Rice WOX11 promotes crown root development by interacting with a histone acetylation complex to induce permissive chromatin of gene expression.
Abstract
Shoot-borne crown roots are the major root system in cereals. Previous work has shown that the Wuschel-related homeobox gene WOX11 is necessary and sufficient to promote rice (Oryza sativa) crown root emergence and elongation. Here, we show that WOX11 recruits the ADA2-GCN5 histone acetyltransferase module to activate downstream target genes in crown root meristem. Rice ADA2 and GCN5 genes are highly expressed in root meristem and are shown to be essential for cell division and growth. WOX11 and ADA2-GCN5 commonly target and regulate a set of root-specific genes involved in energy metabolism, cell wall biosynthesis, and hormone response, some of which are known to be important for root development. The results indicate that the recruitment of ADA2-GCN5 by WOX11 establishes gene expression programs of crown root meristem cell division and suggest that permissive chromatin modification involving histone acetylation is a strategy for WOX11 to stimulate root meristem development.
INTRODUCTION
Roots represent a vital organ for plants, responsible for absorption of water and mineral nutrients in addition to anchoring plants in the soil. Unlike dicot plants, cereals such as rice (Oryza sativa) and maize (Zea mays) have an extensive postembryonic shoot-borne root system. Shoot-borne roots are initiated from stem nodes or coleoptile sections and are termed adventitious roots or crown roots (Marcon et al., 2013). Rice growth is mainly reliant on a crown root system that is continuously replenished and reshaped by production and elongation of new adventitious roots throughout the growth period. Specific developmental stages of rice crown roots including initiation, emergence, and elongation can be distinguished (Itoh et al., 2005; Coudert et al., 2010; Kitomi et al., 2011; Wang et al., 2011). In the past years, several key genes such as CROWN ROOTLESS1 (CRL1)/ADVENTITIOUS ROOTLESS1, CRL4, OsGNOM1, and WUSCHEL (WUS)-RELATED HOMEOBOX11 (WOX11) have been identified and characterized to function in the regulation of different stages of crown root development (Inukai et al., 2005; Liu et al., 2005, 2009; Kitomi et al., 2008; Zhao et al., 2009). Some of the identified regulators control crown root development by responding to and/or regulating in auxin or cytokinin signaling (Zhao et al., 2009, 2015; Coudert et al., 2011, 2015). For instance, WOX11 is expressed throughout the root meristem region and stimulates crown root emergence and elongation by regulating genes of both auxin and cytokinin signaling (Zhao et al., 2009, 2015).
WUS is the founding member of the plant-specific WOX gene family, which has three major phylogenetic branches: an ancient clade consisting of WOX13-related genes, present in some green algae and in all land plant genomes; the WUS clade; and the intermediate WOX9 clade, to which the rice WOX11 belongs (van der Graaff et al., 2009). The different clades share a characteristic N-terminal homeodomain but contain distinct conserved sequence motifs in the C termini. For instance, members of the WUS clade can contain any of three conserved sequence motifs at their C-terminal ends: a WUS-box, an EAR domain, and an acidic domain (Haecker et al., 2004). The canonical WUS-box is restricted to the WUS clade and has been shown to interact with TOPLESS (TPL)-type corepressors (Zhang et al., 2014; Pi et al., 2015; Dolzblasz et al., 2016). The WUS clade member WOX5, a regulator of the root stem cell niche, interacts with TPL and a histone deacetylase to repress cell differentiation programs in Arabidopsis thaliana root meristem (Pi et al., 2015), suggesting that utilization of chromatin-mediated repression to silence differentiation programs is a strategy for root stem cell maintenance. Consistent with the cell division-promoting role of WOX11 in the rice crown root meristem, WOX9 clade members do not function in stem cell maintenance (Dolzblasz et al., 2016). However, how WOX11 establishes gene expression programs of rice crown root meristem cell proliferation remains unclear.
Histone modifications such as lysine acetylation and lysine methylation are important chromatin modifications in gene expression reprogramming during plant development (Servet et al., 2010; Chen and Zhou, 2013). Acetylation of histone lysine residues induces a permissive chromatin structure for gene activation. The acetylation level of histone lysine residues is a balance between the action of histone acetyltransferases (HATs) and histone deacetylases. In yeast, Gcn5 was identified as the first transcription-related HAT (Brownell et al., 1996). Gcn5 is integrated into the SAGA (Spt-Ada-Gcn5 acetyltransferase) complex, which is an evolutionarily conserved transcriptional coactivator (Grant et al., 1997; Weake and Workman, 2012; Wang and Dent, 2014) that has several modules. The association of Gcn5 with the Ada proteins and Sgf29 in the complex enables Gcn5 to acetylate multiple lysine residues on nucleosomal histone H3 and is important for in vivo HAT activity (Grant et al., 1999; Balasubramanian et al., 2002; Bian et al., 2011). While SAGA is not essential for viability in Saccharomyces cerevisiae, mutations in SAGA subunits in Drosophila melanogaster and in mice (Mus musculus) cause severe developmental defects that are ultimately lethal (Koutelou et al., 2010; Weake and Workman, 2012). Orthologs of Gcn5 and ADA2 have been characterized in Arabidopsis; T-DNA mutants of AtGCN5 and AtADA2b show developmental defects including dwarf size, deformed flowers, and decreased root length (Vlachonasios et al., 2003), suggesting that specific plant developmental pathways may be particularly sensitive to loss of GCN5 function.
In this work, we show that rice WOX11 recruits the ADA2-GCN5 HAT module to regulate crown root cell proliferation. WOX11 and the HAT module commonly target and regulate genes involved in carbon metabolism related to energy production and cell component biosynthesis and in auxin transport and response. The results indicate a permissive chromatin state is a basis for root meristem cell proliferation. Thus, histone acetylation and deacetylation are used by WOX proteins to establish gene expression programs to respectively regulate cell proliferation and stem cell maintenance of root meristem.
RESULTS
Rice WOX11 Interacts with the Histone Acetyltransferase Complex ADA2-GCN5
To study the molecular mechanism of WOX11 function in rice crown root development, we performed yeast two-hybrid screening of a rice root cDNA library to find WOX11-interacting proteins. One of the candidate interacting proteins was homologous to the ADA2 proteins (Vlachonasios et al., 2003; Lee and Workman, 2007). Different from Arabidopsis, which has two genes (AtADA2a and AtADA2b) (Vlachonasios et al., 2003), only one gene encoding the ADA2 homolog was found in rice genome (Supplemental Figure 1A).
In other species, ADA2 and GCN5 form a histone acetyltransferase complex (Kleff et al., 1995; Brownell et al., 1996; Carrozza et al., 2003; Vlachonasios et al., 2003). To confirm the interaction between rice ADA2 and WOX11 and between ADA2 and GCN5, we cloned the full-length cDNAs of ADA2 and GCN5 and performed yeast two-hybrid assays. As shown in Figure 1A, interaction between ADA2 and WOX11 and between ADA2 and GCN5 occurred in yeast cells. However, no direct interaction between GCN5 and WOX11 was detected in the assays. Analysis with truncated WOX11 and ADA2 revealed that the homeobox domain of WOX11 and the middle region of ADA2 (named as ADA2-2) interacted with each other. The ADA2-2 region was also found to interact with GCN5 (Supplemental Figure 1B).
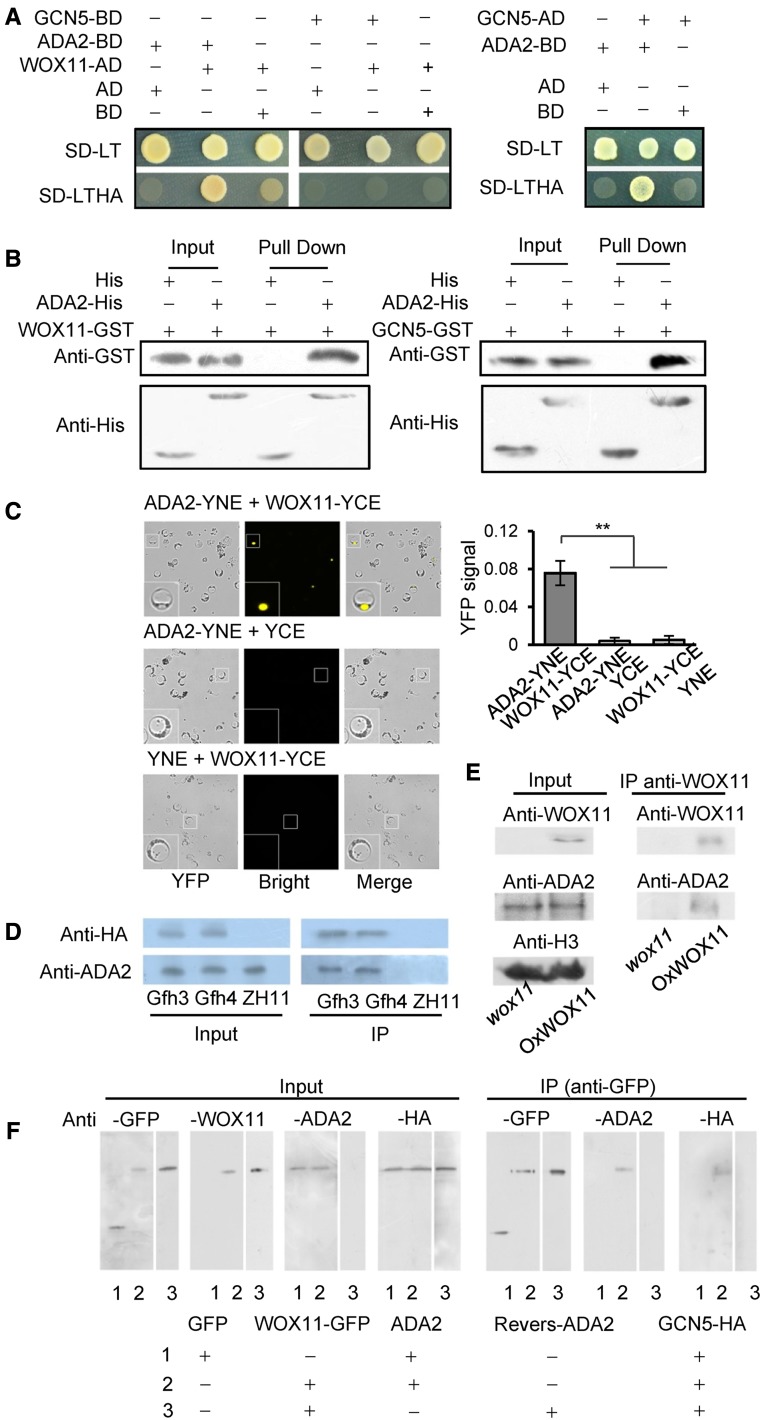
Figure 1.
Rice GCN5 Interacts with WOX11 through ADA2 in Vitro and in Vivo.
(A) Yeast two-hybrid assay of ADA2 with WOX11 and GCN5. Full-length cDNAs of GCN5, ADA2, and WOX11 were cloned into AD (the prey plasmid pGADT7) or BD (the bait plasmid pGBKT7). Yeast cells transformed with the indicated plasmids were grown on control SD-LT medium and selective SD-LTHA medium.
(B) Pull-down assay of ADA2 interaction with WOX11 and GCN5. ADA2-His fusion protein or His alone (from FET32a vector) was tested for the ability to pull down WOX11-GST (left) or GCN5-GST fusion protein (right).
(C) Confirmation of interaction of ADA2 with WOX11 in rice protoplast nuclei by BiFC. ADA2-YNE (YFP N-terminal element) and WOX11-YCE (YFP C-terminal element) interaction produced a yellow signal in the nucleus. Protoplasts transformed with ADA2-YNE and YCE, and YNE and WOX11-YCE are negative controls. Ratios of YFP signal in each group of cells are shown as histograms; significant difference between test and control groups (Student’s t tests, P value < 0.01) is denoted by double asterisks.
(D) Coimmunoprecipitation assays of ADA2 and GCN5 interaction in vivo. Immunoprecipitations were performed with GCN5-flag-HA lines (Gfh3 and Gfh4) and the wild type (ZH11) using anti-HA antibody. The precipitated proteins were analyzed by immunoblots using anti-HA and anti-ADA2 as indicated.
(E) Coimmunoprecipitation assays of sADA2 and WOX11 interaction in vivo in crown root. Immunoprecipitations were performed with WOX11 overexpression lines (OxWOX11) and wox11 mutant with anti-WOX11 antibody. The precipitated proteins were analyzed by immunoblots with anti-WOX11 and anti-ADA2.
(F) Detection of WOX11-ADA2-GCN5 complex in rice cells by coimmunoprecipitation assays. Plasmids containing WOX11-GFP or GFP alone were transformed together with those containing GCN5-2XHA and ADA2 or reverse ADA2 cDNA as a control into rice protoplasts. Immunoprecipitation assays were performed using GFP-Trap agarose beads. Samples of input and immunoprecipitation products were analyzed by immunoblots using anti-GFP, anti-ADA2, anti-HA, and/or anti-WOX11 antibody as indicated. Combinations 1, 2, and 3 of plasmids used for protoplast transformation are indicated on the bottom.
To validate the yeast two-hybrid results, both in vitro and in vivo protein interaction assays were performed. Pull-down assays using Escherichia coli-produced ADA2-6XHis-tag and WOX11-GST or GCN5-GST fusion proteins showed that ADA2-6×His, but not 6×His alone, could retain the WOX11 protein (Figure 1B). Interactions were further validated by bimolecular fluorescence complementation (BiFC) assays, in which YFP was reconstituted in the nucleus when ADA2-YNE and WOX11-YCE were coexpressed (Figure 1C). By contrast, coexpression of ADA2-YNE (YFP N terminus) and YCE (YFP C terminus), or YNE and WOX11-YCE did not produce any YFP signal.
To study in vivo interaction between rice ADA2 and GCN5, we constructed 35S:GCN5-2xflag-2xHA (GCN5-fh) transgenic plants and prepared polyclonal antibodies for the ADA2 protein (Supplemental Figure 2). Protein extracts of two lines of GCN5-fh transgenic plants and of the wild type were first immunoprecipitated with anti-HA antibody and then analyzed by immunoblotting with anti-HA and anti-ADA2 antibodies. The GCN5-fh fusion protein was coimmunoprecipitated with ADA2 (Figure 1D). To confirm the interaction, we extracted nuclear proteins from wox11 mutant and WOX11 overexpression lines and used them for immunoprecipitations with anti-WOX11 antibody, followed by immunoblotting with anti-WOX11 and anti-ADA2 antibodies. As shown in Figure 1E, ADA2 could be immunoprecipitated by anti-WOX11 in the overexpression lines but not in the wox11 mutant.
To study whether WOX11, ADA2, and GCN5 could form a complex in vivo, we cotransformed rice protoplasts with 35S:WOX11-GFP or 35S:GFP constructs together with 35S:ADA2 and 35S:GCN5-HA. A construct with ADA2 in the reverse orientation was used as a control. Protein extracts from the transfected protoplasts were first immunoprecipitated by anti-GFP antibody and then analyzed by immunoblotting with anti-GFP, anti-HA, and anti-ADA2 antibodies, respectively. As shown in Figure 1F, ADA2 and GCN5 were coimmunoprecipitated with WOX1-GFP, but not with GFP alone, nor with the control vector, indicating that these three proteins could form a complex in rice cells.
Rice ADA2 and GCN5 Are Highly Expressed in Root Meristem, and GCN5 Has a General Histone Acetyltransferase Activity
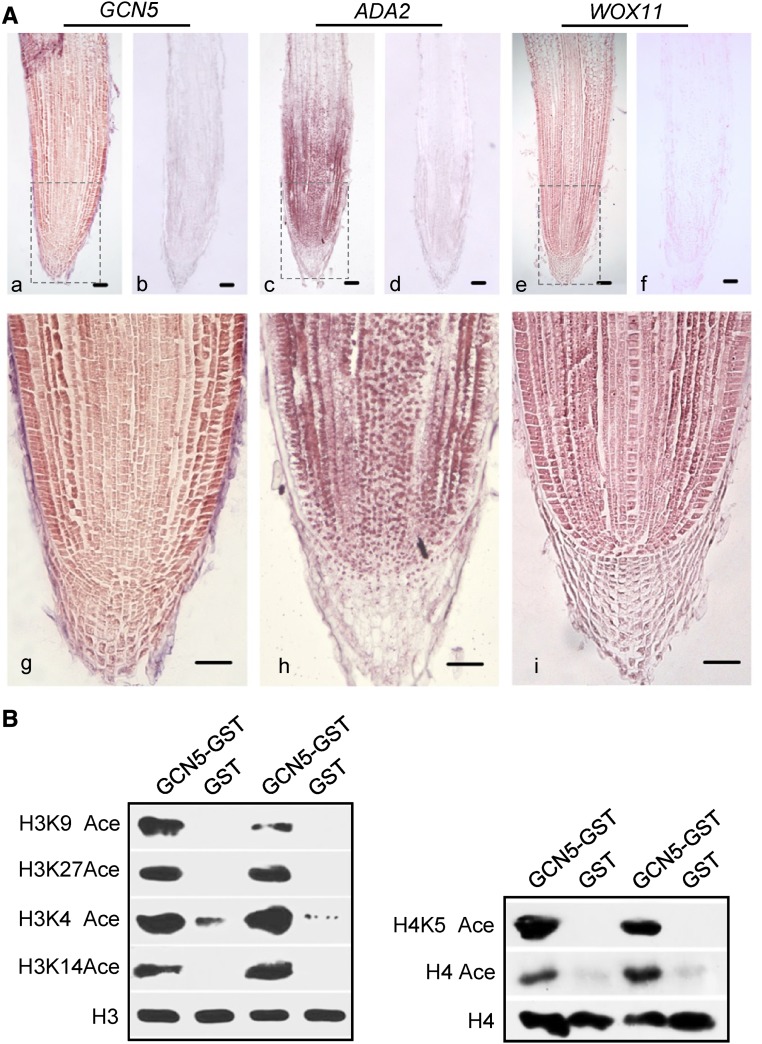
GCN5 and ADA2 displayed similar expression profiles and were broadly expressed in different rice tissues/organs (Supplemental Figure 1C). In situ hybridization indicated that GCN5, ADA2, and WOX11 transcripts were present in all cell types of the crown root tip (although ADA2 appeared to be slightly less expressed in root cap cells), indicating that the expression of these genes overlapped in the root meristem (Figure 2A). This coexpression and the interactions between GCN5, ADA2, and WOX11 in root meristem cells suggested the possibility of their cooperative function in root meristem development.
Figure 2.
The ADA2 and GCN5 HAT Module Is Highly Expressed in Crown Root Meristem.
(A) In situ hybridization of GCN5, ADA2, and WOX11 transcripts in crown root tip. Antisense (a, c, and e) and sense (b, d, and f) probes were used. Close-up views of boxed areas in a, c, and e are shown in g, h, and i, respectively. Bars = 25 μm.
(B) In vitro assay of GCN5 acetylation activity. Recombinant histone H3 or H4 was incubated with purified GCN5-GST protein or GST alone and analyzed by immunoblot using the antibodies indicated on the left. Ace, acetylated. Anti-H3 and anti H4 were used as controls.
To test for histone acetyltransferase activity of GCN5, E. coli-expressed and affinity-purified GCN5-GST protein was used for in vitro histone acetyltransferase assays. Consistent with previous studies in S. cerevisiae and mammals, GCN5 could acetylate histone H3 K4, K9, K14, and K27 sites, as well as H4 (Figure 2B).
Knockdown of Rice GCN5 and ADA2 Affects Crown Root Initiation and Elongation and Mature Plant Height
To study whether GCN5 and ADA2 were required for rice crown root initiation and development, we attempted to knockout the genes by CRISPR-Case 9 technology. However, we could not obtain homozygous knockout plants, suggesting that the genes may be essential for plant regeneration from tissue culture. Instead, we produced GCN5 and ADA2 RNAi transgenic lines. Four RNAi transgenic lines with significantly decreased expression (5-fold) of GCN5 (RG lines) or ADA2 (RA lines) were selected for analysis (Figures 3A and 3B). The ADA2 protein levels in the ADA2 RNAi lines checked by immunoblots using ADA2 antibody (Supplemental Figure 2B) confirmed the downregulation of the gene in the RNAi transgenic lines.
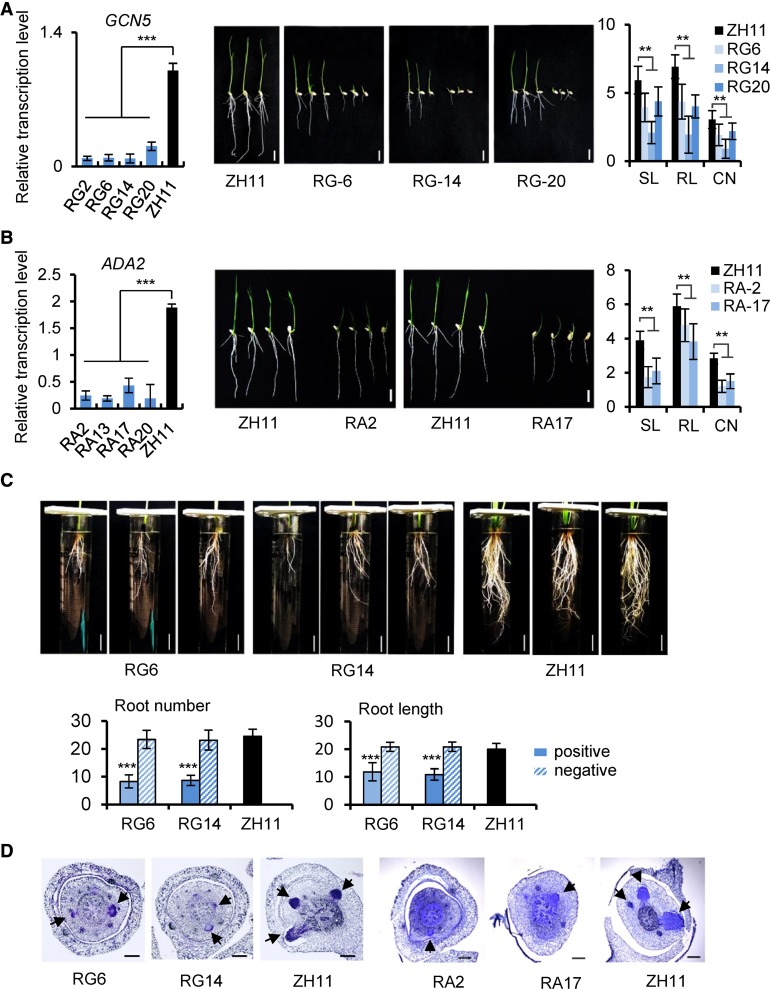
Figure 3.
Characterization of Rice GCN5 and ADA2 RNAi Lines.
(A) and (B) GCN5 (A) and ADA2 (B) RNAi lines. Left, transcripts in four transgenic RNAi lines and the wild type. Values are relative to ACTIN1 transcripts. Bars represent means ± sd from three biological replicates. Each replicate was performed with independent mRNA extractions from different 10-d-old seedlings for each genotype. Significant differences calculated from the three biological replicates are indicated by triple asterisks (P value < 0.001). Middle, 7-d-old seedling phenotype; bars = 1 cm. Right, shoot (SL) and root (RL) lengths, and crown root number (CN) of the RNAi lines compared with the wild type (ZH11). Data are from three GCN5 or ADA2 RNAi lines. Bars are means ± sd of 30 plants. Significant difference between transgenic lines and the wild type (Student’s t tests, P value < 0.01) are marked by double asterisks.
(C) Upper panels: Comparison of root number and length of 45-d-old hydroponic GCN5 RNAi lines and the wild type. Bars = 3 cm. Lower panels: Statistical data of root numbers and root lengths of RG6 and RG14 positive and negative (without the transgene) segregants from T3 generation and wild-type plants. Error bars of histogram denote ± sd of 12 plants. Significant difference between the positive RNAi lines and the wild type (Student’s t tests, P value < 0.001) was marked by triple asterisks.
(D) Toluidine blue-stained cross sections of coleoptilar nodes of GCN5 (3 d old) and ADA2 (5 d old) RNAi lines and corresponding wild type. Arrows indicate emerging crown root initials. Bar = 100 μm.
Inspecting the phenotypes of 7-d-old seedlings revealed that both the GCN5 and ADA2 RNAi lines produced fewer crown roots and showed reduced primary root length and shoot height compared with the wild type (Student’s t test, P value < 0.01) (Figures 3A and 3B). After 45 d of hydroponic culture, the GCN5 RNAi plants (T3) showed significant decreases in crown root number and length compared with negative segregants (T3) or wild-type plants (Figure 3C). To exclude the possibility that the root phenotypes observed in GCN5 and ADA2 RNAi plants were caused by delayed shoot growth, we germinated GCN5 and ADA2 RNAi seeds 1.5 d earlier than the wild type to have about the same shoot lengths and then compared root phenotypes between the RNAi and wild-type plants. As shown in Supplemental Figure 3, with the same shoot lengths, the RNAi plants produced fewer crown roots and reduced root lengths compared with the wild type. To check whether downregulation of GCN5 and ADA2 affected crown root initiation, cross sections of 3-d-old coleoptilar nodes were stained with toluidine blue. The staining revealed clear delays of crown root primordium formation in GCN5 and ADA2 RNAi plants compared with the wild type. The phenotypes of the RNAi plants were similar to those of the wox11 mutant (Zhao et al., 2009). At maturity, GCN5 RNAi and ADA2 RNAi plants were semidwarf with much reduced internode lengths (Supplemental Figures 4A and 4B). Histological analysis of internode tissues revealed no difference in cell length between the GCN5 RNAi lines and the wild type (Supplemental Figure 4C), suggesting that the shorter internodes of the RNAi plants may be caused by reduced cell division rates. By contrast, GCN5 overexpression lines did not show any obvious difference in root development or plant growth (Supplemental Figure 5), suggesting that GCN5 levels were not limiting in wild-type plants.
GCN5 Regulates Meristem Size and Cell Division of Crown Roots
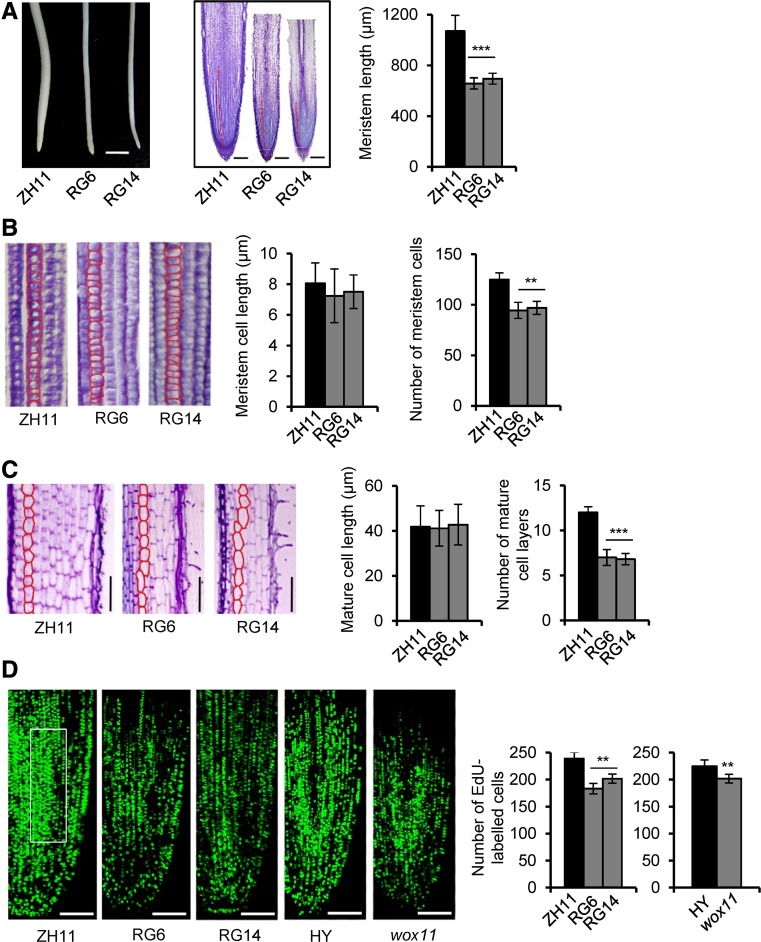
To further understand the effect of GCN5 RNAi on crown root development, longitudinal root sections of RNAi and wild-type crown roots of hydroponic culture were inspected by toluidine blue staining. The RNAi crown roots were thinner and showed smaller meristem zones compared with the wild type (Figure 4). Examination of cell number and longitudinal length in the meristem zone revealed significantly fewer cells in the meristem with no obvious difference of cell lengths in the RNAi root meristem zones (Figure 4B) (see Methods). Longitudinal sections of differentiated zones indicated that the RNAi line had about only half as many cortical cell layers as in the wild type (Figure 4C). These data indicated that the smaller roots of the RNAi plants were due to reduced cell numbers and cell layers, suggesting that GCN5 is required for both transversal and longitudinal divisions of meristem cells. To examine whether the cell division rate was affected in GCN5 RNAi roots, 5-ethynyl-20-deoxyuridine (EdU), which labels newly replicated DNA, was used to stain crown root tips. The EdU signal was noticeably decreased in the RNAi lines compared with the wild type. A decrease of staining was also observed in the wox11 mutant compared with its wild type (HY) (Figure 4D). These data suggested that GCN5 and WOX11 may modulate crown root development by regulation of cell division in root meristem.
Figure 4.
GCN5 RNAi Lines Have a Reduced Rice Crown Root Meristem Cell Division Rate.
(A) Root phenotype of GCN5 RNAi lines and the wild type (ZH11). Left: GCN5 RNAi lines show smaller root tips. Bars = 3 mm. Middle: Median longitudinal sections through root tips of GCN5 RNAi and the wild type; red lines delimit the meristem size (from the quiescent center to the transition zone). Bars = 200 μm. Right: Statistical data for meristem lengths from two RNAi lines (RG6 and RG14) and the wild type. Bars represent means ± sd of 10 roots. Significant differences between transgenic lines and the wild type (Student’s t test, P value < 0.001) are marked by triple asterisks.
(B) Cell size and number within the same length of root meristem cell files of GCN5 RNAi and the wild type. Statistical data for meristem cell length and number from each material are shown on the right. Bars show means ± sd of 10 roots. Significant differences with P < 0.01 (Student’s t tests) between RNAi and the wild type are marked by two asterisks.
(C) Cell size and layer number in the differentiated zone of GCN5 RNAi and wild-type roots. Bars = 100 μm. Statistical analysis showed significant difference (Student’s t test, P < 0.001) between the RNAi lines and the wild type. Bars show means ± sd.
(D) Longitudinal view of EdU-labeled cells in the 4-d-old seedling root meristems of GCN5 RNAi and its wild type (ZH11), or wox11 mutant and its wild type (HY). Bars = 100 μm. Numbers of EdU-labeled cells in an arbitrary area of root tip (white box) were counted. Error bars show means ± sd of five roots. Significant differences between the knockdown/knockout lines and the corresponding wild type are indicated by double asterisks (Student’s t test P value<0.01).
GCN5 RNAi and the wox11 Mutant Display Attenuated Auxin Accumulation in Root
Polar auxin transport and an auxin maximum near the quiescent center (QC) are vital for maintaining neighboring meristem cell division (Overvoorde et al., 2010). To study whether GCN5 RNAi and the wox11 mutation affected auxin accumulation in crown roots, the auxin reporter line DR5-GUS was crossed with GCN5 RNAi lines and the wox11 mutant. In GCN5 RNAi and wox11 mutant backgrounds, the expression of DR5-GUS in regions around the root cap and stele was much weaker compared with the wild type (Supplemental Figure 6A). Histological analysis suggested that the auxin accumulation was much weaker in the crown root cap, QC, and the epidermal region of the meristem zone in 5-d-old GCN5 RNAi and wox11 mutant plants (Supplemental Figure 6A). These observations suggested that GCN5 and WOX11 may be required for auxin transport or responses in the root meristem to stimulate cell division.
GCN5 RNAi Preferentially Reduces the Expression of Root-Specific Genes Related to Metabolism, Cell Components, and Stress
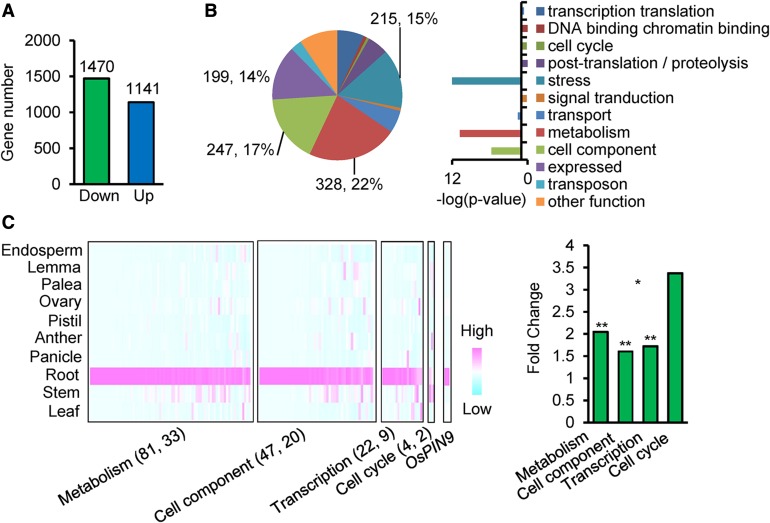
To evaluate the effect of GCN5 RNAi on gene expression in roots, we analyzed the transcriptome of 5-d-old roots (a mixture of crown roots and the primary root) of wild-type (ZH11) and RNAi (lines 6, 14, and 20 combined) plants by RNA-seq. Two biological repeats with mRNA isolated from different plants were performed. Trimmomatic (version 0.32), TopHat (version 2.0.13), and Cufflink pipelined software were used to identify differentially expressed genes; >99.5% raw tags were of high quality and were kept for further alignment. In total, >85% clean tags could be aligned to the reference rice genome. These parameters confirmed the high quality of the sequencing data (Supplemental Table 1). Pairwise scatterplots reflected the highly repeatability of the biological replicates (Supplemental Figure 7A). Compared with the wild type, 1470 down- and 1141 upregulated (P value < 0.05, fold change > 2) genes were identified in GCN5 RNAi roots (Figure 5A).
Figure 5.
RNA-Seq Analysis of GCN5 RNAi Roots.
(A) Histogram shows numbers of up- and downregulated genes (>2-fold, P < 0.05) in 5-d-old roots of three combined GCN5 RNAi lines compared with the wild type.
(B) Distribution of functional categories of downregulated genes in GCN5 RNAi roots. Gene numbers and percentages are indicated. Histograms indicate P values of the enriched functional categories.
(C) Many downregulated metabolism-, cell component-, transcription-, and cell cycle-related genes in GCN5 RNAi plants are specifically expressed in roots. Left panel: Expression heat map of downregulated genes of the specific categories. Numbers in parentheses are genes specifically expressed in roots and genes containing the WOX binding motif. Right panel: Relative enrichments of root specific genes of the different categories. Fold changes are differences relative to the observed frequencies of specific subsets in the genome. Significant enrichments (Fisher’s tests) are indicated: *P value < 0.05 and **P value < 0.01.
Given the function of histone acetyltransferases in gene activation, we reasoned that the downstream genes of GCN5 were likely to be among the downregulated genes. Of those downregulated genes, 22% (328), 17% (247), and 15% (215) were respectively related to the significantly enriched categories metabolism, cell component, and stress (Figure 5B). Genes involved in energy metabolism (such as glucan, glucose, and lipid metabolic/transport processes), cell component (such as cell wall organization/cellulose catabolic processes) were particularly enriched (Supplemental Table 2). In addition, 98 downregulated genes were related to gene transcription/translation and 9 were related to cell cycle. Among the downregulated genes, 81 metabolism-, 47 cell component-, 22 transcription-, and 4 cell cycle-related genes showed root-specific expression (see Methods) and were significantly enriched compared with their genomic frequencies (Figure 5C). These observations suggest that GCN5 may be involved in root-specific gene expression. In addition, among the downregulated root-specific genes, 33 metabolism, 20 cell component, 9 transcription, and 2 cell cycle genes were found to contain the WOX binding motif in the 1-kb promoter region or in the gene body (Figure 5C). In addition, consistent with the defects in auxin transport or responses in wox11 and GCN5 RNAi root meristems (Supplemental Figure 6A), the putative auxin efflux transporter gene OsPIN9 was also found to be among the downregulated root-specific genes in GCN5 RNAi and to contain the WOX binding motif (Figure 5C). OsPIN9 is mostly closely related to Arabidopsis AtPIN1 and AtPIN4, which are reported to play a key role in auxin transport from the stele to the QC (Blilou et al., 2005). The downregulation of OsPIN9 might be related to the decrease of auxin concentration in the root meristems of the wox11 mutant and GCN5 RNAi lines. In addition, we identified 47 auxin-inducible genes among downregulated genes in GCN5 RNAi roots, some of which were also downregulated in the wox11 mutant (Supplemental Figure 6B). These data suggested that GCN5 and WOX11 might be required for the expression of auxin transport- and response-related genes in roots.
Important Cell Metabolic and Hormonal Genes Are Common Targets of ADA2-GCN5 and WOX11 in Crown Root Cells
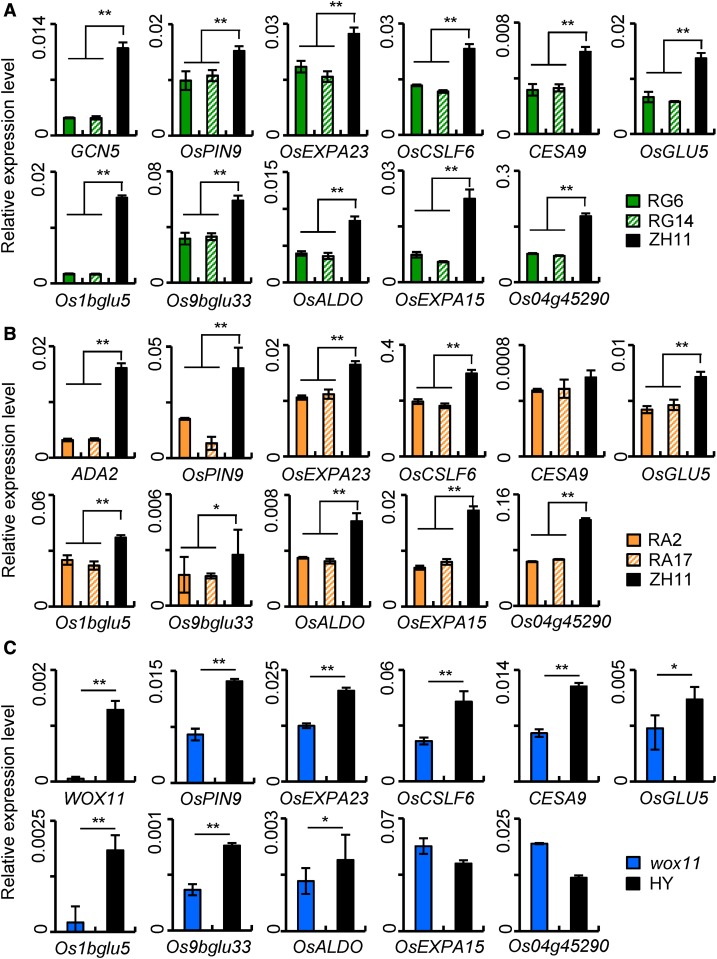
About 30% (128/434) downregulated genes in wox11 root tips overlapped with those downregulated in GCN5 RNAi (Supplemental Figure 7B) (Jiang et al., 2017). These genes might be potential common targets of WOX11 and GCN5 in rice roots. From those that contain the WOX binding site and were downregulated in both GCN5 RNAi and wox11 mutant, we selected eight genes from representative categories (including metabolism, cell component, and hormone) for experimental validation. These genes included auxin transport-related OsPIN9, cell wall biosynthesis-related CESA9 (encoding cellulose synthase), CSLF6 (cellulose synthase-like; Jin et al., 2015), and OsEXPA23 (expansin precursor), and energy metabolism-related OsGLU5 (1,4 β-endoglucanase; Zhang et al., 2012), Os1bglu5 (β-glucosidase), Os9bglu33 (β-glucosidase; Rouyi et al., 2014), and OsALDO (fructose-bisphosphate aldolase, a key enzyme of glycolysis). qRT-PCR analysis indicated that all of these genes were indeed downregulated in crown roots of GCN5 and ADA2 RNAi and wox11 mutant plants compared with the respective wild type (Figures 6A to 6C). OsEXPA15 and Os04g45290, which do not contain the WOX binding site and were downregulated only in GCN5 RNAi, were used as controls. In situ hybridization assays detected the transcripts of OsPIN9 and OsCSLF6 in all cell types of the wild-type crown root tips, and GCN5 RNAi and the wox11 mutation reduced the overall hybridization signals (Supplemental Figure 8), consistent with GCN5 and WOX11 expression pattern in the root tip (Figure 2A).
Figure 6.
Analysis of GCN5 and WOX11 Common Target Genes.
Transcript levels of putative common target genes in root of two GCN5 RNAi lines (RG6 and RG14) compared with the wild type (ZH11) (A), two ADA2 RNAi lines (RA2 and RA17) compared with the wild type (ZH11) (B), and of wox11 mutant compared with its wild type (HY) (C). Values are relative to ACTIN1 transcripts. Bars indicate means ± sd from three biological replicates. Each replicate was performed with independent mRNA extraction from crown roots of different plants for each genotype. Significance of differences between indicated samples was calculated from three biological replicates (**P value < 0.01 and *P value < 0.05).
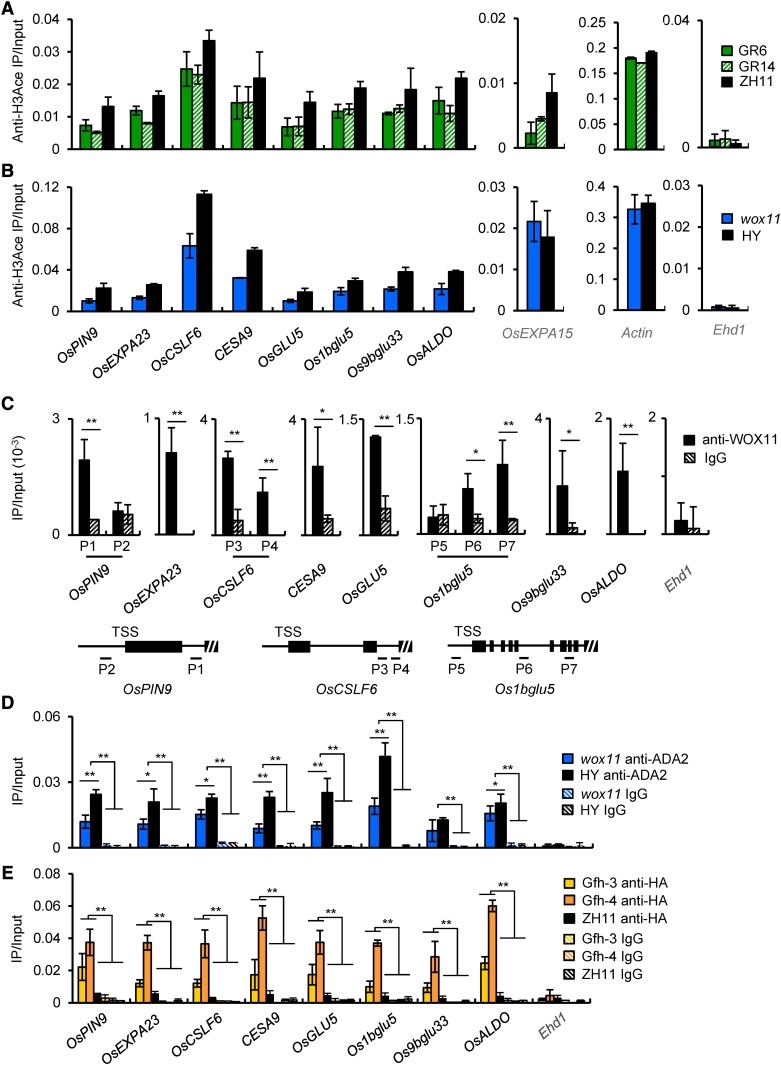
In addition, histone acetylation levels at the promoter region of the eight genes together with three control genes (ACTIN1, OsEXPA15, and Ehd1, which had no transcript detected in root tips in our RNA-seq data) were analyzed by chromatin immunoprecipitation (ChIP) assays using antibody of acetylated H3 and 5-d-old roots (a mixture of and crown roots and the primary root). The precipitated chromatin fragments were analyzed by qPCR using primer sets corresponding to the promoter region (from −300 bp to +1 bp relative to the transcription start site of the genes). Decreased histone H3 acetylation was observed in the promoters of the eight genes in both the GCN5 RNAi lines and the wox11 mutant. A decrease of acetylation in the control gene OsEXPA15 was detected only in the GCN5 RNAi plants but not in wox11, while no decrease was observed at ACTIN1 in either genotype, and only background acetylation level was detected in Ehd1 (Figures 7A and 7B; Supplemental Figure 9). Notably, the acetylation data corresponded well with the expression levels of the tested genes in the different genotypes (Figure 6).
Figure 7.
ADA2-GCN5 and WOX11 Directly Bind to Common Downstream Genes and Regulate Histone H3 Acetylation in Crown Root.
(A) and (B) GCN5 and WOX11 regulate histone H3 acetylation of eight common downstream genes. Histograms on left are ChIP assays with anti H3ace of eight common target genes in roots of two GCN5 RNAi lines compared with the wild type (ZH11) (A) and wox11 mutant compared with the wild type (HY) (B). Histograms on side are tests of OsEXPA15 and ACTIN loci as controls. Bars represent means ± sd from three technical repeats of one biological replicate. The other two biological replicates are shown in Supplemental Figure 9. Each biological replicate was performed using crown roots collected from independently germinated seedlings of each genotype.
(C) WOX11 binds directly to common target genes. ChIP assays were performed using anti-WOX11 antibody with roots of wild-type plants. Anti-IgG was used as a control. Three of the eight genes have more than one putative WOX binding site. Primer sets corresponding to each of the binding sites were used (see schematic below the graphs).
(D) and (E) ADA2 and GCN5 associate directly with the downstream genes. ChIP assays were performed using polyclonal anti-ADA2 (D) or anti-HA (E). Anti-IgG was used as a control. ChIP assays with anti-ADA2 or anti-HA antibodies were performed with root chromatin fragments of wox11 mutant and wild-type (HY) plants and of GCN5-falg-HA and wild-type (ZH11) plants, respectively.
Bars in (C) to (E) are means ± sd from three biological replicates. Each replicate was performed using crown roots harvested from independently germinated seedlings for each genotype. Significant enrichments (Student’s t tests) calculated from three biological replicates are indicated: *P value < 0.05 and **P value < 0.01.
To examine whether WOX11, ADA2, and GCN5 directly associated with these genes, we first performed ChIP analysis of wild-type roots using anti-WOX11, with IgG as control. The precipitated chromatin fragments were analyzed by qPCR using the primer sets covering the WOX11 binding motifs in the downstream genes. Among the tested genes, OsPIN9, OsCSLF6, and Os1bglu5 contain more than one putative WOX binding site, while the other genes each have only one in the gene body. At least one putative WOX binding site was found to be associated with WOX11 in each of the tested genes (Figure 7C). To study whether ADA2 bound to the genes, the same root chromatin fragments were precipitated using anti-ADA2 and analyzed by qPCR with the same primer sets as used in anti-WOX11 ChIP assays (i.e., P1 for OsPIN9, P3 for OsCSLF6, and P6 for Os1bglu5). The analysis revealed a clear association of ADA2 with the genes in wild-type roots. However, the ADA2 binding was much reduced in wox11 roots, suggesting that WOX11 was required for efficient recruitment of ADA2 to the target genes (Figure 7D). Next, to study whether GCN5 also associated with the genes, we analyzed two lines of GCN5-fh transgenic roots together with the wild type by ChIP assays using anti-HA, with IgG as control. The same primer sets for examining ADA2 binding were used for qPCR. The analysis with anti-HA revealed noticeably higher signals in the transgenic roots than in the wild type or than with IgG, suggesting that GCN5 also bound to the genes in root cells (Figure 7E). The negative control, Ehd1, was not bound by WOX11, ADA2, or GCN5 (Figures 7C to 7E). By contrast, in GCN5 RNAi roots, the binding of WOX11 to the common target genes was not clearly affected (Supplemental Figure 10), suggesting that GCN5-mediated histone acetylation is not a prerequisite of WOX11 binding to target loci. Together, the data indicated that the binding of WOX11 is required for the recruitment of the ADA2-GCN5 HAT module to the common target genes.
DISCUSSION
WOX11 Recruits ADA2/GCN5 to Establish Gene Expression Programs of Crown Root Meristem Cell Proliferation
Our previous and present data, showing that the rice WOX11 is required to stimulate crown root initiation and elongation by stimulating cell division in the meristem (Zhao et al., 2009, 2015), suggest that WOX11 may activate specific gene expression programs required for crown root meristem cell proliferation. In Arabidopsis, WOX11 and WOX12 directly activate WOX5 and WOX7 to promote root primordium initiation and organogenesis (Liu et al., 2014; Hu and Xu, 2016). However, it remains unclear whether WOX11 has a similar function as Arabidopsis WOX11/12 to promote the formation of crown root primordia, considering that developmental functions of WOX family members between rice and Arabidopsis are not always conserved (Somssich et al., 2016). Arabidopsis WUS clade members WUS, WOX4, and WOX5 are regulators of stem cell maintenance in the shoot floral meristems, cambium, and root meristem, respectively. WOX5, which regulates the root stem cell niche in the quiescent center, represses transcription of CDF4, which encodes a cell differentiation-promoting transcription factor, by recruiting TPL/TPR corepressors and the histone deacetylase HDA19, and consequently inducing histone deacetylation at the CDF4 regulatory region (Pi et al., 2015). The WOX9 clade includes WOX8, 9, 11, and 12. Members of the WOX9 and the ancient WOX13 clades are nonfunctional in stem cell maintenance (Dolzblasz et al., 2016), perhaps due to the absence of a canonical WUS box (and also an EAR domain). It is thus likely that the members of these clades do not act via recruitment of TPL/TPR proteins for gene repression. These data showing protein interaction and overlapping expression patterns between rice WOX11, ADA2, and GCN5 in the crown root meristem suggest that WOX11-dependent transcription involves permissive chromatin modifying activity to establish gene activation programs required for root meristem cell proliferation during crown root initiation and elongation. The results indicate that intermediate clade WOX transcription factor WOX11 directly activates target genes through recruiting the histone acetyltransferase complex. Thus, WOX proteins recruit histone acetylation and deacetylation enzymes to regulate different aspects of plant meristem development.
Our results establish a model in which WOX11 recruits the ADA2-GCN5 HAT module to target root-specific genes involved in diverse processes, including auxin transport and response, transcription, cell cycle, and metabolic processes such as energy metabolism and biosynthesis of cell wall components. Indeed, some of the common target genes have been shown to be essential for root growth in rice. For instance, CSLF6, a putative cellulose-like synthase family member, is required for rice primary and adventitious root development through a phosphate accumulation-dependent pathway (Jin et al., 2015). Rice expansin genes, including OsEXPA8, OsEXPA17, and OsEXPB5, are reported to modulate root development (Won et al., 2010; ZhiMing et al., 2011; Ma et al., 2013). In addition, CESAs (cellulose synthases) are delivered to cell plate and deposit cellulose via phragmoplast-associated vesicles during cell division in Arabidopsis root (Miart et al., 2014). Their altered expression may be related to the root phenotypes observed in wox11 and GCN5 RNAi plants.
Function of the ADA2-GCN5 HAT Module in Root Meristem Development
Although there are a large number of predicted targets for SAGA-like activities in plants, experimental evidence implies that many key regulatory genes of cell proliferation and cell identity are particularly regulated by plant SAGA complexes and their misexpression in SAGA mutants causes specific developmental defects (Bertrand et al., 2003; Vlachonasios et al., 2003; Benhamed et al., 2006; Kornet and Scheres, 2009; Anzola et al., 2010; Servet et al., 2010). Our finding that auxin-responsive genes are enriched among downregulated genes in rice GCN5 RNAi roots is consistent with previous results that GCN5 and ADA2b are required for auxin signaling gene expression in the Arabidopsis root meristem (Aida et al., 2004; Kornet and Scheres, 2009). These results suggest that plant ADA2/GCN5 HAT modules might stimulate root meristem cell division by activating auxin signaling genes. Interaction with WOX11 may allow specific targeting of the HAT complex to auxin genes for transcriptional activation in the rice crown root meristem. This work thus establishes a link between control of cell proliferation, auxin signaling, and transcriptional regulation involving histone acetylation enzymes.
The observation that genes involved in carbon and energy metabolism are enriched among downregulated genes in GCN5 RNAi root tips suggests that high carbon metabolic activity that supplies catabolic precursors of cell component synthesis and energy may be essential for root meristem development. The observations that ADA2-GCN5 directly targeted to OsEXPA23, CESA9, CSLF6, and OsGlu5 (involved in cellulose synthesis and breakdown) and Os1bglu5, Os9bglu33, and OsALDO (involved in energy metabolism) and that the targeting required the presence of WOX11 (Figure 7), indicate that a cooperation between this HAT complex and WOX11 is necessary for expression of the genes in rice roots. Previous results indicate that glucose is a main energy source for generating ATP and also serves as an important signaling factor in accelerating cell division in Arabidopsis root meristem (Gibson, 2005; Mishra et al., 2009; Xiong et al., 2013). Similarly, activation of carbon metabolic genes is observed during the transition from shoot to inflorescence meristem in rice (Liu et al., 2015). Likely, higher carbon and energy metabolism is required to support rapid cell division and meristem growth. In addition, HAT enzymes use acetyl-CoA to acetylate histones. Perhaps a higher energy metabolic activity may be required to sustain the acetyl-CoA pool required for ADA2-GCN5 HAT-mediated histone acetylation in crown root meristem cells, which highlights the interplay between histone modification and metabolism for plant developmental regulation (Shen et al., 2015, 2016).
Plant ADA2 Proteins Function as a Scaffold for HAT Interaction with Transcription Factors
The SAGA complex is a highly conserved transcriptional coactivator that is involved in the transcription of nearly all active genes in yeast and in plants (Huisinga and Pugh, 2004; Benhamed et al., 2008; Koutelou et al., 2010; Bonnet et al., 2014). In the rice genome, there are single genes encoding ADA2 and GCN5, while there seems to be no ADA3 in plants (Vlachonasios et al., 2003). The severe phenotypes of atgcn5 or atada2b T-DNA mutants (which are not complete loss-of-function mutants) and our failure to recover loss-of-function mutants of the rice genes suggest that the ADA2-GCN5 HAT module is likely to be essential in plants. Our data indicating an important role for the HAT module of the rice SAGA-like complex in cell division of crown root meristem lead us to hypothesize that specific developmental genes might be particularly sensitive to loss of SAGA activities, which has important implications for specific recruitment of SAGA to target genes in particular cell types.
There are different ways to recruit HAT modules of SAGA to gene promoters. HAT complexes can interact with active chromatin marks such as acetylated histone lysine residues and trimethylated H3 lysine 4 (H3K4me3) or with the transcriptional machinery components (e.g., Sanchez et al., 2014). Interaction between HAT and specific DNA binding transcription factors would increase the specificity and durability of target gene activation, which is important for regulation of specific developmental processes. These data together with previous results indicate that in plants, ADA2 proteins are able to interact with DNA binding transcription factors, which provides a mechanism to recruit HAT-containing SAGA-like complexes to specific genes. For instance, Arabidopsis ADA2b interacts with the AP2 transcription factor CBF1, which promotes expression of several cold-responsive genes (Mao et al., 2006), whereas ADA2 from maize binds the basic leucine zipper (bZIP) factor OPAQUE2 (Bhat et al., 2004), which is involved in regulating seed storage genes during maize endosperm development. These observations suggest that plant ADA2 proteins may serve as a scaffold for interaction between transcription factors and GCN5-containing HAT complexes and may play an important role in signal-induced or tissue-specific gene expression programs.
METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa spp japonica) material for transformation of GCN5 RNAi and overexpression vectors used in this study was from the ‘ZhongHua11’ (ZH11) background. The wox11 mutant was described previously (Zhao et al., 2009). Transgenic plants of T3 segregating populations were used for genotypic and phenotypic analysis. For germination culture, seeds were surface-sterilized and germinated in medium containing 0.8% agar supplemented with 3% (w/v) sucrose at 28°C (in light) and 24°C (in dark) with a 14-h-light (white, 10,000 lx)/10-h-dark cycle. Seedling ages were counted in days after germination for phenotype inspection. For growth in field, germinated rice seedlings were transplanted in the field at the beginning of May in the Wuhan area; seeds were harvested in September.
Yeast Two-Hybrid Screening
Yeast two-hybrid screening followed the protocol provided by Clontech. In brief, WOX11 was cloned into the bait plasmid pGBKT7, and cDNA library of rice root was built into prey plasmid pGADT7 (Clontech). Yeast containing the bait and prey plasmids were grown on SD-LTHA medium (solid)-Leu/-Trp/-His/-Ade medium). Colony PCR and sequencing were performed to eliminate frame shifts and verify prey plasmid sequences.
Pull-Down Assays
His alone and ADA2-His, WOX11-GST, and GCN5-GST fusion proteins were expressed in Escherichia coli. Equal volumes of His alone protein or ADA2-His fusion protein were incubated with WOX11-GST or GCN5-GST protein in total 1.2 mL volume of pull-down buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5% Igepal CA-630, and protease inhibitor) at 4°C with gentle upside down rotation overnight. His beads (40 μL) (Promega; REF V8500) were added, followed by six more hours of incubation. After extensive washing, the pulled down proteins were eluted with His elution buffer provided by the kit and denatured for 12% SDS-PAGE. Immunoblots were performed using anti-His antibody (abcam; ab9180, lot GR248369-2) and anti-GST antibody (abcam; ab19256, lot GR1093331-1).
Coimmunoprecipitation Assay
Plasmids containing 35S:GCN5-HA:NOS-terminator, 35S:ADA2:NOS-terminator (with ADA2 cDNA in either sense or antisense direction), 35:GFP:NOS-terminator, or 35S:WOX11-GFP:NOS-terminator fragments were transformed via polyethylene glycol method into rice shoot cell protoplasts prepared from 10-d-old wild-type seedlings as described (Zong et al., 2016). After 16 h cultivation, protoplasts were harvested and broken by centrifugal force; GFP-Trap beads (ChromoTek; gta-20) were incubated with supernatant of the protoplasts overnight at 4°C. After extensive washing, beads were denatured with SDS loading buffer at 95°C for 10 min. Immunoblots were performed with anti-GFP (Abmart; 7G9, lot 264174), anti-HA (Abmart; 26D11, lot 264066), anti-ADA2, and anti-WOX11 (Zhao et al., 2015).
BiFC Assay
Rice ADA2 full-length cDNA was cloned into the pVYNE vector for expression of ADA2 fused to the N-terminal 1 to 155 amino acids of YFP, and WOX11 full-length cDNA was cloned into the pVYCE vector for expression of WOX11 fused to the C-terminal 156 to 239 amino acids of YFP (Waadt et al., 2008). Combinations (10 μg for each plasmid) were transformed via the polyethylene glycol method and transiently expressed in shoot cells of 10-d-old (after germination) seedlings. The fluorescence was detected by confocal microscopy (Leica). To exclude false positive signals (arising from the tendency of split fragments to spontaneously reassemble autonomously if unhindered; Xing et al., 2016), cell ratios of YFP signal in test group and control groups were calculated.
In Vitro Acetyltransferase Activity Assay
Affinity-purified GCN5-GST fusion or GST alone protein was incubated with 10 μg histone H3 or H4 in acetylation buffer (40 mM Tris-HCl, pH 8.0, 0.1 M NaCl, 10% glycerol, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 5 mM sodium butyrate, and 0.1 mM acetyl-CoA). After 4 h incubation at 30°C, the reaction was terminated by boiling. Samples were analyzed by electrophoresis in 12% SDS-PAGE gels followed by immunoblotting.
In Situ Hybridization
The hybridization and immunological detection were performed as described by Zhao et al. (2009, 2015). Gene-specific regions of GCN5, ADA2, OsPIN9, and OsCSLF6 were cloned into the pGEM-T vector (Promega A1360) (Supplemental Data Set 1). WOX11-pGEM-T was previously described (Zhao et al., 2009). In vitro transcription either from the T7 or SP6 promoter was performed to generate sense and antisense probe using the Digoxigenin RNA labeling kit (Roche). RNase-free slices of rice root tip were hybridized to digoxigenin-labeled probes and used for subsequent immunological detection.
Meristem Size Measurement
Root meristem size was determined by measuring the length from the quiescent center to the first elongated cell in the 6th cortex layer. When length of a cortex cell was twice of that of the neighbor cell, it was considered to be the first elongated cortex cell.
EdU Staining
EdU staining was performed as described by Li et al. (2015) using an EdU kit (C10310; Apollo 488) from Ribobio. Briefly, crown roots of 5-d-old (after germination) seedling were immersed in 50 μM EdU solution for 3 h. After fixation for 30 min in 4% paraformaldehyde, longitudinal vibration section was obtained and followed by treatment with Apollo. The florescence was detected with confocal microscopy.
GUS Staining and Sectioning
GCN5 RNAi and wox11 mutants were crossed with DR5:GUS transgenic lines (Zhao et al., 2009; Wang et al., 2014); homozygotes in the wox11 mutant and wild-type backgrounds were segregated from self-pollinating crossed plants. Crown roots from different backgrounds were stained with GUS staining buffer (2 mM N,N-dimethylformamide, 50 mM NaH2PO4, 50 mM Na2HPO4, 10 mM Na2EDTA, 0.1% Triton X-100, and 0.5 mM K3[Fe(CN)6]) for 4 h followed by photography. For semithin sections, 8-h stained crown roots were embedded in resin using Technovit 7100 kit (Heraeus Kulzer).
RNA-Seq and Analysis
Roots of 5-d-old (after germination) seedlings were harvested. Extracted high-quality total RNA was used for RNA-seq library construction. Briefly, 2 μg of total RNA was used for mRNA purification and cDNA synthesis. Double-stranded cDNA was generated by reverse transcription and DNA-dependent DNA synthesis. A single “A” nucleotide was added the 3′ end of the blunt fragment of double-stranded DNA followed by indexing adapter alignments. The amplified DNA fragments were sequenced with Illumina HiSeq 3000 system.
For data analysis, Trimmomatic (version 0.32) (Bolger et al., 2014) was used to remove low-quality tags, and TopHat (version 2.0.13) was used to map tags to the reference genome (RGAP version 7.0). The Cufflinks suite was used to splice transcripts and find different expressed genes (P value < 0.05, fold change > 2). Different expressed genes were automatically annotated and manually categorized according to their putative or demonstrated function based on gene ontology as described by Coudert et al. (2015). Gene Ontology enrichment analysis was performed at http://www.ricearray.org/. For tissue specificity analysis, genome-wide expression profiles were downloaded from RiceXPro website (http://ricexpro.dna.affrc.go.jp/). Transcript ratios (root versus summation of all other tissues) larger than 0.5-fold were considered to reflect root-specific gene expression.
RT-qPCR
Total RNA was extracted from tissues using TRIzol reagent followed by reverse transcription using reagent (cat. no. 12183555) from Invitrogen and according to the manufacturer’s protocol. qRT-PCR was performed using the gene-specific primers listed in Supplemental Data Set 1 and a real-time PCR 7500 system (Applied Biosystems). Data were collected using the ABI PRISM 7500 sequence detection system. Three biological replicates with independent mRNA isolations were performed, each with three technical repeats.
ChIP-qPCR
About 0.5 to 1 g root material was cross-linked in 1% formaldehyde under vacuum. Chromatin was extracted and fragmented via ultrasound to 200 to 800 bp; 5 μL antibody (anti-H3Ace, Millipore, 06-599, lot 2469593; anti-HA, Abmart, 26D11, lot 264066; anti-ADA2 and anti-WOX11, polyclonal antibody) was incubated with 40 μL protein A/G beads (Invitrogen) at 4°C for more than 4 h. After bead washing, 100 μL fragmented chromatin suspension was added, followed by incubation at 4°C overnight. After extensive washing, immunoprecipitated chromatin was de-cross-linked and retrieved for qPCR. Primers for ChIP-qPCR are listed in Supplemental Data Set 1. Three biological replicates were performed using crown roots harvested from three independent cultures. Each biological replicate was tested with three technical repeats.
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project website (http://rice.plantbiology.msu.edu/) under the following accession numbers: GCN5, LOC_Os10g28040; ADA2, LOC_Os03g53960; OsPIN9, LOC_Os01g58860; OsEXPA23, LOC_Os02g16809; OsCSLF6, LOC_Os08g06380; CESA9, LOC_Os09g25490; OsGLU5, LOC_Os01g12070; Osbglu5, LOC_Os01g70520; Os9bglu33, LOC_Os09g33710; OsALDO, LOC_Os11g07020; OsEXPA15, LOC_Os03g06020; and Ehd1, LOC_Os10g32600. The RNA-seq data described in this article have been deposited into the Gene Expression Omnibus database (accession number GSE95481).
Supplemental Data
Supplemental Figure 1. Mapping the interaction regions of ADA2, GCN5, and WOX11 by yeast two-hybrid assays and comparison of their expression profiles in rice tissues/organs.
Supplemental Figure 2. Protein levels of GCN5 and ADA2 in transgenic plants.
Supplemental Figure 3. Root phenotypes of GCN5 and ADA2 RNAi seedlings with the same shoot length as the wild type.
Supplemental Figure 4. Phenotypes of GCN5 and ADA2 RNAi lines at mature stages.
Supplemental Figure 5. Production and phenotype of GCN5 overexpression lines.
Supplemental Figure 6. GCN5 and WOX11 are required for auxin accumulation or response in crown root tip.
Supplemental Figure 7. RNA-seq analysis of GCN5 RNAi plants and comparison with downregulated genes in wox11 root tips.
Supplemental Figure 8. In situ hybridization detection of OsPIN9 and OsCSLF6 transcripts in crown root tip of two GCN5 RNAi lines, wox11 mutant, and their corresponding wild types.
Supplemental Figure 9. The other two biological replicates of the H3 acetylation ChIP-qPCR assays shown in Figures 7A and 7B.
Supplemental Figure 10. GCN5 RNAi does not affect WOX11 binding to target genes.
Supplemental Table 1. RNA-seq reads and analysis data.
Supplemental Table 2. Gene Ontology analysis of downregulated genes by GCN5 RNAi.
Supplemental Data Set 1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Qinghua Zhang for high-throughput sequencing and Qinglu Zhang and Xianghua Li for help in field experiments and management. We also thank Jun Yang for help in protoplast transformation, and Yue Lu and Qiutao Xu for OsGCN5 knockout plant construction with CRIPSR-Case 9 technology. This research was supported by grants from the National Key Research and Development Program of China (2016YFD0100903 and 2016YFD0100802) and the National Natural Science Foundation of China (31371468 and 31571256), and supported by the Huazhong Agricultural University Scientific and Technological Self-Innovation Foundation (program no. 2016RC003).
AUTHOR CONTRIBUTIONS
S.Z., W.J., F.L., W.Y., S.C., and Y.Z. performed research. S.Z. and D.-X.Z. analyzed data. D.-X.Z. and S.Z. wrote the article.
Glossary
- HAT
histone acetyltransferase
- BiFC
bimolecular fluorescence complementation
- QC
quiescent center
- ChIP
chromatin immunoprecipitation
- EdU
5-ethynyl-20-deoxyuridine
Footnotes
Articles can be viewed without a subscription.
References
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Anzola J.M., Sieberer T., Ortbauer M., Butt H., Korbei B., Weinhofer I., Müllner A.E., Luschnig C. (2010). Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc. Natl. Acad. Sci. USA 107: 10308–10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R., Pray-Grant M.G., Selleck W., Grant P.A., Tan S. (2002). Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277: 7989–7995. [DOI] [PubMed] [Google Scholar]
- Benhamed M., Bertrand C., Servet C., Zhou D.X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M., et al. (2008). Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 56: 493–504. [DOI] [PubMed] [Google Scholar]
- Bertrand C., Bergounioux C., Domenichini S., Delarue M., Zhou D.X. (2003). Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278: 28246–28251. [DOI] [PubMed] [Google Scholar]
- Bhat R.A., Borst J.W., Riehl M., Thompson R.D. (2004). Interaction of maize Opaque-2 and the transcriptional co-activators GCN5 and ADA2, in the modulation of transcriptional activity. Plant Mol. Biol. 55: 239–252. [DOI] [PubMed] [Google Scholar]
- Bian C., et al. (2011). Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30: 2829–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J., Wang C.Y., Baptista T., Vincent S.D., Hsiao W.C., Stierle M., Kao C.F., Tora L., Devys D. (2014). The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 28: 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J.E., Zhou J., Ranalli T., Kobayashi R., Edmondson D.G., Roth S.Y., Allis C.D. (1996). Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Utley R.T., Workman J.L., Côté J. (2003). The diverse functions of histone acetyltransferase complexes. Trends Genet. 19: 321–329. [DOI] [PubMed] [Google Scholar]
- Chen X., Zhou D.X. (2013). Rice epigenomics and epigenetics: challenges and opportunities. Curr. Opin. Plant Biol. 16: 164–169. [DOI] [PubMed] [Google Scholar]
- Coudert Y., Périn C., Courtois B., Khong N.G., Gantet P. (2010). Genetic control of root development in rice, the model cereal. Trends Plant Sci. 15: 219–226. [DOI] [PubMed] [Google Scholar]
- Coudert Y., Bès M., Le T.V., Pré M., Guiderdoni E., Gantet P. (2011). Transcript profiling of crown rootless1 mutant stem base reveals new elements associated with crown root development in rice. BMC Genomics 12: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudert Y., Le V.A., Adam H., Bès M., Vignols F., Jouannic S., Guiderdoni E., Gantet P. (2015). Identification of CROWN ROOTLESS1-regulated genes in rice reveals specific and conserved elements of postembryonic root formation. New Phytol. 206: 243–254. [DOI] [PubMed] [Google Scholar]
- Dolzblasz A., Nardmann J., Clerici E., Causier B., van der Graaff E., Chen J., Davies B., Werr W., Laux T. (2016). Stem cell regulation by Arabidopsis WOX genes. Mol. Plant 9: 1028–1039. [DOI] [PubMed] [Google Scholar]
- Gibson S.I. (2005). Control of plant development and gene expression by sugar signaling. Curr. Opin. Plant Biol. 8: 93–102. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Eberharter A., John S., Cook R.G., Turner B.M., Workman J.L. (1999). Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274: 5895–5900. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Duggan L., Côté J., Roberts S.M., Brownell J.E., Candau R., Ohba R., Owen-Hughes T., Allis C.D., Winston F., Berger S.L., Workman J.L. (1997). Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11: 1640–1650. [DOI] [PubMed] [Google Scholar]
- Haecker A., Gross-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668. [DOI] [PubMed] [Google Scholar]
- Hu X., Xu L. (2016). Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 172: 2363–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K.L., Pugh B.F. (2004). A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13: 573–585. [DOI] [PubMed] [Google Scholar]
- Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Shibata Y., Gomi K., Umemura I., Hasegawa Y., Ashikari M., Kitano H., Matsuoka M. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46: 23–47. [DOI] [PubMed] [Google Scholar]
- Jiang W., Zhou S., Zhang Q., Song H., Zhou D.X., Zhao Y. (2017). Transcriptional regulatory network of WOX11 involved in the control of crown root development, cytokinin signals, and redox in rice. J. Exp. Bot., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Fang C., Yuan H., Wang S., Wu Y., Liu X., Zhang Y., Luo J. (2015). Interaction between carbon metabolism and phosphate accumulation is revealed by a mutation of a cellulose synthase-like protein, CSLF6. J. Exp. Bot. 66: 2557–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi Y., Kitano H., Inukai Y. (2011). Molecular mechanism of crown root initiation and the different mechanisms between crown root and radicle in rice. Plant Signal. Behav. 6: 1270–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi Y., Ogawa A., Kitano H., Inukai Y. (2008). CRL4 regulates crown root formation through auxin transport in rice. Plant Root 2: 19–28. [Google Scholar]
- Kleff S., Andrulis E.D., Anderson C.W., Sternglanz R. (1995). Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem. 270: 24674–24677. [DOI] [PubMed] [Google Scholar]
- Kornet N., Scheres B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21: 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E., Hirsch C.L., Dent S.Y. (2010). Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 22: 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.K., Workman J.L. (2007). Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 8: 284–295. [DOI] [PubMed] [Google Scholar]
- Li J., Zhao Y., Chu H., Wang L., Fu Y., Liu P., Upadhyaya N., Chen C., Mou T., Feng Y., Kumar P., Xu J. (2015). SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation. PLoS Genet. 11: e1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang S., Yu X., Yu J., He X., Zhang S., Shou H., Wu P. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43: 47–56. [DOI] [PubMed] [Google Scholar]
- Liu J., Sheng L., Xu Y., Li J., Yang Z., Huang H., Xu L. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Wang J., Wang L., Wang X., Xue Y., Wu P., Shou H. (2009). Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 19: 1110–1119. [DOI] [PubMed] [Google Scholar]
- Liu X., Zhou S., Wang W., Ye Y., Zhao Y., Xu Q., Zhou C., Tan F., Cheng S., Zhou D.X. (2015). Regulation of histone methylation and reprogramming of gene expression in the rice inflorescence meristem. Plant Cell 27: 1428–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Wang Y., Qiu S., Kang Z., Che S., Wang G., Huang J. (2013). Overexpression of OsEXPA8, a root-specific gene, improves rice growth and root system architecture by facilitating cell extension. PLoS One 8: e75997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Pavangadkar K.A., Thomashow M.F., Triezenberg S.J. (2006). Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim. Biophys. Acta 1759: 69–79. [DOI] [PubMed] [Google Scholar]
- Marcon C., Paschold A., Hochholdinger F. (2013). Genetic control of root organogenesis in cereals. Methods Mol. Biol. 959: 69–81. [DOI] [PubMed] [Google Scholar]
- Miart F., Desprez T., Biot E., Morin H., Belcram K., Höfte H., Gonneau M., Vernhettes S. (2014). Spatio-temporal analysis of cellulose synthesis during cell plate formation in Arabidopsis. Plant J. 77: 71–84. [DOI] [PubMed] [Google Scholar]
- Mishra B.S., Singh M., Aggrawal P., Laxmi A. (2009). Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS One 4: e4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P., Fukaki H., Beeckman T. (2010). Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi L., Aichinger E., van der Graaff E., Llavata-Peris C.I., Weijers D., Hennig L., Groot E., Laux T. (2015). Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 33: 576–588. [DOI] [PubMed] [Google Scholar]
- Rouyi C., Baiya S., Lee S.K., Mahong B., Jeon J.S., Ketudat-Cairns J.R., Ketudat-Cairns M. (2014). Recombinant expression and characterization of the cytoplasmic rice β-glucosidase Os1BGlu4. PLoS One 9: e96712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R., Meslamani J., Zhou M.M. (2014). The bromodomain: from epigenome reader to druggable target. Biochim. Biophys. Acta 1839: 676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servet C., Conde e Silva N., Zhou D.X. (2010). Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol. Plant 3: 670–677. [DOI] [PubMed] [Google Scholar]
- Shen Y., Wei W., Zhou D.X. (2015). Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 20: 614–621. [DOI] [PubMed] [Google Scholar]
- Shen Y., Issakidis-Bourguet E., Zhou D.X. (2016). Perspectives on the interactions between metabolism, redox, and epigenetics in plants. J. Exp. Bot. 67: 5291–5300. [DOI] [PubMed] [Google Scholar]
- Somssich M., Je B.I., Simon R., Jackson D. (2016). CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143: 3238–3248. [DOI] [PubMed] [Google Scholar]
- van der Graaff E., Laux T., Rensing S.A. (2009). The WUS homeobox-containing (WOX) protein family. Genome Biol. 10: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachonasios K.E., Thomashow M.F., Triezenberg S.J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15: 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Wang L., Dent S.Y. (2014). Functions of SAGA in development and disease. Epigenomics 6: 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Chu H., Li Z., Wang J., Li J., Qiao Y., Fu Y., Mou T., Chen C., Xu J. (2014). Origin and development of the root cap in rice. Plant Physiol. 166: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.F., He F.F., Ma X.X., Mao C.Z., Hodgman C., Lu C.G., Wu P. (2011). OsCAND1 is required for crown root emergence in rice. Mol. Plant 4: 289–299. [DOI] [PubMed] [Google Scholar]
- Weake V.M., Workman J.L. (2012). SAGA function in tissue-specific gene expression. Trends Cell Biol. 22: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S.K., Choi S.B., Kumari S., Cho M., Lee S.H., Cho H.T. (2010). Root hair-specific EXPANSIN B genes have been selected for Graminaceae root hairs. Mol. Cells 30: 369–376. [DOI] [PubMed] [Google Scholar]
- Xing S., Wallmeroth N., Berendzen K.W., Grefen C. (2016). Techniques for the analysis of protein-protein interactions in vivo. Plant Physiol. 171: 727–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., McCormack M., Li L., Hall Q., Xiang C., Sheen J. (2013). Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang Y., Li G., Tang Y., Kramer E.M., Tadege M. (2014). STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula. Plant Cell 26: 650–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.W., Xu L., Wu Y.R., Chen X.A., Liu Y., Zhu S.H., Ding W.N., Wu P., Yi K.K. (2012). OsGLU3, a putative membrane-bound endo-1,4-beta-glucanase, is required for root cell elongation and division in rice (Oryza sativa L.). Mol. Plant 5: 176–186. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hu Y., Dai M., Huang L., Zhou D.X. (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21: 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Cheng S., Song Y., Huang Y., Zhou S., Liu X., Zhou D.X. (2015). The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 27: 2469–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZhiMing Y., Bo K., XiaoWei H., ShaoLei L., YouHuang B., WoNa D., Ming C., Hyung-Taeg C., Ping W. (2011). Root hair-specific expansins modulate root hair elongation in rice. Plant J. 66: 725–734. [DOI] [PubMed] [Google Scholar]
- Zong W., Tang N., Yang J., Peng L., Ma S., Xu Y., Li G., Xiong L. (2016). Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 171: 2810–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.