Abstract
Background: Electronic health and administrative data are increasingly being used for identifying surgical site infections (SSI). We found an unexpectedly high number of patients who could not be classified definitively as having an infection or not. To further explore this, we present an electronic classification algorithm for conservative case finding and identify alterations that would adapt the method for other purposes.
Methods: Two computer algorithms were created to identify SSI. One model used a strict National Healthcare Safety Network (NHSN) based SSI algorithm, which was applied to all discharges from 443,284 all discharges from four hospitals in Manhattan, NY, 2009 through 2012. The second model used discharges that only had NHSN-defined SSI procedures during the same period.
Results: The strict SSI algorithm was able to classify SSI status for 27.3% of discharges; there was a high number of indeterminate cases. In contrast, the modified, less strict model, classified 97.2% of discharges with NHSN-approved SSI procedures.
Conclusion: Electronic records provide several options for aiding with the identification of infections in healthcare settings and can be tailored to suit specific uses. While algorithms for SSI classification should reflect the NHSN definition, our research emphasizes how variations of model building can affect the number of indeterminate cases that may necessitate manual review.
Keywords: : hospital-acquired infection, surgical site infection
Manual surveillance of health-care–associated infections (HAIs) is notoriously labor intensive and lacking in inter-rater reliability [1]. To mitigate the time burden of HAI surveillance and improve its accuracy, a growing number of hospitals are turning to electronic methods. Electronic health and administrative data are increasingly available for this purpose, although their use in classification algorithms has been shown to have low positive predictive value, among other challenges [2,3]. The validity of electronic surveillance varies considerably by infection type, granting this method greater utility for certain kinds of reportable infections [4]. Outcomes such as blood stream infections (BSI), for example, are more readily identified via electronic methods while others, particularly surgical site infections (SSIs), require more nuanced review.
As part of a federally funded study (Health Information Technology to Reduce Healthcare-Associated Infections, NR010822), we developed computerized classification algorithms to identify four types of HAIs using electronically available data [5]. Electronic data were suitable for identifying pneumonia, BSI, and urinary tract infection (UTI), but for SSIs, we found a high number of patients who could not be classified definitively as having an infection or not. To further explore this limitation and optimize SSI detection for specific uses, we present an electronic classification algorithm for conservative case finding and identify alterations that would adapt the method for other purposes.
Methods
We developed a data mart that included electronic health and administrative records for patients discharged from four academically affiliated acute care hospitals in Manhattan, NY [5]. All patients discharged from 2009 through 2012 were included. The four hospitals are part of the same network and share information technology systems including a commercially available electronic medical record and charting system, an admission-discharge-transfer (ADT) system, and a clinical data warehouse storing information from several smaller sources such as clinical laboratory records.
Based on HAI surveillance guidelines published by the Centers for Disease Control and Prevention's (CDC) National Healthcare Safety Network (NHSN) [6], electronic algorithms for identifying BSI, SSI, UTI, and pneumonia were developed by an interdisciplinary team that included an infectious disease physician, an infection prevention nurse, an epidemiologist, a programmer/data manager, and an information technology systems manager with expertise in the use of hospital administrative data. The algorithms, described in Table 1, use a combination of International Classification of Diseases, 9th Revision, Clinical Modification (ICD-9-CM) procedure and diagnosis codes, date and time-stamped clinical microbiology culture results, and admission and discharge dates. The classification algorithms were designed to be conservative in their identification of patients who likely had infections and those who likely did not have infections by creating an “indeterminate” category for those whose records contained incomplete, insufficient, or conflicting information.
Table 1.
Classification Algorithms for Identifying Health-Care–Associated Infections
| Infection type | Infection | No infection | Indeterminate |
|---|---|---|---|
| Blood stream | (+) blood culture AND no other (+) culture with same organism at another body site in previous 14 d | [No (+) blood culture] OR [<2 (+) cultures with common skin contaminant within 2 d period] | [ICD-9-CM code for sepsis AND no (+) blood culture] OR [(+) culture with same organism at another body site in previous 14 d] |
| Pneumonia | ICD-9-CM code for pneumonia AND (+) respiratory culture | No ICD-9-CM code for pneumonia AND no (+) respiratory culture | [ICD-9-CM code for pneumonia AND no (+) respiratory culture] OR [no ICD-9-CM code for pneumonia AND (+) respiratory culture] OR [no ICD-9-CM code for pneumonia AND (+) urine streptococcal antigen] |
| Surgical site | ICD-9-CM code for NHSN procedure AND (+) wound culture within 30 d | ICD-9-CM code for NHSN procedure AND no wound culture performed within 30 d AND no ICD-9-CM code for post-operative infection | [No ICD-9-CM code for NHSN procedure] OR [NHSN procedure AND (-) wound culture within 30 d] OR [NHSN procedure AND no wound culture performed AND ICD-9-CM code for post operative infection] |
| Urinary tract | [(+) urine culture of ≥105 CFU/mL with <2 other species] OR [(+) urine culture of 103–105 CFU/mL with <2 other species AND pyuria (≥3 WBC per high power field in urine microscopy) within ±48 h of culture] | No (+) urine culture AND no ICD-9-CM code for urinary tract infection | ICD-9-CM code for urinary tract infection AND no (+) urine culture |
ICD-9-CM = International Classification of Diseases, 9th Revision, Clinical Modification; NHSN = National Healthcare Safety Network; CFU = colony forming unit.
The completed algorithms were applied to all patient discharges occurring during the study period. For each infection type, we counted the number and percent of patient discharges that were classified as indeterminate. For SSIs, we used a flow diagram to determine the number and percent of patient discharges that were classified as having an infection, not having an infection, and having indeterminate status at each stage of the algorithm.
Results
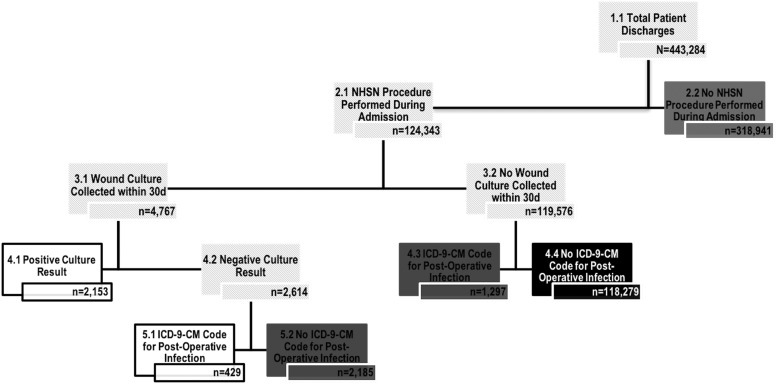
A total of 443,284 discharges occurred during the four-year study period and were included in our analyses. Our algorithms were able to classify BSI status for 97.5%, UTI status for 95.8%, and pneumonia status for 94.4% of discharges (n = 10,930, n = 18,501, and n = 24,980 indeterminate discharges, respectively). When applying the strict NHSN guidelines, SSI status was classified for 27.3% of discharges (n = 322,423 indeterminate discharges). Figure 1 illustrates the number of indeterminate discharges resulting from each step of the SSI classification algorithm. Some discharges were indeterminate because at least one post-operative incision culture was taken, but all results were negative, and no clinical diagnosis of post-operative infection was recorded in the discharge codes (0.7%, n = 2,185). A smaller percentage (0.3%, n = 1,297) was indeterminate, because there was a discrepancy between the microbiology record and ICD-9-CM diagnosis codes (i.e., a post-operative infection was documented but no incision culture was taken).
FIG. 1.
Classification algorithm for identifying surgical site infections (SSI) using electronic data using strict NHSN definitions. White boxes represent the number of patient discharges identified as having an SSI. Black boxes represent the number of patient discharges identified as not having an SSI. Dark gray boxes represent the number of patient discharges who could not be definitively classified using the algorithm.
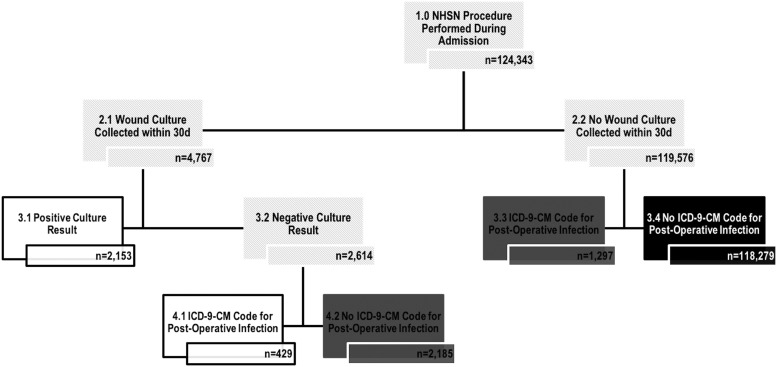
Figure 2 illustrates the number of indeterminate discharges resulting from each step of the SSI classification algorithm when the NHSN definition is modified to only consider patients who underwent a NHSN-defined SSI procedure (n = 124,343). Using the modified algorithm, the indeterminate numbers were reduced substantially (n = 3,482).
FIG. 2.
Classification algorithm for identifying surgical site infections (SSI) using electronic data using a modified NHSN definition only to consider patients that underwent a NHSN defined SSI procedure. White boxes represent the number of patient discharges identified as having an SSI. Black boxes represent the number of patient discharges identified as not having an SSI. Dark gray boxes represent the number of patient discharges who could not be definitively classified using the algorithm.
Discussion
Electronic records provide several options for aiding with the identification of infections in healthcare settings and can be tailored to suit specific uses [2]. Nevertheless, work is still needed to assess and improve the validity and reliability of electronic administrative data and identify the most useful, parsimonious, and accurate data elements for SSI surveillance.
Administrative data have been used for surveillance of various types of infections with mixed results to date. A decade ago, researchers reported a positive predictive value of 20% for HAIs identified by administrative data as compared with 100% identified through active surveillance by an experienced infection prevention professional [7]. More recently, Snyders et al. [8] found that electronic algorithms to identify central line associated BSI required adjustment for various populations, and others have noted discrepancies in diagnosing SSIs, depending on definitions used [9,10]. Singh et al. [11] found a significant discrepancy between SSI rates reported to a national United Kingdom surveillance system when compared with rates identified by a retrospective review of electronic medical records, with higher rates identified electronically. It was not possible to discern, however, whether the discrepancy was attributable to under-reporting or to under-identification of cases.
Others have evaluated the use of ICD-CM (currently ICD-10-CM) codes for post-operative infection, although there are limitations to relying solely on discharge diagnosis codes because sensitivity and specificity may not be adequate [12]. Another method would be to identify patients who had incision cultures performed during their admission. Because incision cultures may be collected for reasons unrelated to SSI or may not be collected at all, however, this approach would still yield a large proportion of patient discharges for whom manual chart review would be required and the positive predictive value of this added step would likely be low.
In a recent systematic review of 57 studies using electronic surveillance for HAIs, sensitivities and predictive values were highly variable, and the studies were characterized by considerable methodologic heterogeneity. Hence, the authors recommended careful use of such data and continued work to improve algorithms [13]. In January 2016, the CDC's NHSN Patient Safety Component Manual published updated guidelines that provide more specificity for identifying and monitoring SSI and re-emphasized the importance of using epidemiologically sound infection definitions and effective surveillance methods [14]. The European Centre for Disease Prevention and Control also publishes an annual epidemiologic report on SSIs [15] using slightly different case definitions [16].
Our study adds to the extant and burgeoning literature on use of administrative data for case finding and identification of SSIs. The algorithm presented in this study was initially developed for the purpose of a research study in which the goal was to match patients who had infections with those who did not. Therefore, our classification schema was designed to be conservative in the identification of infected and non-infected patient discharges to minimize false positives and false negatives. The objectives of electronic case finding for the of purpose institutional surveillance, however, are different. Infection control staff members need to apply a more inclusive case definition, one that identifies definitive versus possible infections to flag those that would require manual chart review and clinical adjudication. The ultimate goal of surveillance methods is to provide valid and reliable, efficient, real-time SSI data that would minimize the resources required for manual record searches and other surveillance activities that now require a large proportion of time from infection prevention and control staff [17,18].
In 2014, Woeltje et al. [19] published recommended data elements for effective electronic surveillance of selected HAIs and the possible complications associated with each. For SSI surveillance, a combination of microbiologic cultures, procedure and diagnosis billing codes, and ADT data were identified as key elements. Ultimately, electronic surveillance that allows use of data from multiple sources including surgical, laboratory, radiologic and medication records, physician and nursing notes as well as ICD-10-CM codes shows great promise for more precise and efficient case finding.
Like all studies involving electronic record review, our investigation has some limitations. Although the classifications were thoroughly reviewed by several members of our institution's infection control team and an initial validation study was performed [20], further validation studies with larger and more geographically representative samples are in order. Finally, the largest proportion of SSIs generally occur after the patient's discharge from the hospital, and hence those identified among inpatients represent only a small proportion of the total incidence. Our aim, however, was not to assess incidence of SSI but rather to apply the NHSN definitions of SSI and determine the extent to which those infections that do manifest among inpatients can be detected using electronically collected data.
Even when essential data elements are available, reliance on electronic review has several limitations because of the complexity of clinical presentation and diagnostic criteria for SSIs, and further work is needed to take maximum advantage of the large volumes of electronic data now available. Despite limitations of electronic monitoring for SSI, progress has been made in data mining, programming, and standardization of definitions. Hence, electronic monitoring is useful for identifying patients likely to have an SSI so that follow-up by infection prevention and control staff is expedited. Ultimately, increasingly sensitive and sophisticated surveillance and reporting methods will free up more time for other prevention activities such as staff and patient education.
Acknowledgments
This study was funded in part by a grant from National Institute of Nursing Research, R01 NR010822-07S1.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Freeman R, Moore LS, García Álvarez L, et al. Advances in electronic surveillance for healthcare-associated infections in the 21st Century: A systematic review. J Hosp Infect 2013;84:106–119 [DOI] [PubMed] [Google Scholar]
- 2.Klompas M, Yokoe DS. Automated surveillance of health care-associated infections. Clin Infect Dis 2009;48:1268–1275 [DOI] [PubMed] [Google Scholar]
- 3.Cohen B, Vawdrey DK, Liu J, et al. Challenges associated with using large data sets for quality assessment and research in clinical settings. Policy Polit Nurs Pract 2015;16:117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tokars JI, Richards C, Andrus M, et al. The changing face of surveillance for health care-associated infections. Clin Infect Dis 2004;39:1347–1352 [DOI] [PubMed] [Google Scholar]
- 5.Apte M, Neidell M, Furuya EY, et al. Using electronically available inpatient hospital data for research. Clin Transl Sci 2011;4:338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 2008;36:309–332 [DOI] [PubMed] [Google Scholar]
- 7.Sherman ER, Heydon KH, St John KH, et al. Administrative data fail to accurately identify cases of healthcare-associated infections. Infect Control Hosp Epidemiol 2006;27:332–337 [DOI] [PubMed] [Google Scholar]
- 8.Snyders RE, Goris AJ, Gase KA, et al. Increasing reliability of fully automated surveillance for central line-associated bloodstream infections. Infect Control Hosp Epidemiol 2015;36:1396–1400 [DOI] [PubMed] [Google Scholar]
- 9.Taylor JS, Marten C, Potts K, et al. What is the real rate of surgical-site infection? In: Proceedings of the ASCO Annual Meeting, Chicago, Illinois, June3–7, 2016 2016;34(Suppl 7):171) [Google Scholar]
- 10.Ju MH, Ko CY, Hall BL, et al. A comparison of 2 surgical site infection monitoring systems. JAMA Surg 2015;150:51–7 [DOI] [PubMed] [Google Scholar]
- 11.Singh S, Davies J, Sabou S, et al. Challenges in reporting surgical site infections to the national surgical site infection surveillance and suggestions for improvement. Ann R Coll Surg Engl 2015;97:460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schweizer ML, Eber MR, Laxminarayan R, et al. Validity of ICD-9-CM coding for identifying incident methicillin-resistant Staphylococcus aureus (MRSA) infections: Is MRSA infection coded as a chronic disease? Infect Control Hosp Epidemiol 2011;32:148–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Mourik MS, van Duijn PJ, Moons KG, et al. Accuracy of administrative date for surveillance of healthcare-associated infections: A systematic review. BMJ Open 2015;5:e008424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disesease Control and Prevention. National Healthcare Safety Network (NHSN) Patient Safety Component Manual. 2016. www.cdc.gov/nhsn/pdfs/pscmanual/pcsmanual_current.pdf Last accessed March15, 2017
- 15.European Centre for Disease Prevention and Control. Annual epidemiologic report. Surgical site infections. ecdc.europa.eu/en/healthtopics/Healthcare-associated_infections/surgical/site-infections/Pages/Annual-epidemiological-report-2016.aspx Last accessed March15, 2017
- 16.European Commission. Commission Implementing Decision amending Decision 2002/253/EC laying down case definitions for reporting communicable diseases to the Community network under Decision No 2119/98/EC of the European Parliament and of the Council (2012/506/EU). Off J Eur Union 2012;L262:1–57 [Google Scholar]
- 17.Grota PG, Stone PW, Jordan S, et al. Electronic surveillance systems in infection prevention: Organizational support, program characteristics, and user satisfaction. Am J Infect Control 2010;38:509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stone PW, Dick A, Pogorzelska M, et al. Staffing and structure of infection prevention and control programs. Am J Infect Control 2009;37:351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woeltje KF, Lin MY, Klompas M, et al. Data requirements for electronic surveillance of healthcare-associated infections. Infect Control Hosp Epidemiol 2014;35:1083–1091 [DOI] [PubMed] [Google Scholar]
- 20.Apte M, Landers T, Furuya Y, et al. Comparison of two computer algorithms to identify surgical site infections. Surg Infect (Larchmt) 2011;12:459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]