Abstract
The parvovirus minute virus of mice (MVM) packages predominantly negative-sense single strands, while its close relative LuIII encapsidates strands of both polarities with equal efficiency. Using genomic chimeras and mutagenesis, we show that the ability to package positive strands maps not, as originally postulated, to divergent untranslated regions downstream of the capsid gene but to the viral hairpins and predominantly to the nick site of OriR, the right-end replication origin. In MVM, the sequence of this site is 5′-CTAT▾TCA-3′, while in LuIII a two-base insertion (underlined) changes it to 5′-CTATAT▾TCA-3′. Matched LuIII genomes differing only at this position (designated LuIII and LuΔ2) packaged 47 and <8% positive-sense strands, respectively. OriR sequences from these viruses were both able to support NS1-mediated nicking in vitro, but initiation efficiency was consistently two- to threefold higher for LuΔ2 derivatives, suggesting that LuIII's ability to package positive strands is determined by a suboptimal right-end origin rather than by strand-specific packaging sequences. These observations support a mathematical “kinetic hairpin transfer” model, previously described by Chen and colleagues (K. C. Chen, J. J. Tyson, M. Lederman, E. R. Stout, and R. C. Bates, J. Mol. Biol. 208:283-296, 1989), that postulates that preferential excision of particular strands is solely responsible for packaging specificity. By analyzing replicative-form (RF) DNA generated in vivo during LuIII and LuΔ2 infections, we extend this model, showing that positive-sense strands do accumulate in LuΔ2 infections as part of duplex RF DNA, but these do not support packaging. However, replication is biphasic, so that accumulation of positive-sense strands is ultimately suppressed, probably because the onset of packaging removes newly displaced single strands from the replicating pool.
The family Parvoviridae consists of small (250 to 280 Å in diameter) animal viruses that package a single copy of their linear single-stranded 5-kb DNA genome into preformed protein capsids. They replicate through a series of intracellular duplex replicative-form (RF) DNA intermediates, and while most can encapsidate DNA strands of either polarity with equal efficiency, members of some genera, including Parvovirus, vary in the ability to package DNA that is positive in sense with regard to transcription (22). Thus, while most of the members of this genus, including the type species minute virus of mice (MVM), encapsidate predominantly negative-sense DNA, one virus, LuIII, which is structurally very similar to and shares 73% sequence identity with MVM, packages negative- and positive-sense strands with equal efficiency (2, 17).
How parvovirus DNA is selected for encapsidation remains poorly understood. Viral genomes contain a long (∼4.8-kb), single-stranded coding region bracketed by small imperfect terminal palindromes that fold back on themselves to create duplex hairpin telomeres. These hairpins, together with a few adjacent nucleotides, contain all of the cis-acting information required for efficient genome replication and can also support packaging (11, 22), although their ability to control strand selection has not been explored. While the genomic organization and sequence of LuIII generally resemble those of the other viruses, its genome does contain a unique 47-nucleotide AT-rich element located at map unit 89 that was originally thought to be responsible for its different encapsidation pattern (17). However, in this paper we show that substituting the LuIII AT-rich element for equivalent sequences in MVM has no influence on MVM's inability to package positive-sense DNA.
Whereas most of the members of the family Parvoviridae have terminal repeat sequences, and thus have identical origins at both genomic termini, viruses in the genus Parvovirus have unique termini. Hairpins at the left and right ends of these genomes are around 121 and 248 nucleotides, respectively, and differ markedly in sequence and secondary structure. Moreover, while sequences at the right end function as replication origins in their hairpin configuration, those at the left end do not but instead give rise to a single competent duplex initiation site when this hairpin is extended and copied to form the junction sequence that bridges adjacent genomes in dimer RF (10). This junction is then resolved asymmetrically by a repetitive form of hairpin transfer that is sometimes, but not invariably, linked to a second, slower, reaction that appears to involve nicking preexposed single-stranded DNA and that generates positive-sense strands with an alternate terminal sequence orientation (16). Mechanistic differences in the processing of these two disparate origins could therefore result in the excision of the two ends of the genome from concatemeric RF DNA in different conformations and at different rates. Moreover, if the efficiency of the two types of origins differed between virus species, this could alter the availability of excised positive- and negative-sense unit length (5-kb) DNA strands in the infected cell and, if all excised strands were packaged with equal efficiency, would generate apparent differences in packaging specificity.
Some years ago a mathematical model, called the “kinetic hairpin transfer” model, proposed essentially this type of mechanism to explain why different parvoviruses package different forms of the genome (4, 29), but the idea was not tested experimentally. The data we present here substantiate this model, showing that the sequences controlling strand selection map to the viral hairpins, and predominantly to the nick site of oriR, the right-end replication origin, where they control origin efficiency. Using matched genomes that differ only by the presence or absence of a two-nucleotide deletion in the right-end origin of LuIII, we explored the RF DNAs that are generated as infection progresses. This revealed the biphasic nature of viral replication and allowed us insight into the structure of the packaging substrates.
MATERIALS AND METHODS
Plasmids.
The infectious plasmid clone of LuIII, pGLU833, contains a full-length LuIII genome cloned into the BamHI site of pUC19 (17). An infectious clone of the prototype strain of MVM, pdBMVp, has been described previously (20). Chimeric plasmid pMVAT was generated by using PCR to copy the entire untranslated region (UTR) sequence from LuIII (Fig. 1A) with an upstream primer that bound to both genomes but contained a BstAPI site unique to the MVM sequence (nucleotide 4510) and a downstream primer that covered the PshAI site at MVM nucleotide 4913. The product was then digested with the above enzymes and used to replace the corresponding fragment in pdBMVp, creating pMVAT.
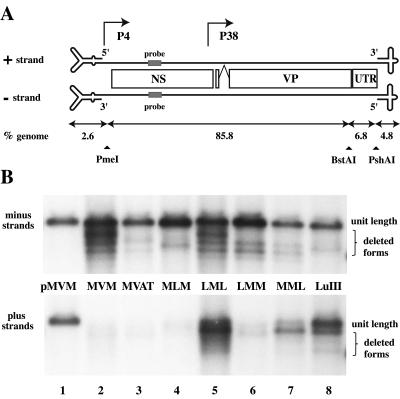
FIG. 1.
Packaging strand bias is controlled predominantly by sequences in the right hairpin. (A) Diagrammatic representation of the MVM and LuIII genomes with the structure of their terminal hairpins enlarged ∼20-fold. Nonstructural (NS) and capsid (VP) coding sequences are shown together with the 350-bp 3′ UTR. Sequences used for strand-specific Southern probes are shaded (MVM nucleotides 1165 to 1190). Restriction endonuclease sites used to create the chimeras are indicated, together with the proportion of the genome present in each fragment. (B) Southern transfers of DNA from micrococcal nuclease-digested virions analyzed on denaturing gels and probed to detect negative- and positive-sense genomes. pMVM is the duplex viral genome excised from pdBMVp with restriction enzymes and used here as a standard for determining the size and concentration of individual packaged strands. Chimeric viruses are described in the text. Full-length genomes are indicated, together with a series of deleted forms that are a common feature of preparations generated in NB324K cells.
Double-hairpin chimeras MLM and LML were obtained by exchanging viral sequences between the shared unique PmeI site (MVM nucleotide 134) and a PshAI site (at MVM nucleotide 4913). Single-hairpin chimeras were similarly generated by exchanges between one of these two sites and a unique AhdI site in vector sequences.
pLU1Nde, an infectious clone of LuIII used as the parent for sequence exchanges within the right hairpin, was generated by destroying first the NdeI site in the plasmid backbone of pGLU833 and then the NdeI site in the outer arm of the viral hairpin while also deleting the remainder of the outboard arm between this site and the polylinker. The resulting plasmid has a single remaining NdeI site, in the inboard arm of the hairpin, allowing facile replacement of the LuIII sequence from the stem of the hairpin with the corresponding mutagenized sequence between the NdeI and PshAI sites (LuIII nucleotides 4966 and 4891, respectively). Mutant sequences were generated by PCR for pLLM5L on an MVM template with a mutagenic primer that created the required NdeI site and for pLuΔ2 on a LuIII template with a mutant primer that introduced the necessary two-base deletion at the nick site.
PCR2.1 constructs (Invitrogen, San Diego, Calif.) containing the 134-bp pLuΔ2 and 136-bp LuIII minimal right-end origins were generated by cloning PCR fragments corresponding to LuIII nucleotides 4887 to 5022 from the shorter, inboard (so-called Hha) arm of the appropriate infectious clones. Larger origin constructs, which included most of the adjacent UTR, were derived directly from the infectious clones by deleting all XbaI fragments between the polylinker at the left end of the viral sequence and LuIII nucleotide 4523, leaving 564 bp of viral sequence from the right end of the genome.
Virus stocks.
324K cells (5 × 105) were seeded (at 0.25× confluence) on 60-mm-diameter dishes and transfected with 2 μg of infectious plasmid DNA with Superfect (QIAGEN, Valencia, Calif.) in accordance with the manufacturer's instructions. Cells were subcultured 1:4 the next day and harvested 3 days later, when they showed conspicuous cytopathic effects from the virus. Cell pellets were resuspended in 50 mM Tris-HCl pH 8.7-0.5 mM EDTA, and virus was liberated by freezing and thawing three times. Residual cell debris was removed by centrifugation, and packaged DNA was assessed in the supernatant by digestion with micrococcal nuclease, electrophoresis through denaturing alkaline agarose gels, and Southern transfer (18). Viral DNA was detected with 5′-32P-labeled oligonucleotide probes NScPOS and NScMIN, which hybridize perfectly to both MVM and LuIII positive- and negative-sense DNAs, respectively (MVM nucleotides 1165 to 1190), and quantitated by comparison with genome length restriction fragments excised from the infectious plasmids, with a Molecular Dynamics PhosphorImager SI.
Replication assays.
Replication assays were carried out at 37°C as previously described (7) with extracts from uninfected murine A9 cells prepared as described by Wobbe and Mitra (33). Assay mixtures (20 μl) contained template plasmid DNA (10 μg/ml), histidine-tagged NS1 (20 μg/ml) prepared in HeLa cells from recombinant vaccinia virus and purified as described by Nuesch et al. (23), deoxynucleotides, MgCl2, ATP, an ATP-regenerating system, and one 32P-labeled deoxynucleoside triphosphate. Reactions were terminated by addition of sodium dodecyl sulfate (SDS; to 0.5%) and EDTA (to 10 mM), and the products were processed as previously described (9) for electrophoresis by digestion with proteinase K and ScaI, which linearizes plasmid DNA by cutting once in the vector sequence, or for immunoprecipitation with rabbit antiserum directed against NS1, followed by ScaI and proteinase K digestion. Reaction products were analyzed on neutral agarose gels, which were then dried, and samples were quantitated by phosphorimager (PI) analysis.
In vivo replication products.
324K cells (1.5 × 106) were plated in 100-mm-diameter dishes (at 0.25× confluence) and infected with LuIII or LuΔ2 virus (2,500 genomes per cell) for 3 h at 37°C. One-step growth conditions were then imposed by adding neuraminidase (0.05 U/ml) to destroy virus receptors and hence block readsorption of released virions, and the incubation was continued for 30 min. Inocula were removed, cells were washed, and fresh medium containing neuraminidase was added. After incubation at 37°C for 14, 18, or 22 h, cells and medium were harvested separately and viral DNA was extracted from the cell pellet by a modified Hirt procedure (27). Samples were quantitated by electrophoresis through denaturing gels, Southern transfer, hybridization with strand-specific probes and PI analysis as described previously (15). Replication products and intermediates were also analyzed after electrophoresis through neutral agarose gels, with or without the addition of 0.2% SDS to both gel and electrophoresis buffer as specified in the figure legends, and by two-dimensional (2D) gel electrophoresis (15), in which lanes from neutral agarose gels were rotated through 90° and re-electrophoresed under denaturing conditions.
RESULTS
The unique AT-rich element at map unit 89 in LuIII does not contain a positive-strand-specific packaging element.
Members of the genus Parvovirus have a 350-bp UTR located between the capsid-coding sequences and the right-end hairpin (Fig. 1A), which differs in sequence and organization in MVM and LuIII (nucleotides 4563 to 4924 and 4558 to 4905, respectively). In this region, LuIII has a 47-bp AT-rich sequence absent from MVM (LuIII nucleotides 4558 to 4604), which begins 5 nucleotides downstream of the termination codon for the capsid proteins. In MVM, the UTR contains downstream elements from a complex group of sequences that has been referred to as an “internal replication sequence,” MVM nucleotides ∼4520 to 4631 (3), that have been reported to bind unidentified cellular proteins and to function as a cis-acting replication element in some extensively deleted forms of the genome (25, 26). Equivalent sequences in LuIII have not been examined for such activity. Finally, in the prototype strain of MVM used here (MVMp), one 65-bp sequence is duplicated (nucleotides 4720 to 4784 and 4785 to 4849).
Previously, it has been suggested that sequences in these disparate UTRs might mediate strand selection (6, 17). To address this question directly, we copied the UTR from the infectious plasmid clone of LuIII and used it to replace the equivalent sequence in MVMp clone pdBMVp, yielding hybrid plasmid pMVAT. We then recovered a chimeric virus, MVAT, by transfecting plasmid DNA into NB324K cells, which serve as productive hosts for both MVM and LuIII.
Virion DNA from this infection was analyzed for strand specificity by Southern transfer and hybridization with strand-specific oligonucleotide probes NScPOS and NScMIN, derived from a region of the NS gene (MVM nucleotides 1165 to 1190) that is identical in the two viruses. Control duplex genomes excised in vitro from infectious plasmid clones of each virus with restriction enzymes were used as standards for genome size and quantitation (e.g., Fig. 1B, lane 1). Whereas LuIII virions were estimated to package approximately 52% positive-sense strands in this analysis (Fig. 1B, lane 8), the MVM and MVAT genomes contained little positive-sense DNA (Fig. 1B, lanes 2 and 3). Estimates made by PI analysis suggest that these viruses packaged 6 and 8% positive strands, respectively, although these are probably overestimates due, at least in part, to weak cross-hybridization between this probe and reiterated regions of the abundant negative-sense genomes present in this region of the blot. This tendency to overestimate low concentrations of a particular strand was also compounded by our need to compare DNA concentrations in relatively large areas of gel in order to included both full-length genomes of each virus and a variety of slightly deleted forms (as seen in Fig. 1) that are a common feature of virus stocks grown in 324K cells. The structures of these deleted forms will be explored elsewhere (S. F. Cotmore and P. Tattersall, unpublished data). We conclude from this analysis that the presence of the entire UTR from the right end of LuIII has no influence on the negative-strand bias shown by MVM.
The ability to package positive-sense DNA maps to the viral hairpins, predominantly to the right-end telomere.
The left-end hairpins present in the published sequences of MVM and LuIII extend to nucleotide 118 and can effectively be excised from duplex forms of the genome by digestion with PmeI (nucleotide 134), giving left-end fragments that comprise 2.6% of each genome (Fig. 1A). Right-end hairpins of both viruses can be excised by digestion with PshAI (nucleotide 4912 in MVM), which cuts eight nucleotides inboard of the right-end nick site, giving a fragment that accounts for 4.8% of the genome. When the remaining body sequence of LuIII (92.6% of the total) was placed between MVM termini (in the virus MLM, Fig. 1B, lane 4), virion populations contained predominantly negative-sense genomes (estimated by PI analysis to be 93% of the total), whereas when the body region of MVM was sandwiched between LuIII termini (in the virus LML, lane 5), approximately 52% of the packaged DNA in this region of the gel hybridized with the plus strand probe.
We then analyzed the influence of these two termini separately in the context of the MVM body sequence. PI estimates suggest that virus LMM, which contained just the left-end hairpin from LuIII, packaged approximately 14% positive-sense strands (Fig. 1B, lane 6), whereas MML, carrying the LuIII right terminus, packaged a much greater proportion (37%) of positive-sense DNA (Fig. 1B, lane 7). We conclude that the ability to package positive-sense strands maps to the LuIII hairpins, with the right-end hairpin having by far the greater influence.
The OriR nick site controls the virus's ability to package positive-sense DNA.
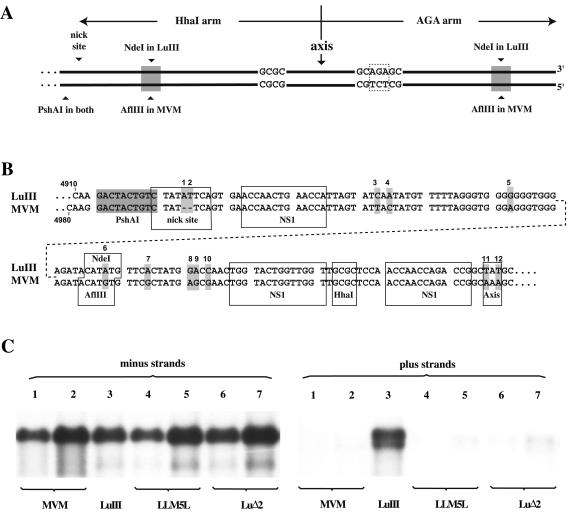
The right-end hairpins of both MVM and LuIII are 248 nucleotides in length and are expressed in two sequence orientations, flip and flop, that are inverted complements of one another. These sequences can be folded into almost perfect duplex hairpins, with just three unpaired bases at the axis of the hairpin and a single mismatched region in the stem, where a three-nucleotide insertion (AGA or TCT) on one strand interrupts the sequence 5′-GCGC-3′ on the other. In extended duplex copies of the hairpin (as illustrated in Fig. 2A), the GCGC sequence creates an HhaI restriction site in one arm, which we consequently term the Hha arm, while the other is referred to as the AGA arm, deriving its name from the triplet that interrupts the HhaI site. Both the LuIII and MVMp infectious clones, pGLU833 and pdBMVp, have this right-end palindrome in the flip orientation, such that the Hha arm is inboard of the axis. The sequences of this inboard arm in pGLU833 and pdBMVp are shown in Fig. 2B, from the flanking PshAI site to the axis. Complementary changes are also present at each of these positions in the outboard arm. As indicated, there are 10 sequence changes scattered throughout each arm, together with 2 at the axis. One change, positioned in the middle of the hairpin stem, converts an AflIII site in MVM into an NdeI site in LuIII. Duplex genomes that are truncated at the right end but extend beyond the trinucleotide asymmetry (AGA) in the outboard arm yield infectious progeny, presumably because deleted outboard sequences can be repaired by copying the inboard arm. Thus, to facilitate manipulation of mutant sequences we deleted the outboard arm of pGLU833 from the position of the NdeI site, making the equivalent site in the inboard arm unique within viral sequences. This construct, referred to as pLu1Nde, served as the parent for subsequent modifications.
FIG. 2.
The ability to package positive-sense DNA maps to the nick site of the right origin. (A) Diagram of the right-end palindromes of MVM and LuIII in the extended configuration, showing sequence asymmetries and the positions of PshAI and NdeI cloning sites used to introduce stem sequences from the inboard arm of MVM into LuIII. (B) Sequence comparison of the inboard arm of the LuIII and MVM right hairpins (presented in two parts because of size constraints) showing differences between the two genomes (indicated by shading and numbered). Change 6 in LuIII destroys the AflIII site (ACATGT) present in MVM but creates an NdeI site (CATATG) in its place. The nick site consensus sequence, NS1 binding sites, unpaired axis nucleotides, and restriction endonuclease recognition sites are boxed. (C) Southern transfer showing virion DNA probed to detect negative- and positive-sense genomes. LuIII (lane 3) is a wild-type virus derived from a truncated LuIII genome in which the terminal half of the outer hairpin arm was deleted to facilitate cloning of mutant sequences (described in the text). LLM5L (shown at two concentrations in lanes 4 and 5) is a virus derived from a LuIII genome carrying MVM changes 1 to 5 from panel B; LuΔ2 is a similar construct that has just two bases from the LuIII nick site deleted (lanes 6 and 7, carrying MVM changes 1 and 2 in panel B).
MVM sequences corresponding to those in LuIII between the PshAI site and the remaining NdeI site were obtained by PCR with a primer containing a new NdeI site and introduced into pLu1Nde, leaving the outer part of the hairpin stem and the axis as LuIII sequence. This created a LuIII construct, pLLM5L, with five nucleotide changes (three changes and a two-base deletion at the nick site) inserted into the stem of the inboard arm. Virus derived from pLLM5L, shown at two different concentrations in Fig. 2C, lanes 4 and 5, packaged <7% positive-sense DNA, indicating that some or all of these five MVM-specific nucleotides contribute to the control element in MVM that suppresses positive-strand packaging. We then created a second pLu1Nde-based construct, pLuΔ2, that simply contained the two-base deletion at the nick site found in MVM. LuIII virus containing this minimally altered nick site packaged approximately 8% positive strands in the analysis shown in Fig. 2C, lanes 6 and 7. Thus, comparison of lane 3 with lane 6 or 7 demonstrates that the critical element in LuIII that allows it to package DNA strands of both senses is the two-base insertion in the consensus nick site of OriR.
Efficiency of the LuIII and LuΔ2 OriR sequences in vitro.
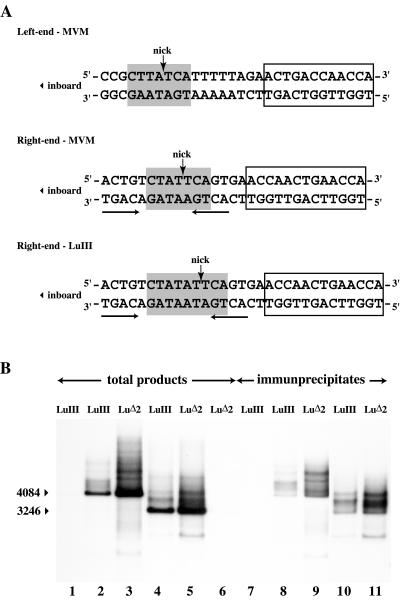
Such an insertion might well be expected to influence origin efficiency. The consensus nick site for MVM, 5′-CTWWTCA-3′, was derived simply by comparing the somewhat disparate sequences present in the origins at the two ends of the genome, and an equivalent sequence is present at the left end of LuIII (Fig. 3A). While it is known that this entire consensus element cannot be deleted (8) or inverted (5), the importance of individual nucleotides has not been determined experimentally. The exact position of the nick within this consensus appears slightly variable in vivo, plus or minus one nucleotide (13, 17), but analysis of cleavage products generated in vitro from 5′-labeled substrates derived from OriL suggests that the position indicated in Fig. 3A is preferred (5). The LuIII right-end sequence from pGLU833, also shown Fig. 3A, contains a TA dinucleotide inserted within this consensus immediately upstream of the presumed nick site.
FIG. 3.
In vitro replication assays. (A) Sequences of the MVM left-end and MVM and LuIII right-end nick sites, together with flanking nucleotides and the nearby NS1 binding site. Consensus nick sites are shaded, the position of the nick is indicated with an arrow, and NS1 binding sites are boxed. Palindromic tetranucleotide sequences in the right origin are underlined. These could potentially anneal to create a minimal stem-loop structure with the nick site exposed on one side of the loop, as commonly found in rolling-circle replication initiation sites. (B) Autoradiograph of a neutral agarose gel showing ScaI-digested, 32P-labeled in vitro replication products generated from substrates containing minimal viral origin sequences of 136 bp from the LuIII genome (lane 1 to 2) and 134 bp from the LuΔ2 genome (lane 3) and substrates carrying minimal origins plus additional UTR sequences (563 to 565 bp total, lanes 4 to 6 as indicated), generated in the presence (lanes 2 to 5) or absence (lane 1 and 6) of NS1. The arrowheads at the left indicate the positions of linearized forms of the input substrate (4084 and 3246 bp, respectively, for the minimal and UTR-containing constructs). Relative origin activity was determined by PI quantitation of 32P incorporation into DNA species migrating in each lane throughout the entire gel region shown here. Lanes 7 to 11 contain ScaI-digested, 32P-labeled products from samples equivalent to those shown in lanes 1 to 5, following immunoprecipitation with anti-NS1 antibodies.
Initially we used in vitro nicking assays to assess origin efficiency. In these assays, 32P-labeled, hairpinned LuIII and LuΔ2 origin sequences were incubated with recombinant NS1 and HMG1 in the presence of ATP (14). NS1 and HMG1 concentrations were titrated against both substrates to make sure the assays were linear with respect to these components, and several time points were investigated without revealing any significant difference in the efficiency of nicking at the two origins (data not shown). However, such assays are not very sensitive to variations in template concentration, so that we suspected that their failure to discriminate between the two sequences could simply reflect this shortcoming in the assay.
We therefore went on to explore the efficiency with which LuIII and LuΔ2 origin sequences were able to support NS1-dependent replication in vitro with an assay that measures 32P-nucleotide incorporation into newly synthesized DNA and therefore monitors a combination of nicking efficiency, the establishment of a competent replication fork, and limited processive synthesis. Two different types of substrates were used in these assays. The first contained a predicted minimal linear origin sequence, based on the minimal MVM origin (8), of 134 and 136 bp for LuΔ2 and LuIII, respectively (LuIII nucleotides 4887 to 5022), cloned into a plasmid vector. The second set of constructs contained the origin plus extensive LuIII UTR sequences (LuIII nucleotides 4523 to 4987), in case this UTR influences the minimal origin in a way not seen before. Origin efficiency was assessed in vitro in cell extracts supplemented with purified recombinant NS1, an ATP-regenerating system, and a mixture of unlabeled and 32P-labeled deoxynucleoside triphosphates. Assay duration was limited to 1 h in order to emphasize the influence of initiation efficiency over processive synthesis, and murine cell extracts were used since these support only limited processive synthesis compared to their human HeLa and 293 cell counterparts (9). In these assays, both forms of LuΔ2-based origin (Fig. 3B, lanes 3 and 5) consistently supported two- to threefold more NS1-dependent DNA synthesis than did their LuIII counterparts (lanes 2 and 4). Thus, there appeared to be a marked difference in efficiency between the LuIII and LuΔ2 origins in this assay.
NS1 initiates replication via a transesterification reaction that leaves its tyrosine residue at position 210 (Y210) covalently attached to the 5′ end of the DNA at the nick site (23). Authentic initiation can therefore be confirmed by demonstrating that a proportion of the in vitro reaction products remain covalently associated with NS1. These can then be immunoprecipitated with NS1-specific antiserum, as shown in Fig. 3B, lanes 7 to 11. In this analysis, the LuΔ2 origin was again seen to support two- to threefold more NS1-associated synthesis than its LuIII counterpart.
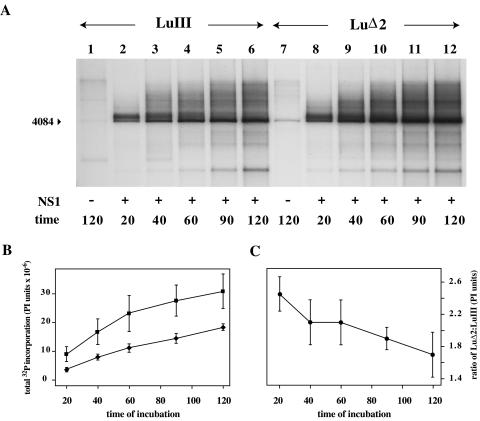
The LuΔ2 and LuIII substrates used here were so closely matched that we considered it unlikely that processive DNA synthesis would occur at different rates on the two templates once an initiating fork had been established. However, to differentiate between these two events, we analyzed NS1-dependent DNA synthesis as a function of time (Fig. 4). In these assays, LuIII-based origins were found to be impaired relative to their LuΔ2 counterparts at all times during the reaction (Fig. 4A and B), but this divergence was most extreme at the earliest time points (Fig. 4C), indicating that an early event was specifically inhibited. Thus, the data indicate that LuΔ2 and LuIII origins support different rates of an early event, such as initiation, rather than different rates of processive synthesis. Since the specific nicking assays failed to discriminate between these two substrates, as mentioned previously, it could be argued that fork assembly at the LuIII origin, rather than nicking, was impaired. However, we believe this would be an overinterpretation of the data since the reaction conditions used for nicking assays are less physiologically robust than those used in cell extract-based replication assays. It is also easier to rationalize how a two-base insertion could influence nicking efficiency rather than fork assembly since we know that a specific consensus sequence is required for nicking, whereas there is no evidence that fork assembly at a base-paired 3′ hydroxyl shows sequence specificity. Even if this were the case, the three nucleotides at the 3′ end of the primer strands exposed in LuIII and LuΔ2 are identical, so that longer-range interactions would have to be postulated. Thus, we conclude that the different initiation rates observed here reflect differences in nicking efficiency rather than fork assembly, even though our assay results appear to suggest otherwise.
FIG. 4.
In vitro replication time course. (A) ScaI-digested, 32P-labeled in vitro replication products generated from minimal origin substrates following incubation for the indicated time in the presence (lanes 2 to 6 and 8 to 12) and absence (lanes 1 and 7) of NS1. (B) 32P incorporation (PI units) determined by PI analysis of DNA species migrating in each lane throughout the entire gel region shown in panel A. (C) Changes in relative 32P incorporation into LuΔ2 and LuIII products with time.
Kinetics of unit length strand generation from replicative intermediates in vivo.
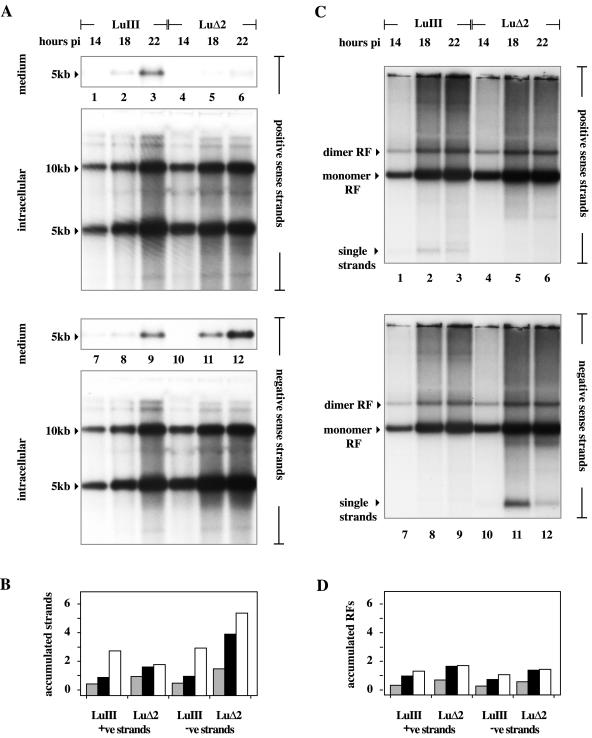
Removal of the additional AT dinucleotide from the consensus nick site in the right-end origin of LuIII thus enhanced its ability to support replication initiation in vitro. If a similar change in OriR efficiency occurred in vivo, we should be able to detect this because it would be likely to influence the distribution of amplified replication intermediates generated during infection. The kinetic hairpin transfer model proposed by Chen and colleagues (4) postulated that changing the relative efficiencies of the origins at the two ends of the genome might indirectly modulate strand selection for encapsidation by modifying both the distribution of different forms of replicative intermediates and the efficiency with which these forms generated unit length single-stranded DNAs of each polarity, which would then be packaged with equal efficiency. To determine whether packaged DNA truly reflects the polarity of all of the unit length strands present in RF—an idea that seemed to us unlikely—we explored this model by infecting 324K cells with LuIII or LuΔ2 under one-step growth conditions. Medium and cells were harvested separately 14, 18, and 22 h postinfection, and viral DNA was extracted from cell pellets by a modified Hirt procedure. RF DNAs were then quantitated following electrophoresis through denaturing and neutral agarose gels by Southern transfer and hybridization with strand-specific probes.
Remarkably, 5-kb positive-sense DNA strands accumulated rapidly in both LuIII- and LuΔ2-infected cells, and at early time points the distributions of unit length RF intermediates in the two infections appeared very similar. When analyzed under denaturing conditions, as shown in Fig. 5A, at each individual time point equivalent fractions from LuIII infections contained similar numbers of positive- and negative-sense monomer and dimer length (5- and 10-kb) strands, and amplification of these forms progressed throughout the duration of the study (Fig. 5A, lanes 1 to 3 and 7 to 9, and B), although by the final time point (22 h) relatively little packaged virus containing either strand had been released into the medium. In contrast, amplification of both positive- and negative-sense 5- and 10-kb strands was initially faster in LuΔ2 infections (Fig. 5A, lanes 4 to 6 and 10 to 12, and B), but negative-sense strands accumulated to progressively higher levels than positive-sense strands and continued to accumulate throughout the duration of the study, whereas positive-sense DNA levels reached a plateau at around 18 h. As expected, only negative-sense LuΔ2 DNA was packaged into virions and released into the medium. While there are differences in the kinetics with which positive and negative LuΔ2 strands accumulate in the cell, particularly late in infection, if all 5-kb strands were packaged with equal efficiency, then positive-sense strands should account for approximately 30% of the DNA being packaged at the 18-h time point and 25% of the DNA being packaged at 22 h, which they do not.
FIG. 5.
Accumulation of unit length viral DNA in cells infected with the LuIII and LuΔ2 viruses. (A) Southern transfers of total viral DNA either released into the medium or retained in cells 14, 18, and 22 h postinfection (pi) following electrophoresis through denaturing gels, transfer, and incubation with strand-specific probes. Total unit length DNA was quantitated by PI analysis against standard DNAs run in the same gel (as detailed in the text). (B) Accumulation of unit length (5- plus 10-kb) DNA strands of each polarity with time, expressed on the y axis as micrograms of DNA quantitated for each strand following electrophoresis through denaturing gels. Individual histogram blocks shaded gray or black or unshaded represent DNA harvested at 14, 18, and 22 h postinfection, respectively. (C) Southern transfers of total viral DNA from cells 14, 18, and 22 h postinfection, following electrophoresis through neutral agarose gels. The migration positions of monomer RF, dimer RF, and progeny single-stranded DNAs are indicated. (D) Accumulation of unit length duplex DNA (5 plus 10 kb) with time, expressed on the y axis as micrograms of DNA quantitated for each strand following electrophoresis through neutral gels. Individual histogram blocks shaded gray or black or unshaded represent DNA harvested at 14, 18, or 22 h postinfection, respectively. +ve, positive; −ve, negative.
We conclude that the balance of origin efficiency is an essential element in determining packaging specificity but that it does not operate simply by determining which unit length DNA strands are excised. Rather, it appears that the molecular form in which individual strands are expressed may be significant. Encapsidation of parvovirus DNA is thought to require ongoing DNA replication (24, 34), although it is not clear if the substrate must first be released as a free single strand or if packaging is somehow coordinated with displacement from duplex monomeric or concatemeric templates, as suggested for adeno-associated virus (21).
Since the positive-sense unit length LuΔ2 strands observed here did not serve as substrates for encapsidation, this suggests that they were sequestered in some RF that could not serve as a packaging substrate, at least for positive-sense DNA. To explore this possibility, strand accumulation was also analyzed on nondenaturing gels. Once again, accumulation of both monomer and dimer RFs appeared faster in LuΔ2 infections than in the LuIII samples, but the overall patterns of these replication intermediates looked similar in the two infections (Fig. 5C and D). Moreover, by comparing the histograms in Fig. 5B and D it becomes apparent that, at all times, the total accumulation of positive-sense LuΔ2 DNA in monomer and dimer duplex RFs (as seen in neutral gels) was very similar to the total accumulation of monomer and dimer length positive-sense single strands seen in denaturing gels. Since this DNA strand is not packaged, we conclude that these monomer and dimer duplex RFs do not serve as packaging substrates for positive-sense strands. A similar close correlation between the accumulation of 5- and 10-kb single strands and monomer and dimer RF concentrations also exists at early time points (14 and 18 h) for both strands of LuIII DNA and at 14 h for negative strands of LuΔ2, but at later times the correlation disappeared and all of these DNA species started to accumulate rapidly in some form that was not a monomer or dimer RF. Thus, by 22 h only approximately half of the unit length positive and negative LuIII DNA in the cell was in the form of unit length duplexes, while for negative-sense LuΔ2 DNA at 18 h only 36% and at 22 h only 27% of the unit length single strands were part of such duplexes. Thus, as each infection proceeded, unit length single strands of the sense(s) that were destined to be packaged progressively accumulated in the cell in a form that was not a discrete monomer or dimer duplex RF, suggesting that these RFs may not be packaging substrates for any DNA species. Rather, it appears that infection is biphasic, with monomer and dimer RFs (containing unit length positive- and negative-sense LuΔ2 strands) accumulating at early times, followed by a switch to predominantly negative-strand synthesis later in infection. It seems likely that this switch may result from the onset of packaging.
Structure and distribution of viral replication intermediates in vivo.
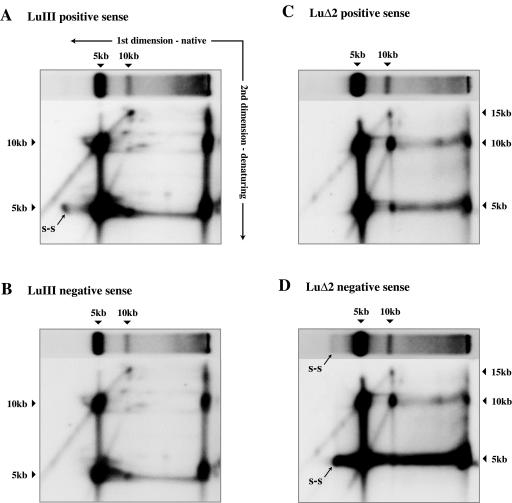
Some of the excess unit length DNA seen in the denaturing gels cannot be accounted for in the native gels. Certainly some single strands did appear as intracellular packaged virions (probably accounting for most of the DNA migrating as single strands in Fig. 5C). Packaged single-stranded DNA was more apparent in LuΔ2 negative-sense samples (Fig. 5C, lanes 11 to 12) than in LuIII infections, but this apparent discrepancy between viruses may simply reflect reannealing of positive- and negative-sense LuIII single strands during sample preparation. However, as infection progressed more of the unaccounted for DNA appeared to be represented by a dense smear of low-mobility complexes that migrated between the position of unit length replicative intermediates and the well of the gel. This suggests that in the Hirt extracts the excess single-stranded DNA may exist as part of a series of progressively more intricate complexes, the most elaborate of which may simply not enter the neutral gel (Fig. 5C). Subjecting samples from the 22-h time point to 2D electrophoresis, first through native and then through denaturing gels (Fig. 6), confirmed that much of this retarded material is unit length DNA, predominantly of the packaged sense(s), rather than covalently continuous concatemers. Samples shown in Fig. 5C were subjected to extensive digestion with proteinase K in SDS prior to electrophoresis. Equivalent samples analyzed on neutral gels containing 0.2% SDS (as in the first-dimension gels shown in Fig. 6) gave essentially similar profiles in this upper region of the gel as did samples in which incubation (and hence proteolysis) was strictly limited in an attempt to stop free positive and negative strands from annealing. Thus, it seems unlikely that these highly retarded structures merely reflect the presence of residual undigested protein.
FIG. 6.
Structure of LuIII and LuΔ2 replication intermediates. Southern transfers of 2D agarose gels in which LuIII and LuΔ2 Hirt extracts harvested 22 h postinfection were subjected to electrophoresis first through a neutral agarose gel (as indicated by the horizontal arrow in panel A) in the presence of 0.2% SDS. An equivalent lane was turned through 90° and re-electrophoresed under denaturing conditions (in the direction indicate by the vertical arrow in panel A). Transfers were probed consecutively for positive- and negative-sense strands. DNA migrating as free single strands under native conditions is labeled s-s.
2D gel analysis, such as that shown in Fig. 6, also allowed us to explore the structure of the various RFs. This showed that at all times, in all infections, duplex 5-kb monomer RF molecules contain both 5- and 10-kb single strands, thus confirming that such RFs do indeed contain discrete 5-kb positive-sense single strands, whether or not these strands are destined to be packaged. A second striking observation was that LuIII dimer RF contained a very different, and much more complex, mixture of DNA fragments than did LuΔ2 dimers. Thus, whereas LuΔ2 dimers predominantly comprised 10-kb single strands, with rather fewer 5- and 15-kb species, LuIII dimers predominantly contained 5- and 15-kb species, with few unit length 10-kb molecules but a spectrum of discrete species of around this size. We subsequently reexamined dimer structure on neutral gels following digestion with single-cutter restriction enzymes that allow discrimination between right-end-right-end and left-end-left-end dimer junctions. In LuIII infections, these two forms of dimer junction appeared equimolar, suggesting that the right- and left-end junction fragments are resolved with equal efficiency and that the disparate mixture of 5-, 10-, and 15-kb species seen in the 2D gel (Fig. 6A and B) represents processing intermediates from two different (right-end-right-end and left-end-left-end) substrates. In contrast, in extracts from LuΔ2 infections digestion gave a discrete band, indicating that left-end-left-end forms of the dimer predominate, presumably because they are resolved more slowly than their counterparts from the right end. Accordingly, in the 2D gels (Fig. 6C and D) LuΔ2 dimers were much simpler and were either unprocessed forms of the left-end bridge or showed evidence of having been nicked once, presumably at the active OriLTC nick site. This difference in the structure of the dimer intermediates thus supports our previous in vitro observations, since it suggests that the LuΔ2 mutation alters the efficiency of one origin in vivo, thus shifting the distribution of replication intermediates that accumulate. Finally, the 2D gels show that retarded complexes, both in the wells of the native gel and smearing between the wells and the unit length RFs, contained a high proportion of 5- and 10-kb unit length strands. Remarkably, in the LuΔ2 infections negative-sense strands outnumbered positive-sense strands by three- to fourfold in these retarded complexes, suggesting either that these excess unit length negative strands formed complexes with a component not detected in this blot (such as protein, RNA, or cellular DNA) or that multiple unit length negative strands were associated with each positive-sense strand, presumably in the form of complex, branched structures.
DISCUSSION
In this report, we show that the differential ability of the parvoviruses MVM and LuIII to package positive-sense DNA is controlled by the viral hairpins, and in particular by the nick site in OriR, the right-end replication origin. In LuIII, this site is modified by insertion of a dinucleotide, TA, that impairs its ability to support NS1-mediated replication initiation in vitro and induces a shift in the spectrum of replication intermediates generated in vivo. While LuIII normally packages approximately equimolar positive- and negative-sense DNA strands, removal of this dinucleotide generates a virus, LuΔ2, that packages in excess of 90% negative-sense DNA. Similarly, if the hairpins of MVM are replaced with their LuIII counterparts, MVM shifts from packaging predominantly negative-sense DNA to packaging around 50% positive-sense strands.
These findings indicate that encapsidation of positive-sense DNA does not require specific packaging signals, but rather they lend support to a unified mathematical scheme postulated some years ago by Chen and colleagues (4), called the kinetic hairpin transfer model. This scheme explains the distribution of the strands and terminal conformations of packaged genomes of all known parvoviruses in terms of the efficiency with which each individual type of terminus can undergo a series of reactions, collectively called “hairpin transfer” by the authors (as discussed below), that allow the terminus to be copied and reformed. Parvovirus hairpins are slightly imperfect palindromes and are generally present in two different sequence orientations, termed flip and flop, that are inverted complements of one another. Chen and colleagues argued that selective encapsidation could not be responsible for the ratios of terminal sequence orientations observed in virion DNA, because the same ratios of flip to flop forms were found in the intracellular RF population that gave rise to them. Instead, they postulated that the balance between the efficiency of hairpin transfer reactions at the two genomic termini must determine the distribution of amplified replication intermediates generated during the infectious cycle and, ultimately, the efficiency with which single-stranded DNAs of characteristic polarity and terminal orientation were excised, which would then be packaged with equal efficiency. Surprisingly, this simple model has never been tested experimentally, but the data presented here clearly support its central tenets since we demonstrate that changing the efficiency of one origin generates a different pattern of dimer replication intermediates in vivo and ultimately modifies strand selection. The mechanism of selection, however, appears slightly more complex than we originally imagined, since in infections in which only a single sense strand will be packaged, monomer length strands of both senses do accumulate.
If relative origin efficiency can modulate packaging specificity, it follows that minor sequence differences between the left-end hairpins of LuIII and MVM could cause the chimeric virus MML to encapsidate slightly more positive-sense DNA (14%) than its MVM parent (6%), as shown in Fig. 1. The left-end hairpins of these viruses differ by four point mutations: (i) a C-to-T transition at MVM nucleotide 13 and its complement on the inboard arm at nucleotide 104, which form part of the NS1 binding site and would, perhaps, be predicted to result in NS1 binding to the LuIII origin with slightly higher efficiency; (ii) an A-to-T transversion at MVM nucleotide 24 and its complement at residue 93, which abut the bubble asymmetry but would not be predicted to have any detectable effect on nicking efficiency; (iii) a C-to-G change at MVM nucleotide 53 that enhances the asymmetry at the tip of one ear, potentially making the LuIII hairpin slightly less stable than its MVM counterpart; and (iv) an A-to-G change at nucleotide 90 that forms part of the bubble but would not be predicted to influence origin efficiency. To accord with the model, the slight increase in encapsidation of positive-sense DNA by the LMM chimera should correlate with enhanced processing of the LuIII sequence relative to its MVM counterpart, but we do not know whether one or several of these nucleotide changes contribute to the observed effect. The original model also postulates that protein sequence changes between the NS1 molecules encoded by these viruses might influence strand selection by favoring mechanisms needed for initiation at one or the other origin. While this remains a plausible scenario, the coding sequences of LuIII and MVM in the LML and MLM chimeras described here did not influence strand selection in either direction.
In the original kinetic hairpin model, the term hairpin transfer was used as a catchall phrase to encompass all of the steps needed to activate, copy, and reform a functional origin at either end of the genome. In MVM and LuIII, this is even more complicated than it first appeared because the mechanisms of resolution and reformation associated with the two ends of the genome are substantially different (8, 10). At the critical right terminus, monomer duplexes in the hairpin configuration are nicked by NS1 and a replication fork is assembled that unfolds and copies the hairpin, generating an extended-form duplex palindrome. This is then melted out, and the two strands are allowed to anneal back on themselves in a reaction called “rabbit ear formation,” that pairs the exposed 3′ nucleotide with an internal base and thus creates a primer for synthesis of a new complementary strand (12, 28). Changes in the efficiency of any one of these steps could theoretically influence the speed with which right termini were processed and consequently modulate the pattern of replication intermediates generated in vivo. However, the ability to package positive-sense DNA maps directly to the nick site of the hairpin, rather than to other regions in this telomere that are known to be involved in NS1-mediated conformation changes (31). This suggests that initiation, rather than subsequent modulation of the duplex structure, is critical in this particular case. This probability is supported by the in vitro replication data, which show that removal of the TA dinucleotide from the LuIII nick site in the virus LuΔ2 results in a detectable, albeit surprisingly modest, shift in origin efficiency. A similar shift also appears to occur in vivo, causing LuIII and LuΔ2 to generate dimer RF intermediates with very different structures.
We show here that monomer length positive-sense strands of LuΔ2 exist predominantly as part of duplex RF DNA that is apparently unable to function as a packaging substrate, at least for positive-sense DNA. We also show that for both viruses replication is biphasic. As infection progresses, a switch in the replication pattern occurs and unit length strands of the sense(s) that will be packaged rapidly come to predominate. However, this new packaging sense DNA is not present in a duplex unit length RF, even in virus infections in which both strands are packaged, but rather accumulates as part of a spectrum of complex intermediates that smear throughout the upper regions of a native gel. Although encapsidation appears to require ongoing DNA replication (19, 34), it has never been clear whether the substrate is a newly displaced unit length strand or if packaging is coordinated with displacement synthesis on a duplex template (21). The present data argue for the former scenario, suggesting that a unit length RF is not a functional precursor and, by default, that fully displaced single strands are packaged. If correct, this would provide a mechanism for the biphasic nature of DNA replication seen in LuΔ2 infections, since the observed shift from synthesizing strands of both senses to generating predominantly negative-sense DNA can readily be explained if newly displaced single strands are prevented from re-entering the replicating RF pool by the onset of packaging, as discussed below.
In the present study, we did not further characterize the “packaging phase” complexes that migrate throughout the upper region of the native gel because we were concerned that the high-salt precipitation step of the Hirt extraction procedure may have led to artifactual aggregation of replication and packaging complexes. Late in infection, however, unit length DNA strands of the packaged sense(s) progressively accumulate within this continuum of complexes, suggesting that it likely contains packaging intermediates, in addition to DNA that is clearly being actively replicated. Evidence for this comes from 2D gel electrophoresis following digestion with EcoRI or PmlI, which showed that around 75% of the unit length negative strands in these complexes were single stranded at the right end, whereas less than 50% were single stranded at the left end (data not shown). Thus, a significant proportion of these strands were in the process of being displaced from RF in a right-to-left (5′-to-3′) direction, compatible with displacement during replication, rather than in the 3′-to-5′ direction that would indicate ongoing encapsidation (21; Cotmore and Tattersall, unpublished). Accumulation of unit length, packaging sense DNA in these high-molecular-weight complexes could thus be interpreted as suggesting that packaging and replication occur on the same, highly branched, DNA substrates. However, we suspect that this inference is incorrect. Rather, we suggest that in vivo DNA molecules in packaging and replication complexes are separate, but the complexes are physically clustered in space because of interactions between the mediating NS1 complexes. This seems particularly likely because in order to prevent initiation of DNA synthesis on the 3′ ends of newly displaced strands, and their subsequent diversion into the replication pool, packaging must initiate immediately when displaced strands are released from their parental duplexes. Thus, we suggest that NS1 complexes associated with replicating duplex molecules direct the newly displaced 5-kb strands directly into particles, resulting in the creation of heterogeneous, protein-mediated complexes in which multiple displaced strands are being packaged while other, predominantly duplex, molecules remain actively involved in replication. Following partial removal of proteins such as NS1 and RPA by proteolysis, we suggest that protracted exposure to a high salt concentration at a low temperature during the Hirt extraction procedure could promote short-range invasion and annealing between strands involved in these alternate pathways, resulting in the smear of complexes observed on neutral gels. This would also explain how such structures could contain three to four full-length negative-sense strands for every positive-sense strand, a ratio that is hard to explain by successive waves of strand displacement synthesis alone. Thus, we suspect that the complexes observed in these extracts under nondenaturing conditions represent spurious interactions between replication and packaging DNA intermediates that occur following their release from protein-mediated supercomplexes in which newly displaced single strands are packaged while they remain physically associated with the replicating DNA. However, further evaluation of such intermediates requires the use of alternative isolation techniques.
We suggest that the switch from duplex DNA amplification to the preferential synthesis of strands of the packaged sense(s) occurs simply because the packaging machinery sequesters newly released single strands. This argument hinges on the fact that the MVM and LuIII genomes are highly asymmetric in two critical ways. First, the right and left termini of monomer RFs are structurally and functionally very different, and while right termini in the hairpin configuration can be nicked by NS1, origin sequences from the left end can only be nicked when this structure has been unfolded and copied, to form the fully base-paired junction region that bridges adjacent genomes in dimer RF (1, 10). This means that monomer duplexes with left termini in the hairpin configuration must be cycled through dimer intermediates before they can be nicked and, as a consequence, that positive-sense single strands are predominantly released by displacement synthesis from dimeric, rather than monomeric, intermediates.
The second critical asymmetry involves the ability of duplex extended-form termini to melt out and reconfigure into the rabbit ear structures that are capable of priming further rounds of displacement synthesis. It has been demonstrated in vitro that extended forms of the right end of MVM are substantially more likely to undergo NS1-mediated rabbit ear formation than their left-end counterparts (1, 30, 32), probably because the axial region of the right telomere is better adapted to support such transitions. Thus, while both hairpins have NS1 binding sites that position the nuclease over the nick site of the origin, extended forms of the right terminus also have high-affinity NS1 binding sites immediately flanking the axis of symmetry that mediate the melting-out process via a reaction that involves ATP hydrolysis (31). In consequence, extended-form monomer RF molecules generally undergo rabbit ear formation at the right end, as depicted in Fig. 7, step 1, leading to the establishment of leftward forks that displace negative-sense single strands and leave a hairpin structure at the right end of the parental duplex (Fig. 7, steps 2 and 3). Since this right-end hairpin functions as an origin (Fig. 7, step 3), it can (in viruses like LuΔ2) immediately support a new round of initiation, regenerating the parental extended-form monomer RF molecule (Fig. 7, step 4).
FIG. 7.
Removal of displaced negative-sense LuΔ2 strands by encapsidation effectively precludes dimer formation and hence displacement synthesis of positive-sense DNA. The viral genome is represented by a continuous line, with its 3′ end indicated by an arrowhead. The grey sphere represents an NS1 molecule. The letters L and R represent left- and right-end palindromic sequences, respectively. Upper- and lowercase letters represent flip and flop versions of these sequences, respectively, which are inverted complements of one another. Shaded hexagons represent preassembled capsids, into which are packaged displaced strands, vectorially in a 3′-to-5′ direction. Differences in the efficiency of rabbit ear formation at the left and right termini of extended-form monomer RF DNA promote the preferential synthesis and displacement of negative-sense DNA (steps 1 and 2). Since hairpin forms of the right end serve as origins, these support repeated waves of negative-sense strand displacement (steps 1 through 5). Removal of these displaced strands by encapsidation limits synthesis to negative-sense DNA (step 6A), but replication of the displaced strands, during the amplification phase of the reaction, generates dimer intermediates that can support displacement of both positive- and negative-sense DNAs (steps 6B through 12).
Thus, in viruses that have efficient right-end origins, extended-form monomer RF molecules would be expected to predominantly support successive waves of strand displacement synthesis from the right end of the genome, leading to the synthesis of negative-sense DNA and the displacement of negative-sense single strands. We suggest that this is the only reaction that proceeds efficiently during the packaging phase of the LuΔ2 infections reported here and that this mode predominates because the displaced negative-sense strands are sequestered by the packaging apparatus (Fig. 7, steps 6A and 7A) and are thus prevented from entering the pool of replicating DNA. In contrast, during the earlier “amplification” phase, when approximately equimolar copies of positive- and negative-sense DNA are generated, displaced negative-sense strands would be expected to feed into the replicating pool (Fig. 7, step 6B) and be converted into dimer intermediates that can support displacement of positive-sense DNA. In this case, the base-paired 3′ (left-end) hairpin of the newly displaced negative-sense strand would prime complementary strand synthesis (Fig. 7, step 7B), generating monomer RF molecules with a covalently closed left-end hairpin and an extended-form right terminus. The left-end hairpin of such an RF cannot be nicked, but its right terminus can undergo rabbit ear formation (Fig. 7, step 8), leading to the synthesis of a dimer intermediate (Fig. 7, step 9) containing two active origins: a hairpin form of the right end (OriR), which would allow displacement synthesis of negative-sense DNA, and OriLTC, the active arm of the dimer bridge, which would support read-through displacement synthesis of a positive-sense strand (16), as shown in Fig. 7, steps 10 through 12. Thus, during the amplification phase of LuΔ2 replication, rolling-hairpin replication of displaced strands through a dimer intermediate would allow the synthesis of both positive- and negative-sense DNAs, and relative initiation efficiency at OriR and OriLTC would then determine whether just positive-sense unit length strands or 5-kb strands of both senses were released by displacement synthesis from the dimer.
According to this scenario, the switch to predominantly negative-sense DNA synthesis during the packaging phase fails to occur during LuIII infections because the suboptimal right-end origin effectively dampens the frequency of strand displacement synthesis from the right end of the genome. This would favor the occurrence of rabbit ear formation at the left end, leading to the synthesis of positive-sense DNA by displacement of a strand that is still in the hairpin configuration at its right end, which would be unfolded to generate a right-end-right-end dimer intermediate. This suggestion is supported by the observation that while such junctions were relatively rare in LuΔ2 infections, in LuIII-infected cells they are as common as left-end-left-end junctions (data not shown). Since this first dimer would still lack a strong nick site, the fork could have time to unfold and copy the left-end hairpin, generating a tetrameric duplex that would contain suboptimal right-end origins but the strong left-end origin, OriLTC. Accordingly, which strands were subsequently displaced would be determined by the relative efficiency of the competing origin sequences, given the obvious caveat that both origins must function sufficiently well for amplified DNA to be efficiently converted into unit length genomes for packaging. This concept, which we call the “relative single-strand displacement” scheme, thus extends the kinetic hairpin transfer model of Chen et al. (4, 29) by taking into account the fact that only certain types of molecule, we suggest specifically newly displaced single strands, can serve as packaging substrates. In consequence, unit length strands of the nonpackaged polarity can accumulate in vivo as part of duplex RF DNA, perhaps even at a low level during the packaging phase of the reaction, but they will not be packaged unless they are subsequently released by displacement synthesis.
Thus, in members of the Parvovirus genus, preferential rabbit ear formation at the right end of the genome creates a situation that favors the synthesis and displacement of negative-sense DNA. However, in LuIII this is tempered by the quality of the right-end origin because the efficiency of this nick site effectively determines how often the left-end origin will be regenerated and hence whether progeny single strands will be displaced predominantly from the right or left telomere. We therefore propose that the diminished efficiency of the LuIII right-end origin not only leads to the relatively slower genome amplification kinetics of LuIII observed in vivo but also readily explains the shift in dimer RF structure and ultimately the observed differences in packaging specificity.
Acknowledgments
We thank Nanette Diffoot for providing pGLU833, the infectious plasmid clone of LuIII.
This work was supported by Public Health Service grants AI26109 and CA29303 from the National Institutes of Health.
REFERENCES
- 1.Baldauf, A. Q., K. Willwand, E. Mumtsidu, J. P. Nuesch, and J. Rommelaere. 1997. Specific initiation of replication at the right-end telomere of the closed species of minute virus of mice replicative-form DNA. J. Virol. 71:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, R. C., C. E. Snyder, P. T. Banerjee, and S. Mitra. 1984. Autonomous parvovirus LuIII encapsidates equal amounts of plus and minus DNA strands. J. Virol. 49:319-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunstein, J., and C. R. Astell. 1997. Analysis of the internal replication sequence indicates that there are three elements required for efficient replication of minute virus of mice minigenomes. J. Virol. 71:9087-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, K. C., J. J. Tyson, M. Lederman, E. R. Stout, and R. C. Bates. 1989. A kinetic hairpin transfer model for parvoviral DNA replication. J. Mol. Biol. 208:283-296. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corsini, J., J. O. Carlson, F. Maxwell, and I. H. Maxwell. 1995. Symmetric-strand packaging of recombinant parvovirus LuIII genomes that retain only the terminal regions. J. Virol. 69:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corsini, J., S. F. Cotmore, P. Tattersall, and E. Winocour. 2001. The left-end and right-end origins of minute virus of mice DNA differ in their capacity to direct episomal amplification and integration in vivo. Virology 288:154-163. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore, S. F., J. Christensen, and P. Tattersall. 2000. Two widely spaced initiator binding sites create an HMG1-dependent parvovirus rolling-hairpin replication origin. J. Virol. 74:1332-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotmore, S. F., J. P. Nuesch, and P. Tattersall. 1993. Asymmetric resolution of a parvovirus palindrome in vitro. J. Virol. 67:1579-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotmore, S. F., and P. Tattersall. 1994. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 13:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotmore, S. F., and P. Tattersall. 1996. Parvovirus DNA replication, p. 799-813. In M. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Cotmore, S. F., and P. Tattersall. 1995. DNA replication in the autonomous parvoviruses. Semin. Virol. 6:271-281. [Google Scholar]
- 13.Cotmore, S. F., and P. Tattersall. 1989. A genome-linked copy of the NS-1 polypeptide is located on the outside of infectious parvovirus particles. J. Virol. 63:3902-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotmore, S. F., and P. Tattersall. 1998. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J. Virol. 72:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotmore, S. F., and P. Tattersall. 1992. In vivo resolution of circular plasmids containing concatemer junction fragments from minute virus of mice DNA and their subsequent replication as linear molecules. J. Virol. 66:420-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore, S. F., and P. Tattersall. 2003. Resolution of parvovirus dimer junctions proceeds through a novel heterocruciform intermediate. J. Virol. 77:6245-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diffoot, N., K. C. Chen, R. C. Bates, and M. Lederman. 1993. The complete nucleotide sequence of parvovirus LuIII and localization of a unique sequence possibly responsible for its encapsidation pattern. Virology 192:339-345. [DOI] [PubMed] [Google Scholar]
- 18.Farr, G. A., and P. Tattersall. 2004. A conserved leucine that constricts the pore through the capsid fivefold cylinder plays a central role in parvoviral infection. Virology 323:243-256. [DOI] [PubMed] [Google Scholar]
- 19.Hardt, N., C. Dinsart, S. Spadari, N. G. Pedrali, and J. Rommelaere. 1983. Interrelation between viral and cellular DNA synthesis in mouse cells infected with the parvovirus minute virus of mice. J. Gen. Virol. 64:1991-1998. [DOI] [PubMed] [Google Scholar]
- 20.Kestler, J., B. Neeb, S. Struyf, J. Van Damme, S. F. Cotmore, A. D'Abramo, P. Tattersall, J. Rommelaere, C. Dinsart, and J. J. Cornelis. 1999. cis requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum. Gene Ther. 10:1619-1632. [DOI] [PubMed] [Google Scholar]
- 21.King, J. A., R. Dubielzig, D. Grimm, and J. A. Kleinschmidt. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2359. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Nuesch, J. P., S. F. Cotmore, and P. Tattersall. 1995. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology 209:122-135. [DOI] [PubMed] [Google Scholar]
- 24.Richards, R., P. Linser, and R. W. Armentrout. 1977. Kinetics of assembly of a parvovirus, minute virus of mice, in synchronized rat brain cells. J. Virol. 22:778-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam, P., and C. R. Astell. 1994. Multiple cellular factors bind to cis-regulatory elements found inboard of the 5′ palindrome of minute virus of mice. J. Virol. 68:2840-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam, P., and C. R. Astell. 1993. Replication of minute virus of mice minigenomes: novel replication elements required for MVM DNA replication. Virology 193:812-824. [DOI] [PubMed] [Google Scholar]
- 27.Tattersall, P., L. V. Crawford, and A. J. Shatkin. 1973. Replication of the parvovirus MVM. II. Isolation and characterization of intermediates in the replication of the viral deoxyribonucleic acid. J. Virol. 12:1446-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tattersall, P., and D. C. Ward. 1976. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature 263:106-109. [DOI] [PubMed] [Google Scholar]
- 29.Tyson, J. J., K. C. Chen, M. Lederman, and R. C. Bates. 1990. Analysis of the kinetic hairpin transfer model for parvoviral DNA replication. J. Theor. Biol. 144:155-169. [DOI] [PubMed] [Google Scholar]
- 30.Willwand, K., A. Q. Baldauf, L. Deleu, E. Mumtsidu, E. Costello, P. Beard, and J. Rommelaere. 1997. The minute virus of mice (MVM) nonstructural protein NS1 induces nicking of MVM DNA at a unique site of the right-end telomere in both hairpin and duplex conformations in vitro. J. Gen. Virol. 78:2647-2655. [DOI] [PubMed] [Google Scholar]
- 31.Willwand, K., A. Moroianu, R. Horlein, W. Stremmel, and J. Rommelaere. 2002. Specific interaction of the nonstructural protein NS1 of minute virus of mice (MVM) with [ACCA]2 motifs in the centre of the right-end MVM DNA palindrome induces hairpin-primed viral DNA replication. J. Gen. Virol. 83:1659-1664. [DOI] [PubMed] [Google Scholar]
- 32.Willwand, K., E. Mumtsidu, G. Kuntz-Simon, and J. Rommelaere. 1998. Initiation of DNA replication at palindromic telomeres is mediated by a duplex-to-hairpin transition induced by the minute virus of mice nonstructural protein NS1. J. Biol. Chem. 273:1165-1174. [DOI] [PubMed] [Google Scholar]
- 33.Wobbe, C. R., and S. Mitra. 1985. Proteins tightly associated with the termini of replicative form DNA of Kilham rat virus, an autonomous parvovirus. Proc. Natl. Acad. Sci. USA 82:8335-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou, X., and N. Muzyczka. 1998. In vitro packaging of adeno-associated virus DNA. J. Virol. 72:3241-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]