Abstract
Placental abnormalities can cause Pregnancy-Associated Disorders, including preeclampsia, intrauterine growth restriction, and placental insufficiency, resulting in complications for both the mother and fetus. Trophoblast cells within the labyrinthine layer of the placenta facilitate the exchange of nutrients, gases, and waste between mother and fetus; therefore, the development of this cell layer is critical for fetal development. As trophoblast cells differentiate, it is assumed their metabolism changes with their energy requirements. We hypothesize that proper regulation of trophoblast metabolism is a key component of normal placental development; therefore, we examined the role of AMP-activated kinase (AMPK, PRKAA1/2), a sensor of cellular energy status. Our previous studies have shown that AMPK knockdown alters both trophoblast differentiation and nutrient transport. In this study, AMPKα1/2 shRNA was used to investigate the metabolic effects of AMPK knockdown on SM10 placental labyrinthine progenitor cells before and after differentiation. Extracellular flux analysis confirmed that AMPK knockdown was sufficient to reduce trophoblast glycolysis, mitochondrial respiration, and ATP coupling efficiency. A reduction in AMPK in differentiated trophoblasts also resulted in increased mitochondrial volume. These data indicate that a reduction in AMPK disrupts cellular metabolism in both progenitors and differentiated placental trophoblasts. This disruption correlates to abortive trophoblast differentiation that may contribute to the development of Pregnancy-Associated Disorders.
Keywords: : AMP-activated protein kinase, placenta, trophoblast, differentiation, mitochondria, glycolysis
Introduction
The placenta is an essential and complex organ that develops exclusively during pregnancy, and abnormal placental development has been implicated in a number of Pregnancy-Associated Disorders, which can have detrimental effects on fetal and maternal health [1–3]. Placental development is a delicate balance between trophoblast stem cell proliferation and differentiation into distinct placental cell types (lineages). The rodent placenta, which shares numerous functional and morphological features with the human placenta, is composed of three predominant cell lineages that are important for invasion, progenitor propagation, and nutrient transport [4,5]. In this study, we focused on the labyrinthine trophoblast cells, which are primarily responsible for the transport of nutrients, gases, and wastes between the mother and fetus [6,7].
As trophoblasts differentiate, their rates of energy production are assumed to change in response to their energy requirements. We hypothesize that proper regulation of trophoblast metabolism is a key component of normal placental development. To evaluate trophoblast metabolism, we examined the role of AMP-activated kinase (AMPK, PRKAA1/2) in labyrinthine trophoblast differentiation. AMPK (AMP-activated protein kinase, PRKAA1/2, or hydroxymethylglutaryl-CoA reductase NADPH kinase) is a sensor of cellular energy and regulates cell metabolism [8–10]. AMPK is a ubiquitously expressed heterotrimeric serine/threonine kinase composed of alpha, beta, and gamma subunits [11–13]. The catalytic alpha subunit exists in two isoforms (AMPKα1 and AMPKα2). Recent reports have suggested that placental trophoblast differentiation may be regulated by AMPK [14,15].
As proper placental development is necessary for a healthy pregnancy, alterations in AMPK signaling and trophoblast differentiation may result in Pregnancy-Associated Disorders and impair fetal growth and/or viability.
In humans, AMPKα1 genotypic changes are associated with irregular birth weight, oxygen regulation, and metabolic homeostasis, and are implicated in intrauterine growth restriction [16]. In addition, AMPK is present in the placenta of humans and mice, is increased under hypoxia, and facilitates uterine artery blood flow [17]. Furthermore, the role of AMPK in the pathogenesis of preeclampsia has been reported, using a reduced uteroplacental perfusion pressure model. In this system, administration of an AMPK activator was able to prevent the development of hypertension and normalize angiogenesis [18].
However, the role of AMPK to directly effect trophoblast differentiation has only recently been established. Our recent study showed that simultaneous knockdown of AMPKα1/2 isoforms prevents appropriate placental differentiation with significant alterations in glucose and amino acid transport, as well as morphology in the placental labyrinthine progenitor cell line, SM10 [14]. Together, these observations led us to hypothesize that AMPK may play a key role in regulating metabolic stability in placental trophoblasts and that its dysregulation could therefore negatively impact successful differentiation of these cells.
In this study, we used stable clones of SM10 mouse trophoblast progenitor cells that expressed a scrambled (Control) shRNA or AMPK shRNA (specific for both AMPKα1 and AMPKα2 subunits) to investigate the metabolic effects of AMPKα1/α2 knockdown on labyrinthine trophoblast differentiation. Our data indicate that a reduction in AMPK leads to reduced glycolysis, inhibition of mitochondrial respiration, and an increase in total mitochondrial volume, and results in the failure of placental labyrinthine trophoblast cells to fully differentiate.
Experimental Procedures
Materials
RPMI-1640/l-glutamine media (SH30027.01), 50 μM β-mercaptoethanol (35602), 1% antibiotic–antimycotic (SV30079.01), and Hoechst (62249) were purchased from Thermo Scientific. Fetal bovine serum (S01520) was obtained from Biowest. Sodium pyruvate (25-000-CI) and blasticidin (ant-bl-1) were purchased from Mediatech, Inc. and InvivoGen, respectively. Polyclonal rabbit TOM-20 antibody (sc-11415) was purchased from Santa Cruz. Mouse monoclonal anti-alpha 1 sodium/potassium ATPase antibody (ab7671) was obtained from Abcam. Rhodamine Red x-conjugated AffiniPure donkey anti-rabbit IgG (711-295-152) was purchased from Jackson Labs. Alexa-Fluor-488 goat anti-mouse secondary antibody (A10667) was obtained from Molecular Probes and TGF-β2 was a kind gift from Dr. Steve Ledbetter (Genzyme, Inc.).
Cell culture
The SM10 trophoblast cell line has been previously characterized as trophoblast progenitors that differentiate into the labyrinthine, nutrient transport, lineage [19–23]. SM10 cells were cultured in RPMI-1640/l-glutamine medium supplemented with 10% nonheat-inactivated fetal bovine serum, 1% antibiotic–antimycotic, 1 mM sodium pyruvate, and 50 μM β-mercaptoethanol. SM10 shRNA Control (clone B5) and SM10 AMPKα1/2 shRNA knockdown (clone F6) were cultured as previously described and passaged at 80%–90% confluence [14]. Stable expression of the scrambled Control shRNA and AMPKα1/2 shRNA clones was maintained in media containing 12 μg/mL of blasticidin [14]. SM10 cells were differentiated by the addition of 5 ng/mL of TGF-β2 for 72 h, as previously described [14,21,23]. After 3 days of TGF-β induction they are terminally differentiated [14], except for AMPK KD clones, which are unable to fully differentiate.
Cell staining
Cells were plated on sterilized Superfrost Plus slides in 100-mm tissue culture dishes. The following day, the cells were treated with 5 ng/mL TGF-β2 for 72 h to induce differentiation. Media were removed and slides were washed in PBS, fixed with 4% paraformaldehyde for 10 min at room temperature, and then simultaneously incubated with the polyclonal IgG rabbit TOM-20 antibody (FL-145), a mitochondrial marker (sc-11415, 1:200; Santa Cruz), mouse monoclonal anti-α1 sodium/potassium ATPase antibody (464.6), and a plasma membrane marker (ab7671, 1:200; Abcam) overnight at 4°C. The secondary antibodies, Rhodamine Red x-conjugated AffiniPure donkey anti-rabbit IgG (1:200) and Alexa Fluor-488 goat anti-mouse secondary (1:500), were applied for 2 h at room temperature followed by 5 min of Hoechst staining (1 μg/mL).
Image data acquisition with fluorescence microscopy
Fifteen SM10 cells from each group were analyzed (five cells from each replicate experiment) and images were acquired using a DeltaVision microscope system on an inverted Olympus IX71 microscope with an oil immersion, 1.4 N.A. objective, 60 × magnification, with a CoolSNAP HQ CCD camera. Z-sections with a z-step of 0.25 μm and focal depth of 0.28 μm were taken through each cell. Exposure conditions were Texas Red channel (0.05-s exposure, 50% transmission, 617/72 nm emission, and 555/28 nm excitation) for TOM-20 stain; DAPI channel (0.05-s exposure, 100% transmission, 457/50 nm emission, and 360/40 nm excitation) for Hoechst staining; and Alexa-Fluor-488 FITC channel (1.0-s exposure, 100% transmission, 528/38 nm emission, and 490/20 nm excitation) for cytoplasmic staining. Each set of z-sections was deconvolved using SoftWoRx software (Applied Precision).
Image data analysis
IMARIS XT (version 7.7.1; Bitplane, Zurich, Switzerland) was used to perform three-dimensional (3D) reconstruction of SM10 cell z-sections and to quantify mitochondrial, nuclear, and cell volumes [24,25]. Using IMARIS software, surfaces were placed around the nuclei (filtered to one object, smoothing function enabled) and mitochondria (background contrast subtraction applied, unsmoothed). The cytoplasmic staining channel was used to determine cell volume by increasing the signal gain to fill cell boundaries and then applying a surface (smoothing function enabled). The volume and surface area functions in IMARIS were then applied to these surfaces.
Metabolic assays (Seahorse)
SM10 cells (1 × 104 in 100 μL media) were plated in XF24 V7 cell culture microplates (n = 5 wells/condition). Cells were seeded in a two-step process to promote even seeding. They were first incubated in culture media at room temperature for 1 h before being transferred to an incubator at 37°C/5% CO2 for 1 h as per the manufacturer's instructions. After adding 150 μL of media, the cells were then allowed to grow for 24 h in a humidified incubator (37°C with 5% CO2/air). The following day, cells were treated with 5 ng/mL TGF-β2 for 72 h to induce differentiation. To determine glycolysis and mitochondrial respiration, extracellular acidification (ECAR) and oxygen consumption rates (OCR) were monitored using an Extracellular Flux (XF) 24 Analyzer (Agilent Technologies, Santa Clara, CA). SM10 growth media were replaced with 525 μL assay media, as described below.
Glycolysis stress test assay media consisted of XF Base Medium Minimal DMEM (Part No. 103193-100; Agilent Technologies) supplemented with 2.05 mM l-glutamine (Cat. No. SH30034.02; Hyclone). Mitochondrial stress test assay media consisted of XF Base Medium Minimal DMEM (Part No. 103193-100; Agilent Technologies) supplemented with 2.05 mM l-glutamine (Cat. No. SH30034.02; Hyclone), 1 mM sodium pyruvate (S8636; Sigma Life Science), and 25 mM d-glucose (Cat. No. G7528-250G; Sigma Life Science). Assay media were adjusted to pH 7.35 with NaOH at 37°C and were sterile filtered. The cells in assay media were incubated for 1 h in a CO2-free incubator at 37°C to deplete bicarbonate buffering before performing the assays.
Glycolysis and mitochondrial stress tests used mix/wait/measure times of 3/3/3 min, 75 μL drug injections, and three replicate measurements were analyzed after each condition change. Measured rates were normalized to total cell number. The Seahorse XF Glycolysis Stress Test Kit (Part No. 103020-100; Agilent Technologies) used sequential injections of 10 mM d-glucose, 1 μM oligomycin A, and 100 mM 2-deoxy-d-glucose (2-DG) following baseline ECAR measurements. The Seahorse Mito Stress Test Kit (Part No. 103015-100; Agilent Technologies) used sequential injections of 1 μM oligomycin A, 1 μM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), and 1 μM rotenone/antimycin A after basal respiration OCR measurements.
Statistics
For imaging experiments, 5 cells from each condition were imaged for each of three replicates for a total of 15 cells in each condition and 60 cells imaged in total. Average values for mitochondrial volumes, estimated cell volumes, cell surface area, mitochondrial to cell volume ratios, and surface area to cell volume ratios were calculated for each condition and compared using two-way analysis of variance (ANOVA) followed by Tukey's post hoc multiple comparisons test. For Seahorse experiments, groups were an n ≥ 4. Cells were plated in each condition simultaneously and experiments were independently repeated at least three times. Data were analyzed using Microsoft Excel 2011 and two-way ANOVA with Tukey's post hoc multiple comparisons tests and were calculated using PRISM 6 software.
Results
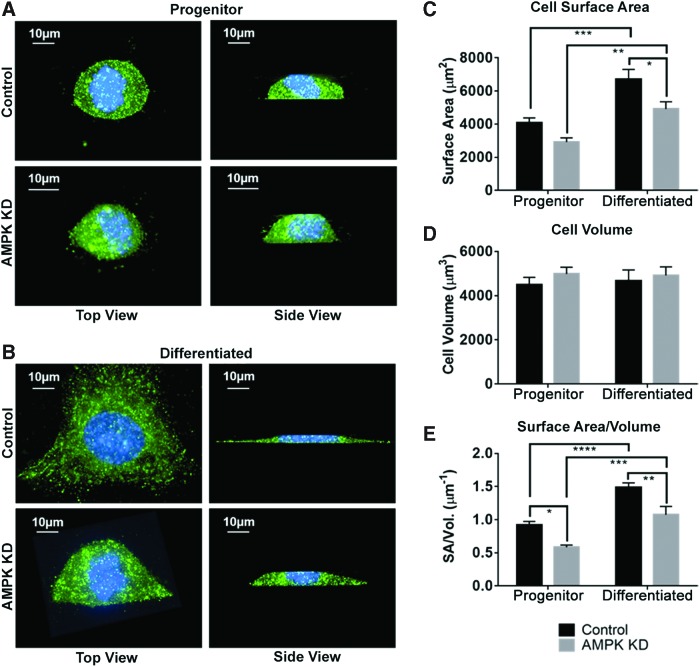
Our previous studies have shown that SM10 progenitor cells can be induced to differentiate on addition of TGF-β, resulting in cell aggregation and colony formation [21,23]. In addition, AMPKα1/2 knockdown via shRNA was shown to inhibit TGF-β-induced differentiation and reduce nutrient transport [14]. To more closely examine the effect of AMPK knockdown on cellular morphology, SM10 cells stably transduced with scrambled shRNA (Control) or AMPKα1/2 shRNA (AMPK KD) were treated with TGF-β or left untreated for 72 h (Fig. 1). SM10 cells were analyzed by immunocytochemistry to visualize cell boundaries using the sodium/potassium ATPase antibody, as it is a highly expressed membrane-bound protein. Z-stack images of individual cells were captured and a 3D composite was rendered (Fig. 1). Cell morphology was assessed by inspection and by measuring surface area and cell volume, followed by calculation of surface area to volume ratios.
FIG. 1.
AMPK knockdown reduces cell surface area. Cells were visualized using the anti-alpha 1 sodium/potassium ATPase antibody (green) and DAPI (blue) in conjunction with deconvolving microscopy and IMARIS software. (A) Representative progenitor AMPK KD and Control SM10 trophoblast cells. (B) Representative TGF-β-treated, differentiated (Differentiated) AMPK KD, and Control SM10 cells. Mean ± SEM for (C) cell surface area and (D) cell volume, calculated in IMARIS from user placed surfaces. (E) Surface area to volume ratio (n = 15 for each group). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 two-way ANOVA with Tukey's multiple comparisons test. ANOVA, analysis of variance. Color images available online at www.liebertpub.com/scd
Control progenitor and differentiated SM10 cell morphology
Treatment with TGF-β causes cells to undergo characteristic differentiation-like behavior [21,23]. In Control SM10 trophoblasts, progenitor cells display a rounded morphology (Fig. 1A). After differentiation, the surface area but not the cellular volume of these cells significantly increased, which led to a subsequent increase in surface area to volume ratio compared to progenitor cells (Fig. 1A–E). Differentiation of SM10 cells using TGF-β resulted in a significant increase in cellular surface area and surface area to volume ratio compared to progenitor cells (Fig. 1C, E).
AMPK knockdown—progenitor and differentiated SM10 cell morphology
AMPKα1/2 knockdown (AMPK KD) significantly reduced surface area to volume ratio in progenitor cells (Fig. 1A, E). Following differentiation, both surface area and the surface area to volume ratio were significantly lower in AMPK KD cells compared to Controls, consistent with compromised differentiation and reduced cell flattening (Fig. 1B, E). Cell volume was not significantly different across all groups (Fig. 1D).
Control progenitor and differentiated SM10 cell glycolysis
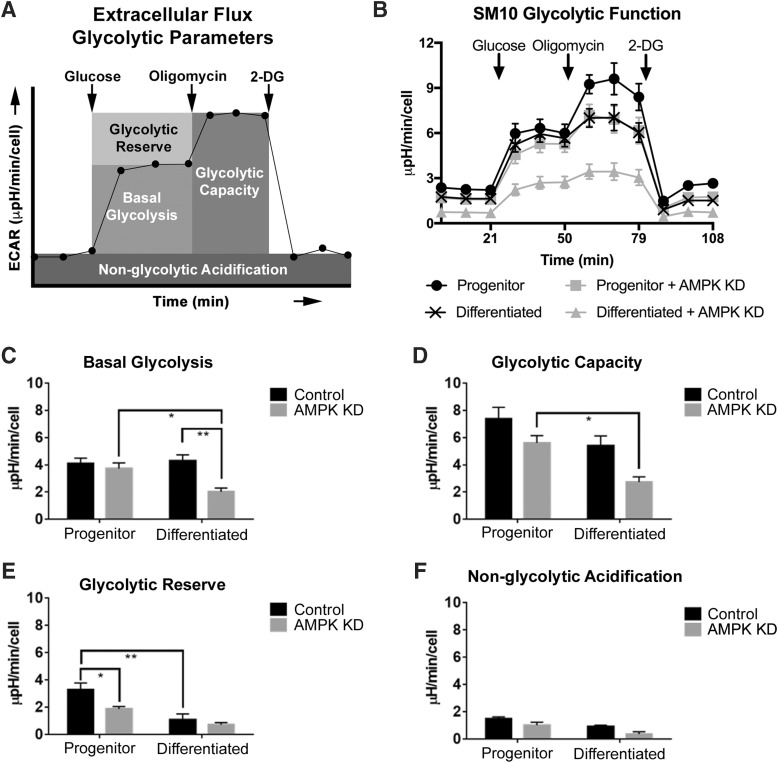
SM10 glycolysis was examined using extracellular flux analysis with the Seahorse Glycolysis Stress Test. Extracellular flux analysis measures oxygen consumption and extracellular pH to analyze mitochondrial and glycolytic responses. A representative diagram of the Extracellular Flux Glycolytic Parameters used in this assay is shown in Fig. 2A. No significant differences were noted between Control progenitor and differentiated SM10 cells for glycolysis, glycolytic capacity, and nonglycolytic acidification (Fig. 2B–D, F). However, a significant reduction in the glycolytic reserve of terminally differentiated Control cells was observed compared to Control progenitor cells (Fig. 2B, E).
FIG. 2.
AMPK knockdown inhibits glycolytic capability. Extracellular Flux Glycolytic Parameters are diagramed in (A) and line graph of the data in (B) are quantified in (C–F). Extracellular flux analysis of basal glycolysis (C), glycolytic capacity (D), glycolytic reserve (E), and nonglycolytic acidification (F) in Control and AMPK KD progenitor and differentiated cells. Extracellular acidification rates (ECAR, μpH/min/cell) are expressed as mean ± SEM and are normalized to number of cells seeded. *P ≤ 0.05, **P ≤ 0.01, two-way ANOVA with Tukey's multiple comparisons test.
AMPK knockdown—progenitor and differentiated SM10 cell glycolysis
AMPK is a major sensor of cellular energy and regulator of metabolism [26]. When activated, AMPK has been shown to increase glycolysis, and our previous studies have shown that AMPK knockdown decreases glucose transport in SM10 cells [14]. To further our understanding of glycolytic regulation by AMPK knockdown, we investigated the ECAR (μpH/min/cell) in AMPK knockdown or Control progenitor and differentiated SM10 cells (Fig. 2).
Resting glycolysis, also known as basal glycolysis, is the glycolytic rate of a cell at rest. Progenitor SM10 cells showed no difference in resting glycolysis in Control versus AMPK KD cells (Fig. 2B, C). However, differentiated AMPK KD cells had significantly lower glycolysis than both progenitor AMPK KD cells and differentiated Control cells (Fig. 2B, C).
The glycolytic capacity of a cell is defined as the highest rate of glycolysis the cell can achieve after ATP synthase is inhibited using oligomycin. No significant change in glycolytic capacity was observed between progenitor cells in Control versus AMPK KD; however, the glycolytic capacity of differentiated AMPK KD cells was significantly reduced compared to progenitor AMPK KD cells (Fig. 2B, D).
During periods of energetic demand, cells can increase glycolysis to produce more ATP. The glycolytic reserve is a measure of the cellular ability to respond to this energetic demand. To determine the effect of AMPK knockdown on SM10 glycolytic reserve in progenitor and differentiated cells, we measured the glycolytic rate, after ATP synthase inhibition with oligomycin. Progenitor AMPK KD cells had a significantly lower glycolytic reserve than progenitor Control cells (Fig. 2B, E). However, this difference in glycolytic reserve is no longer present after treatment with TGF-β (Differentiated) (Fig. 2B, E).
To determine whether the ECAR identified are solely related to glycolysis, nonglycolytic acidification was also measured by inhibiting glycolysis using 2-deoxy-d-glucose (2-DG). No significant changes in nonglycolytic extracellular acidification rates were noted in either progenitor or differentiated SM10 cells in the Control or AMPK KD groups (Fig. 2B, F).
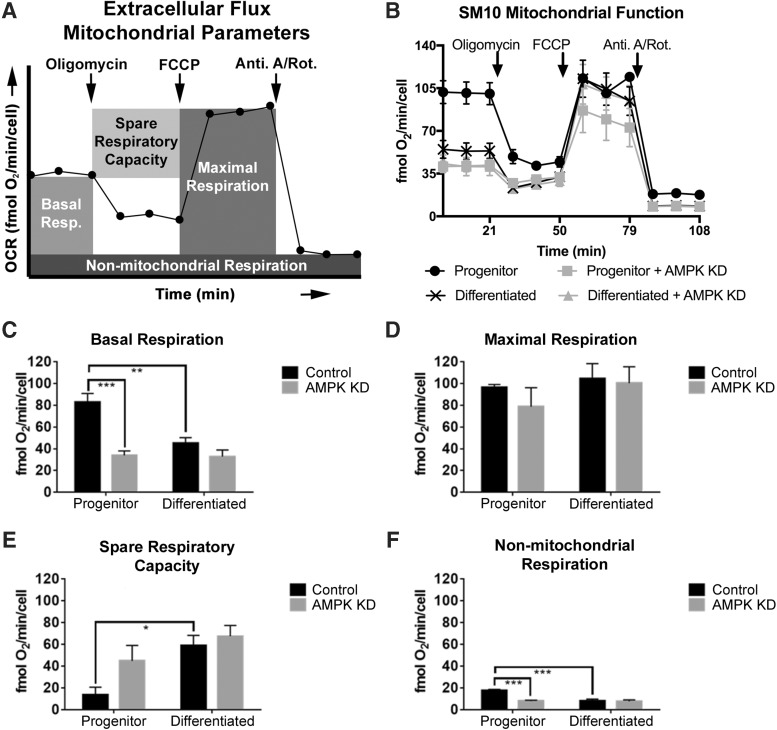
Control progenitor and differentiated SM10 cell mitochondrial respiration
In addition to glycolysis, cells can produce energy via mitochondrial respiration. To investigate the impact of SM10 differentiation on mitochondrial respiration, oxygen consumption was measured via extracellular flux analysis (Figs. 3 and 4). A representative diagram of the Extracellular Flux Mitochondrial Parameters is shown in Figs. 3A and 4A. Resting oxygen consumption (ie, basal respiration) was significantly lower in Control differentiated cells when compared to progenitor cells (Fig. 3B, C).
FIG. 3.
AMPK knockdown alters mitochondrial respiration. Extracellular Flux Mitochondrial Parameters are diagramed in (A) and line graph of the data in (B) are quantified in (C–F). Extracellular Flux Analysis of basal respiration (C), maximal respiration (D), spare respiratory capacity (E), and nonmitochondrial respiration (F) in Control and AMPK KD progenitor and differentiated cells. Oxygen consumption rates (OCR, fmol O2/min/cell) are expressed as mean ± SEM and are normalized to number of cells seeded. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, two-way ANOVA with Tukey's multiple comparisons test.
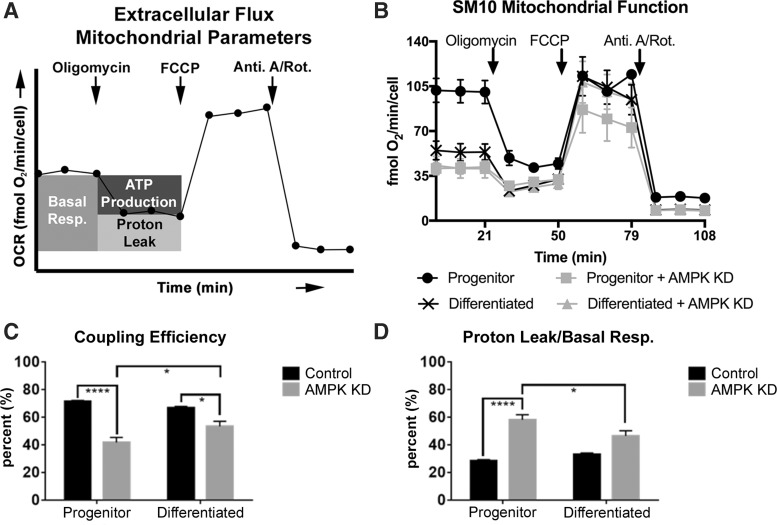
FIG. 4.
AMPK knockdown reduces ATP coupling and increases proton leak. Extracellular Flux Mitochondrial Parameters are diagramed in (A) and line graph of the data in (B) are quantified in (C, D). Ratio of ATP production to basal respiration (Coupling Efficiency) (C). Ratio of proton leak to basal respiration in Control and AMPK KD progenitor and differentiated cells (D). *P ≤ 0.05, ****P ≤ 0.0001 two-way ANOVA with Tukey's multiple comparisons test.
To test the maximal respiration rate that SM10 cells could achieve, the electron transport chain was uncoupled from oxidative phosphorylation using FCCP. No significant difference in maximal respiration was present between Control progenitors or differentiated cells (Fig. 3B, D). We also calculated the capability of the cells to respond to increased energy demands, termed the spare respiratory capacity. This parameter describes how close basal respiration is to the maximal respiratory rate. Differentiated Control cells had a significantly higher spare respiratory capacity than progenitor Control cells (Fig. 3B, E).
Cells can consume oxygen utilizing a variety of nonmitochondrial processes. To demonstrate that the OCR identified are specific to the mitochondria, mitochondrial respiration was assessed by inhibiting Complex I and Complex III with rotenone and antimycin A (Fig. 3B, F). Nonmitochondrial respiration was found to be lower in Control differentiated cells when compared to Control progenitors (Fig. 3B, F).
AMPK knockdown—progenitor and differentiated SM10 cell mitochondrial respiration
Resting oxygen consumption (ie, basal respiration) was significantly lower in progenitor AMPK KD cells compared to progenitor Controls; however, no significant difference was observed in the basal respiration of AMPK KD progenitors compared to AMPK KD differentiated cells (Fig. 3B, C). AMPK knockdown did not result in any differences in maximal respiration or spare respiratory capacity (Fig. 3B, D, E). Progenitor AMPK KD cells exhibited significantly lower nonmitochondrial respiration compared to progenitor Control cells; however, no significant differences were noted between progenitor and differentiated AMPK KD cells (Fig. 3B, F).
Control progenitor and differentiated SM10 cell coupling efficiency and proton leak
Oxygen consumed to establish a proton gradient across the inner mitochondrial membrane supports two processes, ATP production and proton leak. Coupling efficiency refers to the fraction of basal respiration associated with ATP production (Fig. 4A, B). Coupling efficiency was not different in Control progenitors compared to differentiated Control cells (Fig. 4B, C).
Coupling efficiency is impacted by the amount of proton leak from the inner membrane space back into the mitochondrial matrix. Basal respiration includes proton leak; therefore, quantitation of proton leak can be reported as a fraction of proton leak/basal respiration. An increase in proton leak can be a sign of mitochondrial damage and dysfunction. Differentiation did not significantly affect the amount of basal respiration contributed to proton leak in progenitor versus differentiated Control SM10 cells (Fig. 4B, D).
AMPK knockdown—progenitor and differentiated SM10 cell coupling efficiency and proton leak
AMPK knockdown significantly decreased the coupling efficiency compared to Control cells in both progenitor and differentiated conditions (Fig. 4B, C). However, differentiated AMPK KD cells had a significantly higher coupling efficiency compared to progenitor AMPK KD cells (Fig. 4B, C). In progenitor cells, AMPK knockdown resulted in a significantly larger portion of basal respiration to be attributed to proton leak compared to Control cells; however, the portion of proton leak decreased in differentiated AMPK KD cells compared to AMPK KD progenitors (Fig. 4B, D).
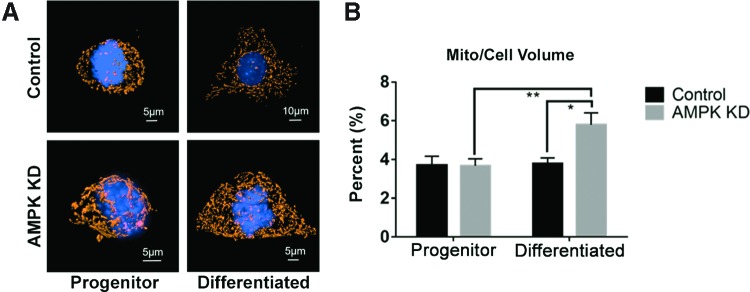
Control progenitor and differentiated SM10 cell mitochondrial volume
To determine whether the mitochondrial changes noted between Control progenitor cells and Control differentiated cells were due to alterations in mitochondrial volume within the cells, we examined the mitochondrial marker, TOM20, using immunocytochemistry. There was no significant difference in mitochondrial volume between Control progenitor and differentiated SM10 cells (Fig. 5A, B).
FIG. 5.
AMPK knockdown increases mitochondrial cell volume. (A) Representative images of SM10 cells in Control and AMPK KD progenitor and differentiated cells. Mitochondria were visualized using TOM-20 antibody (orange) and nuclei were stained with DAPI (blue). (B) Mitochondrial volume as a percentage of cell volume ± SEM. *P ≤ 0.05, **P ≤ 0.01, n = 15 cells per group, two-way ANOVA with Tukey's multiple comparisons test. Color images available online at www.liebertpub.com/scd
AMPK knockdown progenitor and differentiated SM10 cell mitochondrial volume
The reduced glycolysis and mitochondrial respiration with AMPK knockdown suggest that AMPK is an integral component controlling energy production in SM10 placental cells. To further investigate the effect of AMPK knockdown on mitochondria, we assessed mitochondrial volume. Mitochondrial volume increased significantly in differentiated AMPK KD cells compared to progenitor AMPK KD cells or differentiated Control cells (Fig. 5A, B).
Discussion
The SM10 cell line is derived from the labyrinthine layer of the murine placenta, which is involved in the transport of nutrients, gases, and wastes between the mother and fetus [7,19,20]. Abnormal differentiation and development of the placental labyrinth can result in improper nutrient transport and have been implicated in Pregnancy-Associated Disorders [7,14,27]. In this study, we propose that metabolic changes induced by AMPK knockdown alter SM10 trophoblast differentiation. To examine this question, we have first shown that simultaneous knockdown of the AMPKα1 and AMPKα2 subunits in progenitor SM10 trophoblast cells does indeed alter cellular metabolism, resulting in the restriction of critical glycolytic and mitochondrial energy resources (Figs. 2 and 3).
To further understand the role of AMPK in trophoblast differentiation, we analyzed individual cell volumes and cell surface areas. TGF-β induces SM10 cell differentiation, which can be noted morphologically [21]. When trophoblast cells differentiate, they increase their surface area, aggregate, and form colonies with neighboring differentiating cells, thus facilitating proper nutrient transport. We found that TGF-β-induced differentiation resulted in increased surface area, and AMPK knockdown suppressed this effect (Fig. 1). Similar to previous findings [14], AMPK knockdown reduces the ability of SM10 cells to differentiate following TGF-β treatment. Interestingly, knockdown of AMPK resulted in decreased surface area to volume ratio in both progenitor cells and differentiated cells compared to Controls.
These data along with the previously reported decreased transport in differentiated AMPK knockdown cells suggest that the affected cells have a decreased interface for nutrient transport. Moreover, when AMPK was reduced, the placental cells were not able to form colonies and aggregate properly, suggesting a problem with terminal differentiation. In this study, we propose that metabolic changes induced by AMPK knockdown alter SM10 trophoblast differentiation. To test this hypothesis, we have first shown that simultaneous knockdown of the AMPKα1 and AMPKα2 subunits in progenitor SM10 trophoblast cells alter cellular metabolism, resulting in the restriction of critical glycolytic and mitochondrial energy resources.
Cells produce energy by two predominant means: glycolysis and oxidative phosphorylation. Glycolysis can rapidly produce ATP to meet cellular demands, and activation of AMPK increases glucose transport and glycolysis while inhibiting some anabolic processes in response to perceived energetic deficit [28]. Our studies indicate that AMPK knockdown significantly limited the glycolytic reserve of progenitor SM10 cells. Since differentiation is assumed to be an energetically demanding process, the lower glycolytic reserve induced by AMPK knockdown may not allow the cells to meet energy requirements for differentiation.
A recent review has highlighted the importance of mitochondria in embryonic stem cell differentiation—with proliferation and differentiation being dependent on the relative amount of mitochondrial biogenesis [29]. Although embryonic and trophoblast cells are distinctly different, a correlative parallel may exist. During differentiation, when the energy demands of the cell increase, the inability to meet the energy demand may halt the differentiation process. In this study, AMPK knockdown limited trophoblast progenitor cell metabolism and differentiation. We found that AMPK knockdown significantly decreased basal respiration in SM10 trophoblast progenitor cells (Fig. 3B, C). Since AMPK KD progenitor cells exhibit similar basal respiration to differentiated Control cells, the AMPK KD cells may not be able to reduce their basal respiration further. It is unknown whether reduction of basal respiration is required for terminal differentiation. TGF-β-induced differentiation may not be able to further reduce basal respiration, which was observed in differentiated Control cells (Fig. 3B, C). Low basal respiration does not necessarily indicate defective mitochondria; however, cellular changes, including the combination of lower respiration, decreased coupling efficiency, and increased proton leak, as seen in AMPK knockdown cells, are indicative of abnormal mitochondrial function (Figs. 3 and 4). Interestingly, each experimental group had similar maximal respiration rates, indicating that the mitochondrial respiration potential did not change after AMPK knockdown or due to differentiation. However, the ability of the cells to regulate mitochondrial respiration was effected; and therefore, the failure of the AMPK knockdown cells to differentiate may be due to this change in regulation and not overall ability.
AMPK has been previously shown to play a role in mitophagy and mitochondrial biogenesis, as well as mitochondrial fission and fusion [30–32]. Specifically, AMPK activation increases mitochondrial turnover by stimulating mitophagy via ULK1 and increasing mitochondrial biogenesis through PGC1α [30–32]. Our studies have found an increase in mitochondrial volume after AMPK knockdown in differentiated cells that was not present in differentiated Control or progenitor AMPK knockdown cells (Fig. 5). Increased mitochondrial volume was therefore a result of both the TGF-β-induced differentiation and AMPK knockdown. This increased mitochondrial volume is not likely a result of increased mitochondrial biogenesis because both AMPK knockdown and TGF-β signaling have been reported to depress the expression of PGC1α and transcription factors targeted by PGC1α activation [33]. One possible explanation may be that knockdown of AMPK results in defective mitophagy. Thus, these cells accumulate mitochondria that are not functioning optimally, as shown by similar maximal respiration regardless of AMPK knockdown or differentiation status.
Overall, our studies indicate that AMPK knockdown results in metabolic alterations and incomplete differentiation. Previous data have shown that AMPK knockdown inhibits glucose transport and alters amino acid transport, while preventing terminal differentiation [14]. Our data identify AMPK as an important regulator of trophoblast differentiation and placental development and suggest that AMPK knockdown decreases the ability of SM10 cells to sense energetic deficits, thereby resulting in insufficient activation of AMPK and the inability of SM10 cells to meet energy requirements necessary to fully differentiate. As the placenta must balance its energy needs with the needs of the growing fetus, the inability of the placenta to properly regulate metabolism may result in fetal growth restriction or a loss of viability. Future research into the role of AMPK and cellular metabolism in placental differentiation will provide new insights into the development of Pregnancy-Associated Disorders.
Acknowledgments
The authors thank Dr. Paula A. Bubulya for use of the DeltaVision microscope system and Dr. David R. Ladle for use of the IMARIS imaging station and software. This work was supported, in part, by funding from the Biomedical Sciences PhD Program (REA), the Wright State University Graduate Council Scholarship (REA), the Wright State University Endowment for Research on Pregnancy-Associated Disorders (www.wright.edu/give/pregnancyassociateddisorders), and The National Institutes of Health NICHD-R01 HD059969-(T.L.B.), R01 HL091836 (C.N.W.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Caniggia I, Winter J, Lye SJ. and Post M. (2000). Oxygen and placental development during the first trimester: implications for the pathophysiology of pre-eclampsia. Placenta 21 (Suppl. A):S25–S30 [DOI] [PubMed] [Google Scholar]
- 2.Chaddha V, Viero S, Huppertz B. and Kingdom J. (2004). Developmental biology of the placenta and the origins of placental insufficiency. Semin Fetal Neonatal Med 9:357–369 [DOI] [PubMed] [Google Scholar]
- 3.Ilekis JV, Tsilou E, Fisher S, Abrahams VM, Soares MJ, Cross JC, Zamudio S, Illsley NP, Myatt L, et al. (2016). Placental origins of adverse pregnancy outcomes: potential molecular targets: an Executive Workshop Summary of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Am J Obstet Gynecol 215 (1 Suppl.):S1–S46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmons DG. and Cross JC. (2005). Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev Biol 28:12–24 [DOI] [PubMed] [Google Scholar]
- 5.Natale DR, Starovic M. and Cross JC. (2006). Phenotypic analysis of the mouse placenta. Methods Mol Med 121:275–293 [DOI] [PubMed] [Google Scholar]
- 6.Jansson T. and Powell TL. (2013). Role of placental nutrient sensing in developmental programming. Clin Obstet Gynecol 56:591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lager S. and Powell TL. (2012). Regulation of nutrient transport across the placenta. J Pregnancy 2012:179827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Y, Awonuga A, Liu J, Rings E, Puscheck EE. and Rappolee DA. (2013). Stress induces AMP-dependent loss of potency factors ID2 and Cdx2 in early embryos and stem cells. Stem Cells Dev 22:1564–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardie DG. (2011). AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev 25:1895–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rafalski VA, Mancini E. and Brunet A. (2012). Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. J Cell Sci 125:5597–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mihaylova MM. and Shaw RJ. (2011). The AMPK signaling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 13:1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steinberg GR. and Kemp BE. (2009). AMPK in health and disease. Physiol Rev 89:1025–1078 [DOI] [PubMed] [Google Scholar]
- 13.Viollet B, Athea Y, Mounier R, Guigas B, Zarrinpashneh E, Horman S, Lantier L, Hebrard S, Devin-Leclerc J, et al. (2009). AMPK: lessons from transgenic and knockout animals. Front Biosci 14:19–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey EAK, Albers RE, Doliboa SR, Hughes M, Wyatt CN, Natale DRC. and Brown TL. (2014). AMPK knockdown in placental trophoblast cells results in altered morphology and function. Stem Cells Dev 23:2921–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong W, Xie Y, Abdallah M, Awonuga AO, Slater JA, Sipahi L, Puscheck EE. and Rappolee DA. (2010). Cellular stress causes reversible, PRKAA1/2-, and proteasome-dependent ID2 protein loss in trophoblast stem cells. Reproduction 140:921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigham AW, Julian CG, Wilson MJ, Vargas E, Browne VA, Shriver MD. and Moore LG. (2014). Maternal PRKAA1 and EDNRA genotypes are associated with birth weight, and PRKAA1 with uterine artery diameter and metabolic homeostasis at high altitude. Physiol Genomics 46:687–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skeffington KL, Higgins JS, Mahmoud AD, Evans AM, Sferruzzi-Perri AN, Fowden AL, Yung HW, Burton GJ, Giussani DA. and Moore LG. (2016). Hypoxia, AMPK activation and uterine artery vasoreactivity. J Physiol 594:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banek CT, Bauer AJ, Needham KM, Dreyer HC. and Gilbert JS. (2013). AICAR administration ameliorates hypertension and angiogenic imbalance in a model of preeclampsia in the rat. Am J Physiol Heart Circ Physiol 304:H1159–H1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma RK. (1998). Mouse trophoblastic cell lines: I—relationship between invasive potential and TGF-beta 1. In Vivo 12:431–440 [PubMed] [Google Scholar]
- 20.Selesniemi KL, Reedy MA, Gultice AD. and Brown TL. (2005). Identification of committed placental stem cell lines for studies of differentiation. Stem Cells Dev 14:535–547 [DOI] [PubMed] [Google Scholar]
- 21.Selesniemi K, Reedy M, Gultice A, Guilbert LJ. and Brown TL. (2005). Transforming growth factor-beta induces differentiation of the labyrinthine trophoblast stem cell line SM10. Stem Cells Dev 14:697–711 [DOI] [PubMed] [Google Scholar]
- 22.Hemberger M, Yang W, Natale D, Brown TL, Dunk C, Gargett CE. and Tanaka S. (2008). Stem cells from fetal membranes—a workshop report. Placenta 29 (Suppl. A):S17–S19 [DOI] [PubMed] [Google Scholar]
- 23.Selesniemi KL, Albers RE. and Brown TL. (2016). Id2 mediates differentiation of labyrinthine placental progenitor cell line, SM10. Stem Cells Dev 25:959–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ausdenmoore BD, Markwell ZA. and Ladle DR. (2011). Localization of presynaptic Inputs on dendrites of individually labeled neurons in three dimensional space using a center distance algorithm. J Neurosci Methods 200:129–143 [DOI] [PubMed] [Google Scholar]
- 25.Fogarty MJ, Hammond LA, Kanjhan R, Bellingham MC. and Noakes PG. (2013). A method for the three-dimensional reconstruction of Neurobiotin-filled neurons and the location of their synaptic inputs. Front Neural Circuits 7:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng J, Zhang T, Ji H, Tao K, Guo J. and Wei W. (2016). Functional characterization of AMP-activated protein kinase signaling in tumorigenesis. Biochim Biophys Acta 1866:232–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Illsley NP, Caniggia I. and Zamudio S. (2010). Placental metabolic reprogramming: do changes in the mix of energy-generating substrates modulate fetal growth? Int J Dev Biol 54:409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardie DG, Ross FA. and Hawley SA. (2012). AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13:251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folmes CDL, Dzeja PP, Nelson TJ. and Terzic A. (2012). Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11:596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, et al. (2011). Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331:456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toyama EQ, Herzig S, Courchet J, Lewis TL, Jr., Losón OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC. and Shaw RJ. (2016). Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 351:275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Kundu M, Viollet B. and Guan KL. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13:132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sohn EJ, Kim J, Hwang Y, Im S, Moon Y. and Kang DM. (2012). TGF-β suppresses the expression of genes related to mitochondrial function in lung A549 cells. Cell Mol Biol 58:OL1763–OL1767 [PubMed] [Google Scholar]