A Kelch domain-containing F-box protein mediates degradation of chalcone synthase, by which it acts as a proteolytic regulator coordinately tuning flavonoid biosynthesis.
Abstract
Flavonoids represent a large family of specialized metabolites involved in plant growth, development, and adaptation. Chalcone synthase (CHS) catalyzes the first step of flavonoid biosynthesis by directing carbon flux from general phenylpropanoid metabolism to flavonoid pathway. Despite extensive characterization of its function and transcriptional regulation, the molecular basis governing its posttranslational modification is enigmatic. Here, we report the discovery of a proteolytic regulator of CHS, namely, KFBCHS, a Kelch domain-containing F-box protein in Arabidopsis thaliana. KFBCHS physically interacts with CHS and specifically mediates its ubiquitination and degradation. KFBCHS exhibits developmental expression patterns in Arabidopsis leaves, stems, and siliques and strongly responds to the dark-to-light (or the light-to-dark) switch, the blue, red, and far-red light signals, and UV-B irradiation. Alteration of KFBCHS expression negatively correlates to the cellular concentration of CHS and the production of flavonoids. Our study suggests that KFBCHS serves as a crucial negative regulator, via mediating CHS degradation, coordinately controlling flavonoid biosynthesis in response to the developmental cues and environmental stimuli.
INTRODUCTION
As a large family of (poly)phenolics, flavonoids are ubiquitously distributed throughout the plant kingdom. Structurally they are classified as flavones, flavonols, anthocyanins, proanthocyanidins, and isoflavones (Winkel-Shirley, 2001; Grotewold, 2006; Saito et al., 2013). Flavonoids play important roles in the biology of plants; for example, they are the major photoprotectant in plants, conferring flower pigmentation and protecting plants from UV irradiation. Flavonoids also function as antimicrobial compounds (phytoalexins) or insect repellents defending against phytopathogens and herbivores. Some of flavonoid metabolites have pharmacological activities (Grotewold, 2006).
The biosynthesis of flavonoids branches from the general phenylpropanoid pathway via the rate-limiting enzyme chalcone synthase (CHS; EC 2.3.1.74) (Grotewold, 2006; Saito et al., 2013). CHS catalyzes the stepwise condensation of three acetate residues from malonyl-CoA with phenylpropanoid biosynthetic intermediate p-coumaroyl CoA to form naringenin chalcone, which leads to the synthesis of a variety of flavonoid derivatives (Winkel-Shirley, 2001; Grotewold, 2006). As the first committed enzyme of flavonoid biosynthesis, CHS is rigorously controlled in response to a wide range of environmental and developmental stimuli (Feinbaum and Ausubel, 1988; Schmid et al., 1990; Christie and Jenkins, 1996; Dao et al., 2011). When plant cells were treated with UV light or other abiotic elicitors, CHS exhibits a transient burst of de novo enzyme synthesis, followed by a decay of active enzyme (Schröder and Schäfer, 1980; Chappell and Hahlbrock, 1984; Ryder et al., 1984). The induction of its de novo synthesis was demonstrated as the result of a rapid but transient increases of CHS transcripts in response to the elicitation (Bell et al., 1984; Chappell and Hahlbrock, 1984; Ryder et al., 1984; Feinbaum and Ausubel, 1988), and the decline of CHS activity in the induced cell cultures was postulated to be caused by the inactivation and/or degradation of CHS proteins (Schröder and Schäfer, 1980). These early investigations suggest that CHS in plant cells is regulated at both the transcriptional and posttranslational levels. The transcriptional regulation of CHS has been investigated intensively. Studies on the CHS promoter in Arabidopsis thaliana and many other species have led to the identification of several consensus cis-elements such as the G-box (GACGTG) and/or H-box (CCTACC) that are important for transcriptional activation of CHS in response to abiotic elicitation or environmental stimuli (Schulze-Lefert et al., 1989; Staiger et al., 1989; van der Meer et al., 1990; Harrison et al., 1991; Loake et al., 1992; Hartmann et al., 1998). The cis-elements found in the defined minimal Arabidopsis CHS promoter are bound by basic region helix-loop-helix (bHLH) and R2R3-MYB-type transcription factors (Hartmann et al., 2005). Several R2R3-MYB and bHLH transcription factors were found to interact with the WD40-containing protein TRANSPARENT TESTA GLABRA1, to form a regulatory complex controlling multiple enzymatic steps of flavonoid biosynthetic pathway (Broun, 2005).
Among various environmental stimuli, light is one of the most important factors triggering flavonoid biosynthesis. The induction of light-responsive gene expression and flavonoid biosynthesis depends on photoperiod, light intensity, direction, and quality (wavelength) (Zoratti et al., 2014). The light induction process is transcriptionally controlled via a core signaling pathway, in which the CONSTITUTIVE PHOTOMORPHOGENIC1/SUPPRESOR OF PHYTOCHROME A-105 (COP1/SPA) ubiquitin ligase acts as the central regulator (Lau and Deng, 2012). In the darkness, the nuclear-localized COP1/SPA targets positive regulators, such as the bZIP transcription factor ELONGATED HYPOCOTYL5 (HY5) and R2R3-MYB transcription factors for ubiquitination and subsequent protein degradation through 26S proteasome pathway. Under the visible light condition, the activity of COP1/SPA is directly inhibited by the light-activated photoreceptors such as phytochromes, cryptochromes, and phototropins via their physical interaction, which leads to the stabilization of HY5 and R2R3-MYBs, subsequently activating a series of flavonoid/anthocyanin biosynthetic genes including CHS (Maier et al., 2013; Maier and Hoecker, 2015). Upon UV-B exposure, the induction of gene expression and flavonoid biosynthesis in Arabidopsis is achieved through both UV-B-specific and nonspecific signaling pathways (Jenkins, 2009; Hideg et al., 2013). At low doses of UV-B radiation, UV RESISTANCE LOCUS8 (UVR8) serves as the UV absorbing photoreceptor and mediates the rapid expression of HY5 transcription factor and the closely related HY5 HOMOLOG (HYH) (Brown et al., 2005). In this process, the dimeric form of UVR8 undergoes instantaneous monomerization. The monomeric UVR8 interacts with COP1, forming a protein complex that accumulates in the nucleus of the cells, which leads to the transcriptional activation and posttranslational stabilization of HY5/HYH; in turn, HY5/HYH activates a family of R2R3-MYB transcription factors and promotes expression of flavonoid biosynthetic genes including CHS (Brown et al., 2005; Favory et al., 2009; Stracke et al., 2010; Heijde and Ulm, 2012). With high doses of UV-B radiation, however, it induces DNA damage, defense, and wound signaling pathways, in which many signaling components, including reactive oxygen species, MAP kinases, jasmonic acid, salicylic acid, nitric oxide, ethylene, abscisic acid, etc., have been implicated in UV-B-induced gene expression (Jenkins, 2009; Hideg et al., 2013).
While our knowledge on the transcriptional regulation of CHS expression and flavonoid biosynthesis is relatively abundant, little is known about the molecular basis underlying the potential posttranslational regulation of CHS. Ubiquitination is one of the prevalent posttranslational modifications. In most cases, this type of modification leads to protein degradation via the 26S proteasome system (Vierstra, 2012). Whereas the less target-specific ubiquitination occurs as part of a quality control mechanism to remove misfolded and aggregated polypeptides, the programmed ubiquitination of specific target proteins can serve as a sophisticated regulatory mechanism controlling a wide spectrum of plant cellular processes (Smalle and Vierstra, 2004; Vierstra, 2009). The key player of protein ubiquitination is the E3 ubiquitin-protein ligase that specifies appropriate target proteins for modification. One of the best characterized classes of E3 ligases is the Skp1-Cullin-F-box (SCF) complex. Within this complex, Cullin interacts both with SKP1 and RBX1, forming a scaffold on which the F-box protein assembles (del Pozo and Estelle, 2000; Lechner et al., 2006). The specificity of the SCF complex is conferred by the F-box proteins that selectively interact with target proteins (del Pozo and Estelle, 2000). Nearly 700 F-box proteins are predicted in Arabidopsis (Gagne et al., 2002). They are classified into different subfamilies according to the presence of additional protein-protein interaction domains near their C terminus. One subfamily was designated as the Kelch domain (repeat)-containing F-box (KFB) proteins (Bork and Doolittle, 1994), which is composed of ∼103 members in Arabidopsis with only a few that have been characterized (Xu et al., 2009; Schumann et al., 2011). Among them, we recently demonstrated that Arabidopsis KFB01, 20, 39, and 50 (collectively named KFBPALs) interact with phenylalanine ammonia-lyase (PAL), the first rate-limiting enzyme of phenylpropanoid biosynthetic pathway, and mediate its ubiquitination and subsequent degradation, by which the FKBPALs act as negative regulators controlling the synthesis of the entire class of phenylpropanoid metabolites (Zhang et al., 2013, 2015). Given the large number of KFBs found in Arabidopsis, we deduced that the KFB-mediated degradation of the key metabolic enzymes could be a prevalent regulatory mechanism governing the synthesis of different classes of specialized metabolites. In this work, through the targeted yeast two-hybrid screening, we identify a KFB that specifically interacts with CHS, the flavonoid-branch key enzyme, and mediates its ubiquitination and degradation. Moreover, we found that this KFB serves as an essential proteolytic regulator, coordinating with transcriptional regulation mechanism, negatively controlling the production of flavonoids during plant growth and development and in the processes responding to environmental stimuli, in particular to different light signals and UV-B radiation.
RESULTS
The At1g23390 Encoded KFB Protein Physically Interacts with CHS
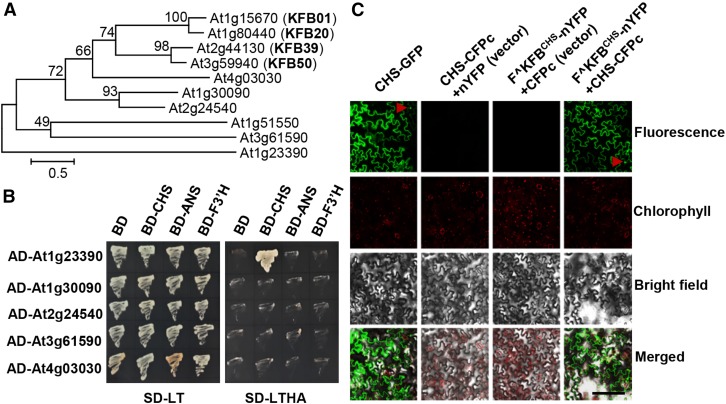
In an established phylogenetic tree of the entire set of Arabidopsis putative KFB polypeptides (Supplemental Figure 1), we noticed that several KFB members were closely clustered with the previously characterized KFBPALs (KFB01, 20, 39, and 50) (Figure 1A) sharing more than 30% sequence similarity at their amino acid levels. The concurrence of some of those KFB members with KFBPALs in response to the carbon deprival or supply was revealed in a previous cDNA microarray study (Osuna et al., 2007). Based on this, we deduced that some of those evolutionarily related KFB proteins may also have biochemical and biological functions that are potentially analogous to those of KFBPALs in mediating the turnover of proteins or enzymes in particular metabolic pathways. Being interested in comprehensively identifying KFBs involved in phenylpropanoid biosynthesis, we selected the members of the cluster (except for At1g51550, for which we did not successfully obtain a cDNA) as the initial targets to examine their potential protein interactions with phenylpropanoid biosynthetic enzymes via conventional yeast two-hybrid (Y2H) assay. A set of monolignol and flavonoid biosynthetic enzymes were used as “baits” and coexpressed with the selected KFBs in the assays (Figure 1B; Supplemental Figure 2). Of 55 pairs with lignin biosynthetic enzymes, none were able to sustain yeast cell growth on the selective medium lacking His, Ade, Trp, and Leu supplements (Supplemental Figure 2), indicating no physical interaction occurs between the paired proteins. However, intriguingly, of the 20 pairwise assays using flavonoid biosynthetic enzymes, CHS, flavonoid 3′-hydroxylase (F3′H), and anthocyanin synthase (ANS), the yeast cells harboring CHS and one KFB encoded by At1g23390 survived well on the stringent selective medium (-His, -Ade, -Trp, and -Leu) (Figure 1B), suggesting that the KFB encoded by At1g23390 physically interacts with CHS in vivo (thereafter, it is designated as KFBCHS). The deduced polypeptide of KFBCHS consists of 394 amino acid residues. As the conserved features of kelch domain-containing F-box protein, its N terminus contains a typical F-box motif but its C-terminal domain only consists of one predicted kelch motif, which is distinct from the characterized KFBPALs that encompass two to approximately four predicted kelch repeats (Supplemental Figure 3).
Figure 1.
Interaction of Arabidopsis Flavonoids Biosynthetic Enzymes with KFB Proteins.
(A) Phylogeny of KFBs clustered with KFB01, 20, 39, and 50 (marked as bold). Scale shows the length of branch that represents an amount amino acid change of 0.5.
(B) Pairwise Y2H assays of the “bait” pDEST-GBKT7-CHS, -ANS or -F3′H with the “prey” pDEST-GADT7-At1g23390, -At1g30090, -At2g24540, -At3g61590, and -At4g03030. Yeast harboring the pairwise expression cassettes were grown on both the growth medium lacking Leu and Trp (SD-LT) and on the selective medium lacking Leu, Trp, His, and adenine (SD-LTHA) for 3 d.
(C) BiFC assays for the CHS-CFPc fusion with the truncated KFBCHS that had a deletion of its F-box motif and was fused with nYFP (F^KFBCHS-nYFP). The expression constructs were coinfiltrated into tobacco leaves according to the indicated combinations on the top of panels. The fluorescence distribution of CHS-full length of GFP fusion is shown in the left panel. Arrowheads indicate the nucleus. Bar = 200 μm.
To validate the detected interaction between CHS and KFBCHS in planta, we conducted, a bimolecular fluorescence complementation (BiFC) assay by coexpression of CHS and KFBCHS tagged with complementary half-molecule fragments of cyan or yellow fluorescent protein (CFP or YFP). Since the interaction between the full length of F-box protein and its target in plant cells could potentially lead to the turnover of the substrate protein, thus preventing the detection of in vivo interaction (Zhang et al., 2013), the truncated KFBCHS with a deletion of its F-box domain was created and used in the study; this was designated F^KFBCHS. Both CHS and F^KFBCHS that were fused with the complementary half-molecule of CFP or YFP were coexpressed in tobacco (Nicotiana tabacum) leaf cells. The CHS full-length GFP fusion was used as a positive control to illustrate its subcellular localization. The data revealed that the fluorescence of the CHS-GFP fusion was predominantly distributed in the cytosol; meanwhile, nuclear localization was also occasionally observed in a portion of leaf cells (Figure 1C). This observation is consistent with previous reports that Arabidopsis CHS is a cytoplasmic and nuclear-localized enzyme for local synthesis of flavonoids (Hrazdina and Jensen, 1992; Saslowsky and Winkel-Shirley, 2001). While coexpression of F^KFBCHS-nYFP with CFPc, or coexpression of CHS-CFPc with nYFP in the tobacco leaves did not yield any chimeric fluorescence, coexpressing F^KFBCHS-nYFP with CHS-CFPc generated strong fluorescence signals in the cytosol and nucleus, similar to the CHS-full length GFP fusion (Figure 1C). These results confirm the spatial proximity or the physical interaction of CHS and KFBCHS in planta.
CHS Is Ubiquitinated and Degraded through the Ubiquitin/26S Proteasome Pathway
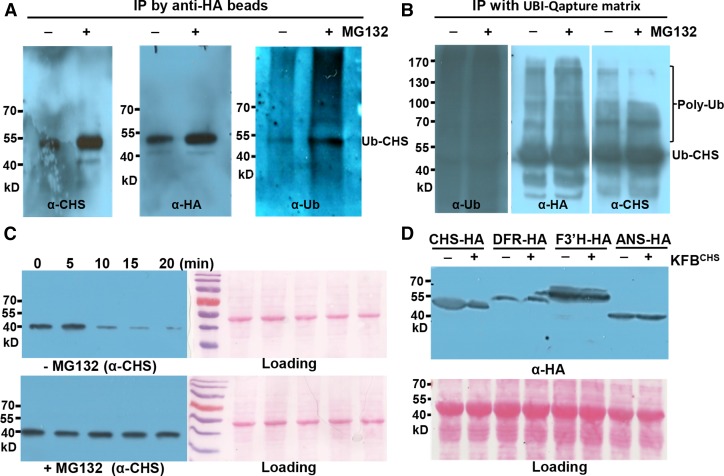
Since KFBCHS is an annotated structural component of the SCF type ubiquitin-ligase complex (Xu et al., 2009; Schumann et al., 2011), its physical interaction with CHS indicates that CHS might be ubiquitinated and degraded through the ubiquitin/26S proteasome pathway in planta. To test this hypothesis, HA-tagged CHS was transiently expressed in tobacco leaves and examined for potential ubiquitination. Two sets of reciprocal affinity binding experiments were conducted. First, after 3 d transfection with Agrobacterium tumefaciens harboring the AtCHS-HA expression cassette, equal amounts of tobacco leaves were ground in ATP-containing extraction buffer in the presence or absence of the specific 26S proteasome inhibitor MG132 (Lyzenga et al., 2012). The AtCHS-HA proteins were then immunoprecipitated with anti-HA antibody and detected with antiubiquitin, anti-AtCHS, and anti-HA antibodies, respectively. As depicted in Figure 2A, a prominent band was detected in all three blots probed with antiubiquitin, anti-AtCHS, or anti-HA antibody, with an apparent molecular mass of ∼51 kD, referenced with the prestained protein molecular markers (Figure 2A; Supplemental Figure 4A). In addition to this primary band of ∼51 kD, both anti-AtCHS and anti-HA probes also detected a weaker signal with apparent molecular mass slightly above the 40-kD prestained protein marker, which is coincident with the predicted molecular mass of the AtCHS-HA polypeptide (∼43 kD). Notably, this weak band was not detected with antiubiquitin antibody (Figure 2A; Supplemental Figure 4A) therefore likely representing the nonubiquitinated AtCHS-HA species. Considering the molecular mass of ubiquitin at ∼8.5 kD, it is possible that the 51-kD band visualized with all three antibodies might represent the monoubiquitinated form of AtCHS-HA. Furthermore, the smear of signals above the detected 51-kD band was readily observed with the antiubiquitin antibody (Figure 2A; Supplemental Figure 4A) and such species were actually also detectable with anti-CHS (or anti-HA) antibody in the immunoblot with prolonged signal detection or exposure process (Supplemental Figure 4B). These data suggest that the expressed AtCHS-HA in tobacco leaves might be polyubiquitinated, besides its predominant monoubiquitinated form. Since the polyubiquitinated species yield more abundant ubiquitin epitopes than the antigen protein itself, they can be visualized more easily by probing with antiubiquitin antibody than with the anti-HA or anti-CHS antibody with the same detection method. Therefore, the detected smear signals were more obvious in the blot with the antiubiquitin antibody. Since antiubiquitin and anti-AtCHS (or anti-HA) probes essentially detect different abundant epitopes, the smear patterns visualized with two different probes were also slightly different (Figure 2A; Supplemental Figure 4).
Figure 2.
CHS Protein Is Subject to Ubiquitination and 26S Proteasome-Mediated Degradation.
(A) Detection of the ubiquitinated AtCHS-HA proteins. Transiently expressed AtCHS-HA was IP-enriched by anti-HA beads from tobacco leaves in the presence or absence of 50 μM MG132 and detected with anti-AtCHS (left), anti-HA (middle), and antiubiquitin antibodies (right), respectively. The images of original blots with full molecular markers are presented in Supplemental Figure 4.
(B) Enrichment of the ubiquitinated CHS-HA proteins. Total ubiquitinated proteins were enriched by ubiquitin affinity binding matrix (UBI-Qapture matrix) from tobacco leaves with transiently expressed AtCHS-HA and preinfiltrated with or without 40 μM MG132. The enriched proteins were then detected with antiubiquitin (left), anti-HA (middle), and anti-AtCHS (right) antibodies, respectively; the smear/bands above the primary ubiquitinated CHS (Ub-CHS) band represent the polyubiquitinated CHS species (Poly-Ub).
(C) Cell-free degradation of the endogenous CHS enzyme from Arabidopsis. Total proteins from 5-DAG Arabidopsis seedlings were incubated in the degradation buffer at 37°C in the presence or absence of 50 μM MG132. At the indicated time points, the level of CHS protein was monitored by immunoblot using anti-AtCHS antibody. Ponceau S staining serves as the loading control for the amount of total protein.
(D) Immunoblots with anti-HA antibody for AtCHS-HA, AtDFR-HA, AtF3′H-HA, and AtANS-HA fusion proteins coexpressed with KFBCHS (pGWB402-KFBCHS) or the empty vector (pGWB402) in tobacco leaves. Ponceau S staining serves as the loading control for the amount of total proteins. Note that both AtCHS-HA and AtDFR-HA exhibited the larger molecular sizes than their predicted molecular masses.
However, it is also possible that the detected smear of ubiquitinated species might be derived from the potential interaction partners of CHS or nonspecific proteins that coimmunoprecipitated with AtCHS-HA. To further verify the polyubiquitination of AtCHS-HA, we further conducted a reciprocal affinity binding assay, in which the total ubiquitinated proteins were first enriched by incubating the crude protein extracts from the tobacco leaves transiently expressing AtCHS-HA with Ubi-Qapture-Q matrix (Enzo life Science), the ubiquitin-affinity resin showing superior capacity for efficiently isolating both mono- and polyubiquitinated proteins from cells (Schwertman et al., 2013). The enriched ubiquitinated proteins were then probed with antiubiquitin, anti-HA, and anti-CHS antibodies, respectively. The data showed that, while antiubiquitin probe delivered an intensive smear immunosignals in the blot, indicating the effective enrichment of different sizes of ubiquitinated proteins, a profound immunoband was detected either by anti-HA or by anti-AtCHS antibody with its migration between the 40 and 55 kD of the prestained protein markers (Figure 2B; Supplemental Figure 5), representing the monoubiquitinated AtCHS-HA species. In addition, the obvious larger molecular mass bands and/or smear species were monitored by anti-HA and anti-AtCHS antibodies (Figure 2B; Supplemental Figure 5), confirming the polyubiquitination of the expressed AtCHS-HA.
Comparing the samples incubated with and without MG132 in immunoprecipitation experiments (Figures 2A and 2B), it is noticeable that the intensity of AtCHS-HA immunosignals monitored with any of the applied antibodies was much stronger in the samples treated with MG132. These data suggest that the 26S proteasome inhibitor stabilizes CHS-HA proteins, implying the ubiquitinated CHS is degraded via the 26S proteasome system.
The 26S proteasome-mediated turnover of CHS was further examined using a cell-free degradation assay for Arabidopsis endogenous proteins. The crude proteins extracted from Arabidopsis seedlings at 5 d after germination (DAG) were incubated with degradation assay buffer containing 10 mM ATP in the presence or absence of MG132. The endogenous AtCHS, when blotted with anti-AtCHS antibody, routinely showed a signal around 41 kD, compared with the prestained protein markers (Figure 2C). Occasionally an additional weaker band with higher molecular mass (∼50 kD) could be detected (e.g., in the crude proteins from the dark-treated plants; Supplemental Figure 6), which apparently is consistent with the molecular size of its ubiquitinated species. These data indicate that the dominant form of endogenous CHS existing in Arabidopsis differs from that of AtCHS overexpressed in tobacco leaves and its prominent form in Arabidopsis is represented with the nonubiquitinated species. Over the 20-min incubation period, the detected CHS showed a fairly stable signal in the samples with MG132 treatment, whereas obvious turnover was observed after 10 min processing in the samples without MG132 (Figure 2C). These data confirm that CHS in Arabidopsis is indeed degraded through the ubiquitin/26S proteasome pathway.
To examine whether KFBCHS mediates CHS degradation, we coexpressed CHS-HA with KFBCHS in tobacco leaf cells. Crude protein extracts were blotted with anti-HA instead of anti-AtCHS antibody to avoid potential cross-reaction with the endogenous CHS from tobacco. The data showed that coexpression of KFBCHS substantially reduced or abolished the level of overexpressed AtCHS-HA (Figure 2D; Supplemental Figure 7), but the control leaves harboring the AtCHS-HA expression cassette and the empty vector for KFBCHS maintained strong CHS signals, indicating KFBCHS is the crucial component mediating CHS degradation (Figure 2D; Supplemental Figure 7). In contrast, when KFBCHS was coexpressed with other flavonoid biosynthetic enzyme dihydroflavonol 4-reductase (DFR), F3′H, or ANS, no effects were observed on the stability of the expressed proteins, in comparison with their corresponding control sets (Figure 2D). Interestingly, among the other three expressed flavonoid enzymes, AtDFR-HA, like AtCHS-HA, also exhibited a band with larger molecular mass (∼55 kD) than its predicted molecular size in the blot, indicating that the potential modification, likely ubiquitination, might also occur on this protein. However, coexpressing it with KFBCHS did not affect its stability (Figure 2D). These results indicate that KFBCHS specifically triggers CHS degradation.
Manipulating KFBCHS Expression in Arabidopsis Alters CHS Cellular Concentration
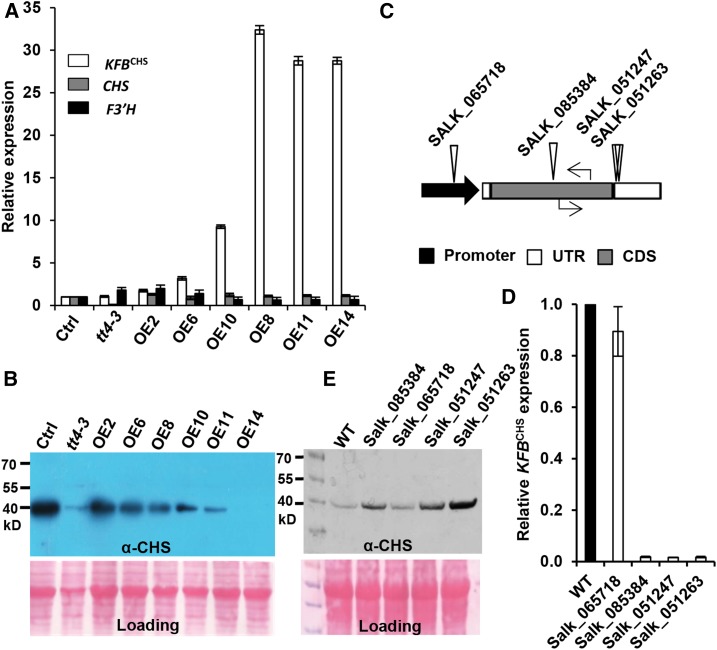
The in vitro degradation assays showed that KFBCHS affects CHS stability. Therefore, we speculated that manipulating KFBCHS expression in planta should affect CHS cellular abundance, thus disturbing flavonoid production. To investigate this hypothesis, KFBCHS was overexpressed in Arabidopsis (Col-0), driven by a 35S promoter. RT-qPCR revealed that the expression level of the KFBCHS transgene varied in a set of established transgenic lines, with ∼10- to ∼30-fold increase in the lines of KFBCHS-OE10, 11, and 14, compared with that in the control (Figure 3A). Conversely, the CHS protein levels in 5-DAG seedlings of T2 transgenic lines exhibited a profound reduction, particularly in the KFBCHS high expression lines (Figure 3B); notably, CHS in the line OE14 was barely detectable (Figure 3B). To exclude the possibility that lowering CHS cellular concentration was due to the unexpected effects on flavonoid biosynthetic gene expression, the transcript levels of CHS and F3′H genes were examined in the KFBCHS overexpression lines. Both genes showed similar expression levels as those in the control plants (Figure 3A), indicating that overexpression of KFBCHS does not significantly affect the intrinsic flavonoid biosynthetic gene expression. Therefore, the attenuation of CHS cellular concentration in planta appears to result from the direct effect of KFBCHS-mediated proteolysis.
Figure 3.
Disturbing KFBCHS Expression Alters CHS Cellular Concentration in Arabidopsis Seedlings.
The plants were grown under white light with a fluence rate of 104 ± 5 µmol·m−2·s−1 and a 16-h-light/8-h-night regime at 22°C.
(A) The relative expression levels of KFBCHS, CHS, and F3′H in the independent T2 KFBCHS overexpression (OEs), the empty vector control (Ctrl.), and tt4-3 (CS66119) mutant lines, detected by RT-qPCR. Total RNA was extracted from the mixed 5-DAG agar plate-grown seedlings. Data represent means ± sd of three biological replicates; each replicate represents the mixed seedlings (0.1 g FW) from one agar plate. The expression level of each gene in the empty vector control lines was set as “1.”
(B) Immunoblot of CHS protein probed with anti-AtCHS antibody in the KFBCHS overexpression, the empty vector control, and the tt4-3 mutant lines from the same set of materials as in (A). Total protein stained with Ponceau S serves as the loading controls.
(C) Diagram of T-DNA insertion mutations (triangles) of KFBCHS gene, and the positions of RT-qPCR primers (arrows) used for detecting gene expression. Black arrow represents the promoter region; empty bars represent untranslated regions; gray bar represents coding sequence. No intron exists in the KFBCHS gene.
(D) The expression levels of KFBCHS in T-DNA insertion mutant lines, detected by RT-qPCR. The expression level in the wild type was set as “1.” Data represent means ± sd of three biological replicates; and each replicate represents the pooled seedlings (0.1 g FW) from one agar plate.
(E) Immunoblot detection of CHS protein in kfbchs homozygous mutant lines. An equal amount of total proteins (10 µg) from 5-DAG seedlings of each line was developed by 12% SDS-PAGE gel and transferred to nitrocellulose membrane. CHS was detected using anti-AtCHS antibody. Ponceau S staining serves as the loading control for the amount of total protein.
To further examine the effect of KFBCHS on CHS stability, we also analyzed T-DNA insertion mutant lines of KFBCHS. In three mutant alleles, Salk_085384, Salk_051247, and Salk_051263, with T-DNA insertions in the coding sequence or 3′ untranslated regions of At1g23390, KFBCHS expression was completely disrupted; whereas the line Salk_065718 with insertion in the promoter region of the gene exhibited only minor effect on KFBCHS gene expression (Figures 3C and 3D). Correspondingly, stronger CHS signals were observed in three knockout kfpchs mutants, whereas only a slight or no increase of CHS levels was detected in the line Salk_065718 in comparison with that in the wild type (Figure 3E). These data further suggest that KFBCHS dominates the turnover of CHS proteins in vivo.
Manipulating KFBCHS Expression in Arabidopsis Disturbs Flavonoid Synthesis
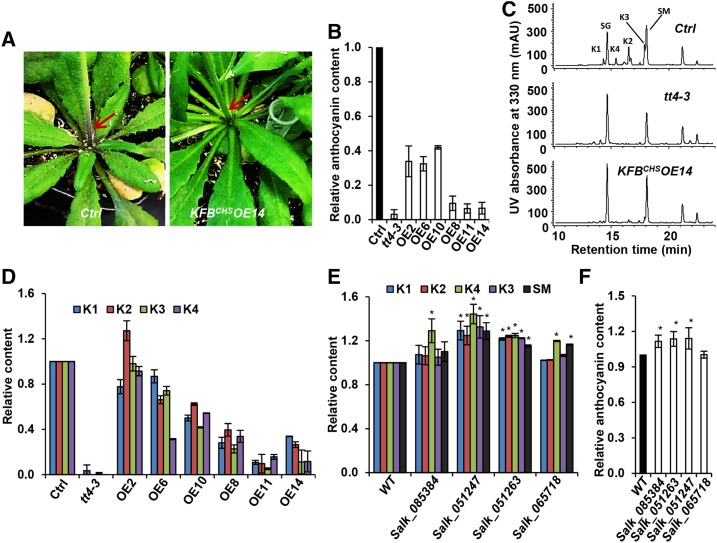
Under normal growth conditions, the 6-week-old control plants (with empty vector) exhibited obvious purple pigmentation at their basal stems and petioles of rosette leaves; by contrast, some of the KFBCHS overexpression lines nearly abolished the pigmentation at the same position of stem or petioles (Figure 4A). Quantifying the level of accumulated anthocyanin pigments in rosette leaves revealed that the contents of anthocyanin in a series of overexpression lines exhibited a 60 to ∼90% reduction, compared with that of the control. In particular, the transgenic lines OE11 and OE14 with high levels of KFBCHS expression showed a near negligible amount of anthocyanin as did the tt4-3, a chs mutant line that was created by zinc finger nuclease-mediated target mutagenesis (Zhang et al., 2010) and that is not a complete null mutant of CHS (Figure 4B). Similarly, profiling other soluble phenolics with liquid chromatography-mass spectrometry revealed that a set of flavonol glycoconjugates that were readily detected in the control plants almost completely disappeared in the transgenic lines OE11 and OE14, which resulted in a nearly identical phenolic profile to that of tt4-3 (Figure 4C). Quantification of the major soluble phenolics indicated that flavonol glycoconjugate contents were substantially reduced in most of the generated overexpression lines and, in general, the higher KFBCHS expression, the more severe the suppression of flavonol biosynthesis (Figures 3A and 4D). However, the levels of sinapate derivatives that are derived from monolignol biosynthetic pathway appeared less affected or even slightly increased (for its glucose derivative) in the lines with high expression of KFBCHS transgene, compared with those of the control plants (Supplemental Figure 8A). These data indicate that KFBCHS negatively affects flavonol and anthocyanin biosynthesis. However, when proanthocyanidin (PA) content in mature seeds was measured, only a mild reduction was observed in the KFBCHS transgenic lines compared with the control plants (Supplemental Figure 8B). This result is probably due to the large pool size of PA polymers in seed coats; thus, the effect of the reduction caused by KFBCHS is less pronounced. Alternatively, the observed weak effect of KFBCHS expression on PA accumulation may be just due to the less efficient expression of the 35S promoter in the seed coat, as previously reported (Chen et al., 2014; Dutt et al., 2014), where PA is synthesized locally.
Figure 4.
Overexpression of KFBCHS in Arabidopsis Impairs Flavonoid Production.
Both 6-week-old-plants and 5-DAG seedlings were grown under white light with a fluence rate of 104 ± 5 µmol·m−2·s−1 and a 16-h-light/8-h-night regime at 22°C.
(A) Visible pigmentation at the basal stem and petioles of rosette leaves of the control and KFBCHS overexpression line #14. Note the reduction of pigmentation in the overexpression line (indicated with arrow).
(B) The relative accumulation levels of anthocyanin in the 6-week-old rosette leaves of KFBCHS OE lines. The content in the control line (50 µg/g FW cyanidin 3-O-glucoside) was set as “1.” Data represents mean ± sd from three biological repeats; each repeat represents the mixed rosette leaves from nine individual plants.
(C) Representative UV-HPLC profiles of methanolic soluble phenolics in KFBCHS OE14 and tt4-3 mutant lines. K1, Kaempferol 3-O-[6′′-O-(rhamnosyl)glucoside] 7-O-rhamnoside; K2, kaempferol 3-O-glucoside 7-O-rhamnoside; K3, kaempferol 3-O-rhamnoside 7-O-rhamnoside; K4, kaempferol 3-O-rhamnoside 7-O-glucoside. Metabolites were extracted from 0.1 g FW of 5-DAG seedlings with 1 mL of 80% methanol, and 10-μL samples of each extract were injected for HPLC profiling.
(D) Relative content of soluble phenolics in the 5-DAG seedlings of KFBCHS OE lines. Peak areas for each identified compound from the control lines were set as “1.” Data represent means ± sd from three biological replicates; each repeat represents the mixed 0.1 g FW seedlings from one agar plate. For clarity, the statistical indicators were not shown.
(E) Relative content of flavonoids and sinapoyl esters accumulated in the flowers and buds of 6-week-old kfbCHS mutant lines. Metabolites were extracted from 0.1 g FW of flower and buds tissue with 1 mL of 80% methanol, and 10-μL extracts were injected for HPLC profiling. Peak areas for each identified compound from the wild type were set as “1.” Data represent means ± sd of three biological replicates. The mixed materials from three individual plants were defined as a replicate. K1, K2, K3, and K4 are same as in (C). SM, sinapoyl malate. *P < 0.05 (Student’s t test) compared with the corresponding compound in the wild type.
(F) Relative content of anthocyanin in the flowers and buds of 6-week-old kfbCHS mutant lines. Absorbance of A530 to A700 from the wild-type extract was set as “1.” Data represent means ± sd of three biological replicates as the defined in (E). *P < 0.05 (Student’s t test) compared with the wild type.
Conversely, examining the accumulation of flavonoids in the kfbchs mutant lines, where CHS level showed a substantial increase, we found that the contents of flavonol derivatives and anthocyanin in flower and buds tissues of the mutant lines were also increased, although the levels of increase were less substantial (Figures 4E and 4F). These data indicate that KFBCHS indeed negatively affects flavonoid biosynthesis. Meanwhile, the results likely implicate additional control factors might exist for regulating either CHS activity or the downstream flavonoid biosynthetic activity.
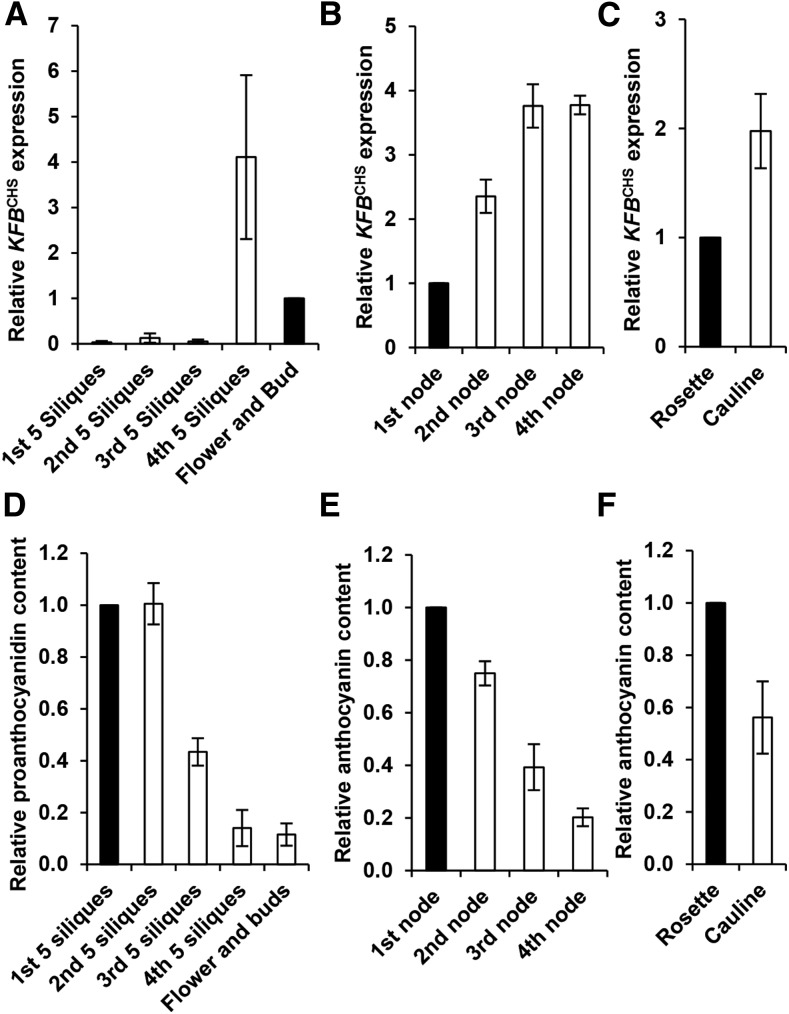
KFBCHS Expression in Different Tissues
Examining KFBCHS expression in different tissues of 6-week-old Arabidopsis, we found that KFBCHS was mostly abundant in the early developing siliques or flowers/buds but nearly absent in the developed siliques (Figure 5A). In different internodes of inflorescence stem, the transcript abundance of KFBCHS was about 4-fold higher in the young stem internodes than in the basal stem (the first nodes) (Figure 5B). Similarly, its expression level in cauline leaves was about 2-fold higher than that in rosette leaves (Figure 5C). These data indicate that KFBCHS exhibits appreciable developmental or tissue-preferential expression patterns. The developmental programming of KFBCHS expression in general was reciprocally correlated with the accumulation of flavonoids in the corresponding tissues under distinct developmental stages. As depicted in Figure 5, PAs accumulated abundantly in the mature siliques/seeds but barely detectable in the early developing siliques or flowers/buds (Figure 5D); similarly, in different stem internodes, the level of anthocyanin was high in the basal nodes but low in young stem nodes (Figure 5E). In the leaf tissues, the rosette leaves accumulated ∼2-fold higher amounts of anthocyanin than did the cauline leaves (Figure 5F). The negative correlation of KFBCHS expression and the developmental production of flavonoids suggest the involvement of KFBCHS in regulating developmental programming of flavonoid biosynthesis.
Figure 5.
Developmental Expression of KFBCHS and the Accumulation Levels of Anthocyanins and/or Proanthocyanidins in Different Tissues.
Six-week-old plants were grown under white light with a fluence rate of 104 ± 5 µmol·m−2·s−1 and a 16-h-light/8-h-night regime at 22°C.
(A) to (C) RT-qPCR analyses of the expression levels of KFBCHS in developing siliques and flowers/buds (A), in stem internodes from the bottom (1st node) to the top (4th node) of the inflorescence (B), and in rosette and cauline leaves (C). Every fifth silique from bottom to top of the inflorescence was grouped together, representing one growth stage in (A). The expression levels detected in the flowers/buds (A), the first basal stem internode (B), and the rosette leaves (C) were set as “1.” Data represent means ± sd of three biological replicates. Each replicate represents the mixed materials of nine 6-week-old plants.
(D) Proanthocyanidin contents in the developing siliques and flowers/buds. The average absorbance at 550 nm in the first group of siliques (the oldest) was set as “1.” Data represent means ± sd from three biological replicates. The biological replicate was defined as in (A).
(E) and (F) Anthocyanin contents in the different stem internodes (E) and in the rosette and cauline leaves (F). The contents quantified in the rosette leaves (∼43 µg/g FW cyanidin 3-O-glucosides) and in the first stem internode (∼55 µg/g FW) were set as “1,” respectively. Data represent means ± sd of three biological replicates. The biological replicate was defined as in (B) and (C).
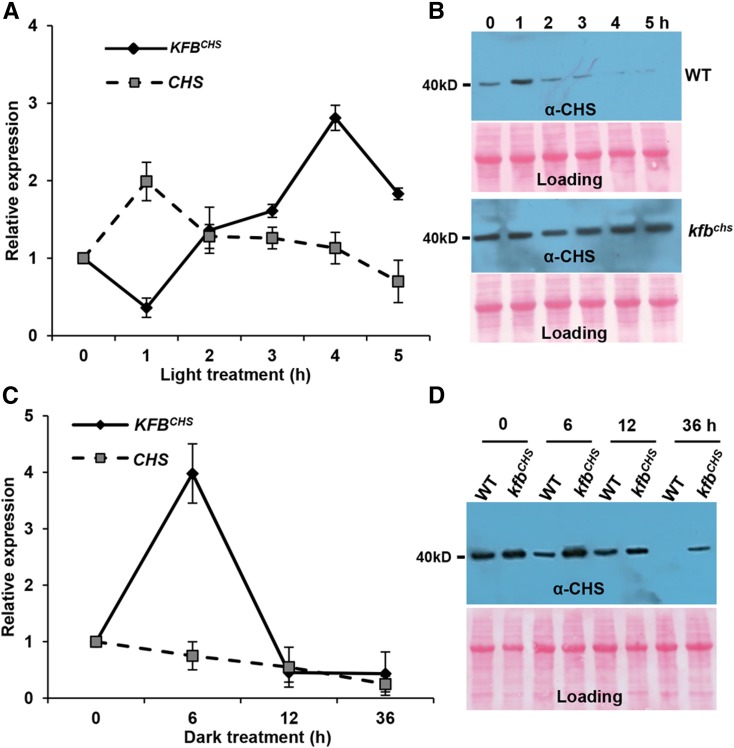
KFBCHS Responds to the Switch from Dark to Light and from Light to Dark
CHS gene expression and flavonoid biosynthesis are induced by a range of environment stimuli; in particular, the inducible synthesis of flavonoids plays a critical role in plant light responsiveness and UV damage protection (Ryder et al., 1984; Bell et al., 1986; Kaulen et al., 1986; Weisshaar et al., 1991; Thain et al., 2002). To explore whether the KFBCHS-mediated proteolysis of CHS also functions in the light- and/or stress-responsive synthesis of flavonoids, we first examined the transcriptional alteration of KFBCHS in the plants treated with white light. Prior to the light treatment, the 5-DAG Arabidopsis seedlings were kept in the dark for 12 h, which exhausts the intricate CHS protein or its activity according to previous reports (Schröder and Schäfer, 1980; Kaulen et al., 1986). The seedlings were then placed under white light with a fluence rate of 104 ± 5 µmol·m−2·s−1 and sampled hourly. RT-qPCR and immunoblotting with anti-AtCHS antibody revealed that both the transcripts and cellular concentration of CHS were concomitantly surged in the early period of the dark-to-light switch (Figures 6A and 6B), which is consistent with the previous observation that high-intensity light treatment induces CHS mRNA synthesis and the corresponding CHS activity (Feinbaum and Ausubel, 1988). Interestingly, within the early period of dark-to-light transition, the expression of KFBCHS was substantially decreased, presenting an opposite pattern to CHS (Figures 6A and 6B). Following the initial suppression, KFBCHS expression then significantly increased, while CHS transcripts began to decline. Correspondingly, the peak level of CHS protein sharply decreased to near detection limit after 4 h of illumination (Figures 6A and 6B). At this point, KFBCHS expression was resuppressed, approaching its basal level (Figure 6A). In contrast to the obvious fluctuation of CHS protein levels in wild-type plants in response to the light treatment, kfbchs mutants under the same treatment condition maintained a constant yet high cellular concentration of CHS (Figure 6B), suggesting the substantial role of KFBCHS in coordinately tuning cellular CHS levels in light responsiveness.
Figure 6.
KFBCHS Is Responsive to Dark-to-Light and Light-to-Dark Transitions.
For dark-to-light treatment, the 5-DAG Arabidopsis seedlings were grown under normal growth conditions as described in Methods and first transferred to the dark, then kept for 12 h, and finally switched to white light (fluence rate 104 ± 5 µmol·m−2·s−1) at 22°C for 5 h ([A] and [B]). For light-to-dark treatment, the 5-DAG seedlings under continuous white light (fluence rate 64 ± 2 µmol·m−2·s−1) at 22°C were switched to the dark for 36 h ([C] and [D]). The total RNAs and proteins were extracted from the collected samples at the indicated time points.
(A) RT-qPCR analyses of the expression levels of KFBCHS and CHS over the course of 5 h after seedlings switching from dark to light. Data represent means ± sd of three biological replicates; each replicate represents the mixed seedlings (0.1 g FW) from one Petri disk plate.
(B) Immunoblots with anti-AtCHS antibody against the total proteins from wild-type (upper) and kfbchs mutant (Salk_085384) seedlings (lower) with the same treatments as in (A). Total protein was stained with Ponceau S as the loading control.
(C) RT-qPCR analyses of the expression levels of KFBCHS and CHS over the course of 36 h after seedlings switching from light to dark. Data represent means ± sd of three biological replicates; each replicate represents the mixed seedlings (0.1 g FW) from one Petri dish plate.
(D) Immunoblots with anti-AtCHS antibody against the total proteins from wild-type and kfbchs mutant (Salk_085384) seedlings. Total protein was stained with Ponceau S as the loading control.
Conversely, when 5-DAG Arabidopsis seedlings were transferred from the continuous white light (64 ± 2 µmol·m−2·s−1) condition to the dark and were maintained for 36 h, we found that CHS expression slowly but steadily decreased from onset of the dark treatment, whereas the expression of KFBCHS showed a drastic increase within the first 6 h of darkness and then sharply decreased (Figure 6C). Correspondingly, the level of CHS proteins in the wild-type seedlings in the dark exhibited a substantial decrease when the treatment proceeded for 6 h and became hardly detectable when the treatment approached 36 h. In contrast, in the kfbchs mutant line under the same treatment, CHS level remains nearly unchanged at the first 6 h of darkness and showed only a partial decrease at 36 h (Figure 6D).
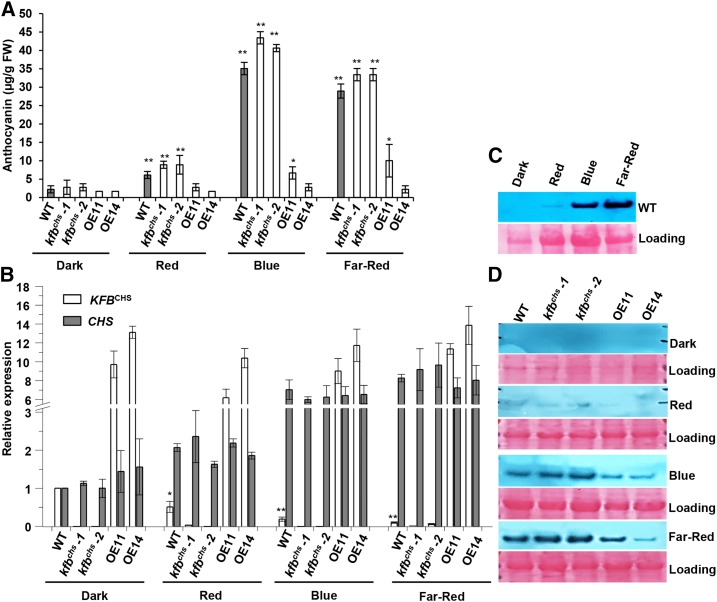
KFBCHS Responds to Different Light Signals
To further assess the roles of KFBCHS in coordinating flavonoid production of Arabidopsis seedlings during photomorphogenesis in response to different light signals, Arabidopsis seeds were illuminated with blue (36 µmol·m−2·s−1), red (40 µmol·m−2·s−1), or far-red (20 µmol·m−2·s−1) light after synchronized germination with the short exposure of red light (30 µmol·m−2·s−1) and maintained under the same light conditions during seedling growth. The 9-d-old seedlings were harvested and examined. Consistent with a previous study (Ahmad and Cashmore, 1997), the blue and far-red light treatment induced abundant production of anthocyanin, showing a >30-fold increase compared with the trace level of the seedlings grown in the dark. By contrast, the induction of anthocyanin synthesis by the illumination with red light was less pronounced, exhibiting about a 5-fold increase compared with the level of the seedlings in the dark (Figure 7A). While the blue and far-red light strongly induced CHS expression, the same treatments profoundly suppressed KFBCHS expression, resulting in 80 to ∼90% reduction of its transcripts in the wild-type seedlings under blue and far-red illumination and an ∼50% decrease with red light treatment, compared with the level in the dark-grown seedlings (Figure 7B). As a consequence of the opposite alterations of CHS and KFBCHS transcripts, the AtCHS protein was readily detected in the wild-type seedlings illuminated with either red, blue, or far-red light, in contrast to the barely detectable level of CHS in the seedlings grown in dark (Figure 7C).
Figure 7.
KFBCHS Is Responsive to Different Qualities of Light.
Seedlings of the wild type (Col-0), kfbchs mutant lines (kfbchs -1, Salk_085384; kfbchs -2, Salk_051263), and overexpression lines OE11 and OE14 were germinated as described in Methods and transferred to blue (36 µmol·m−2·s−1), red (40 µmol·m−2·s−1), or far-red (20 µmol·m−2·s−1) light for 9 d at 22°C. Dark: the seedlings remained in the total darkness for 9 d.
(A) The accumulated levels of anthocyanin in the seedlings grown under different light conditions. Data represent means ± sd of three biological replicates. The seedlings harvested from each agar plate were regarded as one biological replicate. *P < 0.05 and **P < 0.01 (Student’s t test) compared with the corresponding control in the dark.
(B) Relative expression levels of KFBCHS and CHS in the seedlings of the wild type, kfbchs mutant, and overexpression lines under dark, or red, blue, and far-red light. Each gene’s expression level in the wild type in the dark was set as “1.” Data represent ± sd of three biological replicates as the defined in (A). For clarity, statistic indicators were only presented for KFBCHS in the wild type. *P < 0.05 and **P < 0.01 (Student’s t test) compared with the sample in the dark.
(C) Immunoblot of CHS from wild-type seedlings under dark, or red, blue, and far-red light. Ten micrograms of total proteins for each sample were resolved on 12% SDS-PAGE gel and probed with anti-AtCHS antibody (upper). Total protein stained with Ponceau S serves as the loading controls (lower); note that the amount of Rubisco in the loading proteins also varied in the plants under different light conditions.
(D) Immunoblot of CHS from the seedlings of the wild type, kfbchs mutant, and overexpression lines under different qualities of light. Total protein stained with Ponceau S serves as loading controls.
Under the same light treatment, disruption of KFBCHS in general did not affect the light-induced CHS gene expression (Figure 7C) but further enhanced the amounts of CHS protein (Figure 7D) and the anthocyanin accumulation (Figure 7A). Conversely, overexpression of KFBCHS substantially depleted the light-induced accumulation of both CHS and anthocyanin. All these data together substantiate that KFBCHS is strongly responsive to different light signals and contributes to the regulation of the light-induced photomorphogenic anthocyanin accumulation.
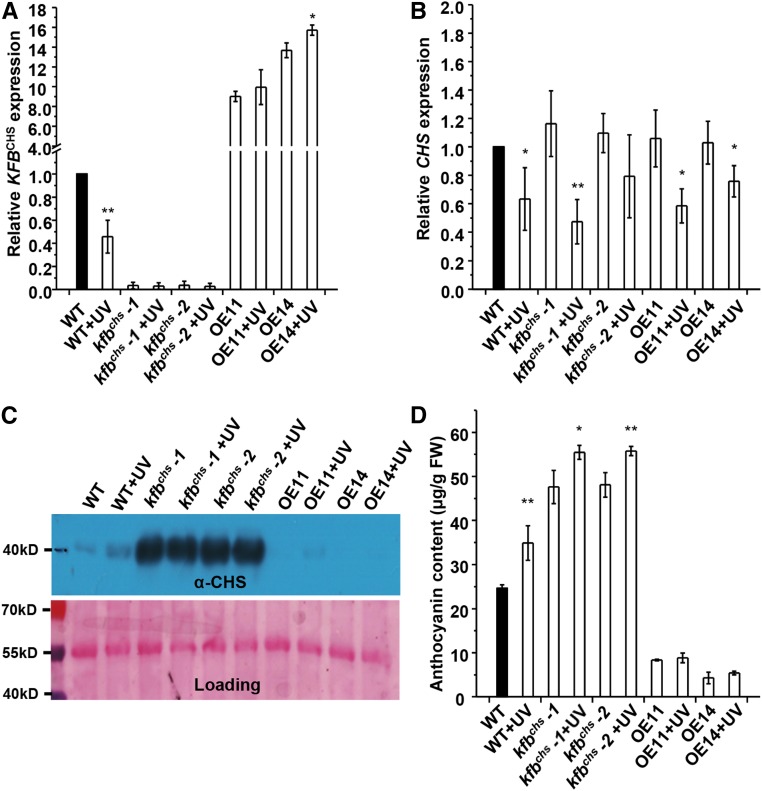
KFBCHS Responds to UV-B Stress
Hyperaccumulation of flavonoids is a primary defense response mechanism of plants to protect from UV irradiation (Li et al., 1993). To examine the potential responsiveness of KFBCHS to UV stress, 4-week-old Arabidopsis plants grown under normal light conditions (i.e., 16/8-h light/night regime, with a fluence rate of light at 104 ± 5 µmol·m−2·s−1) were transferred to the dark and supplemented with UV-B light (fluence rate 3.86 µmol·m−2·s−1) for 15 h. Compared with the control plants that were set in the dark without UV-B supplement, the exposure of wild-type plants to the UV-B resulted in >50% reduction of KFBCHS expression (Figure 8A). Although CHS transcript accumulation after such a long period of onset of UV-B irradiation showed a discernible suppression (Figure 8B), the level of CHS protein in UV-B irradiated rosette leaves was more abundant than that in the plants without UV treatment (Figure 8C). Consequently, anthocyanin content in the plants exposed to UV-B light increased ∼40% (Figure 8D). Disruption of KFBCHS resulted in higher accumulation levels of CHS and anthocyanin in kfbchs mutant lines, with or without UV irradiation. By contrast, overexpression of KFBCHS largely abolished CHS and anthocyanin accumulation in either UV-B treated or nontreated plants (Figures 8C and 8D). These data indicate that the UV stress-induced suppression of KFBCHS expression contributes to the surge of CHS cellular concentration and anthocyanin accumulation in the plants under UV-B irradiation.
Figure 8.
UV Light Irradiation Suppresses KFBCHS Expression.
The 4-week-old wild type, kfbchs mutant lines (kfbchs -1, Salk_085384; kfbchs -2, Salk_051263), and OE11 and OE14 plants (before flowering) were transferred to the dark and supplemented with UV-B light (0.2 J⋅m−2⋅s−1) from a UV-B-FS20 fluorescent lamp for 15 h. The control sets were kept in the dark without UV-B light supplement. The rosette leaves were harvested for RNA and protein extraction.
(A) and (B) Relative expression levels of KFBCHS (A) and CHS (B). Data represent means ± sd of three biological replicates and each replicate is a mixture of rosette leaves from nine individual plants. *P < 0.05 and **P < 0.01 (Student’s t test) compared with the corresponding controls without UV treatment.
(C) Immunoblot of CHS from the same treated materials as in (A) (upper). Total protein stained with Ponceau S serves as the loading control (lower).
(D) Anthocyanins accumulated in the same set of UV-treated rosette leaves as in (A). Data represent means ± sd from three biological replicates and each replicate is a mixture of rosette leaves from nine individual plants. *P < 0.05 and **P < 0.01 (Student’s t test) compared with the corresponding controls without UV treatment.
DISCUSSION
Flavonoid biosynthesis is a precisely controlled process that is activated in response to the developmental cues and different environmental challenges (Weisshaar and Jenkins, 1998; Winkel-Shirley, 2002; Taylor and Grotewold, 2005; Dao et al., 2011). The regulation of this biological process primarily occurs at the transcriptional level, with a set of transcription factors controlling the expression of pathway structural genes, including CHS (Holton and Cornish, 1995; Hartmann et al., 1998, 2005; Broun, 2005). Although an early study speculated that posttranslational mechanisms such as inactivation and/or sequential degradation of CHS might play roles in attenuating CHS activity, thus balancing flavonoid biosynthesis in the plant cells under UV irradiation (Schröder and Schäfer, 1980), the underlying molecular basis and the mechanism governing CHS modification and the pertinent biological significance remain enigmatic. Two genetic studies have observed that alteration of KFB gene expression could affect flavonoid accumulation in rice (Oryza sativa) hull furrows or muskmelon (Cucumis melo) fruit (Shao et al., 2012; Feder et al., 2015). However, the underlying mechanism by which the KFB protein functions was not yet resolved. The data in this work establish that the KFB encoded by At1g23390 in Arabidopsis functions as a negative regulator specifically controlling flavonoid biosynthesis via mediating CHS ubiquitination and subsequent degradation.
CHS Undergoes Selective Degradation via the Ubiquitin/26S Proteasome System
Although previous proteomic studies did not reveal any ubiquitinated forms of particular flavonoid biosynthetic enzymes (Saracco et al., 2009; Kim et al., 2013), our exploration showed that CHS is polyubiquitinated in planta and its stability is governed by the ubiquitin/26S proteasome system. This conclusion can be drawn from several lines of experimental evidence: First, the CHS purified from plant cells cross-reacted with antiubiquitin antibody (Figure 2A; Supplemental Figure 4), indicating that the proteins are tagged with ubiquitin epitopes; conversely, when the total ubiquitinated proteins were enriched from tobacco cells harboring an AtCHS-HA expression cassette, the CHS species among the enriched proteins were detected by anti-AtCHS antibody (Figure 2B), further confirming the ubiquitination of CHS. Moreover, based on the molecular sizes of the detected immunosignals by anti-AtCHS antibody, the ubiquitinated CHS most likely consists of both mono- and polyubiquitinated species, since besides the primary immunosignal of ∼51 kD, the bands/smear of higher molecular mass were cross-detected by antiubiquitin probe in the immunoprecipitated CHS proteins and by anti-AtCHS probe in the enriched ubiquitinated proteins (Figures 2A and 2B; Supplemental Figure 4). Furthermore, both in vitro and in vivo measurements reveal that MG132, a specific inhibitor of 26S proteasome that forms a covalent hemiacetal adduct with proteasome and inhibits the chymotrypsin-like peptidase activity (Lyzenga et al., 2012), substantially slowed down the turnover of either the endogenous or the overexpressed CHS proteins and enhanced the abundance of ubiquitinated CHS species in the extracted total proteins (Figures 2A to 2C). These data suggest that the stability of flavonoid branch enzyme CHS, like those PAL isoforms in the general phenylpropanoid pathway, is controlled by ubiquitin/26S proteasome system.
It is interesting to point out that in this study on CHS and in our previous analyses of PAL isoforms (Zhang et al., 2013, 2015) the detected dominant forms of the overexpressed proteins in tobacco leaf cells differ from the primary forms of the corresponding endogenous proteins existing in Arabidopsis (Figure 2; Supplemental Figure 6). When Arabidopsis CHS and PAL were transiently expressed in tobacco leaf cells, the predominant protein species detected corresponded to their monoubiquitinated forms, whereas the monitored endogenous CHS (Figure 2; Supplemental Figure 6) or PAL (Zhang et al., 2013, 2015) from Arabidopsis appeared with a dominant form coincident with their predicted native molecular sizes, i.e., nonubiquitinated. Although the exact reasons for such a difference remain to be further determined, presumably, overexpression of these key biosynthetic enzymes may tend to provoke or intensify their intrinsic ubiquitination modification and/or the subsequent 26S-proteasome-mediated turnover mechanism. On the other hand, this unique feature of tobacco cells maintaining highly abundant ubiquitinated protein species in fact offers an effective experimental system to detect the ubiquitination modification of the target proteins.
KFBCHS Specifically Mediates CHS Degradation
As one of the large subfamilies of F-box proteins and as the critical structural component of SCF-type E3 ubiquitin-protein ligase complex, KFBs are responsible for recruiting the target proteins into the ligase complex for ubiquitination, thus marking them for subsequent degradation (Schumann et al., 2011). Although the Arabidopsis genome codes for more than 100 KFBs, only a small set of these have been functionally characterized so far. These include ATTENUATED FAR-RED RESPONSE, a positive regulator of phytochrome A-mediated light signaling (Harmon and Kay, 2003); ZEITLUPE, FLAVIN BINDING KELCH-REPEAT F-BOX1, and LOV KELCH PROTEIN2, all involved in photomorphogenesis and regulation of the circadian clock (Somers et al., 2004; Yasuhara et al., 2004; Imaizumi et al., 2005); and KFBPALs (i.e., KFB01, 20, 39, and 50), responsible for the modification of PAL isozymes and as negative regulators controlling general phenylpropanoid synthesis (Zhang et al., 2013, 2015). In this study, among five KFB homologs that we tested pairwise with 14 monolignol and flavonoid biosynthetic enzymes in Y2H assays, the KFBCHS encoded by At1g23390 showed a specific interaction with CHS. Conversely, CHS only interacted with KFBCHS among the five tested KFBs (Figure 1B; Supplemental Figure 2). Such interaction can be validated in planta (Figure 1C). These in vitro and in vivo results suggest that KFBCHS has structurally evolved for specifying CHS modification. Furthermore, coexpression of KFBCHS and CHS in a transient expression system substantially reduces or even totally abolishes CHS protein accumulation (Figure 2D; Supplemental Figure 7). Such an accelerated turnover is not observed for other flavonoid biosynthetic enzymes when their coding sequences were coexpressed with KFBCHS (Figure 2D). Evidently, alteration of KFBCHS expression either via gene disruption or overexpression reciprocally alters the cellular concentration of CHS and/or the accumulation level of flavonoid metabolites (Figures 3 and 6 to 8). These data demonstrate that KFBCHS is a proteolytic mediator specifying the degradation of CHS, the rate-limiting enzyme in flavonoid pathway. As a conserved structural feature of the entire set of F-box proteins, KFBs rely on their C-terminal protein-protein interaction domains to determine their substrate specificity (del Pozo and Estelle, 2000; Lechner et al., 2006). In the established KFB phylogeny, KFBCHS and KFBPALs are clustered within a same clade sharing ∼31 to ∼36% overall sequence similarity. However, their C-terminal domains are quite different, where, as the exception of KFB proteins, KFBCHS only contains one predicted kelch motif, different from KFBPALs. Consistent with this, our analyses reveal that KFBCHS has little propensity to interact with other phenylpropanoid pathway enzymes excepting for CHS (Figure 1; Supplemental Figure 2); vice versa, KFBPALs show no or only very weak physical interactions with CHS in the Y2H assay (Zhang et al., 2015). These data suggest that those KFB members have evolved with high specificity for selectively recruiting the target proteins. On the other hand, it appears that KFBCHS and KFBPALs, the evolutionarily close homologs, serendipitously interact with two sets of metabolically relevant but structurally and functionally distinct key enzymes. It will be intriguing to further explore the sequence and structural features of KFBCHS and KFBPALs, as well as their corresponding substrate targets CHS and PAL to understand the molecular bases governing their substrate specificity and the evolutionary clues for their biological functions.
KFBCHS Integrates Transcriptional Regulation with Posttranslational Modification of CHS in Light and Stress Responsiveness
The ubiquitin/26S proteasome-mediated selective protein degradation allows living organisms to better adapt to changing environments or to effectively meet their developmental needs by providing fast yet precise responses to the intracellular signals (Smalle and Vierstra, 2004). The rapid accumulation of flavonoids with the onset of environmental challenges is paradigmatic of active defense and adaption mechanisms of plants for their terrestrial life. Although early studies discovered that the inducible synthesis of CHS upon the onset of elicitation is the result of de novo transcription of CHS (Ryder et al., 1984; Feinbaum and Ausubel, 1988), this work reveals that KFBCHS, the gene encoding a component of SCF-type E3 ligase mediating CHS ubiquitination and degradation, is substantially responsive to the light and/or stress treatments. Its transcripts, along with CHS expression, displayed dynamic yet coordinated alterations in the plants undergoing the transition from dark to light or, vice versa, light to dark, which conversely correlates with the alterations of cellular concentration of CHS and/or flavonoid accumulation (Figure 6). These data strongly point out that KFBCHS as a proteolytic mediator is an indispensible component in the regulation of CHS and flavonoid biosynthetic activity, which works together with the transcriptional control of CHS gene expression to coordinately balance the homeostasis of cellular concentration of CHS and the production of flavonoids upon environmental stimulation.
In Arabidopsis and many other species, anthocyanin production is a feature of light-grown plants for their photomorphogenesis, and in the dark-grown plants, the synthesis does not occur (Taylor and Briggs, 1990; Kubasek et al., 1992). The involvement of KFBCHS in regulating light-induced flavonoid synthesis is perceived from its substantial alteration in gene expression during the early period of the switch of Arabidopsis seedlings from dark to light or from light to dark (Figure 6). Furthermore, the light responsiveness of KFBCHS is unambiguously evidenced by its profound transcriptional suppression in response to the blue, red, and far-red light signals (Figure 7). Consistent with previous studies (Beggs et al., 1987; Ahmad and Cashmore, 1997), the blue and far-red illumination of Arabidopsis seedlings strongly induced both CHS and anthocyanin accumulation (Figure 7); by contrast, KFBCHS expression was severely suppressed in response to those light signals (Figure 7). This reverse responsive behavior suggests that KFBCHS is capable of directly or indirectly perceiving different light signals and acts as a repressor coordinately regulating photomorphogenic production of anthocyanin pigments in Arabidopsis developing seedlings. In far-red and visible light-induced photomorphogenic production of anthocyanin, COP1/SPA ubiquitin ligase is a central repressor that targets photomorphogenesis, promoting transcription factors including HY5 for 26S proteasome-mediated degradation. The function of COP1/SPA is modulated primarily via the physical interaction with light-activated photoreceptors and their nucleocytoplasmic translocation, which subsequently affects the stability of HY5 and R2R3-MYB transcription factors, thus activating a series of flavonoid/anthocyanin biosynthetic genes (Maier et al., 2013; Maier and Hoecker, 2015). The observed light responsiveness of KFBCHS suggests that KFBCHS is an additional proteolytic regulator recruited in seedling photomorphogenesis for more specifically fine-tuning a flavonoid biosynthetic enzyme and overall pathway activity. Since KFBCHS exhibits profound transcriptional responses to the different light signals, it will be interesting to further determine whether KFBCHS positions in or crosstalks with the COP1-mediated central light signal transduction cascade.
Besides the responsiveness to different light signals, KFBCHS is also suppressed by damaging UV-B light (Figure 8). An early study discovered that the damaging UV-B irradiation of plant cells could trigger significant accumulation of CHS transcripts in just a few hours of the onset of treatment then the overaccumulated transcripts vanished after 12 h of irradiation (Chappell and Hahlbrock, 1984). Consistent with that earlier study, upon the prolonged exposure of 4-week-old Arabidopsis plants to UV-B, the CHS transcripts decreased compared with those in the nontreated control plants (Figure 8); presumably, at the sampling time point, CHS has already passed its peak expression and approached the basal level. At this point, KFBCHS expression showed significant suppression (Figure 8), suggesting its responsiveness to UV-B stress. Evidently such suppression of KFBCHS serves as a crucial regulatory mechanism to maintain the high level of CHS protein and enhanced synthesis of UV-absorbing flavonoids, particularly under the situation of decay in CHS transcripts after prolonged UV-B exposure. The exact reason for decreasing CHS gene expression in the plants with overnight UV exposure is currently unclear; predictably, this could be due to the severe tissue damage thus the loss of a portion of living cells with the prolonged UV treatment. Under such circumstances, suppressing CHS turnover and thus increasing its cellular concentration becomes a crucial means for plants to maintain their active accumulation of UV resistance metabolites. These data exemplify the coordination of both transcriptional and posttranslational regulatory mechanisms of plant cells to fine-tune their biosynthetic processes of specialized metabolites to sustain the cells’ biological activities in response to environmental inputs. Although UV-B light is only a minor component of sunlight, it serves as an important environmental signal that promotes UV acclimation and survival of plants in sunlight. High doses of UV-B radiation act as damaging stimulus triggering distress symptom and inducing stress-related physiological processes, including generation of reactive oxygen species and accumulation of photoprotectant phenolics (Hideg et al., 2013; Jenkins, 2009). This experiment with high-fluence and long-duration UV-B irradiation of 4-week-old Arabidopsis plants primarily measures the stress response of KFBCHS. However, low-intensity UV-B light can act as an informational signal that is perceived and mediated via UVR8-COP1-HY5/HYH signaling pathway in Arabidopsis to regulate genes involved in photomorphogenetic UV-B responses and metabolic accumulation (Heijde and Ulm, 2012). The strong responsiveness of KFBCHS to the far-red and visible light signals and to the damaging UV-B light makes it interesting to further examine whether KFBCHS also connects to the UVR8-mediated photomorphogenetic UV-B response.
Furthermore, KFBCHS exhibits discernible developmental or tissue-preferential expression patterns. Its transcripts are abundant in the early developing tissues, such as the young stem nodes and the early developing siliques, but low in the mature tissues. Its expression patterns in general are inversely related to the developmental accumulation of flavonoids in the corresponding tissues (Figure 5). Such negative correlation hints that KFBCHS-mediated ubiquitination and degradation of CHS is integral to the regulatory cascade developmentally programming flavonoid biosynthesis.
Overall, our study indicates that KFBCHS functions as a proteolytic mediator specific to CHS degradation. Through its transcriptional responses to the developmental cues and environmental challenges, KFBCHS serves as an essential regulatory integrator connecting transcriptional regulation with posttranslational modification to coordinately tune cellular concentrations of CHS, thus controlling flavonoid production. This complex pattern of regulation is essential for plants to achieve their most effective and efficient physiological performance.
METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis thaliana in this study was Columbia-0 (Col-0). The T-DNA insertion lines of KFBCHS used in the study were Salk-065718, Salk_085384, Salk_051247, and Salk_051263. The tt4-3 mutant line was CS66119. All were obtained from the ABRC and genotypically confirmed with the primers listed in Supplemental Table 1.
To generate KFBCHS overexpression lines, a full-length cDNA of KFBCHS was subcloned into the binary vector pMDC32 (Curtis and Grossniklaus, 2003). The construct containing the 35S:KFBCHS cassette was transformed into Arabidopsis wild type (Col-0) using the floral dipping method (Clough and Bent, 1998). The obtained transgenic or mutant plants were maintained in a FLEX series growth chamber (BioChambers) that has combined T5HO fluorescent lamps (Sylvania FP54/841/HO/ECO, Pentron 4100K 54 W 20,906 Hg) and halogen lamps (Topaz 40 W, 130 V A19 Rough Service incandescent bulb) with light intensity of 104 ± 5 µmol·m−2·s−1, 16-h-light/8-h-night regime, at 22 ± 2°C.
Phylogenetic Analysis
The peptide sequences of KFBs were retrieved from TAIR based on previous publications (Xu et al., 2009; Schumann et al., 2011). The full-length amino acid sequences were aligned with the ClustalW program, and phylogenetic analysis was conducted using MEGA6 (Molecular Evolutionary Genetics Analysis Version 6.0) software with the neighbor-joining method. The bootstrap values at branching points were calculated based on 1000 replications.
Pairwise Y2H Assay
cDNAs encoding Arabidopsis flavonoid biosynthetic enzymes CHS, ANS, and F3′H and lignin biosynthetic enzymes C4H, 4CL1, HCT, C3′H, CCOMT1, CCR1, CAD4, F5H, COMT1, and LAC4 were isolated by RT-PCR using the corresponding primers described in Supplemental Table 1 and cloned into the Gateway-compatible shuttle vector PCR8 (Invitrogen), then subcloned into the Gateway-compatible pGBKT7 vector (Clontech) to generate the expression cassettes for the bait-DNA binding domain fusion proteins. Similarly, cDNAs of AT1G23390 (KFBCHS), AT1G30090, AT2G24540, AT3G61590, and AT4G03030, which encode putative KFB proteins, were isolated by RT-PCR using the primers described in Supplemental Table 1 and cloned into the Gateway-compatible pGADT7 vector (Clontech) to generate activation domain fusion proteins. Yeast (AH109; Clontech) transformation and pairwise verification were performed according to the manufacturer’s instructions (Clontech).
BiFC Assay
The BiFC assay was done according to the previously described procedure (Zhang et al., 2013, 2015). Briefly, the truncated KFBCHS (with 156 nucleotides after the ATG, which was removed) was subcloned into pEDST-GWVYNE vector (Gehl et al., 2009) to generate KFBCHS-nYFP (YFP N terminus fragment) fusion protein. A full-length cDNA of CHS was subcloned into the p(MAS)DEST-GWSCYCE (R) vector (Gehl et al., 2009) to generate the CHS-CFPc (CFP C terminus fragment) fusion protein. For CHS-GFP control, the full-length CHS cDNA was subcloned into the pGWB405 vector (Nakagawa et al., 2007) to generate the C-terminal GFP fusion protein. The transient coexpression of CHS-CFPc with F^KFBCHS-nYFP in tobacco (Nicotiana tabacum) leaves was performed via Agrobacterium tumefaciens infiltration as previously described (Zhang et al., 2013, 2015), and observations were conducted after a 3-d infiltration using a Leica SP5 TCS inverted confocal microscope (Leica Microsystems) equipped with an argon ion laser. Samples were excited at 488 nm, and the fluorescence signal was collected between 500 and 530 nm. The chlorophyll signal was collected between 653 and 731 nm. Following the acquisition, brightness and contrast were adjusted using the LCS software (identical parameters were applied to control and fluorescence images).
Enrichment of Ubiquitinated Proteins and Ubiquitination Detection
Arabidopsis CHS was subcloned into the Gateway-compatible binary vector pGWB414 (Nakagawa et al., 2007) (for expressing the CHS-HA fusion protein). Agrobacterium strain GV3101 carrying pGWB414-CHS was infiltrated into tobacco leaves following the protocol described (Sparkes et al., 2006).
For detection of ubiquitination of CHS-HA, the procedure was conducted as previously described (Zhang et al., 2013, 2015). Briefly, total soluble proteins from tobacco leaves expressing CHS-HA were extracted with buffer (2 mL/g fresh weight [FW] leaf powder) containing 25 mM Tris-HCl (pH 7.5), 10 mM NaCl, 10 mM MgCl2, 4 mM PMSF, 14 mM β-mercaptoethanol, 10 mM ATP, and with or without 50 µM MG132 and inoculated with 50 μL of Red Anti-HA Affinity Gel Beads (Sigma-Aldrich; cat. no. E6779) at 4°C for 6 h to precipitate the expressed CHS-HA fusion proteins. The bound proteins were eluted from the beads by adding 2× SDS sample buffer and incubated at 42°C for 10 min. The denatured proteins were separated on 12% SDS-PAGE gel. Immunoblotting was performed using monoclonal antiubiquitin (Agrisera; cat. no. ABIN334561), anti-HA antibodies (Covance; cat. no. MMS-101R), and the anti-Arabidopsis CHS antibody that was developed against with a 20-amino acid peptide from the N-terminal first 50 amino acids of Arabidopsis CHS protein (Santa Cruz; cat. no. sc12620).
To enrich the ubiquitinated proteins from plant cells, the first two leaves of 4-week-old tobacco plants were infiltrated with pGWB414-CHS for transiently expressing CHS-HA fusion protein. The infiltrated leaves were left for 3 d, and each half of individual leaf was infiltrated again with buffer containing with or without 40 μM MG132, and then left for additional 8 h before sampling. The total proteins were extracted from one gram of harvested leaf samples in 2 mL of extraction buffer containing 50 mM Tris-HCl buffer (pH 7.5), 2 mM EDTA, 150 mM NaCl, 10% glycerol, 5 mM DTT, 0.25% Triton X-100, and 1× complete protease inhibitor cocktail. The extracts were then mixed with 40 μL aliquot of UBI-QAPTURE-Q matrix (Enzo; catalog no. BML-UW8995) and kept mixing at 4°C for 2 h. The matrix was collected and washed five times with immunoprecipitation buffer containing 25 mM Tris-HCl (pH 7.5), 1 mM EDTA, 150 mM NaCl, 0.15% Nonidet P-40, and 1× protease inhibitor cocktail. The protein-bound beads were mixed with 50 μL of 2× SDS-PAGE sample buffer and boiled for 5 min. Ten microliters of each sample was separated on a 12% SDS-PAGE gel. Antiubiquitin, anti-HA, and anti-AtCHS antibodies were used for the immunoblots.
In Planta Degradation Assay
Arabidopsis CHS, DFR, F3′H, and ANS genes were subcloned to the Gateway-compatible binary vector pGWB414 (Nakagawa et al., 2007) from their corresponding shuttle vector PCR8 (Invitrogen) to express CHS-HA, DFR-HA, F3′H-HA, and ANS-HA fusion proteins (or to pGWB415 for expressing HA-CHS). Agrobacterium strain GV3101 carrying the binary gene expression vector was mixed with an equal volume of (OD600 = 0.7) Agrobacterium strain carrying pGWB402-KFBCHS or the empty pGWB402 vector and was infiltrated into tobacco leaves following the protocol described (Sparkes et al., 2006). At 3 d after infiltration, the total proteins were extracted with buffers containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 0.1% Nonidet P-40, 14 mM 2-mercaptoethanol, 1× protease inhibitor cocktail, and 40 µM MG132. The ratio of fresh weight to extraction buffer is 1 g/mL. The proteins were examined by immunoblot with anti-HA antibody.
Cell-Free Degradation Assay
Cell-free degradation assay for Arabidopsis endogenous CHS protein was performed according to the procedure described by Lyzenga et al. (2012). Briefly, 5-DAG Col-0 Arabidopsis seedlings were flash frozen in liquid nitrogen and ground to fine powder. One gram (fresh weight) of tissues was extracted with 1 mL of the above-mentioned buffer on ice for the whole procedure. Total soluble proteins at a concentration of 1 mg/mL were divided into two portions and then a final concentration of 50 µM MG132 or DMSO control was added to the corresponding portions. The degradation assays were performed in a water bath at 37°C for 20 min, and 10 μL of the total protein was sampled every 5 min, and then resolved by SDS-PAGE gel. Immunoblotting was performed using anti-Arabidopsis CHS antibody.
Preparation of Developmental Samples and Quantitative Real-Time PCR
To collect samples at different developmental stages, the inflorescence stems of 6-week-old Arabidopsis (Col-0) in the growth chamber were numbered and sampled sequentially from bottom to top for each internode. The internodes from nine individual plants were pooled together, representing one biological replicate. The entire rosette leaves and cauline leaves from the same nine individual plants were respectively collected and pooled as one biological sample. For siliques, every fifth silique in the inflorescence from bottom to top was collected and grouped together to represent one developmental stage; the mixture of 45 siliques from nine individual plants was regarded as one biological replicate. The flowers and unopened buds on the top inflorescence were mixed as one sample. The pooled samples from five plants were considered as one replicate.
For RT-qPCR analysis of gene expression level, the total RNAs were isolated from the tissues of 5-DAG seedlings and the collected rosette leaves, cauline leaves, flowers/buds, siliques, and stem internodes from the bottom to top of 6-week-old plants using TRIzol reagent (Molecular Research Center) and subsequently cleaned up with Qiagen RNA cleanup kit. All the RNAs were treated with DNase I (New England Biolabs) before the use for first-strand cDNA synthesis. First-strand cDNAs were synthesized using Moloney Murine Leukemia Virus reverse transcriptase (Promega). RT-qPCR was performed with SsoAdvanced SYBR Supermix (Bio-Rad) according to the manufacturer’s instructions using primers XB_KFBCHSRT_F and XB_KFBCHSRT_R for KFBCHS, XB_CHS_RTF and XB_CHS_RTR for CHS, and XB_F3′H_RTF and XB_F3′H_RTR for F3′H as listed in Supplemental Table 1. The cycle threshold (Ct) value was calculated by the CFXManager Software v3.0 (Bio-Rad) for each gene. Gene expression was analyzed using the comparative Ct method (i.e., 2−ΔΔCt method) (Livak and Schmittgen, 2001) against the reference gene Arabidopsis UBIQUITIN10 (At4g05320).
Light and Dark Treatments
For light treatment, Arabidopsis Col-0 and Salk_085384 seedlings seeds were surface-sterilized with 70% (v/v) ethanol for 20 min and washed three times with 0.1% Triton X-100 in water. The sterilized seeds were sown on 0.5× Murashige and Skoog medium containing 0.8% agar and 2% sucrose and the agar plates were held at 4°C for 2 d in dark. Plates were then kept in an environmentally controlled growth chamber under growth conditions described above. At 5 DAG, the plates containing seedlings were wrapped with aluminum foil and kept in the growth chamber at 22 ± 2°C for 12 h, then the foil was removed and the seedlings were exposed to the white fluorescent light (fluence rate 104 ± 5 µmol·m−2·s−1) for 5 h. The treated seedlings were sampled every hour. The pooled seedlings (with 0.1 g FW) from one agar plate were treated as one biological replicate. The samples were immediately frozen with liquid N2 and then subjected to RNA and protein extraction as described above. For dark treatment, 5-DAG seedlings grown under continuous light (64 ± 2 µmol·m−2·s−1) at 22 ± 2°C were wrapped with aluminum foil for the time period of 0, 6, 12, and 36 h. The seedlings (with a fresh weight of 0.1 g) from each plate were collected as one biological replicate and flash-frozen in liquid N2 at each time point and used for total RNA and proteins extractions.
For red, blue, and far-red light treatments, the procedure was essentially followed as described (Ahmad and Cashmore, 1997). Briefly, Arabidopsis seeds were surface-sterilized and sown on the solid 0.8% 0.5× Murashige and Skoog medium supplemented with 2% sucrose. The seeds were cold treated at 4°C for 2 d in the dark, then were illuminated with red light (660 nm, 30 ± 5 µmol·m−2·s−1) (from LED, Osram Olson SLL 80 LHCP7P, semicon-ThinInGaAIP) for 15 min to induce germination, followed by an additional 24 h at dark. The plates were then transferred to the blue (470 nm, 36 ± 5 µmol·m−2·s−1) (LED, Osram Olson SLL 80 LBCP7P, semicon-ThinGaN), red (660 nm, 40 ± 5 µmol·m−2·s−1) (from LED, Osram Olson SLL 80 LHCP7P, semicon-ThinInGaAIP), and far-red (730 nm, 20 ± 5 µmol·m−2·s−1) (from LED, Cree XLamp XP-E FARL100000601, semicon-AlGaAs) continuous light for 9 d at 22°C in environmentally controlled growth cabinets. Spectral determinations and photon fluence rates were measured by StellarNet spectroradiometer and visualized with SpectraWiz software (Stellar Net). The seedlings from each plate were collected and weighed in low diffuse white light conditions as one biological repeat and frozen in liquid nitrogen. The frozen samples were used for RNA, protein, and anthocyanin extraction.
UV Light Stress
For UV-B treatment, the 4-week-old soil-grown Col-0, Salk_085384, Salk_051263, and two KFBCHS overexpression plants (before flowering) grown in the normal conditions as described above were transferred to the dark and supplemented with the UV-B light from a UV-B-FS20 fluorescent lamp (Westinghouse) for 15 h with irradiation intensity of 3.86 µmol·m−2·s−1, measured with a Molectron PR200 pyroelectric radiometer. The plants kept in darkness for the same period of time but without supplement of UV-B served as the control. RNAs and proteins were extracted according to the procedures described above. Accordingly, 1 µg of total RNA was used for first-strand cDNA synthesis and followed by RT-qPCR analysis as described above, and 10 μL of total protein with the concentration of 1 mg/mL was resolved by SDS-PAGE gel. Immunoblotting was performed using anti-Arabidopsis CHS antibody.
Phenolic Metabolite Measurement
Soluble phenolic extraction, HPLC-based analysis, anthocyanin measurement, and seed PA measurements were performed as described (Zhang et al., 2013, 2015).
Statistical Analysis
Statistical analysis was performed using Student’s t tests (one-tail distribution and two samples with unequal variances). Statistically significant differences were defined as P < 0.05. Values in graphs were presented as means with sd of at least three biological or technical repeats. Microsoft Excel 2011 was used for data management, statistical analysis, and graph generation.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL database under the following accession numbers: KFBCHS (At1g23390), CHS (At5g13930), DFR (At5g42800), F3′H (At5g07990), and ANS (At4g22880).
Supplemental Data
Supplemental Figure 1. Phylogeny of Arabidopsis Kelch (Repeat) Domain Containing F-Box Protein Family.
Supplemental Figure 2. Pairwise Yeast Two-Hybrid Screening of Arabidopsis Lignin Biosynthesis Enzymes for Potential Interaction with Candidate KFB Proteins.
Supplemental Figure 3. The Predicted F-Box and Kelch Repeat Domains of the KFBCHS and KFBPALs.
Supplemental Figure 4. Detection of Ubiquitinated CHS-HA Proteins.
Supplemental Figure 5. Enrichment of Ubiquitinated Proteins and Visualization of Higher Molecular Mass Ubiquitinated CHS Protein Species.
Supplemental Figure 6. Immunodetection of Arabidopsis Endogenous CHS Protein.
Supplemental Figure 7. Immunoblot of Coexpressed AtCHS-HA or HA-AtCHS Fusion Protein with KFBCHS in Tobacco Leaves.
Supplemental Figure 8. Accumulation of Phenolics in KFBCHS Overexpression Lines.
Supplemental Table 1. Primers Used in This Study.
Supplemental File 1. Text File of Alignment Corresponding to the Phylogenetic Tree in Figure 1.
Supplemental File 2. Text File of Alignment Corresponding to the Phylogenetic Tree in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Mingyu Gou for the help with immunoprecipitation and Paula Bennett for UV-B light source determination. This work was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-SC0012704, specifically through the Physical Biosciences program of the Chemical Sciences, Geosciences, and Biosciences Division and by the National Science Foundation through Grant MCB-1051675 (to C.-J.L.). The use of the confocal microscope in the Center for Nanosciences was supported by the Office of Basic Energy Sciences, U.S. Department of Energy, under Contract DEAC0298CH10886.
AUTHOR CONTRIBUTIONS
X.Z. and C.-J.L. conceived the original research plan and designed the experiments. X.Z. performed the experiments. C.-J.L. supervised the experiments. C.A. and T.A.C. prepared different wavelength light treatment materials. X.Z. and C.-J.L. analyzed data and wrote the article.
Glossary
- Y2H
yeast two-hybrid
- ANS
anthocyanin synthase
- BiFC
bimolecular fluorescence complementation
- DAG
days after germination
- PA
proanthocyanidin
- FW
fresh weight
Footnotes
Articles can be viewed without a subscription.
References
- Ahmad M., Cashmore A.R. (1997). The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J. 11: 421–427. [DOI] [PubMed] [Google Scholar]
- Beggs C.J., Kuhn K., Böcker R., Wellmann E. (1987). Phytochrome-induced flavonoid biosynthesis in mustard (Sinapis alba L.) cotyledons. Enzymic control and differential regulation of anthocyanin and quercetin formation. Planta 172: 121–126. [DOI] [PubMed] [Google Scholar]
- Bell J.N., Ryder T.B., Wingate V.P., Bailey J.A., Lamb C.J. (1986). Differential accumulation of plant defense gene transcripts in a compatible and an incompatible plant-pathogen interaction. Mol. Cell. Biol. 6: 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J.N., Dixon R.A., Bailey J.A., Rowell P.M., Lamb C.J. (1984). Differential induction of chalcone synthase mRNA activity at the onset of phytoalexin accumulation in compatible and incompatible plant-pathogen interactions. Proc. Natl. Acad. Sci. USA 81: 3384–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Doolittle R.F. (1994). Drosophila kelch motif is derived from a common enzyme fold. J. Mol. Biol. 236: 1277–1282. [DOI] [PubMed] [Google Scholar]
- Broun P. (2005). Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8: 272–279. [DOI] [PubMed] [Google Scholar]
- Brown B.A., Cloix C., Jiang G.H., Kaiserli E., Herzyk P., Kliebenstein D.J., Jenkins G.I. (2005). A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA 102: 18225–18230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J., Hahlbrock K. (1984). Transcription of plant defence genes in response to UV light or fungal elicitor. Nature 311: 76–78. [Google Scholar]
- Chen M.-X., Zheng S.-X., Yang Y.-N., Xu C., Liu J.-S., Yang W.-D., Chye M.-L., Li H.-Y. (2014). Strong seed-specific protein expression from the Vigna radiata storage protein 8SGα promoter in transgenic Arabidopsis seeds. J. Biotechnol. 174: 49–56. [DOI] [PubMed] [Google Scholar]
- Christie J.M., Jenkins G.I. (1996). Distinct UV-B and UV-A/blue light signal transduction pathways induce chalcone synthase gene expression in Arabidopsis cells. Plant Cell 8: 1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao T.T.H., Linthorst H.J.M., Verpoorte R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo J.C., Estelle M. (2000). F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol. Biol. 44: 123–128. [DOI] [PubMed] [Google Scholar]
- Dutt M., Dhekney S.A., Soriano L., Kandel R., Grosser J.W. (2014). Temporal and spatial control of gene expression in horticultural crops. Hortic. Res. 1: 14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory J.J., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28: 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder A., et al. (2015). A Kelch domain-containing F-box coding gene negatively regulates flavonoid accumulation in muskmelon. Plant Physiol. 169: 1714–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum R.L., Ausubel F.M. (1988). Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol. Cell. Biol. 8: 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.M., Downes B.P., Shiu S.H., Durski A.M., Vierstra R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C., Waadt R., Kudla J., Mendel R.-R., Hänsch R. (2009). New Gateway vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2: 1051–1058. [DOI] [PubMed] [Google Scholar]
- Grotewold E. (2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57: 761–780. [DOI] [PubMed] [Google Scholar]
- Harmon F.G., Kay S.A. (2003). The F box protein AFR is a positive regulator of phytochrome A-mediated light signaling. Curr. Biol. 13: 2091–2096. [DOI] [PubMed] [Google Scholar]
- Harrison M.J., Lawton M.A., Lamb C.J., Dixon R.A. (1991). Characterization of a nuclear protein that binds to three elements within the silencer region of a bean chalcone synthase gene promoter. Proc. Natl. Acad. Sci. USA 88: 2515–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U., Sagasser M., Mehrtens F., Stracke R., Weisshaar B. (2005). Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57: 155–171. [DOI] [PubMed] [Google Scholar]
- Hartmann U., Valentine W.J., Christie J.M., Hays J., Jenkins G.I., Weisshaar B. (1998). Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol. Biol. 36: 741–754. [DOI] [PubMed] [Google Scholar]
- Heijde M., Ulm R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17: 230–237. [DOI] [PubMed] [Google Scholar]
- Hideg E., Jansen M.A., Strid A. (2013). UV-B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends Plant Sci. 18: 107–115. [DOI] [PubMed] [Google Scholar]
- Holton T.A., Cornish E.C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrazdina G., Jensen R.A. (1992). Spatial organization of enzymes in plant metabolic pathways. Annu. Rev. Plant Biol. 43: 241–267. [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297. [DOI] [PubMed] [Google Scholar]
- Jenkins G.I. (2009). Signal transduction in responses to UV-B radiation. Annu. Rev. Plant Biol. 60: 407–431. [DOI] [PubMed] [Google Scholar]
- Kaulen H., Schell J., Kreuzaler F. (1986). Light-induced expression of the chimeric chalcone synthase-NPTII gene in tobacco cells. EMBO J. 5: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.Y., Scalf M., Smith L.M., Vierstra R.D. (2013). Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubasek W.L., Shirley B.W., McKillop A., Goodman H.M., Briggs W., Ausubel F.M. (1992). Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4: 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593. [DOI] [PubMed] [Google Scholar]
- Lechner E., Achard P., Vansiri A., Potuschak T., Genschik P. (2006). F-box proteins everywhere. Curr. Opin. Plant Biol. 9: 631–638. [DOI] [PubMed] [Google Scholar]
- Li J., Ou-Lee T.M., Raba R., Amundson R.G., Last R.L. (1993). Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Loake G.J., Faktor O., Lamb C.J., Dixon R.A. (1992). Combination of H-box (CCTACC(N7)CT) and G-box (CACGTG)cis-elements is necessary for fedforward stimulation of a chalcone synthase promoter by the phenylpropanoid pathway intermediate p-coumaric acid. Proc. Natl. Acad. Sci. USA 89: 9230–9234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga W.J., Booth J.K., Stone S.L. (2012). The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J. 71: 23–34. [DOI] [PubMed] [Google Scholar]
- Maier A., Hoecker U. (2015). COP1/SPA ubiquitin ligase complexes repress anthocyanin accumulation under low light and high light conditions. Plant Signal. Behav. 10: e970440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A., Schrader A., Kokkelink L., Falke C., Welter B., Iniesto E., Rubio V., Uhrig J.F., Hülskamp M., Hoecker U. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74: 638–651. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Osuna D., Usadel B., Morcuende R., Gibon Y., Bläsing O.E., Höhne M., Günter M., Kamlage B., Trethewey R., Scheible W.R., Stitt M. (2007). Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 49: 463–491. [DOI] [PubMed] [Google Scholar]
- Ryder T.B., Cramer C.L., Bell J.N., Robbins M.P., Dixon R.A., Lamb C.J. (1984). Elicitor rapidly induces chalcone synthase mRNA in Phaseolus vulgaris cells at the onset of the phytoalexin defense response. Proc. Natl. Acad. Sci. USA 81: 5724–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Yonekura-Sakakibara K., Nakabayashi R., Higashi Y., Yamazaki M., Tohge T., Fernie A.R. (2013). The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol. Biochem. 72: 21–34. [DOI] [PubMed] [Google Scholar]
- Saracco S.A., Hansson M., Scalf M., Walker J.M., Smith L.M., Vierstra R.D. (2009). Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 59: 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky D., Winkel-Shirley B. (2001). Localization of flavonoid enzymes in Arabidopsis roots. Plant J. 27: 37–48. [DOI] [PubMed] [Google Scholar]
- Schmid J., Doerner P.W., Clouse S.D., Dixon R.A., Lamb C.J. (1990). Developmental and environmental regulation of a bean chalcone synthase promoter in transgenic tobacco. Plant Cell 2: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder J., Schäfer E. (1980). Radioiodinated antibodies, a tool in studies on the presence and role of inactive enzyme forms: regulation of chalcone synthase in parsley cell suspension cultures. Arch. Biochem. Biophys. 203: 800–808. [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P., Becker-André M., Schulz W., Hahlbrock K., Dangl J.L. (1989). Functional architecture of the light-responsive chalcone synthase promoter from parsley. Plant Cell 1: 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann N., Navarro-Quezada A., Ullrich K., Kuhl C., Quint M. (2011). Molecular evolution and selection patterns of plant F-box proteins with C-terminal kelch repeats. Plant Physiol. 155: 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwertman P., Bezstarosti K., Laffeber C., Vermeulen W., Demmers J.A., Marteijn J.A. (2013). An immunoaffinity purification method for the proteomic analysis of ubiquitinated protein complexes. Anal. Biochem. 440: 227–236. [DOI] [PubMed] [Google Scholar]
- Shao T., Qian Q., Tang D., Chen J., Li M., Cheng Z., Luo Q. (2012). A novel gene IBF1 is required for the inhibition of brown pigment deposition in rice hull furrows. Theor. Appl. Genet. 125: 381–390. [DOI] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590. [DOI] [PubMed] [Google Scholar]
- Somers D.E., Kim W.Y., Geng R. (2004). The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Staiger D., Kaulen H., Schell J. (1989). A CACGTG motif of the Antirrhinum majus chalcone synthase promoter is recognized by an evolutionarily conserved nuclear protein. Proc. Natl. Acad. Sci. USA 86: 6930–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Favory J.J., Gruber H., Bartelniewoehner L., Bartels, S., Binkert M., Funk M., Weisshaar B., Ulm R.(2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Enviro. 33: 88–103. [DOI] [PubMed]
- Taylor L.P., Briggs W.R. (1990). Genetic regulation and photocontrol of anthocyanin accumulation in maize seedlings. Plant Cell 2: 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.P., Grotewold E. (2005). Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 8: 317–323. [DOI] [PubMed] [Google Scholar]
- Thain S.C., Murtas G., Lynn J.R., McGrath R.B., Millar A.J. (2002). The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 130: 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer I.M., Spelt C.E., Mol J.N.M., Stuitje A.R. (1990). Promoter analysis of the chalcone synthase (chsA) gene of Petunia hybrida: a 67 bp promoter region directs flower-specific expression. Plant Mol. Biol. 15: 95–109. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2012). The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 160: 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar B., Jenkins G.I. (1998). Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1: 251–257. [DOI] [PubMed] [Google Scholar]
- Weisshaar B., Armstrong G.A., Block A., da Costa e Silva O., Hahlbrock K. (1991). Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promoter element with functional relevance in light responsiveness. EMBO J. 10: 1777–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5: 218–223. [DOI] [PubMed] [Google Scholar]
- Xu G., Ma H., Nei M., Kong H. (2009). Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc. Natl. Acad. Sci. USA 106: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara M., Mitsui S., Hirano H., Takanabe R., Tokioka Y., Ihara N., Komatsu A., Seki M., Shinozaki K., Kiyosue T. (2004). Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J. Exp. Bot. 55: 2015–2027. [DOI] [PubMed] [Google Scholar]
- Zhang F., Maeder M.L., Unger-Wallace E., Hoshaw J.P., Reyon D., Christian M., Li X., Pierick C.J., Dobbs D., Peterson T., Joung J.K., Voytas D.F. (2010). High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA 107: 12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gou M., Liu C.J. (2013). Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25: 4994–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gou M., Guo C., Yang H., Liu C.J. (2015). Down-regulation of Kelch domain-containing F-box protein in Arabidopsis enhances the production of (poly)phenols and tolerance to ultraviolet radiation. Plant Physiol. 167: 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoratti L., Karppinen K., Luengo Escobar A., Häggman H., Jaakola L. (2014). Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 5: 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.