The lncRNA ELENA1 is positive regulator of plant immune response genes and regulates expression of its target gene, PR1, by promoting MED19a enrichment in the promoter.
Abstract
The plant immune response is a complex process involving transcriptional and posttranscriptional regulation of gene expression. Responses to plant immunity are initiated upon the perception of pathogen-associated molecular patterns, including peptide fragment of bacterial flagellin (flg22) or translation elongation factor Tu (elf18). Here, we identify an Arabidopsis thaliana long-noncoding RNA, designated ELF18-INDUCED LONG-NONCODING RNA1 (ELENA1), as a factor enhancing resistance against Pseudomonas syringe pv tomato DC3000. ELENA1 knockdown plants show decreased expression of PATHOGENESIS-RELATED GENE1 (PR1) and the plants are susceptible to pathogens. By contrast, plants overexpressing ELENA1 show elevated PR1 expression after elf18 treatment and display a pathogen resistance phenotype. RNA-sequencing analysis of ELENA1-overexpressing plants after elf18 treatment confirms increased expression of defense-related genes compared with the wild type. ELENA1 directly interacts with Mediator subunit 19a (MED19a) and affects enrichment of MED19a on the PR1 promoter. These results show that MED19a regulates PR1 expression through ELENA1. Our findings uncover an additional layer of complexity, implicating long-noncoding RNAs in the transcriptional regulation of plant innate immunity.
INTRODUCTION
Plants and animals are confronted with constant risk of infections by various microorganisms in their natural habitats. Like animals, plants have evolved a repertoire of pattern recognition receptors (PRRs) that recognize molecular signatures typical of entire classes of microbial pathogens. Pathogen-associated molecular patterns (PAMPs) include bacterial flagellin, translation elongation factor Tu (EF-Tu), peptidoglycans, lipopolysaccharides, and fungal cell wall-derived chitin fragments (Jones and Dangl, 2006; Boller and Felix, 2009). Perception of different PAMPs by cognate PRRs triggers innate immune responses that restrict pathogen propagation, and this series of signaling events is designated PAMP-triggered immunity (PTI) (Jones and Dangl, 2006). The best-characterized bacterial PAMPs recognized by plants are flg22 and elf18, which are derived from flagellin and EF-Tu, respectively (Kunze et al., 2004). flg22 and elf18 are recognized by FLAGELLIN SENSING2 (FLS2) and EF-Tu RECEPTOR (EFR), respectively, and these receptors belong to the leucine-rich repeat-receptor kinase family XII in Arabidopsis thaliana (Zipfel et al., 2006).
Following pathogen detection by PRRs, plants are able to mount a number of defense responses, including production and secretion of antimicrobial compounds and defense-related proteins (van Loon et al., 2006; Bednarek, 2012). Pathogenesis-related (PR) proteins are an important class of inducible defense-related proteins in various plant species, and they function as key players in an immune surveillance mechanism that protects plants primarily against invasion by microorganisms (van Loon et al., 2006). PR1 expression is highly responsive to salicylic acid (SA) and bacterial pathogens, and its transcriptional regulation has been extensively studied (Pajerowska-Mukhtar et al., 2013). As a key factor in PR1 expression, NONEXPRESSER OF PR GENES1 orchestrates PR1 transcriptional activity in concert with the TGA family of basic region/leucine zipper motif transcription factors (TFs) (Kesarwani et al., 2007; Gatz, 2013) and WRKY TF family (Yu et al., 2001; Eulgem and Somssich, 2007)
TFs typically have effector (activator or repressor) domains that are separated from their DNA binding domains, and they interact with transcriptional regulators. The Mediator complex is a general target of TF effector domains; moreover, as different TFs bind to different Mediator subunits, multiple TFs might bind to the Mediator complex at the same time (Allen and Taatjes, 2015). A basic function of Mediator is to communicate regulatory signals from TFs directly to the RNA polymerase II (Pol II) enzyme. The precise mechanisms by which the Mediator complex regulates Pol II activity remain poorly understood, but they clearly involve extensive protein-protein interactions between Mediator, Pol II, and other general and gene-specific TFs. Through those interactions, the Mediator complex is involved in a broad range of transcriptional events, including transcription initiation, transcript elongation, changes in chromatin architecture, and enhancer-promoter gene looping (Allen and Taatjes, 2015). In plants, quite a number of Mediator subunits have been characterized as important regulators for different signaling networks in response to various developmental as well as environmental changes. So far, at least nine Mediator subunits, MED8, 14, 15, 16, 18, 19, 21, 25, and CDK8, have been implicated in defense signaling (Samanta and Thakur, 2015).
High-resolution RNA sequencing (RNA-seq) analyses in animals and plants have revealed that the transcriptional landscape in eukaryotes is much more complex than previously envisioned and pervasive transcription seems to be a widespread feature of all eukaryotic genomes (Chekanova et al., 2007; Kapranov et al., 2007; Guttman et al., 2009; Liu et al., 2015). Early studies questioned the biological relevance of long-noncoding RNAs (lncRNAs; longer than 200 nucleotides) because of their low expression levels and lack of sequence conservation. But recently, a variety of types and origins of lncRNAs has been identified and shown to play important roles in transcriptional regulation and chromatin modification (Rinn and Chang, 2012; Kung et al., 2013; St Laurent et al., 2015). In plants, the regulatory roles of lncRNAs are only beginning to be recognized, and the molecular basis of lncRNA-mediated gene regulation is still poorly understood. So far, plant lncRNAs have been shown to play key roles in phosphate signaling, flowering time, auxin transport, root organogenesis, and seedling photomorphogenesis (Franco-Zorrilla et al., 2007; Swiezewski et al., 2009; Heo and Sung, 2011; Ariel et al., 2014; Bardou et al., 2014; Wang et al., 2014).
Previously, we performed a custom lncRNA array analysis with elf18-treated Arabidopsis seedlings to screen for PAMP-responsive lncRNAs (Liu et al., 2012). We selected multiple lncRNA candidates that were highly induced by elf18 treatment, designated as ELENAs (ELF18-INDUCED LONG-NONCODING RNAs) for further analysis. Among them, we found that ELENA1 acts as a positive regulator of resistance against bacterial pathogens by analyzing ELENA1 knockdown (KD) and overexpressing (OX) plants. Genome-wide transcriptome analysis of ELENA1 OX plants showed elevated expression of defense-related genes, including PR1. Transcriptional regulation of PR1 by ELENA1 was brought about by interaction with MED19a and its enrichment on the PR1 promoter. Taken together, our results provide evidence that the lncRNA ELENA1 plays a positive role in plant innate immunity by regulating PR1 expression.
RESULTS
ELENA1 Expression Is Induced by elf18 and flg22 in an EFR- and FLS2-Dependent Manner
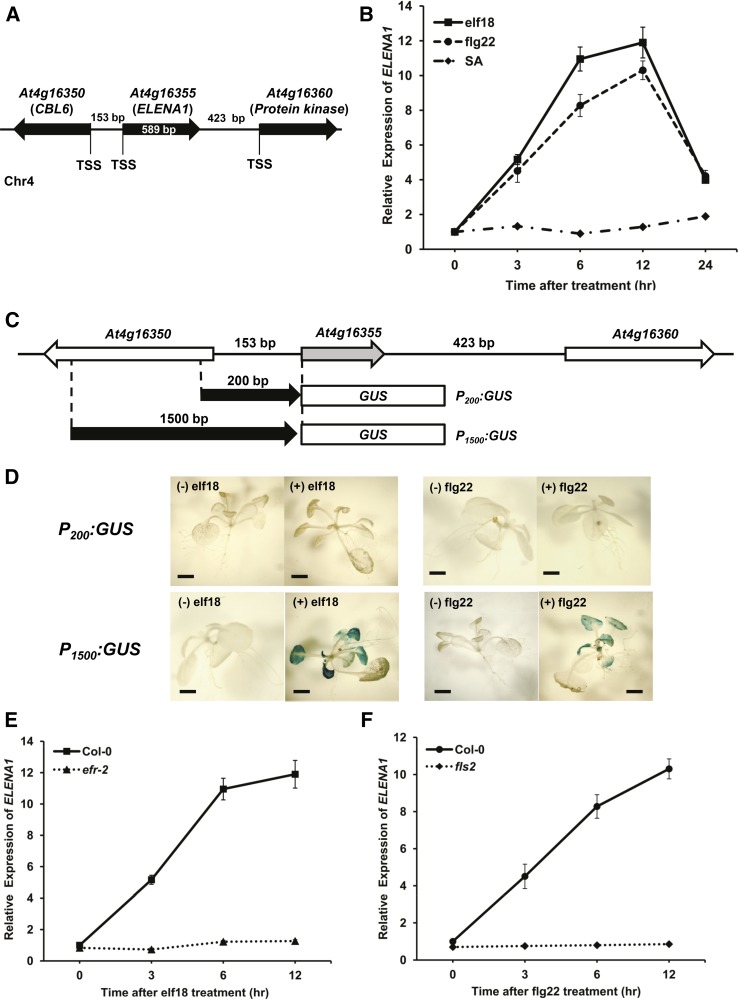
We screened for polyadenylated lncRNAs in response to elf18 treatment (Liu et al., 2012). Among them, ELENA1 (At4g16355) is located between CALCINEURIN B-LIKE6 (At4g16350) and another locus (At4g16360), encoding a putative protein kinase (Figure 1A). The ELENA1 transcript level was induced by both elf18 and flg22 treatments and reached a maximum at 12 h after treatment, but it was not responsive to SA treatment (Figure 1B).
Figure 1.
ELENA1 Transcript Levels Are Induced by elf18 and flg22.
(A) Schematic representation of the ELENA1 genomic location. TSS, transcription start site.
(B) Time-course analysis of ELENA1 expression levels after treatment. Ten-day-old wild-type (Col-0) seedlings were treated with 5 µM elf18, 5 µM flg22, or 1 mM SA.
(C) Schematic representation of PELENA1:GUS constructs. P200 is 200 bp upstream of the transcription start site, and P1500 is 1500 bp upstream.
(D) Histochemical staining of P200:GUS and P1500:GUS transgenic plants with (+) or without (−) elf18 or flg22 treatment for 6 h. Bars = 2 mm.
(E) Time-course analysis of ELENA1 transcript levels in efr-2 mutant after elf18 treatment.
(F) Time-course analysis of ELENA1 transcript levels in fls2 mutant after flg22 treatment. Ten-day-old seedlings were treated with 5 µM elf18 or 5 µM flg22.
For (B), (E), and (F), transcript levels were normalized to ACT2 expression levels. Bars represent average ± sd (n = 3 independent seedling pools).
To define the promoter regions responsible for ELENA1 expression responding to elf18 and flg22, two 5′ upstream fragments of ELENA1, 1.5 kb (P1500) and 0.2 kb (P200), were fused to the GUS coding sequence and transformed into Arabidopsis (Figure 1C). Histochemical GUS staining of transgenic plants showed that elf18- and flg22-responsive GUS activity was detected only in leaves of the P1500:GUS lines, but not in the P200:GUS lines (Figure 1D). These results indicate that the ELENA1 promoter responds to elf18 and flg22 and the region between 0.2 and 1.5 kb upstream of the ELENA1 promoter contains putative cis-acting elements required for elf18 and flg22 responsiveness.
In Arabidopsis, EFR (At5g20480) and FLS2 (At5g46330) function as receptors of elf18 and flg22, respectively (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). To investigate whether ELENA1 transcription was regulated by receptor-dependent pathways, we examined ELENA1 transcript levels in each receptor knockout (KO) mutant, efr-2 and fls2. Transcript induction after elf18 treatment was abolished in efr-2 mutants (Figure 1E), and it was also blocked in fls-2 after flg22 treatment (Figure 1F). These results provide evidence that ELENA1 transcription is regulated through PRR-dependent pathways.
ELENA1 KD and OX Plants Show Altered Responses to Pst DC3000 and Changes in PR1 Expression
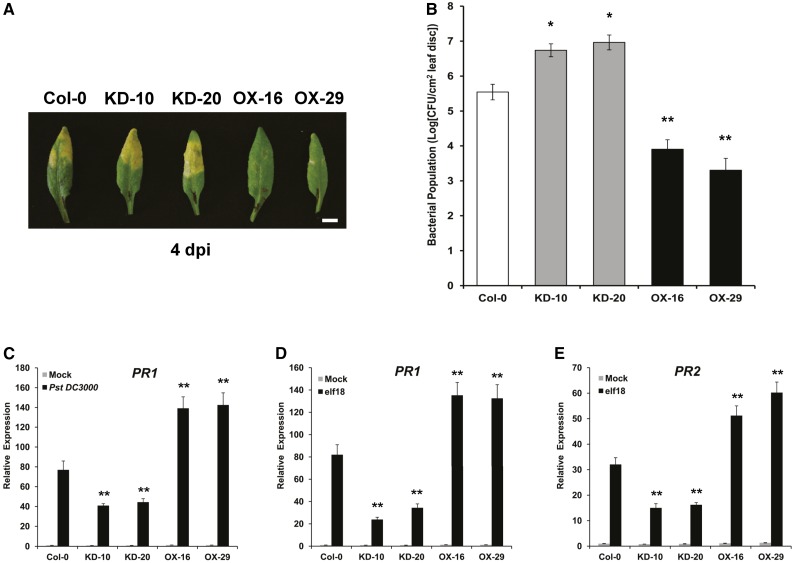
To investigate the function of ELENA1 in plant innate immunity, we generated ELENA1 KD transgenic plants using an artificial miRNA (Niu et al., 2006) (Supplemental Figure 1A) and selected two independent lines (KD-10 and KD-20) showing significantly reduced ELENA1 transcript levels (Supplemental Figure 1B). We also generated ELENA1 OX transgenic plants using a 35S promoter (Supplemental Figure 2A) and selected two independent lines (OX-16 and OX-29) showing high expression of ELENA1 (Supplemental Figure 2B). Selected KD lines and OX lines were inoculated with Pseudomonas syringae pv tomato DC3000 (Pst DC3000) to investigate defense phenotypes. Leaf chlorosis and bacterial growth assay showed that KD lines were more susceptible to Pst DC3000, whereas OX lines were more resistant (Figures 2A and 2B). In addition, PR1 (At2g14610) expression after Pst DC3000 infection was enhanced in OX lines but reduced in KD lines compared with the wild type (Col-0) (Figure 2C). These results indicate that ELENA1 is a positive regulator of resistance against Pst DC3000.
Figure 2.
Defense Phenotypes of ELENA1 KD and OX Plants.
(A) Altered disease susceptibility of plants of various genotypes. Wild-type (left end), ELENA1 KD lines (#10 and #20), and ELENA1 OX lines (#16 and #29) were inoculated with Pst DC3000. Infected leaves were photographed at 4 d postinoculation (dpi). Bar = 5 mm.
(B) Growth of Pst DC3000 in plants of various genotypes. Wild-type, KD lines, and OX lines were inoculated with Pst DC3000 and bacterial population in each was determined at 4 d postinoculation. Bars indicate ± sd (n = 3). CFU, colony-forming units.
(C) Using qRT-PCR, PR1 mRNA levels were determined in ELENA1 transgenic plants at 24 h after infiltration with Pst DC3000. Infiltration with water (gray bars) served as the mock control. All experiments were performed using 4- to 5-week-old leaf tissues and repeated at least three times with similar results.
(D) Real-time PCR analysis of PR1 expression in the absence or presence of 5 µM elf18 for 24 h.
(E) Real-time PCR analysis of PR2 expression in the absence or presence of 5 µM elf18 for 24 h.
For (C) to (E), transcript levels were normalized to ACT2 expression levels. Bars represent average ± sd (n = 3). For (B) to (E), asterisks indicate statistically significant difference compared with the wild type (Col-0). *P < 0.05 and **P < 0.01; two-tailed t test.
We also examined the expression of PR1 and PR2 (At3g57260) in transgenic plants with altered ELENA1 expression after elf18 treatment. PR1 and PR2 expression were reduced in KD lines, but enhanced in OX lines compared with the wild type (Figures 2D and 2E). Overall, our results show that ELENA1 is a positive regulator in PR gene expression and resistance against bacterial pathogen.
ELENA1 Functions as a Bona Fide lncRNA
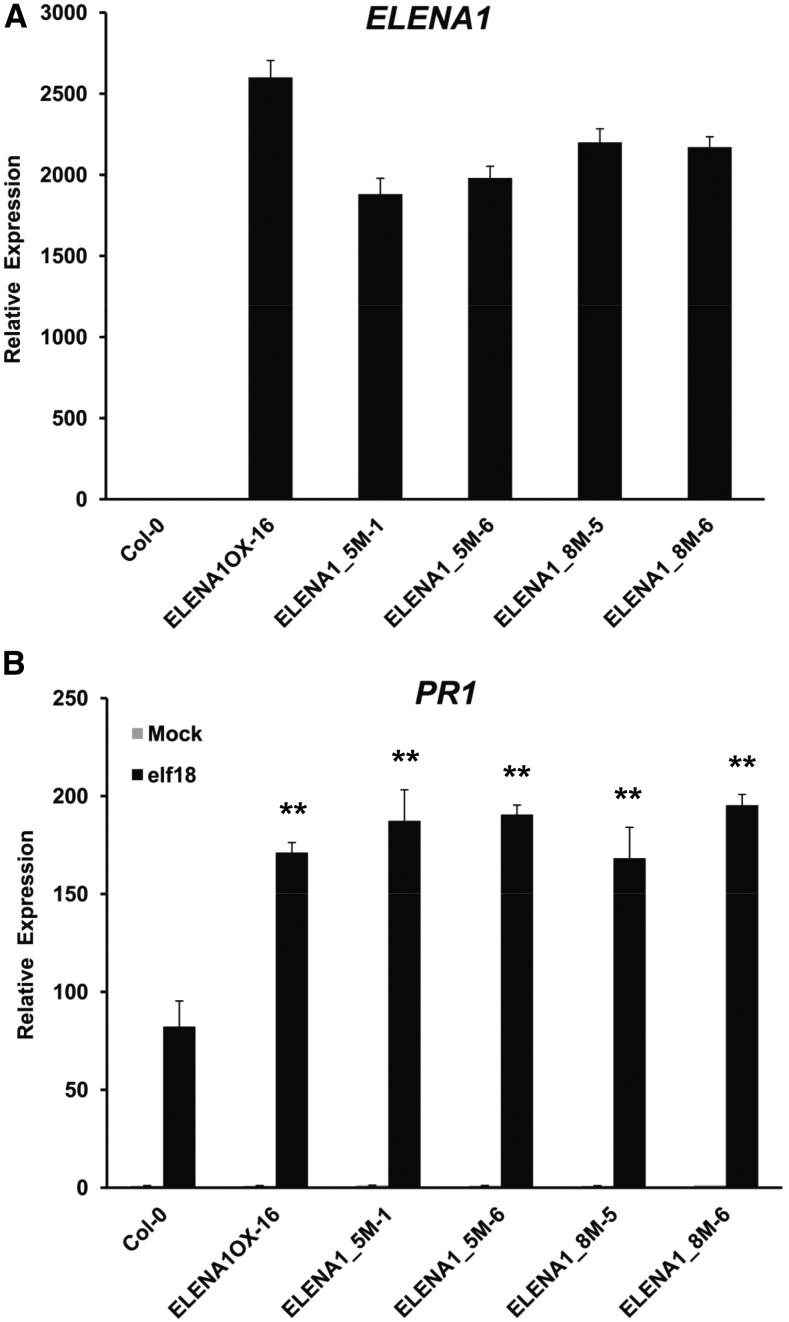
It has been recently reported that several lncRNAs could encode small peptides and function as peptide-coding genes (Anderson et al., 2015; Nelson et al., 2016). In ELENA1 transcripts, a total of eight putative open reading frames (ORFs) are found with expected peptide lengths ranging from 2 to 43 amino acids (Supplemental Figure 3). To test whether ELENA1 functions as a noncoding or coding RNA, we generated two different mutant plants, 35S:5m_ELENA1 (ELENA1_5M) and 35S:8m_ELENA1 (ELENA1_8M). In ELENA1_5M, five start codons of putative ORFs encoding peptides longer than 10 amino acids were mutated to TTG, whereas in ELENA1_8M, every start codon of all putative ORFs was changed to TTG. Notwithstanding some differences in transcript levels (1900–2600-fold) among mutated ELENA1 OX lines, PR1 expression was not significantly affected (Figures 3A and 3B). In the wild type, the maximal induction level of ELENA1 by elf18 is 10 to 20 times compared with the basal level (Figure 1B), but in ELENA1 OX or mutated ELENA1 OX plants, ELENA1 expression levels were over 1000 times compared with the basal level. Furthermore, we have analyzed four different OX lines with varying ELENA1 expression levels (1800–3200-fold) and found that these plants have comparable PR1 expression levels (Supplemental Figure 2). All these results suggested that transcript levels of ELENA1 in the OX plants were already above the saturating levels for PR1 expression, and different ELENA1 expression levels in OX plants were not expected to affect PR1 expression level significantly.
Figure 3.
Elevated PR1 Expression in Transgenic Plants Overexpressing ELENA1 Bearing Multiply Mutated Start Codons.
(A) Real-time PCR analysis of ELENA1 expression with no treatment. Both ELENA1_5M and ELENA1_8M harbor five and eight start codon mutations (ATG to TTG), respectively.
(B) Real-time PCR analysis of PR1 expression in the absence or presence of 5 µM elf18 for 24 h. Asterisks indicate statistically significant difference compared with the wild type (Col-0). **P < 0.01; two-tailed t test.
For (A) and (B), transcript levels were normalized to ACT2 expression levels. Bars represent average ± sd (n = 3 independent seedling pools).
Analyses of elf18-treated transgenic plants overexpressing the mutated ELENA1 showed increased PR1 expression levels compared with those in wild-type plants and comparable PR1 expression levels with WT ELENA1 OX lines (Figure 3A and 3B). These results suggested that ELENA1 functions as an authentic lncRNA rather than a peptide-coding RNA.
ELENA1 Positively Regulates elf18-Induced Immune Response Genes
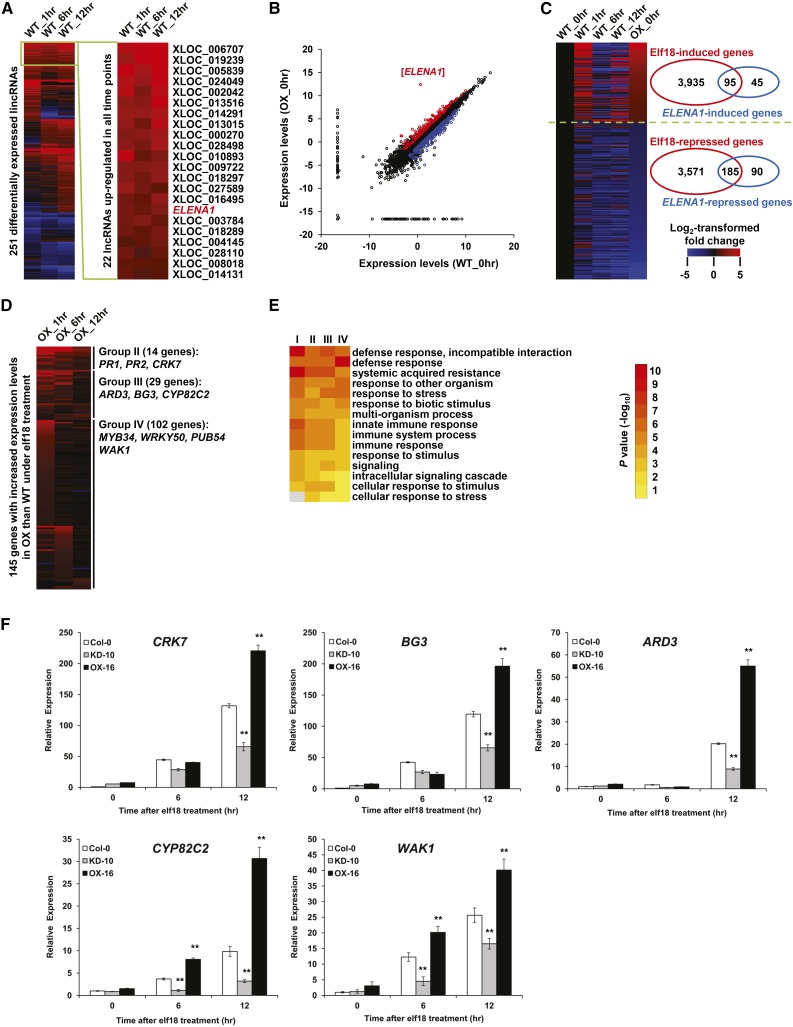
To investigate the role of ELENA1 in regulating the immune response, we performed strand-specific RNA-seq (ssRNA-seq) on wild-type and ELENA1 OX plants grown under normal (0 h) and elf18-treated conditions (1, 6, and 12 h; Supplemental Table 1), and three biological replicates (three independent seedling pools) were used for each condition. Clustering analysis revealed that the replicates of each condition showed gene expression profiles distinct from those of other conditions (Supplemental Figure 4A). Among 22,324 detected genes (TAIR10 annotated), 535 and 603 protein-coding genes were upregulated at all time points in wild-type and OX plants, respectively (fold change ≥ 2, P value < 0.05; Supplemental Figure 4B and Supplemental Data Sets 1 and 2). Gene Ontology (GO) enrichment analysis revealed that the functions of these genes were significantly enriched in biological processes associated with defense response and immune response (Supplemental Figure 4C), indicating that our transcriptome data sets exhibited characteristics of plants under elf18 treatment.
In addition, we confirmed that the ELENA1 transcript level was induced by elf18 because in wild-type plants, ELENA1 was upregulated in all time points following elf18 treatment (Figure 4A; Supplemental Data Set 3). In OX plants, ELENA1 expression was significantly elevated under all conditions compared with wild-type plants (Figure 4B; Supplemental Figure 4D). These results were in agreement with the above real-time PCR analysis (Figure 1B; Supplemental Figure 2).
Figure 4.
Transcriptional Regulation of elf18-Responsive Genes by ELENA1.
(A) Heat map showing fold changes of 251 differentially expressed lncRNAs under 5 µM elf18 treatment compared with normal condition (fold change ≥ 2 or ≤ 0.5, P value < 0.05). Twenty-two lncRNAs that were upregulated in all time points are selectively shown on the right.
(B) Scatterplot showing log2-transformed expression levels (FPKMs) of expressed genes in the wild type (x axis) compared with OX-16 plant (y axis).
(C) Heat map showing fold changes of 280 elf18-responsive genes with altered basal expression levels in OX plants.
(D) Heat map showing fold changes of 145 genes with increased expression levels in OX compared with wild-type plants under elf18 treatment.
(E) GO enrichment analysis of Group I, II, III, and IV genes. Top 15 (with the lowest P values) enriched GO terms of the biological process category are shown.
(F) Time-course analysis of selected target gene expression after elf18 treatment. Transcript levels were normalized to ACT2 expression levels. Bars represent average ± sd (n = 3 independent seedling pools). Asterisks indicate statistically significant difference compared with the wild type (Col-0). **P < 0.01; two-tailed t test.
Correlation analysis (Supplemental Figure 4A) showed that overexpression of ELENA1 did not induce global changes of gene expressions because wild-type and OX samples from the same time points were clustered together. To identify genes specifically regulated by ELENA1, we compared the transcriptomes of OX and wild-type plants under normal and elf18-treated conditions. Detailed analysis showed that the basal expression levels (under normal condition) of 95 elf18-induced (Group I) and 185 elf18-repressed genes increased and decreased, respectively, in OX plants (Figure 4C; Supplemental Data Set 4). In addition, by comparing wild-type and OX plants under elf18 treatment, we identified 145 genes with increased expression levels in OX plants compared with wild-type plants (Supplemental Data Set 4). These genes could be further classified into three additional groups. Group II contained 14 genes that had higher expression levels in all time points (Figure 4D). These genes include previously identified important regulators of defense response such as PR1, PR2, and CRK7 (CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASE7; At4g23150) (Denoux et al., 2008; Oide et al., 2013; Idänheimo et al., 2014). Group III contains 29 genes whose expression levels were increased in two time points. Representative genes of this group were CYP82C2 (CYTOCHROME P450, FAMILY 82, SUBFAMILY C, POLYPEPTIDE2; At4g31970) and BG3 (BETA-1,3-GLUCANASE3; At3g57240), which are known downstream defense-related genes (Dong et al., 1991; Rajniak et al., 2015). Group IV contains 102 genes that were upregulated in one time point and included some genes that have been shown to participate in defense responses, such as PUB54 (At1g01680), WRKY50 (At5g26170), MYB34 (At5g60890), etc. (Gao et al., 2011; Frerigmann and Gigolashvili, 2014). GO enrichment analysis of these genes showed that their functions were significantly enriched in biological processes associated with systemic acquired resistance, immune responses, and defense responses (Figure 4E). Taken together, these results suggested that ELENA1 positively regulates a subset of elf18-induced defense genes.
We verified the expression level of selected genes in ELENA1 KD and OX lines with real-time PCR (Figure 4F). Major candidates of putative ELENA1 target genes, PR1, PR2, CRK7, BG3, and CYP82C2, showed a clear anticorrelation in their expression level between ELENA1 KD and OX plants. These results confirmed that ELENA1 positively regulates expression of selective defense-related genes responsive to elf18.
ELENA1 Interacts with MED19a
Lai et al. (2013) reported that lncRNA may directly associate with Mediator and promote target gene expression in human cells. However, no Mediator subunits with RNA binding motifs have been identified in Arabidopsis yet. Therefore, we examined possible direct interactions between ELENA1 and Mediator subunits. First, we screened a group of Mediator subunits with high probable RNA binding motif using BindN software (Wang and Brown, 2006). Then, we performed in vitro RNA binding assays with maltose binding protein (MBP)-tagged recombinant Mediator subunits and in vitro-transcribed ELENA1 RNA. We found that MED19a (AT5G12230) and MED26b (AT5G05140) were able to bind to ELENA1 in vitro (Supplemental Figure 5). Analyses of KO mutants of mediator subunits showed that med19a (med19a-1 and med19a-2), not med26b plants, displayed a similar phenotype as ELENA1 KD lines in PR1 expression after elf18 treatment (Supplemental Figure 6). The Arabidopsis genome contains MED19b (AT5G19480), a close homolog of MED19a. We found that ELENA1 also associated with MED19b in vitro (Supplemental Figure 7A). However, analysis of the double mutant, MED19b RNAi med19a, showed only a marginal additive effect on PR1 expression compared with med19a single mutant (Supplemental Figure 8). This observation suggests that MED19a is the major regulator for PR1 expression and MED19b played only a minor role in PR1 expression induced by elf18. Therefore, further analysis was performed with MED19a.
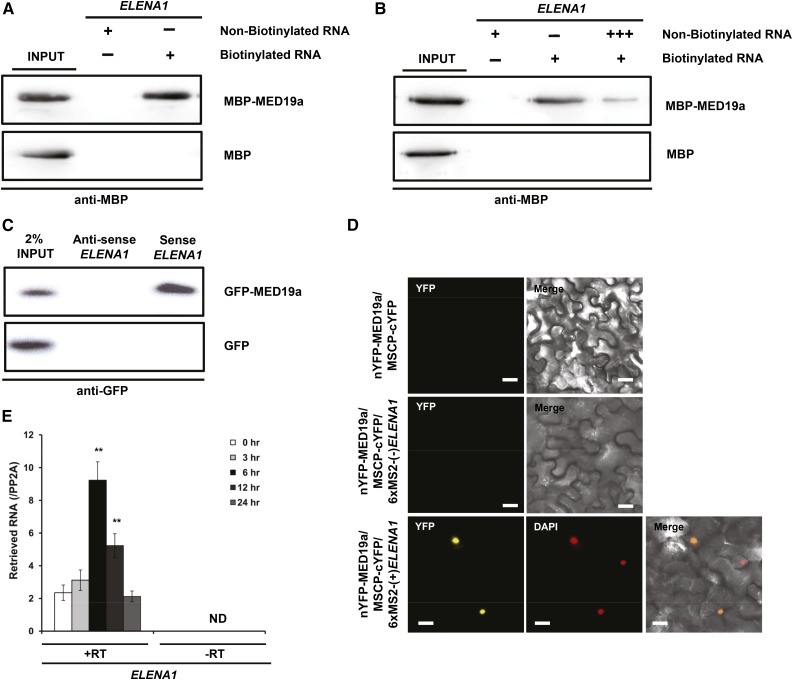
Further binding competition assay with nonbiotinylated RNA confirmed that ELENA1 specifically bound to MED19a (Figures 5A and 5B). However, recombinant MED19a protein also bound to antisense ELENA1 (Supplemental Figure 7B), suggesting non-sequence-specific binding. The non-sequence-specific binding to single-stranded nucleotides (including RNA) by target proteins has been reported previously (Heo and Sung, 2011). To check the specificity of the interaction between ELENA1 and the native MED19a protein, we prepared nuclear extracts from GFP-MED19a transgenic lines to perform RNA binding assay. Contrary to in vitro binding with recombinant MED19a protein purified from Escherichia coli, we detected MED19a association only with the sense strand of ELENA1 (Figure 5C), demonstrating the strand-specific interaction of ELENA1 with native MED19a in vivo. We also checked in vivo association of sense ELENA1 with MED19a using trimolecular fluorescence complementation (TriFC) assay, a modified version of bimolecular fluorescence complementation (BiFC) assay using a MS2 system (Schönberger et al., 2012; Han et al., 2014). We observed that sense strand of ELENA1 associated with MED19a in the nucleus, but antisense of ELENA1 did not (Figure 5D).
Figure 5.
ELENA1 Associates with MED19a.
(A) In vitro binding assay with in vitro-transcribed biotinylated ELENA1 RNA and recombinant MBP-MED19a protein.
(B) In vitro binding competition assay with recombinant MBP-MED19a protein and in vitro-transcribed ELENA1 RNA.
(C) In vitro binding between GFP-MED19a from nuclear extracts and in vitro-transcribed sense or antisense ELENA1 RNA. GFP nuclear extracts from 35S:GFP plants were used as a negative control.
(D) TriFC assay in tobacco leaves. nYFP was fused to MED19a, and cYFP was fused to MSCP. 6xMS2 nucleotide sequences fused to sense or antisense ELENA1. Confocal images were taken 3 d after infiltration. Bars = 20 μm.
(E) RIP assay with 35S:GFP-MED19a line during elf18 treatment. Data (mean ± sd of qPCR; n = 3) are relative to the background level of RNA precipitation (PP2A). +RT, with reverse transcription of precipitates; –RT, without reverse transcription of precipitates. Asterisks indicate statistically significant difference compared with 0 h. **P < 0.01; two-tailed t test.
For (A) and (B), MBP was used as a negative control. For (C), 35S:GFP was used as a negative control.
We also examined in vivo association of ELENA1 with MED19a after elf18 treatment by RNA immunoprecipitation (RIP) assay using the 35S:GFP-MED19a lines. Indeed, we were able to retrieve ELENA1 from GFP-MED19a immunoprecipitates (Figure 5E); more importantly, there was an increased association of ELENA1 with MED19a specifically during elf18 treatment (Figure 5E). Taken together, these results provide further evidence that ELENA1 was specifically associated with MED19a in vivo and their association was enhanced by elf18 treatment.
ELENA1 and MED19a Are Interdependent in PR1 Expression
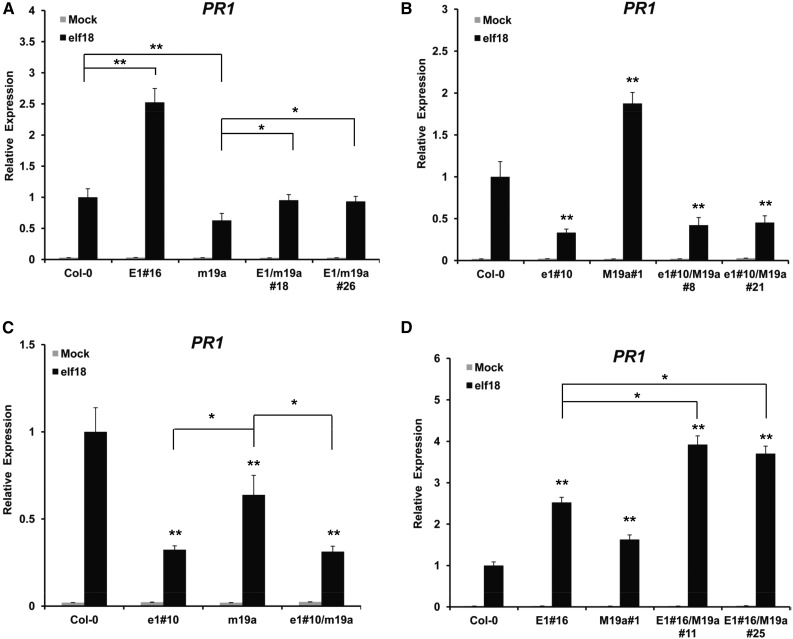
To investigate the relationship between ELENA1 and MED19a in PR1 expression, we generated four different double transgenic lines of ELENA1 and MED19a: 35S:ELENA1/med19a-1 (E1/m19a), ELENA1 KD-10/UBQ:MED19a (e1#10/M19a), ELENA1 KD-10/med19a-1 (e1#10/m19a), and 35S:ELENA1-16/UBQ:MED19a (E1#16/M19a) (Figure 6; Supplemental Figure 9). After selecting two independent lines for each double transgenic combination, we analyzed PR1 expression with elf18 treatment. The E1/m19a lines showed greatly reduced PR1 expression compared with ELENA1 overexpressing plant (E1#16) but showed a small increase of PR1 expression compared with med19a-1 mutant (m19a) (Figure 6A). This result suggested that ELENA1 function in PR1 expression was largely dependent on MED19a, but there were factors other than MED19a associating with ELENA1 to promote PR1 expression. ELENA1 KD mutant showed ∼30% PR1 expression levels compared with the wild type (Figure 6C). In this mutant background, knockout of MED19a (e1#10/m19a) had no further effect, indicating that MED19a action on PR1 expression was dependent on ELENA1. This conclusion is further supported by the results of Figure 6B. In the ELENA1 KD mutant background, overexpression of MED19a (e1#10/M19a) had no significant effect on PR1 expression, whereas in the wild-type background, overexpression of MED19a increased PR1 expression by ∼2-fold (Figure 6B). E1#16/M19a lines showed elevated PR1 expression compared with the overexpressing plants, suggesting that ELENA1 and MED19a can interact to further enhance PR1 expression (Figure 6D). Taken together, these results showed that under elf18 treatment, the promoting effect of MED19a on PR1 expression is dependent on ELENA1; in addition, there was a partial independent role of ELENA1 in PR1 induction upon elf18 treatment.
Figure 6.
PR1 Expression in ELENA1 and MED19a Double Mutants.
(A) PR1 expression in 35S:ELENA1/med19a-1 (E1/m19a) plants. E1#16 is 35S:ELENA1-16, and m19a is med19a-1
(B) PR1 expression in ELENA1 KD-10/UBQ:MED19a (e1#10/M19a) plants. e1#10 is ELENA1 KD-10, and M19a#1 is UBQ:MED19a-1.
(C) PR1 expression in ELENA1 KD-10/med19a-1 (e1#10/m19a) plants.
(D) PR1 expression in 35S:ELENA1-16/UBQ:MED19a (E1#16/M19a) plants.
All gray bars are values for mock treatment (without elf18), and black bars are values for 12 h 5 µM elf18 treatment. Bars represent average ± sd (n = 3 independent seedling pools). Asterisks indicate statistically significant difference compared with the wild type or between indicated values. *P < 0.05 and **P < 0.01; two-tailed t test.
ELENA1 Promotes MED19a Enrichment on the PR1 Promoter
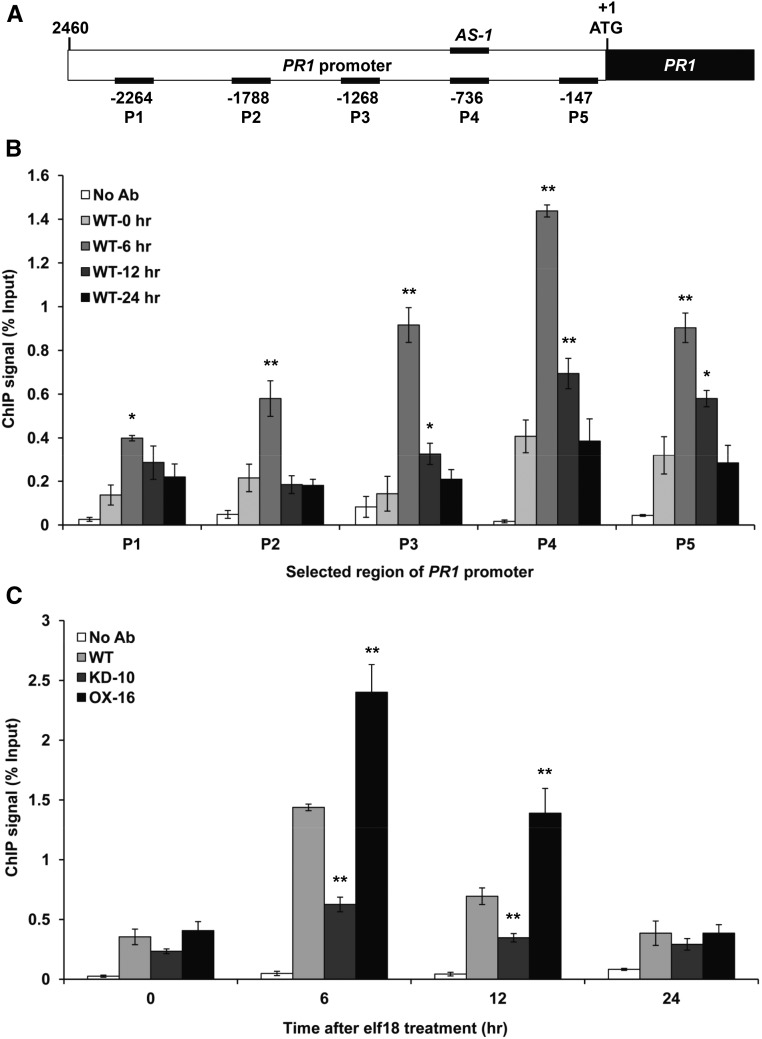
To investigate possible enrichment of MED19a on the PR1 promoter, we performed chromatin immunoprecipitation (ChIP) assays with PMED19a:GFP-MED19a transgenic line. After the ChIP assay, we quantified the enrichment of five different regions (P1 to P5) on the PR1 promoter by qPCR (Figure 7A). MED19a enrichment on the PR1 promoter was enhanced after elf18 treatment. MED19a enrichment was highest at 6 h after elf18 treatment and then reduced over 12 h after elf18 treatment (Figure 7B). Interestingly, MED19a enrichment was highest in the P4 region compared with other regions. Previous reports have identified a TGA binding cis-element, AS-1 like, in this region (Després et al., 2000; Johnson et al., 2003) (Figure 7A). This result demonstrated that MED19a was enriched on the PR1 promoter region, especially including the TGA binding site, during elf18 treatment.
Figure 7.
ELENA1 Promotes MED19a Enrichment on the PR1 Promoter.
(A) Schematic diagrams of PR1 promoter (2460 bp) and five different regions (P1 to P5) for analysis by ChIP assay. P4 region includes AS-1 cis-element.
(B) Time course of ChIP qPCR results of PMED19a:GFP-MED19a line after elf18 treatment. ChIP signal in each region of the PR1 promoter at each time point was quantified by qPCR. The numbers on the x axis indicate the positions of the PCR-amplified sites described in (A).
(C) Relative enrichment of MED19a at P4 region in ELENA1 mutants (KD-10 and OX-16) was shown in different times after 5 µM elf18 treatment. Error bars indicate sd (n = 3). Asterisks indicate statistically significant difference compared with 0 h (B) and the wild type (Col-0) (C). *P < 0.05 and **P < 0.01; two-tailed t test.
We next examined MED19a enrichment in ELENA1 mutants. In ELENA1 KD lines, the enrichment of MED19a on the P4 region of PR1 promoter was greatly reduced compared with the wild type. On the other hand, the enrichment of MED19a in the ELENA1 OX line was increased (Figure 7C). This is consistent with elevated levels of PR1 transcript in ELENA1 OX lines and reduced levels of PR1 transcript in ELENA1 KD lines during elf18 treatment. These results support the view that ELENA1 facilitates the recruitment of MED19a to the PR1 promoter.
DISCUSSION
ELENA1 Is a Positive Regulator of PTI Signaling in Arabidopsis
ELENA1 is an lncRNA induced by PAMP in plants. Like many other lncRNAs, the basal transcript level of ELENA1 was very low, but its expression level increased more than 10 times after elf18 treatment. Abolishment of ELENA1 transcriptional induction in receptor KO mutants (efr and fls2) indicated that ELENA1 transcriptional regulation was controlled by receptor-dependent signal transduction. Additionally, ELENA1 promoter-GUS assays confirmed that its promoter carries cis-acting elements highly responsive to elf18 and flg22. ELENA1 is not early responsive, as its transcript level continuously increased until 12 h after elf18 or flg22 treatment. Moreover, ELENA1 expression did not respond to SA, the key hormone for systemic acquired resistance. This observation suggests that ELENA1 functions downstream of PAMP recognition and by an SA-independent signaling.
In ELENA1 KD and OX plants, the altered defense phenotypes against Pst DC3000 and changes in PR1 expression after elf18 treatment clearly showed that this lncRNA served as a positive regulator of PTI signaling. RNA-seq results also demonstrated that many plant defense-related genes were upregulated in ELENA1 OX plants.
ELENA1 Transcriptionally Regulates Downstream Immune Response Genes, Including PR1
At the initial stage of this study, we assumed that the presumptive target genes of ELENA1 might be located in the nearby genomic region because many lncRNAs are involved in cis-regulatory function (Guil and Esteller, 2012). Two genes are located close to ELENA1. In wild-type plants, CBL6 but not At4g16360 was induced by elf18 treatment (Supplemental Figure 10). Real-time qPCR and RNA-seq results of ELENA1 transgenic plants showed no difference in the transcript levels of neighboring genes including CBL6 (Supplemental Figure 10). These results suggest that ELENA1 likely functions as a trans-acting lncRNA involved in the transcriptional regulation of distant target genes.
We screened candidate target genes of ELENA1 using an RNA-seq data set of ELENA1 OX line. PR1 and PR2 were identified as good candidates because their expression in OX lines was higher compared with the wild type at all time points after elf18 treatment. Moreover, the opposite results were obtained in ELENA1 KD mutants. We tested expression of over 30 candidate target genes screened by RNA-seq, but only a few of these genes showed a clear opposite expression pattern between ELENA1 KD mutant and OX plants. This observation suggested that ELENA1 regulates expression of only a subset of target genes.
RNA-seq and qPCR validation results showed that downstream genes, such as PR1, PR2, BG3, and CYP82C2, were major candidate ELENA1 targets, and these genes clearly showed reduced expression in KD plants but enhanced expression in OX plants. PR2 and BG3 are major β-1,3-glucanases regulating callose accumulation and SA-dependent defense responses (Oide et al., 2013). The recently characterized CYP82C2 is a biosynthetic enzyme generating a cyanogenic metabolite in Arabidopsis required for inducible pathogen defense and innate immunity (Rajniak et al., 2015). There was no difference in expression levels of early and upstream genes in PAMP signaling, e.g., FLS2, EFR, BAK1, BIK1, PBS1, FRK1, and MAPK genes, etc. This is not surprising because the ELENA1 transcript level increased relatively slowly and reached a maximum level between 6 and 12 h. Together, our results suggest that ELENA1 could regulate late and downstream genes rather than early-responsive upstream genes. We propose the major role of ELENA1 in PTI signaling to be enhancement of expression for specific downstream genes involved in plant innate immunity.
ELENA1 Interacts with Mediator Subunit 19a
Two notable classes of lncRNAs, activator RNAs and enhancer RNAs, regulate expression of their targets by interacting with the Mediator complex (Ørom et al., 2010; Lai et al., 2013; Allen and Taatjes, 2015). Inspired by these findings, we examined possible interactions of ELENA1 with selected Mediator subunits carrying a putative RNA binding motif and found that ELENA1 binds to MED19a and MED26b in vitro. However, each mediator KO mutant showed different PR1 gene expression pattern upon elf18 treatment (Supplemental Figure 6). These results raised the possibility that ELENA1 could bind to different sets of Mediator subunits in vivo depending on the signals.
A recent report showed that the fungal effector (HaRxL44) of powdery mildew mediates proteasome-dependent degradation of MED19a and shifts the balance from SA-mediated disease resistance to ethylene/jasmonic acid-mediated transcriptomic changes (Caillaud et al., 2013). This result suggests that MED19a plays an important role in biotrophic pathogen resistance and could be involved in PTI signaling. Our result on PR1 expression in MED19a KO mutants after elf18 treatment also supported this view. Analysis of double mutants of ELENA1 and MED19a showed that the function of MED19a in PR1 expression is closely related to ELENA1. Our results also suggested that ELENA1 can partially promote PR1 expression in a MED19a-independent way (Figure 6). There are two possible explanations for this. First, ELENA1 may bind to other Mediator subunits and partially regulate PR1 expression. We have already shown the possibility that ELENA1 can bind to other Mediator subunits (Supplemental Figure 5). Second, the MED19a homolog, MED19b, also associates with ELENA1 and the complex may regulate PR1 gene expression (Supplemental Figures 7A and 8). In Supplemental Figure 8, we showed that MED19b might play a minor role by interacting with ELENA1 and affect PR1 expression. Therefore, the slightly higher expression of PR1 in E1/m19a compared with that in the med19a single mutant may be caused by MED19b. The e1#10/M19a plants showed almost similar PR1 expression levels as the single ELENA1 KD line because ELENA1 was almost depleted in the e1#10/M19a mutant (Figure 6B). In Figure 6C, the PR1 expression level in med19a-1 was higher than that in the ELENA1 KD plants. Also, the PR1 expression level in the e1#10/m19a line was similar to that in the ELENA1 KD line because ELENA1 expression was already depleted. These observations suggested that MED19b might have redundant function with MED19a. Therefore, all these lines of evidence supported the notion that ELENA1 affects transcription of defense-related target genes through Mediator subunits, including MED19a.
ELENA1 Regulates MED19a Enrichment on the PR1 Promoter
Analysis of transgenic plants with different genotypes of ELENA1 and MED19a suggested that ELENA1 and MED19a were interdependent for PR1 expression induced by elf18 treatment. However, it was not clear whether ELENA1 and MED19a regulate PR1 gene expression directly or indirectly. ChIP-PCR analysis clearly showed that MED19a was enriched on the AS-1-like element-containing region of PR1 promoter. Decreased MED19a enrichment in ELENA1 KD lines and increased MED19a enrichment in ELENA1 OX lines on the PR1 promoter demonstrated that ELENA1 facilitated MED19a enrichment on the PR1 promoter (Supplemental Figure 11). In addition, it was reported that multiple TGA factors have a binding capacity for both positive and negative regulatory AS-1-like cis-elements present on the PR1 promoter (Jupin and Chua, 1996; Després et al., 2000; Pajerowska-Mukhtar et al., 2013). This suggested that transcriptional regulation of PR1 by ELENA1 and MED19a might be closely related with TGA TFs.
Notwithstanding our results, the detailed mechanism of MED19a enrichment on the PR1 promoter by ELENA1 remains to be clarified. Several mechanisms have been proposed for the involvement of trans-acting lncRNA in transcriptional regulation of target genes (Koziol and Rinn, 2010; Vance and Ponting, 2014). One possible model is Triple helix formation between ELENA1 lncRNA and PR1 promoter DNA. Using Triplexator software (Buske et al., 2012), we were unable to find any significant sequence area for triple helix formation between ELENA1 and PR1 promoter sequences. Another possible model is that ELENA1 may interact with other proteins, such as other Mediator subunits, transcription factors, cofactors, or adaptors, thereby recruiting MED19a to the transcription machinery. In this regard, we have shown that ELENA1 could bind to another mediator, MED26b. It is possible that ELENA1 can bind to other Mediator subunits and other transcriptional machinery components. Further study of other interactors will likely provide additional information of the regulatory complexity of this noncoding RNA.
The roles of lncRNA in plant immunity are only beginning to be unraveled. In addition to ELENA1, our custom lncRNA array and RNA-seq data have uncovered hundreds of lncRNAs that are upregulated or downregulated by elf18 treatment, suggesting that many lncRNAs may be involved in the regulation of plant innate immunity. We anticipate that further studies of other lncRNAs may lead to a better understanding of the roles of lncRNAs in transcriptome regulation associated with plant immunity.
METHODS
Plant Materials, Growth Conditions, and Treatment
Arabidopsis thaliana ecotype Columbia (Col-0), efr-2 (SALK_068675), fls2 (SALK_062054), med19a-1 (SALK_037435), med19a-2 (SALK_034955), and med26b-1 (SALK_020870) mutant plants were used. T-DNA insertion lines were obtained from the SALK collection. Homozygous plants for the T-DNA insertion were selected by genotyping progeny plants according to Alonso et al. (2003). Absence of target gene expression in homozygous plants was further confirmed by RT-PCR. Plants of all genotypes were grown on 0.6% agar media containing 0.5× Murashige and Skoog (MS) salts (MP Biomedicals), 1% sucrose (Fisher), and 0.5 g/L MES hydrate (Sigma-Aldrich) in a growth room at 22°C under 16 h light/8 h dark with white fluorescent light (∼100 μmol m−2 s−1). For elf18 (EZBiolab) or flg22 (EZBiolab) treatment, 10-d-old seedlings grown on MS solid medium were transferred to MS liquid medium (pH 5.7) with 1% sucrose and 5 μM elf18 or flg22 and incubated under the same condition and then seedlings were harvested at each time point after peptide application.
Generation of Transgenic Lines
The entry clone of ELENA1 was recombined into pBA-DC (Zhang et al., 2005) to generate overexpressing plants. ELENA1 KD mutants were generated using artificial microRNA (Niu et al., 2006). To produce ELENA1 codon mutants, we made an entry clone of full-length mutated ELENA1 by DNA synthesis (IDT) and recombined entry clone into pBA-DC by LR reaction. The E1/m19a lines were generated by transforming the med19a-1 knockout mutant with pBA-ELENA1. The e1#10/M19a line were generated by transforming ELENA1 KD-10 with pUBQ-MED19a. The e1#10/m19a lines were produced by genetic crossing between ELENA1 KD-10 and med19a-1 knockout mutant. The E1#16/M19a lines were generated by transforming ELENA1 OX-16 with pUBQ-MED19a. PMED19a:GFP-MED19a mutants were generated with pKGWFS7 vector (Karimi et al., 2002). All constructs were verified by sequencing and transformed into Agrobacterium tumefaciens strain GV3101. Wild-type (Col-0) or mutant plants were transformed using the floral dip method (Zhang et al., 2006).
Real-Time RT-PCR Analysis
Total RNA was extracted from Arabidopsis seedlings using RNeasy plant mini kit (Qiagen) including DNase I treatment. Reverse transcription was performed using 2 μg of each total RNA and oligo(dT)20 primers by the SuperScript III reverse transcriptase (Invitrogen). Real-time RT-PCR was performed using SYBR premix Ex Taq (Tli RNaseH plus; TaKaRa) on the Bio-Rad CFX96 real-time system with gene-specific primers. Primer sequences used are listed in Supplemental Table 2.
Histochemical GUS Staining
For promoter-GUS fusion, two promoter fragments of ELENA1 (200 and 1500 bp) were amplified by PCR, cloned into pENTR/D-TOPO vector (Invitrogen), and then recombined into pKGWFS7 vector to obtain the P200:GUS and P1500:GUS fusions. Arabidopsis (Col-0) plants were transformed by the floral dip method and GUS staining was performed as described (Senecoff et al., 1996).
Bacterial Growth Assays
Bacterial growth assays were performed as described (Katagiri et al., 2002) with minor modifications. For leaf assay, an overnight culture of Pseudomonas syringae pv tomato DC3000 was collected by centrifugation, washed, and then resuspended to 5 × 104 colony-forming units/mL in water. Arabidopsis leaves of 4-week-old plants were infiltrated with bacterial suspension using a needleless syringe. Four days after infiltration, leaf disks were ground in 100 μL water, and serial dilutions were plated on King’s B medium. Bacterial colony-forming units were counted 2 d after incubation at 28°C.
RNA Extraction, Library Construction, and Sequencing for ssRNA-Seq
Total RNA was extracted from Arabidopsis 10-d-old seedlings using TRIzol reagent (Ambion), treated with TURBO DNase (Ambion), and purified using RNeasy mini spin column (Qiagen). The quality of purified RNA was assessed using an Agilent 2100 Bioanalyzer. cDNA libraries for ssRNA-seq were prepared using the Illumina TruSeq Stranded mRNA sample preparation kit according to the low sample protocol guidelines. The quality and size of each sample library was assessed using an Agilent High Sensitivity D1K ScreenTape System. The average sizes of the enriched cDNA fragments were between 272 and 300 bp. The three biological replicates for each condition (negative control, 1 h_elf18, 6 h_elf18, and 12 h_elf18) were pooled into one well and sequenced on an Illumina NextSeq High Output SR 75 with 75-cycle single reads per multiplexed sample.
ssRNA-Seq Data Analysis
Strand-specific RNA-seq reads were mapped to the Arabidopsis genome (TAIR10) using TopHat (version 2.0.8) (Kim et al., 2013) with parameters “–library-type=fr-firststrand -i 40 -I 5000 -g 1–segment-length=20.” The mapped reads were assembled using Cufflinks (v2.1.1) (Trapnell et al., 2013) with parameters “–library-type=fr-firststrand -I 5000–min-intron-length=40” and with TAIR10 annotation as the reference (Lamesch et al., 2012). The assembled transcripts of each ssRNA-seq sample were merged and annotated using Cuffcompare (v2.1.1) (Trapnell et al., 2013) with TAIR10 annotations as the reference. The expression level of each gene was then calculated by fragments per kilobase of exons per million fragments mapped (FPKM) using Cuffdiff (v2.1.1) (Trapnell et al., 2013) with parameter “–library-type=fr-firststrand.” A 2-fold variance in FPKM, a P value < 0.05, and an adjusted P value < 0.1 were used as cutoffs to define differentially expressed genes. The P value and adjusted P value were calculated using DESeq2 (Love et al., 2014). All assembled intergenic transcription units were collected as lncRNA candidates. Candidate genes encoding RNA with length ≥200 nucleotides and a predicted ORF ≤100 amino acids were defined as lncRNA (Liu et al., 2012). ORFs were predicted using GenScan with Arabidopsis specific parameters (Burge and Karlin, 1997). GO enrichment analysis was performed using agriGO (Du et al., 2010) with TAIR10 annotation. The smaller the P value is, the more the GO term is significantly enriched; therefore, the top enriched GO terms are those with the smallest P values.
In Vitro RNA Pull-Down Assay and RIP Assay
Biotin-labeled RNAs were in vitro transcribed using the Biotin RNA Labeling Mix (Roche) and T7 RNA polymerase (Roche), treated with RNase-free DNase I (Invitrogen), and purified with RNeasy Mini Kit (Qiagen). Nuclear extract was obtained from seedling samples as described (Heo and Sung, 2011). In addition, recombinant MBP-Mediator subunit proteins were expressed with pMAL-DC and purified using Escherichia coli (BL21) expression system. Two micrograms of biotin-labeled RNAs and nuclear extract from GFP-MED19a transgenic plants or recombinant Mediator proteins were mixed in pull-down buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM DTT, 0.05% Nonidet P-40, and protease inhibitor tablet [Roche]) and incubated for 6 h at 4°C. Thirty microliters of washed streptavidin agarose beads (Roche) were then added to each binding reaction and further incubated for 2 h at 4°C. Beads were washed briefly five times using binding buffer and boiled in SDS buffer, and the supernatant was analyzed by protein gel blot using anti-GFP antibody (Santa Cruz). RIP assay with GFP-MED19a line was performed as previously described (Heo and Sung, 2011)
TriFC Assay
We generated binary gateway BiFC vectors, pBA3136, pBA3134, pBA3132, and pBA3130, by recombining pBA002 binary vector with Gateway cassettes of BiFC vectors, pSAT4-DEST-nEYFP-C1(pE3136), pSAT4(A)-DEST-nEYFP-N1(pE3134), pSAT5-DEST-cEYFP-C1(pE3132), and pSAT5(A)-DEST-cEYFP-N1(pE3130) (ABRC) for transient assay in Nicotiana benthamiana. In the case of TriFC assay, full-length entry clone carrying MED19a was recombined into pBA3136, and entry clone encoding MSCP was recombined into pBA3132 by LR reaction. Entry clone of ELENA1 was recombined into p35S-GW-6xMS2 by LR reaction (Schönberger et al., 2012). All constructs were transformed into Agrobacterium strain GV3101 using the freeze and thaw method. Cultured cells were harvested and resuspended in 10 mM MgCl2 plus 150 μM acetosyringone (Sigma-Aldrich) and then kept at 25°C for at least 3 h without shaking. Agrobacterium suspensions containing 50 μM MG132 were infiltrated into leaves of N. benthamiana with a needleless syringe. Leaf cells were analyzed using LSM 780 confocal laser scanning microscope (Zeiss) 2 to 3 d after infiltration.
ChIP Assay
ChIP assays were performed as described previously (Bowler et al., 2004). After chromatin isolation, immunoprecipitation was performed using anti-GFP (Santa Cruz). Cross-links were reversed by incubation at 65°C for 12 h, and DNA was purified with QIAquick spin columns (Qiagen) and eluted in 50 μL of Tris-EDTA buffer (pH 8.0). Real-time qPCR was used to quantify the enrichment of different fragments on the PR1 promoter. First, relative enrichment of each fragment was calculated based on comparison to qPCR using input control. Second, relative enrichment was calculated by comparison to control region. Real-time PCR reaction was performed on Bio-Rad CFX96 real-time system. Primer sequences are in Supplemental Table 2.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ELENA1 (At4g16355), CALCINEURIN B-LIKE6 (At4g16350), PR1 (At2g14610), PR2 (At3g57260), FLS2 (At5g46330), EFR (At5g20480), PUB54 (At1g01680), WRKY50 (At5g26170), MYB34 (At5g60890), MED19a (AT5G12230), MED19b (AT5G19480), and MED26b (AT5G05140). A total of 24 ssRNA-seq data sets generated in this work have been deposited in the NCBI Gene Expression Omnibus database under accession number GSE93560 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=abehwqeihnsrzohandacc=GSE93560).
Supplemental Data
Supplemental Figure 1. Expression level of selected ELENA1 knock down lines by artificial miRNA.
Supplemental Figure 2. ELENA1 and PR1 expression level in selected ELENA1-overexpressing lines.
Supplemental Figure 3. Predicted ORFs in ELENA1 transcript.
Supplemental Figure 4. Global view of ssRNA-seq results.
Supplemental Figure 5. ELENA1 associates with Mediator subunits in vitro.
Supplemental Figure 6. PR1 expression levels in MED19a and MED26a KO mutants after elf18 treatment.
Supplemental Figure 7. ELENA1 associates with both MED19a and MED19b in vitro.
Supplemental Figure 8. PR1 expression levels in med19a/med19b double mutant plants.
Supplemental Figure 9. ELENA1 and MED19a expression levels in ELENA1 and MED19a double mutants.
Supplemental Figure 10. Time-course expression of genes neighboring ELENA1 after elf18 treatment.
Supplemental Figure 11. A working model of transcriptional regulation of PR1 by ELENA1 in Arabidopsis.
Supplemental Table 1. Statistics of ssRNA-seq reads that could map to the Arabidopsis genome.
Supplemental Table 2. Primers used in this study.
Supplemental Data Set 1. Tabulated data of gene expression levels detected by ssRNA-seq.
Supplemental Data Set 2. Tabulated data of gene expression levels of 535 and 603 protein coding genes that were upregulated at all time points in wild-type and OX plants.
Supplemental Data Set 3. Tabulated data of gene expression levels of 251 differentially expressed lncRNAs.
Supplemental Data Set 4. Tabulated data of gene expression levels of upregulated genes in OX plants.
Supplementary Material
Acknowledgments
We thank Fumiaki Katagiri for Pst DC3000 strain and Jonathan D.G. Jones for MED19a KO and OX seeds. This work was funded in part by Singapore NRF RSSS Grant NRF-RSSS-002.
AUTHOR CONTRIBUTIONS
J.S.S., C.J., and N.-H.C. conceived the research plans. J.S.S. and N.-H.C. designed the experiments. J.S.S., B.S.P., and C.-H.H. performed the experiments. H.-X.S. analyzed the RNA-seq data. J.S.S., H.-X.S., C.J., S.-D.Y., and N.-H.C. wrote the article.
Glossary
- PRR
pattern recognition receptor
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- SA
salicylic acid
- TF
transcription factor
- lncRNA
long-noncoding RNA
- ORF
open reading frame
- ssRNA-seq
strand-specific RNA-seq
- GO
Gene Ontology
- TriFC
trimolecular fluorescence complementation
- BiFC
bimolecular fluorescence complementation
- RIP
RNA immunoprecipitation
- ChIP
chromatin immunoprecipitation
- MS
Murashige and Skoog
- FPKM
fragments per kilobase of exons per million fragments mapped
References
- Allen B.L., Taatjes D.J. (2015). The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., Olson E.N. (2015). A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160: 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F., Jegu T., Latrasse D., Romero-Barrios N., Christ A., Benhamed M., Crespi M. (2014). Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 55: 383–396. [DOI] [PubMed] [Google Scholar]
- Bardou F., Ariel F., Simpson C.G., Romero-Barrios N., Laporte P., Balzergue S., Brown J.W., Crespi M. (2014). Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 30: 166–176. [DOI] [PubMed] [Google Scholar]
- Bednarek P. (2012). Chemical warfare or modulators of defence responses - the function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 15: 407–414. [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A.V., Tariq M., Paszkowski J. (2004). Chromatin techniques for plant cells. Plant J. 39: 776–789. [DOI] [PubMed] [Google Scholar]
- Burge C., Karlin S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Buske F.A., Bauer D.C., Mattick J.S., Bailey T.L. (2012). Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 22: 1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M.C., Asai S., Rallapalli G., Piquerez S., Fabro G., Jones J.D. (2013). A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol. 11: e1001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova J.A., et al. (2007). Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353. [DOI] [PubMed] [Google Scholar]
- Denoux C., Galletti R., Mammarella N., Gopalan S., Werck D., De Lorenzo G., Ferrari S., Ausubel F.M., Dewdney J. (2008). Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1: 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C., DeLong C., Glaze S., Liu E., Fobert P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290. [PMC free article] [PubMed] [Google Scholar]
- Dong X., Mindrinos M., Davis K.R., Ausubel F.M. (1991). Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10: 366–371. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Frerigmann H., Gigolashvili T. (2014). MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 7: 814–828. [DOI] [PubMed] [Google Scholar]
- Gao Q.M., Venugopal S., Navarre D., Kachroo A. (2011). Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155: 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz C. (2013). From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant Microbe Interact. 26: 151–159. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Guil S., Esteller M. (2012). Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol. 19: 1068–1075. [DOI] [PubMed] [Google Scholar]
- Guttman M., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Wang S., Zhang Z., Ma X., Li W., Zhang X., Deng J., Wei H., Li Z., Zhang X.E., Cui Z. (2014). In vivo imaging of protein-protein and RNA-protein interactions using novel far-red fluorescence complementation systems. Nucleic Acids Res. 42: e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J.B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79. [DOI] [PubMed] [Google Scholar]
- Idänheimo N., Gauthier A., Salojärvi J., Siligato R., Brosché M., Kollist H., Mähönen A.P., Kangasjärvi J., Wrzaczek M. (2014). The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Commun. 445: 457–462. [DOI] [PubMed] [Google Scholar]
- Johnson C., Boden E., Arias J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15: 1846–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Jupin I., Chua N.H. (1996). Activation of the CaMV as-1 cis-element by salicylic acid: differential DNA-binding of a factor related to TGA1a. EMBO J. 15: 5679–5689. [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Willingham A.T., Gingeras T.R. (2007). Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8: 413–423. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Katagiri F., Thilmony R., He S.Y. (2002). The Arabidopsis thaliana-Pseudomonas syringae interaction. Arabidopsis Book 1: e0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani M., Yoo J., Dong X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144: 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol M.J., Rinn J.L. (2010). RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 20: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J.T., Colognori D., Lee J.T. (2013). Long noncoding RNAs: past, present, and future. Genetics 193: 651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F., Orom U.A., Cesaroni M., Beringer M., Taatjes D.J., Blobel G.A., Shiekhattar R. (2013). Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang H., Chua N.H. (2015). Long noncoding RNA transcriptome of plants. Plant Biotechnol. J. 13: 319–328. [DOI] [PubMed] [Google Scholar]
- Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L., Arenas-Huertero C., Chua N.H. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.R., et al. (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351: 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q.W., Lin S.S., Reyes J.L., Chen K.C., Wu H.W., Yeh S.D., Chua N.H. (2006). Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Oide S., Bejai S., Staal J., Guan N., Kaliff M., Dixelius C. (2013). A novel role of PR2 in abscisic acid (ABA) mediated, pathogen-induced callose deposition in Arabidopsis thaliana. New Phytol. 200: 1187–1199. [DOI] [PubMed] [Google Scholar]
- Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., Guigo R., Shiekhattar R. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar K.M., Emerine D.K., Mukhtar M.S. (2013). Tell me more: roles of NPRs in plant immunity. Trends Plant Sci. 18: 402–411. [DOI] [PubMed] [Google Scholar]
- Rajniak J., Barco B., Clay N.K., Sattely E.S. (2015). A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature 525: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Chang H.Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S., Thakur J.K. (2015). Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front. Plant Sci. 6: 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönberger J., Hammes U.Z., Dresselhaus T. (2012). In vivo visualization of RNA in plants cells using the λN22 system and a GATEWAY-compatible vector series for candidate RNAs. Plant J. 71: 173–181. [DOI] [PubMed] [Google Scholar]
- Senecoff J.F., McKinney E.C., Meagher R.B. (1996). De novo purine synthesis in Arabidopsis thaliana. II. The PUR7 gene encoding 5′-phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole synthetase is expressed in rapidly dividing tissues. Plant Physiol. 112: 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G., Wahlestedt C., Kapranov P. (2015). The landscape of long noncoding RNA classification. Trends Genet. 31: 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance K.W., Ponting C.P. (2014). Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 30: 348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon L.C., Rep M., Pieterse C.M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44: 135–162. [DOI] [PubMed] [Google Scholar]
- Wang L., Brown S.J. (2006). BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 34: W243–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fan X., Lin F., He G., Terzaghi W., Zhu D., Deng X.W. (2014). Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. USA 111: 10359–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Chen C., Chen Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13: 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N.H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19: 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.