetic conditions (83). In addition, C-type lectin domain family 16 member A (Clec16a) has been implicated in β cell mitophagy through a Parkin-dependent mechanism. Pancreas-specific deletion of Clec16a using Pdx1-Cre leads to accumulation of abnormal mitochondria, impaired glucose tolerance, and insulin secretion in response to glucose challenge (84). Surprisingly, Clec16a-deficient pancreas have more Parkin, which raises questions about the role of Parkin in regulating mitophagy in β cells. Although the role of Parkin in β cell mitophagy remains unclear, these results emphasize that dysfunction in selective mitophagy alone could lead to β cell failure and diabetes.
Liver
Obesity is closely associated with hepatic steatosis. Mice with diet-induced obesity or a genetically induced form of obesity (ob/ob mice) display reduced hepatic autophagy, as assessed by decreased numbers of GFP-LC3 puncta, reduced LC3-II abundance, and accumulation of p62 (85). Liver-specific deletion of Atg7, Vps34 (vacuolar protein sorting 34), or Tfeb results in lipid accumulation and enlarged livers (64, 86, 87). Conversely, overexpression of Atg7 or Tfeb by adenovirus reverses hepatic lipid accumulation, reduces liver size, prevents body weight gain in response to genetic or high-fat diet–induced obesity, and improves glucose tolerance and insulin sensitivity (85, 87). However, contrary to these studies, liver-specific deletion of Atg7 leads to reduced hepatic lipid accumulation (47, 88). Because the diverging phenotypes come from the same genetic model, the discrepancies are presumably due to differences in experimental conditions or analyses.
Starvation is a powerful stimulus that induces autophagy in the liver to control hepatic gluconeogenesis. During the early neonatal period, when the placental nutrient supply is cut off at birth, autophagy-deficient (Atg5−/−) pups die soon after birth from severe hypoglycemia and hypolipidemia (89). Dysregulation of autophagy in a liver-specific fashion in adults leads to a defect in intracellular lipid degradation, enlarged liver, and increased hepatic lipid content under starvation conditions (64, 87). In addition, liver-specific deletion of Atg7 using inducible Mx1-Cre results in hypoglycemia due to reduced gluconeogenesis (90).
Positive regulators of the starvation-induced autophagy include TFEB and PPARα; for instance, liver-specific deletion of Tfeb prevents transcription of various autophagy-related genes and leads to increased hepatic lipid content, increased circulating free fatty acids (FFAs), and impaired FFA oxidation in vitro (87). Similarly, PPARα activates transcription of autophagy components. Pharmacological activation of PPARα induces autophagy in nutrient-replete cells, which mimics a starvation response, whereas the liver of fasted Pparα−/−mice cannot induce autophagy and exhibits increased lipid content (91). On the other hand, farnesoid X receptor (FXR) functions to repress autophagy in the fed state. Pharmacological activation of FXR blunts starvation-induced hepatic autophagy, whereas Fxr−/−mice maintain high amounts of autophagy in the liver even in the fed state (65, 91). It would be important to determine whether the same regulation applies to non–starvation-induced autophagy in other metabolic organs.
Methodologies for Detecting Mitophagy in Adipocytes
Mitophagy structures were initially identified by electron microscopy. Studies from the 1950s described double-membrane vesicles, later termed autophagic vacuoles, which contained recognizable mitochondrial cristae (92). Identification and quantification of mitophagy have been challenging due largely to the substantial overlap with autophagy machinery and an absence of a universally defined marker for mitophagy per se. Despite the current limitations, there are several tools available for monitoring mitophagy as described below.
Colocalization of mitochondria-localized proteins with GFP-LC3
Mitophagy can be assessed on the basis of colocalization of the autophagosome with mitochondria. To this end, GFP-LC3 transgenic mice have been successfully used to visualize autophagosomes in vivo and cultured cells (93). Vital dyes, such as MitoTracker Red are reliable options for labeling mitochondria in vitro. However, these dyes label less than 50% of the existing mitochondria and lose signal after fixation, limitations that preclude their use in vivo (94). An alternative approach to vital dyes is to label mitochondria with mitochondria-localized proteins and assess their sequestration into the autophagosome using GFP-LC3. During the beige-to-white adipocyte conversion, we have observed the colocalization of the mitochondria protein translocase of outer mitochondrial membrane 20 (TOM20) with GFP-LC3; analyzed in conjunction with mitochondria-autophagosome structures observed by electron microscopy, as well as with changes in mitochondrial content, these data suggest that selective mitophagy takes place during the beige-to-white adipocyte conversion (39).
Monitoring mitochondrial turnover: Indirect measurement of mitophagy
Mitophagy is a transient event that cannot be fully examined through snapshots of the mitochondria using mitochondria-localized proteins. MitoTimer is a fluorescent reporter that can measure the kinetics of mitochondrial biogenesis and degradation. A mutant of the red fluorescent protein is attached to the mitochondrial localization sequence of cytochrome c oxidase subunit VIII (COX VIII), which fluoresces to green when the protein is newly synthesized and then gradually transitions to red as the protein matures (95, 96). This system has been used to assess mitochondrial turnover in skeletal muscle and heart (97, 98). Quantifying the loss of red fluorescence determines the kinetics of mitophagy process. This method is an indirect assessment of mitophagy, and as such, a major concern would be that protein degradation is also measured as well as mitophagy flux.
Monitoring delivery of mitochondria to lysosomes
A more direct way of measuring mitophagy is to assess the delivery of mitochondria to lysosomes. A tandem mCherry-GFP fusion protein is attached to the mitochondrial localization sequence of the mitochondrial fission 1 protein (FIS1). Different chemical properties of mCherry and GFP allow the identification of mitochondria that are undergoing lysosomal degradation; GFP is immediately degraded in the acidic lysosomal environment, whereas mCherry persists (99). A transgenic mouse model called mito-QC (quality control), which uses this system, has been generated (100).
A similar strategy for assessing mitophagy directly has been developed using the mt-Keima transgenic mouse system. A tandem repeat of COX VIII tagged with the fluorescent protein Keima is targeted to the mitochondria. Keima fluorescence is pH-dependent; it emits at different wavelengths at neutral or acidic pH, making it possible to determine whether mitochondria are in the cytosol (neutral pH) or lysosomes (acidic pH). In addition, Keima fluorescence is resistant to lysosomal degradation because it is derived from corals (101, 102). The dual fluorescence of this reporter allows direct and quantitative assessment of mitophagic flux using fluorescence-activated cell sorting (FACS), which often is a more sensitive and quantitative tool than imaging analysis (103). At least two potential shortcomings of using mt-Keima system exist: (i) unfixed tissues are required for visualizing changes in Keima fluorescence, and (ii) some spectral overlap of the red and green fluorescence exists (101, 102). The mito-QC system mentioned above could be a better system for imaging analysis because it overcomes these limitations, but it has not yet been optimized for FACS analysis (100). These direct tools of assessing mitophagy could be applied to metabolic organs including adipose tissue.
Future Directions
An intriguing observation from our study is that autophagy-mediated degradation of mitochondria in beige adipocytes maintains the fate of these cells. Whereas the role of the nucleus in regulating mitochondrial mass and function has been examined extensively in the past, the above result indicates, in turn, that mitochondria immensely influence the cellular maintenance of beige adipocytes. What is the molecular signaling that mediates the mitochondria-to-nucleus communication? Mitochondrial clearance could regulate cellular maintenance and function by altering nuclear gene expression by limiting metabolite availability. For instance, several mitochondrial metabolites (such as α-ketoglutarate, succinate, and fumarate) regulate chromatin-modifying enzymes, such as Jumonji demethylases, which control the adipocyte differentiation program (104). In addition, mitochondrial stress in C. elegans activates another type of mitochondrial quality control, UPRmt (unfolded mitochondrial protein response), which alters the chromatin structure, causing persistent changes in gene expression (105). Further studies are needed to uncover the molecular mechanisms and biological roles of the mitochondria-nuclear communication in adipocytes.
Acknowledgments
We would like to thank P. Maretich for constructive comments and edits of the manuscript.
Funding: This work was supported by the NIH (grants DK97441 and DK108822), the Diabetes and Endocrinology Research Center (grant DK63720), the Pew Charitable Trusts, and the Japan Science and Technology Agency (to S.K.). S.A.-K. is supported by fellowships from the American Heart Association (15PRE23050029) and the California Institute of Regenerative Medicine (TG2-01153).
REFERENCES AND NOTES
- 1.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: Origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol. 2009;452:13–23. doi: 10.1016/S0076-6879(08)03602-1. [DOI] [PubMed] [Google Scholar]
- 5.Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 6.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 7.Settembre C, Di Malta C, Polito VA, Arencibia MG, Vetrini F, Erdin S, Uckac Erdin S, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, Settleman J, Stephanopoulos G, Dyson NJ, Zoncu R, Ramaswamy S, Haas W, Bardeesy N. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLOS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 Recruitment and TBK1 activation to promote mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Øvervatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 16.Murakawa T, Yamaguchi O, Hashimoto A, Hikoso S, Takeda T, Oka T, Yasui H, Ueda H, Akazawa Y, Nakayama H, Taneike M, Misaka T, Omiya S, Shah AM, Yamamoto A, Nishida K, Ohsumi Y, Okamoto K, Sakata Y, Otsu K. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun. 2015;6:7527. doi: 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Randall MS, Loyd MR, Dorsey FC, Kundu M, Cleveland JL, Ney PA. Mitochondrial clearance is regulated by Atg7–dependent and –independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortensen M, Ferguson DJP, Edelmann M, Kessler B, Morten KJ, Komatsu M, Simon AK. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci USA. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dötsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 21.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 22.Politi Y, Gal L, Kalifa Y, Ravid L, Elazar Z, Arama E. Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev Cell. 2014;29:305–320. doi: 10.1016/j.devcel.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Thompson WE, Ramalho-Santos J, Sutovsky P. Ubiquitination of prohibitin in mammalian sperm mitochondria: Possible roles in the regulation of mitochondrial inheritance and sperm quality control. Biol Reprod. 2003;69:254–260. doi: 10.1095/biolreprod.102.010975. [DOI] [PubMed] [Google Scholar]
- 24.Song WH, Yi YJ, Sutovsky M, Meyers S, Sutovsky P. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc Natl Acad Sci. 2016;113:E5261–E5270. doi: 10.1073/pnas.1605844113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol Reprod. 2000;63:582–590. doi: 10.1095/biolreprod63.2.582. [DOI] [PubMed] [Google Scholar]
- 26.Sidossis L, Kajimura S. Brown and beige fat in humans: Thermogenic adipocytes that control energy and glucose homeostasis. J Clin Invest. 2015;125:478–486. doi: 10.1172/JCI78362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoneshiro T, Aita S, Matsushita M, Kayahara T, Kameya T, Kawai Y, Iwanaga T, Saito M. Recruited brown adipose tissue as an antiobesity agent in humans. J Clin Invest. 2013;123:3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Lans AAJJ, Hoeks J, Brans B, Vijgen GHEJ, Visser Mariëlle GW, Vosselman MJ, Hansen J, Jörgensen JA, Wu J, Mottaghy FM, Schrauwen P, van Marken Lichtenbelt WD. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J Clin Invest. 2013;123:3395–3403. doi: 10.1172/JCI68993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shinoda K, Luijten IHN, Hasegawa Y, Hong H, Sonne SB, Kim M, Xue R, Chondronikola M, Cypess AM, Tseng YH, Nedergaard J, Sidossis LS, Kajimura S. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat Med. 2015;21:389–394. doi: 10.1038/nm.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp LZ, Shinoda K, Ohno H, Scheel DW, Tomoda E, Ruiz L, Hu H, Wang L, Pavlova Z, Gilsanz V, Kajimura S. Human BAT possesses molecular signatures that resemble beige/brite cells. PLOS ONE. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lidell ME, Betz MJ, Leinhard OD, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, Virtanen KA, Beuschlein F, Persson A, Borga M, Enerbäck S. Evidence for two types of brown adipose tissue in humans. Nat Med. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Boström P, Sparks LM, Ye L, Hyun Choi J, Giang A-H, Khandekar M, Virtanen KA, Nuutila P, Schaart G, Huang K, Tu H, van Marken Lichtenbelt WD, Hoeks J, Enerbäck S, Schrauwen P, Spiegelman BM. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frontini A, Cinti S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell Metab. 2010;11:253–256. doi: 10.1016/j.cmet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor γ (PPARγ) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, Kissig M, Rajakumari S, Huang L, Lim HW, Won KJ, Seale P. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci USA. 2014;111:14466–14471. doi: 10.1073/pnas.1412685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, Schrier D, Falb D, Kirkland JL, Wagers AJ, Tseng YH. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci USA. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenwald M, Perdikari A, Rülicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol. 2013;15:659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- 39.Altshuler-Keylin S, Shinoda K, Hasegawa Y, Ikeda K, Hong H, Kang Q, Yang Y, Perera RM, Debnath J, Kajimura S. Beige adipocyte maintenance is regulated by autophagy-induced mitochondrial clearance. Cell Metab. 2016;24:402–419. doi: 10.1016/j.cmet.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baerga R, Zhang Y, Chen PH, Goldman S, Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Lopez N, Athonvarangkul D, Sahu S, Coletto L, Zong H, Bastie CC, Pessin JE, Schwartz GJ, Singh R. Autophagy in Myf5+ progenitors regulates energy and glucose homeostasis through control of brown fat and skeletal muscle development. EMBO Rep. 2013;14:795–803. doi: 10.1038/embor.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci USA. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller TD, Jun Lee S, Jastroch M, Kabra D, Stemmer K, Aichler M, Abplanalp B, Ananthakrishnan G, Bhardwaj N, Collins S, Divanovic S, Endele M, Finan B, Gao Y, Habegger KM, Hembree J, Heppner KM, Hofmann S, Holland J, Küchler D, Kutschke M, Krishna R, Lehti M, Oelkrug R, Ottaway N, Perez-Tilve D, Raver C, Walch AK, Schriever SC, Speakman J, Tseng Y-H, Diaz-Meco M, Pfluger PT, Moscat J, Tschöp MH. p62 links β-adrenergic input to mitochondrial function and thermogenesis. J Clin Invest. 2013;123:469–478. doi: 10.1172/JCI64209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullican SE, Tomaru T, Gaddis CA, Peed LC, Sundaram A, Lazar MA. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol Endocrinol. 2013;27:127–134. doi: 10.1210/me.2012-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao J, Yang T, Gu Z, Heird WC, Finegold MJ, Lee B, Wakil SJ. aP2-Cre-mediated inactivation of acetyl-CoA carboxylase 1 causes growth retardation and reduced lipid accumulation in adipose tissues. Proc Natl Acad Sci USA. 2009;106:17576–17581. doi: 10.1073/pnas.0909055106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim Y-N, Kim SS, Hoon Kim D, Hur KY, Kim HK, Ko TH, Han J, Lim Kim H, Kim J, Back SH, Komatsu M, Chen H, Chan DC, Konishi M, Itoh N, Choi CS, Lee M-S. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013;19:83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- 48.Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14:143–151. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou Y-S, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S-i, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 51.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Klionsky DJ. The regulation of autophagy – unanswered questions. J Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torres-Quiroz F, Filteau M, Landry CR. Feedback regulation between autophagy and PKA. Autophagy. 2015;11:1181–1183. doi: 10.1080/15548627.2015.1055440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filteau M, Diss G, Torres-Quiroz F, Dubé AK, Schraffl A, Bachmann VA, Gagnon-Arsenault I, Chrétien A, Steunou AL, Dionne U, Côté J, Bisson N, Stefan E, Landry CR. Systematic identification of signal integration by protein kinase A. Proc Natl Acad Sci USA. 2015;112:4501–4506. doi: 10.1073/pnas.1409938112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kajimura S, Saito M. A new era in brown adipose tissue biology: Molecular control of brown fat development and energy homeostasis. Annu Rev Physiol. 2014;76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu D, Bordicchia M, Zhang C, Fang H, Wei W, Li JL, Guilherme A, Guntur K, Czech MP, Collins S. Activation of mTORC1 is essential for β-adrenergic stimulation of adipose browning. J Clin Invest. 2016;126:1704–1716. doi: 10.1172/JCI83532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signaling. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akabane S, Uno M, Tani N, Shimazaki S, Ebara N, Kato H, Kosako H, Oka T. PKA regulates PINK1 stability and Parkin recruitment to damaged mitochondria through phosphorylation of MIC60. Mol Cell. 2016;62:371–384. doi: 10.1016/j.molcel.2016.03.037. [DOI] [PubMed] [Google Scholar]
- 61.Shabalina IG, Petrovic N, de Jong JMA, Kalinovich AV, Cannon B, Nedergaard J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Rep. 2013;5:1196–1203. doi: 10.1016/j.celrep.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 62.Rossmeisl M, Barbatelli G, Flachs P, Brauner P, Zingaretti MC, Marelli M, Janovská P, Horáková M, Syrový I, Cinti S, Kopecký J. Expression of the uncoupling protein 1 from the aP2 gene promoter stimulates mitochondrial biogenesis in unilocular adipocytes in vivo. Eur J Biochem. 2002;269:19–28. doi: 10.1046/j.0014-2956.2002.02627.x. [DOI] [PubMed] [Google Scholar]
- 63.Evans TD, Sergin I, Zhang X, Razani B. Target acquired: Selective autophagy in cardiometabolic disease. Sci Signaling. 2017;10:eaag2298. doi: 10.1126/scisignal.aag2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, Ma J, Kemper B, Kemper JK. Transcriptional regulation of autophagy by an FXR–CREB axis. Nature. 2014;516:108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hernández-Gea V, Ghiassi–Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, Czaja MJ, Friedman SL. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, Cuervo AM, Singh R. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14:173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaushik S, Cuervo AM. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy. 2016;12:432–438. doi: 10.1080/15548627.2015.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cairó M, Villarroya J, Cereijo R, Campderrós L, Giralt M, Villarroya F. Thermogenic activation represses autophagy in brown adipose tissue. Int J Obes Relat Metab Disord. 2016;40:1591–1599. doi: 10.1038/ijo.2016.115. [DOI] [PubMed] [Google Scholar]
- 71.Martinez-Lopez N, Garcia-Macia M, Sahu S, Athonvarangkul D, Liebling E, Merlo P, Cecconi F, Schwartz GJ, Singh R. Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the brown adipose tissue and liver. Cell Metab. 2016;23:113–127. doi: 10.1016/j.cmet.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orava J, Nuutila P, Noponen T, Parkkola R, Viljanen T, Enerbäck S, Rissanen A, Pietiläinen KH, Virtanen KA. Blunted metabolic responses to cold and insulin stimulation in brown adipose tissue of obese humans. Obesity (Silver Spring) 2013;21:2279–2287. doi: 10.1002/oby.20456. [DOI] [PubMed] [Google Scholar]
- 73.Samad F, Yamamoto K, Pandey M, Loskutoff DJ. Elevated expression of transforming growth factor-beta in adipose tissue from obese mice. Mol Med. 1997;3:37–48. [PMC free article] [PubMed] [Google Scholar]
- 74.Yadav H, Quijano C, Kamaraju AK, Gavrilova O, Malek R, Chen W, Zerfas P, Zhigang D, Wright EC, Stuelten C, Sun P, Lonning S, Skarulis M, Sumner AE, Finkel T, Rane SG. Protection from obesity and diabetes by blockade of TGF-β/Smad3 signaling. Cell Metab. 2011;14:67–79. doi: 10.1016/j.cmet.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bi P, Shan T, Liu W, Yue F, Yang X, Liang XR, Wang J, Li J, Carlesso N, Liu X, Kuang S. Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity. Nat Med. 2014;20:911–918. doi: 10.1038/nm.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ost A, Svensson K, Ruishalme I, Brännmark C, Franck N, Krook H, Sandström P, Kjolhede P, Strålfors P. Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol Med. 2010;16:235–246. doi: 10.2119/molmed.2010.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kovsan J, Blüher M, Tarnovscki T, Klöting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schön MR, Greenberg AS, Elazar Z, Bashan N, Rudich A. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011;96:E268–E277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 78.Nuñez CE, Rodrigues VS, Gomes FS, de Moura RF, Victorio SC, Bombassaro B, Chaim EA, Pareja JC, Geloneze B, Velloso LA, Araujo EP. Defective regulation of adipose tissue autophagy in obesity. Int J Obes Relat Metab Disord. 2013;37:1473–1480. doi: 10.1038/ijo.2013.27. [DOI] [PubMed] [Google Scholar]
- 79.Jansen HJ, van Essen P, Koenen T, Joosten LAB, Netea MG, Tack CJ, Stienstra R. Autophagy activity is up-regulated in adipose tissue of obese individuals and modulates proinflammatory cytokine expression. Endocrinology. 2012;153:5866–5874. doi: 10.1210/en.2012-1625. [DOI] [PubMed] [Google Scholar]
- 80.Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, Kawamori R, Fujitani Y, Watada H. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 81.Jung HS, Chung KW, Kim JW, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, Jeong YT, Han MS, Lee MK, Kim KW, Shin J, Lee MS. Loss of autophagy diminishes pancreatic β cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 82.Quan W, Hur KY, Lim Y, Oh SH, Lee JC, Kim KH, Kim GH, Kim SW, Kim HL, Lee MK, Kim KW, Kim J, Komatsu M, Lee MS. Autophagy deficiency in beta cells leads to compromised unfolded protein response and progression from obesity to diabetes in mice. Diabetologia. 2012;55:392–403. doi: 10.1007/s00125-011-2350-y. [DOI] [PubMed] [Google Scholar]
- 83.Hoshino A, Ariyoshi M, Okawa Y, Kaimoto S, Uchihashi M, Fukai K, Iwai-Kanai E, Ikeda K, Ueyama T, Ogata T, Matoba S. Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic β-cell function in diabetes. Proc Natl Acad Sci USA. 2014;111:3116–3121. doi: 10.1073/pnas.1318951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soleimanpour SA, Gupta A, Bakay M, Ferrari AM, Groff DN, Fadista J, Spruce LA, Kushner JA, Groop L, Seeholzer SH, Kaufman BA, Hakonarson H, Stoffers DA. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell. 2014;157:1577–1590. doi: 10.1016/j.cell.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, Zong WX. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci USA. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, Araif H, Tanaka K, Kominami E, Uchiyama Y. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382:419–423. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 89.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 90.Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, Taka H, Fujimura T, Takehana K, Yoshida M, Iwata J, Tanida I, Furuya N, Zheng DM, Tada N, Tanaka K, Kominami E, Ueno T. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient–sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eskelinen EL, Reggiori F, Baba M, Kovács AL, Seglen PO. Seeing is believing: The impact of electron microscopy on autophagy research. Autophagy. 2011;7:935–956. doi: 10.4161/auto.7.9.15760. [DOI] [PubMed] [Google Scholar]
- 93.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Jun Ahn H, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NYO, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai XY, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ETW, Bánhegyi G, Bartholomew CR, Bassham DC, Bast RC, Batoko H, Huat Bay B, Beau I, Béchet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin JP, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahová M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac LAM, Carneiro V, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Jung Chae H, Yin Chai C, Chan DC, Chan EY, Chang RCC, Ming Che C, Chen CC, Chen GC, Chen GQ, Chen M, Chen Q, Chen SSL, Chen WL, Chen X, Chen X, Chen X, Chen YG, Chen Y, Chen Y, Chen YJ, Chen Z, Cheng A, Cheng CHK, Cheng Y, Cheong H, Ho Cheong J, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang HL, Chiarelli R, Chiba T, Shen Chin L, Hwa Chiou S, Chisari FV, Hin Cho C, Hyung Cho D, Choi AMK, Choi DS, Sook Choi K, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Hsien Chuang T, Huei Chueh S, Chun T, Joon Chwae Y, Len Chye M, Ciarcia R, Ciriolo MR, Clague MJ, Clark RSB, Clarke PGH, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronjé MJ, Maria Cuervo A, Cullen JJ, Czaja MJ, D’Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, De Groot JF, De Haan CAM, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LMD, Demarchi F, Zhen Deng Y, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Xing Ding W, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff K, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Tony Eissa N, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Er Eng K, Mart Engelbrecht A, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Liisa Eskelinen E, Espert L, Espina V, Fan H, Fan J, Wen Fan Q, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Claude Farré J, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fésüs L, Feuer R, Figueiredo-Pereira ME, Maria Fimia G, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Biao Gao F, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Ginsberg HN, Golab J, Goligorsky MS, Golstein P, Gomez-Manzano C, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, González-Estévez C, Ana González-Polo R, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Elvira Granato G, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Lin Guan J, Liang Guan K, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson AB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Wade Harper J, Harris AL, Harris J, Harris SD, Hashimoto M, Haspel JA, Ichiro Hayashi S, Hazelhurst LA, He C, Wen He Y, Josée Hébert M, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Höhfeld J, Holyoake TL, Huang Hong M, Hood DA, Hotamisligil GS, Houwerzijl EJ, Høyer-Hansen M, Hu B, Hu CAA, Ming Hu H, Hua Y, Huang C, Huang J, Huang S, Pang Huang W, Huber TB, Ki Huh W, Ho Hung T, Hupp TR, Min Hur G, Hurley JB, Hussain SNA, Hussey PJ, Jin Hwang J, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Ichi Isobe K, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jäättelä M, Jackson GR, Jackson WT, Janji B, Jendrach M, Hong Jeon J, Bae Jeung E, Jiang H, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jiménez A, Jin M, Jin S, Joe CO, Johansen T, Johnson DE, Johnson GVW, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhász G, Juillerat-Jeanneret L, Hwa Jung C, Keun Jung Y, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kagedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Bee Kang K, Kang KI, Kang R, Kang YA, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Yuan Ke P, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JAKW, Kiger AA, Kihara A, Ryong Kim D, Hyung Kim D, Hou Kim D, Kyoung Kim E, Ryong Kim H, Sung Kim J, Hun Kim J, Cheon Kim J, Kim JK, Kim PK, Who Kim S, Sun Kim Y, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BCB, Koch JC, Koga H, Young Koh J, Ho Koh Y, Koike M, Komatsu M, Kominami E, Jeong Kong H, Jia Kong W, Korolchuk VI, Kotake Y, Koukourakis MI, Kouri Flores JB, Kovács AL, Kraft C, Krainc D, Krämer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Krüger R, Krut O, Ktistakis NT, Yi Kuan C, Kucharczyk R, Kumar A, Kumar R, Kumar S, Kundu M, Jien Kung H, Kurz T, Jeong Kwon H, La Spada AR, Lafont F, Lamark T, Landry J, Lane JD, Lapaquette P, Laporte JF, László L, Lavandero S, Lavoie JN, Layfield R, Lazo PA, Le W, Le Cam L, Ledbetter DJ, Lee AJX, Wan Lee B, Min Lee G, Lee J, Hyun Lee J, Lee M, Shik Lee M, Hyung Lee S, Leeuwenburgh C, Legembre P, Legouis R, Lehmann M, Yao Lei H, Ying Lei Q, Leib DA, Leiro J, Lemasters JJ, Lemoine A, Lesniak MS, Lev D, Levenson VV, Levine B, Levy E, Li F, Lin Li J, Li L, Li S, Li W, Jun Li X, Bo Li Y, Ping Li Y, Liang C, Liang Q, Feng Liao Y, Liberski PP, Lieberman A, Lim HJ, Leong Lim K, Lim K, Feng Lin C, Cheng Lin F, Lin J, Lin JD, Lin K, Wan Lin W, Chin Lin W, Ling Lin Y, Linden R, Lingor P, Lippincott-Schwartz J, Lisanti MP, Liton PB, Liu B, Feng Liu C, Liu K, Liu L, Liu QA, Liu W, Chau Liu Y, Liu Y, Lockshin RA, Nam Lok C, Lonial S, Loos B, Lopez-Berestein G, López-Otín C, Lossi L, Lotze MT, Lõw P, Lu B, Lu B, Lu B, Lu Z, Luciano F, Lukacs NW, Lund AH, Lynch-Day MA, Ma Y, Macian F, MacKeigan JP, Macleod KF, Madeo F, Maiuri L, Chiara Maiuri M, Malagoli D, Malicdan MCV, Malorni W, Man N, Maria Mandelkow E, Manon S, Manov I, Mao K, Mao X, Mao Z, Marambaud P, Marazziti D, Marcel YL, Marchbank K, Marchetti P, Marciniak SJ, Marcondes M, Mardi M, Marfe G, Mariño G, Markaki M, Marten MR, Martin SJ, Martinand-Mari C, Martinet W, Martinez-Vicente M, Masini M, Matarrese P, Matsuo S, Matteoni R, Mayer A, Mazure NM, McConkey DJ, McConnell MJ, McDermott C, McDonald C, McInerney GM, McKenna SL, McLaughlin BA, McLean PJ, McMaster CR, Angus McQuibban G, Meijer AJ, Meisler MH, Meléndez A, Melia TJ, Melino G, Mena MA, Menendez JA, Menna-Barreto RFS, Menon MB, Menzies FM, Mercer CA, Merighi A, Merry DE, Meschini S, Meyer CG, Meyer TF, Yu Miao C, Ying Miao J, Michels PAM, Michiels C, Mijaljica D, Milojkovic A, Minucci S, Miracco C, Miranti CK, Mitroulis I, Miyazawa K, Mizushima N, Mograbi B, Mohseni S, Molero X, Mollereau B, Mollinedo F, Momoi T, Monastyrska I, Monick MM, Monteiro MJ, Moore MN, Mora R, Moreau K, Moreira PI, Moriyasu Y, Moscat J, Mostowy S, Mottram JC, Motyl T, Moussa CEH, Müller S, Muller S, Münger K, Münz C, Murphy LO, Murphy ME, Musarò A, Mysorekar I, Nagata E, Nagata K, Nahimana A, Nair U, Nakagawa T, Nakahira K, Nakano H, Nakatogawa H, Nanjundan M, Naqvi NI, Narendra DP, Narita M, Navarro M, Nawrocki ST, Nazarko TY, Nemchenko A, Netea MG, Neufeld TP, Ney PA, Nezis IP, Phuc Nguyen H, Nie D, Nishino I, Nislow C, Nixon RA, Noda T, Noegel AA, Nogalska A, Noguchi S, Notterpek L, Novak I, Nozaki T, Nukina N, Nürnberger T, Nyfeler B, Obara K, Oberley TD, Oddo S, Ogawa M, Ohashi T, Okamoto K, Oleinick NL, Javier Oliver F, Olsen LJ, Olsson S, Opota O, Osborne TF, Ostrander GK, Otsu K, Ou JHJ, Ouimet M, Overholtzer M, Ozpolat B, Paganetti P, Pagnini U, Pallet N, Palmer GE, Palumbo C, Pan T, Panaretakis T, Bhan Pandey U, Papackova Z, Papassideri I, Paris I, Park J, Park OK, Parys JB, Parzych KR, Patschan S, Patterson C, Pattingre S, Pawelek JM, Peng J, Perlmutter DH, Perrotta I, Perry G, Pervaiz S, Peter M, Peters GJ, Petersen M, Petrovski G, Phang JM, Piacentini M, Pierre P, Pierrefite-Carle V, Pierron G, Pinkas-Kramarski R, Piras A, Piri N, Platanias LC, Pöggeler S, Poirot M, Poletti A, Poüs C, Pozuelo-Rubio M, Prætorius-Ibba M, Prasad A, Prescott M, Priault M, Produit-Zengaffinen N, Progulske-Fox A, Proikas-Cezanne T, Przedborski S, Przyklenk K, Puertollano R, Puyal J, Bing Qian S, Qin L, Hong Qin Z, Quaggin SE, Raben N, Rabinowich H, Rabkin SW, Rahman I, Rami A, Ramm G, Randall G, Randow F, Ashutosh Rao V, Rathmell JC, Ravikumar B, Ray SK, Reed BH, Reed JC, Reggiori F, Régnier-Vigouroux A, Reichert AS, Reiners JJ, Reiter RJ, Ren J, Revuelta JL, Rhodes CJ, Ritis K, Rizzo E, Robbins J, Roberge M, Roca H, Roccheri MC, Rocchi S, Peter Rodemann H, Rodríguez De Córdoba S, Rohrer B, Roninson IB, Rosen K, Rost-Roszkowska MM, Rouis M, Rouschop KMA, Rovetta F, Rubin BP, Rubinsztein DC, Ruckdeschel K, Rucker EB, Rudich A, Rudolf E, Ruiz-Opazo N, Russo R, Erik Rusten T, Ryan KM, Ryter SW, Sabatini DM, Sadoshima J, Saha T, Saitoh T, Sakagami H, Sakai Y, Hoseini Salekdeh G, Salomoni P, Salvaterra PM, Salvesen G, Salvioli R, Sanchez AMJ, Sánchez-Alcázar JA, Sánchez-Prieto R, Sandri M, Sankar U, Sansanwal P, Santambrogio L, Saran S, Sarkar S, Sarwal M, Sasakawa C, Sasnauskiene A, Sass M, Sato K, Sato M, Schapira AHV, Scharl M, Schätzl HM, Scheper W, Schiaffino S, Schneider C, Schneider ME, Schneider-Stock R, Schoenlein PV, Schorderet DF, Schüller C, Schwartz GK, Scorrano L, Sealy L, Seglen PO, Segura-Aguilar J, Seiliez I, Seleverstov O, Sell C, Bok Seo J, Separovic D, Setaluri V, Setoguchi T, Settembre C, Shacka JJ, Shanmugam M, Shapiro IM, Shaulian E, Shaw RJ, Shelhamer JH, Ming Shen H, Chiang Shen W, Hang Sheng Z, Shi Y, Shibuya K, Shidoji Y, Jer Shieh J, Ming Shih C, Shimada Y, Shimizu S, Shintani T, Shirihai OS, Shore GC, Sibirny AA, Sidhu SB, Sikorska B, Silva-Zacarin ECM, Simmons A, Katharina Simon A, Uwe Simon H, Simone C, Simonsen A, Sinclair DA, Singh R, Sinha D, Sinicrope FA, Sirko A, Siu PM, Sivridis E, Skop V, Skulachev VP, Slack RS, Smaili SS, Smith DR, Soengas MS, Soldati T, Song X, Sood AK, Wah Soong T, Sotgia F, Spector SA, Spies CD, Springer W, Srinivasula SM, Stefanis L, Steffan JS, Stendel R, Stenmark H, Stephanou A, Stern ST, Sternberg C, Stork B, Strålfors P, Subauste CS, Sui X, Sulzer D, Sun J, Yong Sun S, Jun Sun Z, Sung JJY, Suzuki K, Suzuki T, Swanson MS, Swanton C, Sweeney ST, King Sy L, Szabadkai G, Tabas I, Taegtmeyer H, Tafani M, Takács-Vellai K, Takano Y, Takegawa K, Takemura G, Takeshita F, Talbot NJ, Tan KSW, Tanaka K, Tanaka K, Tang D, Tang D, Tanida I, Tannous BA, Tavernarakis N, Taylor GS, Taylor GA, Paul Taylor J, Terada AS, Terman A, Tettamanti G, Thevissen K, Thompson CB, Thorburn A, Thumm M, Tian FF, Tian Y, Tocchini-Valentini G, Tolkovsky AM, Tomino Y, Tönges L, Tooze SA, Tournier C, Tower J, Towns R, Trajkovic V, Travassos LH, Fen Tsai T, Tschan MP, Tsubata T, Tsung A, Turk B, Turner LS, Tyagi SC, Uchiyama Y, Ueno T, Umekawa M, Umemiya-Shirafuji R, Unni VK, Vaccaro MI, Maria Valente E, Van Den Berghe G, Van Der Klei IJ, Van Doorn WG, Van Dyk LF, Van Egmond M, Van Grunsven LA, Vandenabeele P, Vandenberghe WP, Vanhorebeek I, Vaquero EC, Velasco G, Vellai T, Miguel Vicencio J, Vierstra RD, Vila M, Vindis C, Viola G, Teresa Viscomi M, Voitsekhovskaja OV, Von Haefen C, Votruba M, Wada K, Wade-Martins R, Walker CL, Walsh CM, Walter J, Bo Wan X, Wang A, Wang C, Wang D, Wang F, Wang F, Wang G, Wang H, Gang Wang H, Dar Wang H, Wang J, Wang K, Wang M, Wang RC, Wang X, Wang X, Jan Wang Y, Wang Y, Wang Z, Charles Wang Z, Wang Z, Wansink DG, Ward DM, Watada H, Waters SL, Webster P, Wei L, Weihl CC, Weiss WA, Welford SM, Ping Wen L, Whitehouse CA, Lindsay Whitton J, Whitworth AJ, Wileman T, Wiley JW, Wilkinson S, Willbold D, Williams RL, Williamson PR, Wouters BG, Wu C, Cheng Wu D, Wu WKK, Wyttenbach A, Xavier RJ, Xi Z, Xia P, Xiao G, Xie Z, Xie Z, Zhi Xu D, Xu J, Xu L, Xu X, Yamamoto A, Yamamoto A, Yamashina S, Yamashita M, Yan X, Yanagida M, Sheng Yang D, Yang E, Ming Yang J, Yu Yang S, Yang W, Yuan Yang W, Yang Z, Chao Yao M, Pang Yao T, Yeganeh B, Lien Yen W, Jing Yin J, Ming Yin X, Joon Yoo O, Yoon G, Yong Yoon S, Yorimitsu T, Yoshikawa Y, Yoshimori T, Yoshimoto K, Jin You H, Youle RJ, Younes A, Yu L, Yu L, Woon Yu S, Haung Yu W, Min Yuan Z, Yue Z, Heui Yun C, Yuzaki M, Zabirnyk O, Silva-Zacarin E, David Zacks E, Zacksenhaus L, Zaffaroni N, Zakeri Z, Zeh HJ, Zeitlin SO, Zhang H, Ling Zhang H, Zhang J, Pu Zhang J, Zhang L, Zhang L, Yong Zhang M, Dong Zhang X, Zhao M, Fang Zhao Y, Zhao Y, Zhao ZJ, Zheng X, Zhivotovsky B, Zhong Q, Zhao Zhou C, Zhu C, Guo Zhu W, Feng Zhu X, Zhu X, Zhu Y, Zoladek T, Xing Zong W, Zorzano A, Zschocke J, Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Romanelli E, Sorbara CD, Nikić I, Dagkalis A, Misgeld T, Kerschensteiner M. Cellular, subcellular and functional in vivo labeling of the spinal cord using vital dyes. Nat Protoc. 2013;8:481–490. doi: 10.1038/nprot.2013.022. [DOI] [PubMed] [Google Scholar]
- 95.Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, Weissman I, Siebert P. “Fluorescent timer”: Protein that changes color with time. Science. 2000;290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- 96.Hernandez G, Hernandez G, Thornton C, Stotland A, Lui D, Sin J, Ramil J, Magee N, Andres A, Quarato G, Carreira RS, Richard Sayen M, Wolkowicz R, Gottlieb RA. MitoTimer: A novel tool for monitoring mitochondrial turnover. Autophagy. 2013;9:1852–1861. doi: 10.4161/auto.26501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Laker RC, Xu P, Ryall KA, Sujkowski A, Kenwood BM, Chain KH, Zhang M, Royal MA, Hoehn KL, Dirscoll M, Adler PN, Wessells RJ, Saucerman JJ, Yan Z. A novel MitoTimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J Biol Chem. 2014;289:12005–12015. doi: 10.1074/jbc.M113.530527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stotland A, Gottlieb RA. α-MHC MitoTimer mouse: In vivo mitochondrial turnover model reveals remarkable mitochondrial heterogeneity in the heart. J Mol Cell Cardiol. 2016;90:53–58. doi: 10.1016/j.yjmcc.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Allen GFG, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McWilliams TG, Prescott AR, Allen GFG, Tamjar J, Munson MJ, Thomson C, Muqit MMK, Ganley IG. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. 2016;214:333–345. doi: 10.1083/jcb.201603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 102.Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmström KM, Fergusson MM, Hyun Yoo Y, Combs CA, Finkel T. Measuring in vivo mitophagy. Mol Cell. 2015;60:685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nezich CL, Wang C, Fogel AI, Youle RJ. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J Cell Biol. 2015;210:435–450. doi: 10.1083/jcb.201501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat Rev Mol Cell Biol. 2016;17:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A. Mitochondrial stress induces chromatin reorganization to promote longevity and UPRmt. Cell. 2016;165:1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rodriguez A, Durán A, Selloum M, Champy M-F, Diez-Guerra FJ, María Flores J, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3:211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 107.Kim KY, Stevens MV, Akter MH, Rusk SE, Huang RJ, Cohen A, Noguchi A, Springer D, Bocharov AV, Eggerman TL, Suen DF, Youle RJ, Amar M, Remaley AT, Sack MN. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest. 2011;121:3701–3712. doi: 10.1172/JCI44736. [DOI] [PMC free article] [PubMed] [Google Scholar]