Atomic force microscopy, optical microscopy, and structural modeling reveal extensive macromolecular arrays of cyanobacterial PSI complexes that allow some intertrimer energy migration.
Abstract
Photosystem I (PSI) is the dominant photosystem in cyanobacteria and it plays a pivotal role in cyanobacterial metabolism. Despite its biological importance, the native organization of PSI in cyanobacterial thylakoid membranes is poorly understood. Here, we use atomic force microscopy (AFM) to show that ordered, extensive macromolecular arrays of PSI complexes are present in thylakoids from Thermosynechococcus elongatus, Synechococcus sp PCC 7002, and Synechocystis sp PCC 6803. Hyperspectral confocal fluorescence microscopy and three-dimensional structured illumination microscopy of Synechocystis sp PCC 6803 cells visualize PSI domains within the context of the complete thylakoid system. Crystallographic and AFM data were used to build a structural model of a membrane landscape comprising 96 PSI trimers and 27,648 chlorophyll a molecules. Rather than facilitating intertrimer energy transfer, the close associations between PSI primarily maximize packing efficiency; short-range interactions with Complex I and cytochrome b6f are excluded from these regions of the membrane, so PSI turnover is sustained by long-distance diffusion of the electron donors at the membrane surface. Elsewhere, PSI-photosystem II contact zones provide sites for docking phycobilisomes and the formation of megacomplexes. PSI-enriched domains in cyanobacteria might foreshadow the partitioning of PSI into stromal lamellae in plants, similarly sustained by long-distance diffusion of electron carriers.
INTRODUCTION
Photosystem I (PSI) is one of the two pigment-protein complexes that underpin photosynthesis in cyanobacteria, algae, and plants. PSI and photosystem II (PSII) collect solar energy and use it to reduce electron acceptors, ultimately producing the NADPH required to fix CO2. Cyanobacteria carry out a large proportion of global photosynthesis due to their abundance in the oceans (Ting et al., 2002). In addition to their biological importance, cyanobacteria have provided relatively simple, genetically amenable model systems for dissecting the assembly, structure, and function of photosynthetic complexes (Bryant, 2006). The thermophilic cyanobacteria Thermosynechococcus elongatus and T. vulcanus are particularly significant, as they are the source of purified, stable complexes for a series of x-ray crystallography studies that determined the structures of PSI (Jordan et al., 2001) and PSII (Ferreira et al., 2004; Loll et al., 2005; Umena et al., 2011; Suga et al., 2015). Other cyanobacteria such as Synechococcus sp PCC 7002 (hereafter Synechococcus 7002) and Synechocystis sp PCC 6803 (hereafter Synechocystis) are heavily used models for many areas of research, for example, photosynthesis, gene regulation, and synthetic biology. Given that PSI is the major photosystem complex in many cyanobacteria, this study investigates its membrane organization in T. elongatus, Synechococcus 7002, and Synechocystis using atomic force microscopy (AFM), hyperspectral confocal fluorescence microscopy (HCFM), and three-dimensional structured illumination microscopy (3D-SIM).
The 2.5-Å resolution structure of PSI from T. elongatus (Jordan et al., 2001) shows that it consists of 12 protein subunits (labeled PsaA, PsaB, PsaC, PsaD, PsaE, PsaF, PsaI, PsaJ, PsaK, PsaL, PsaM, and PsaX), 96 chlorophyll a molecules, and 22 β-carotene molecules as well as a number of other cofactors that form an electron transport chain. The core complex of PSI consists of a heterodimer of PsaA and PsaB subunits that bind the so-called P700 special pair of chlorophyll a molecules, located close to the lumenal side of this complex, and the electron transport cofactors, forming two branches that each consist of two chlorophyll a molecules and a phylloquinone (A1) (Jordan et al., 2001). These cofactors act as electron donors to a Fe4S4 cluster (FX), which is located close to the cytoplasmic side of the complex. A third subunit, PsaC, is attached to the cytoplasmic face of the PsaA/PsaB heterodimer and contains two Fe4S4 clusters, FA and FB. Fourth and fifth subunits, PsaD and PsaE, are located either side of PsaC on the cytoplasmic side of the complex and in combination with PsaC provide the binding site for ferredoxin (Sétif et al., 2002). PsaD is also involved in the assembly of PSI and is necessary for the insertion of PsaC, PsaE, and of a sixth subunit, PsaL, into the complex (Xu et al., 2001). PsaL is important for the formation of the PSI trimer and is located in the center of the homotrimer, where it interacts with the PsaL subunits in the other two PSI complexes of the trimeric complex (Chitnis and Chitnis, 1993; Karapetyan et al., 1999). The pigments of subunits PsaL and PsaM play a key role in the excitonic connectivity of PSI monomers within a trimer (Sener et al., 2004). A further subunit, PsaF, is located on the side of the PSI monomer opposite to PsaL; in green algae and plants, the lumenal face of this subunit is involved in binding plastocyanin and cytochrome c6 (Farah et al., 1995), although not in T. elongatus (Mühlenhoff and Chauvat, 1996). The remaining subunits, PsaI, PsaJ, PsaK, PsaM, and PsaX, are small polypeptides believed to be involved with stabilizing PSI; loss of any one of these proteins does not significantly reduce the functionality of the protein complex (Schluchter et al., 1996; Xu et al., 1994; Naithani et al., 2000).
PSI is the dominant photosystem in cyanobacteria and it carries out a pivotal role in cyanobacterial metabolism by supplying the electrons for diverse processes such as pigment biosynthesis; nitrate, nitrite, and sulfite metabolism; and CO2 fixation (Lea-Smith et al., 2015). Despite the biological importance of PSI, the native organization of this complex in cyanobacterial thylakoid membranes is poorly understood; see Supplemental Figure 1 for a schematic diagram of photosynthetic electron flow in cyanobacterial membranes. There is evidence to suggest some degree of spatial heterogeneity in cyanobacterial thylakoids; HCFM of Synechocystis cells has shown that PSI is enriched in thylakoid membranes toward the center of the cell, whereas the greatest levels of PSII fluorescence are detected in thylakoid membranes closer to the plasma membrane (Vermaas et al., 2008). On the other hand, transmission electron microscopy of immunogold-labeled membranes from Synechococcus sp PCC 7942 showed that PSI and ATP synthase are primarily located in the outermost thylakoid membranes that are closest to the plasma membrane, whereas PSII and cytochrome b6f (cytb6f) complexes were found to be evenly distributed through the thylakoid membrane system (Sherman et al., 1994). Although there is no consensus for the membrane distribution of PSI, it does appear that PSI and PSII are to some degree spatially segregated in the thylakoid membrane.
We used AFM to image thylakoid membranes purified from T. elongatus, Synechococcus 7002, and Synechocystis under liquid at room temperature. These images of the native membrane organization reveal ordered macromolecular arrays composed solely of PSI, identified by their characteristic trimeric form; the subnanometer resolution of AFM in the vertical axis allowed the cytoplasmic protrusions of PSI complexes to be identified from their height above the membrane surface and the mica substrate. Other membrane patches showed PSI interspersed among other protein complexes, tentatively identified as PSII. The atomic structure of PSI has been interpolated into membrane topographs to show the placement of PSI electron transfer cofactors in relation to the membrane bilayer; 96 PSI complexes were used to produce a structural model of PSI-only membrane regions. This model shows the true extent of the high packing density of PSI complexes in the thylakoid membrane, which permit no direct interactions with any other membrane complexes, and excitation energy trapping properties were calculated from the positions and orientations of 27,648 chlorophyll molecules. HCFM and 3D-SIM of Synechocystis reveals PSI-enriched membrane domains in whole cells and places the more detailed AFM images of single membranes in a cellular context.
RESULTS
AFM of PSI-Enriched Membrane Patches from T. elongatus
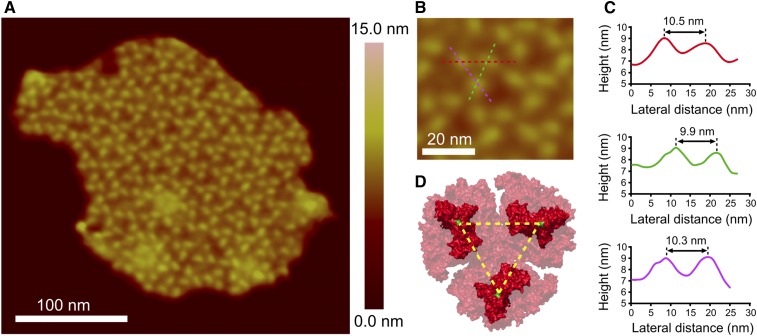
Thylakoid membranes from the cyanobacterium T. elongatus were fractionated on a 30 to 50% (w/w) sucrose density gradient containing 0.1% (w/w) digitonin (Supplemental Figure 2B), which did not affect the absorption spectrum (Supplemental Figures 2C and 2D). As a further check, Clear Native-PAGE (CN-PAGE) was used to analyze the effect of digitonin on photosystem composition and stoichiometry (Supplemental Figure 3). We found that 1.0% (w/w) digitonin, a concentration 10-fold higher than that used to prepare membranes for AFM imaging, did not affect the PSI:PSII ratio, which remained at ∼4 (Supplemental Figure 3D). Membranes from digitonin-free gradients were unsuitable for acquiring high-quality AFM topographs, but Supplemental Figure 4 shows that by using such gradients, plus subsequent drying/rehydration on the mica substrate (see Methods), it was possible to acquire a low-resolution image of a thylakoid membrane housing at least 26 trimeric PSI complexes packed within a small area, although not in a regular array. Thus, PSI-rich membrane domains are a native feature of some areas of the thylakoid membrane in T. elongatus. In order to obtain higher-quality topographs of individual membranes, to allow docking of the PSI trimer structure, a fraction was collected from the middle of the sucrose density gradient containing 0.1% (w/w) digitonin (Supplemental Figure 2B) and imaged by AFM. Figure 1A shows a flat membrane patch that exhibits well-ordered arrays of trimeric complexes. Height sections were taken across one of the trimers, shown in Figure 1B, and the lateral distances between protrusions are 9.9 to 10.5 nm. Taking into account the lateral resolution of our measurements, which is 1 to 2 nm, the distances between protrusions are broadly consistent with the 9.1-nm distance measured from the crystal structure of PSI (PDB:1JB0) between proline 29 on the cytoplasmic side of the PsaC subunit, colored green in Figure 1D. Supplemental Figure 5 shows other examples of membranes where some areas have trimeric complexes as the dominant feature; in Supplemental Figure 5B, a PSI-enriched area is contiguous with another where trimers are interspersed among other complexes, as in Figure 5B.
Figure 1.
AFM Imaging and Analysis of a Thylakoid Membrane Patch from T. elongatus That Contains Ordered Arrays of Trimeric Complexes.
(A) AFM topograph of a membrane patch.
(B) Zoom to a single complex, with the positions of three height profiles shown (red, pink, and green dashed lines), taken across the approximate positions of maximum height.
(C) Height profiles corresponding to the lines in (B), with lateral distances shown.
(D) Structure of the cytoplasmic face of the PSI complex taken from PDB:1JB0. The dashed yellow lines are drawn between proline 29 of the PsaC subunit on each monomer, shown in green, and represent a distance of 9.1 nm.
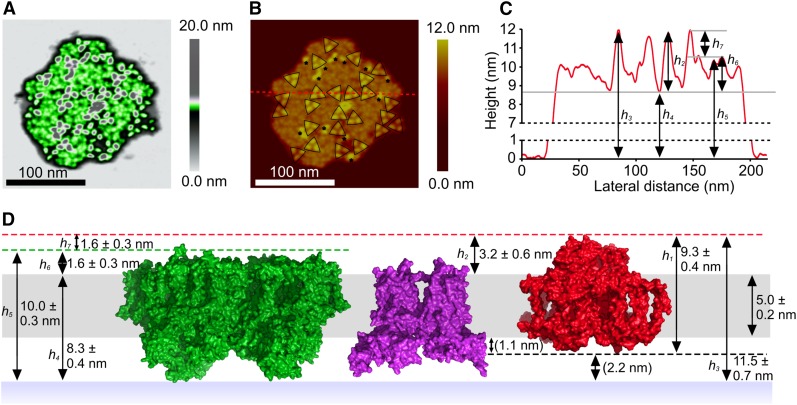
Figure 5.
AFM Imaging of a Membrane Patch from T. elongatus That Contains Trimeric Complexes in a Disordered Arrangement.
(A) Random distribution of trimeric complexes (gray), with intervening complexes in green; the false color representation was used to emphasize the topographic features present in these regions between PSI complexes.
(B) Two-dimensional representation of the AFM data; the dashed red line shows the position of a section used for the height profile in (C). Trimeric PSI complexes are outlined by black triangles, and monomeric and dimeric complexes are indicated by asterisks.
(C) Height profile corresponding to the red dashed line in (B), showing the heights of trimeric complexes with respect to the mica substrate (h3, h4, h5) and in relation to the membrane bilayer (h2). h6 corresponds to the height of a neighboring complex, likely PSII, above the cytoplasmic surface of the membrane.
(D) Diagram showing the heights of PSII (green), cytb6f (purple), and PSI (red) complexes in the thylakoid membrane bilayer, with the vertical positioning of the PSII and PSI complexes, and the thickness of the bilayer, assigned on the basis of AFM measurements. The red and green dashed lines define the respective apices of the PSI and PSII complexes on the cytoplasmic face of the thylakoid membrane. The black dashed line shows the apex of the PSI complex on the lumenal face of the thylakoid membrane which has a height of 1.1 nm. The PSI complex is suspended above the mica surface by 2.2 nm by the lumenal protrusions of the PSII and cytb6f complexes; these distances are shown in parentheses as they were not directly measured but rather calculated from the AFM data and the PSI crystal structure PDB:1JB0. The PSII structure shown is taken from PDB:3WU2 (Umena et al., 2011). See Supplemental Figure 4 for gallery of disordered membrane patches.
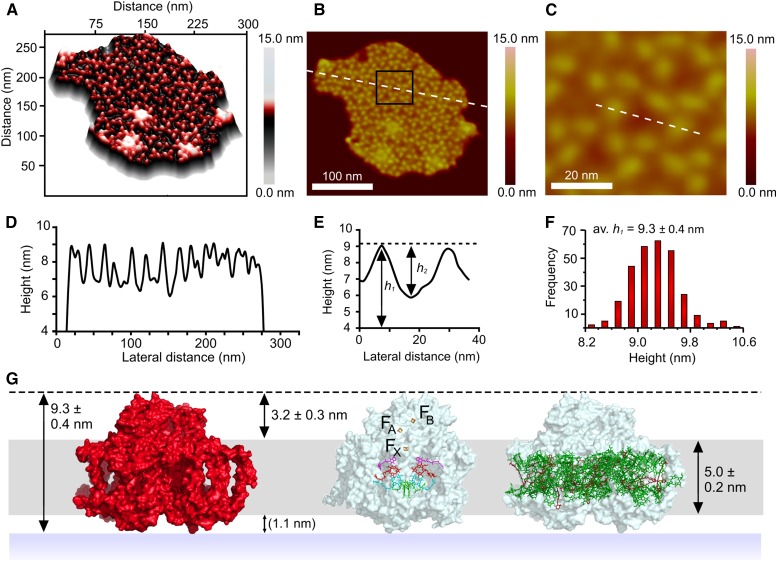
The precision of AFM is higher when vertical dimensions are measured; Figure 2A shows a three-dimensional plot of the same membrane patch as in Figure 1A, which emphasizes the uniformity of the heights of these trimeric complexes. A profile, indicated by the white dashed line in Figure 2B, was taken across the whole membrane, and the data are displayed in Figure 2D as the height above the mica surface. The average height h1 of complexes from the mica substrate, shown in the histogram in Figure 2F, was measured at 9.3 ± 0.4 nm, which fits well with the crystal structure of PSI (Figure 2G). Figure 2C shows a more detailed view of a region of the membrane, corresponding to the black box in Figure 2B. The section shown by the dashed white line in Figure 2C crosses a small area of “empty” membrane adjacent to two PSI complexes and allows measurement of the heights of the protrusions in relation to surface of the membrane; h2 in Figure 2E is 3.2 ± 0.3 nm. This figure is consistent with the value of 3.5 ± 0.2 nm obtained by AFM analysis of 2D crystals reconstituted from detergent-solubilized PSI complexes (Fotiadis et al., 1998). Separately, membranes devoid of protein were imaged to establish the thickness of the membrane bilayer in T. elongatus (Supplemental Figure 6), which is 5.0 ± 0.2 nm. The membrane bilayer is represented in gray in Figure 2G. The other face of the membrane has not been imaged so the heights of the lumenal features of PSI in relation to the membrane bilayer, the site of docking of the plastocyanin electron donor, were calculated to be ∼1.1 nm. On the basis of the vertical dimensions of the complex, as well as the correspondence of surface protrusions with the PSI trimer structure (see also Figure 3), we assign the trimeric features in our AFM topographs to PSI complexes.
Figure 2.
Analysis of the Heights of Topological Features in Relation to the Mica Substrate and the Thylakoid Membrane Surface.
(A) Three-dimensional representation of a membrane patch from T. elongatus that contains ordered arrays of trimeric complexes.
(B) Two-dimensional representation of the AFM data; the dashed white line shows the position of a section used for the height profile in (D). The black box represents the area shown in (C).
(C) Detailed view of an area of membrane showing the position of a section taken across two trimers and an intervening membrane bilayer. The dashed white line shows the position of section used for the height profile in (E).
(D) Height profile corresponding to the white dashed line in panel (B), showing the heights of the trimeric complexes with respect to the membrane bilayer.
(E) Profile showing an approximate height of 3.2 nm from the membrane surface to the top of the protein features.
(F) Histogram of height data compiled from an analysis of 287 complexes; the average height of the complexes above the mica is 9.3 ± 0.4 nm.
(G) Model summarizing the height data, showing their consistency with the height of PSI measured from the crystal structure, and the positions of PSI cofactors in relation to the membrane bilayer (the dashed black line represents the apex of the PSI complex). The 1.1-nm protrusion of the complex from the lumenal surface of the membrane is in parentheses to indicate that it was deduced from the other measurements.
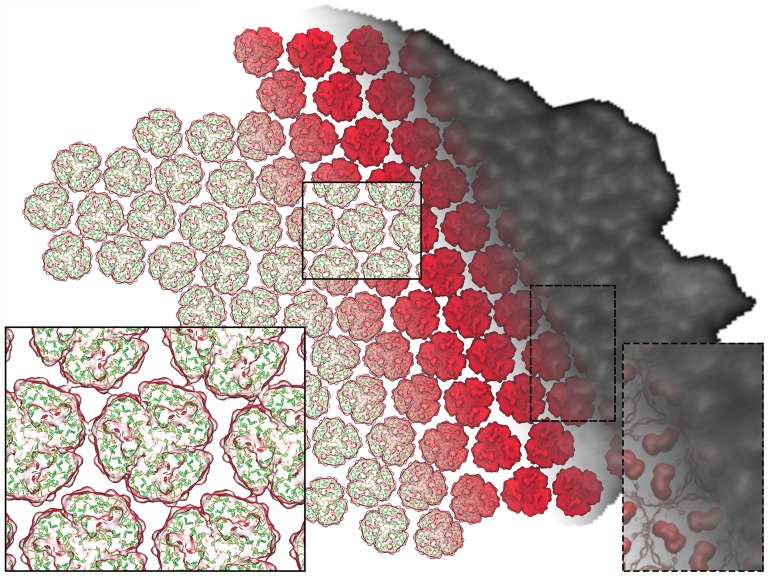
Figure 3.
Structural Model of the Arrangement of Complexes within a PSI-Only Thylakoid Membrane.
The PSI trimers are arranged according to the AFM topograph in Figure 1A, visible as an overlay on the right. The inset on the right shows the correspondence between the PsaC-D-E protrusions of each PSI trimer (red) and corresponding AFM topological features (white). The trimers are shown in surface representation. The pigments can be seen through the transparent surface on the left as the antenna (green) and RC (red) chlorophylls, represented as porphyrin rings, as well as the carotenoids (orange). The inset on the left shows the typical packing pattern around a trimer (see Figure 4). The model features a total of 96 PSI trimers and 27,648 chlorophylls.
The topology measurements summarized in Figure 2 position PSI vertically within the membrane bilayer and provide the first opportunity to define the positions of the PSI energy and electron transfer cofactors in relation to the membrane surface. Figure 2G (middle image) shows the expected intramembrane location of P700, A, A0, B, B0, chlorophylls, as well as the two phylloquinones. The Fx Fe4S4 cluster sits near the cytoplasmic surface of the membrane, and the FA and FB iron-sulfur centers are held outside the bilayer to facilitate electron transfer to the extrinsic acceptor ferredoxin that docks onto the PsaC, PsaD, and PsaE subunits (Sétif et al., 2002). Figure 2G (right) shows the transmembrane location of the antenna chlorophylls and carotenoids. Viewed from the membrane plane, it is clear that a few chlorophylls are held at the extremities of the complex and could mediate energy transfer between PSI trimers. This point was examined in more detail below by interpolating PSI trimers into the AFM topograph.
Computational Model of the PSI Membrane Region
To reveal the packing for the whole membrane array and to determine the excitonic coupling between trimers, we constructed a structural model for the 96 PSI trimers visible in the AFM topograph (Figure 1A). The structural model assigns atomic coordinates based on the crystal structure of PSI (PDB:1JB0) (Jordan et al., 2001) fitted within the AFM density; the inherent resolution of the structural model is limited by the AFM data. The elongated structure of the PsaC-D-E complex on the cytoplasmic face of PSI (Figure 1D) is visualized as an elongated topographic feature in Figures 1A and 1B, which guides the rotation and positioning of PSI structures to fit the overall topographic map of the membrane. The membrane model was built by fitting the protrusion profiles of PsaC-D-E within the trimer given by the crystal structure into the topological features revealed by AFM, employing image recognition methods to determine the position and orientation of each trimer. The resulting model is shown in Figure 3 with the membrane “landscape” viewed from the cytoplasmic side; PSI trimers are shown in surface representation with pigments visible beneath. The gray-scale AFM map on the right hand side of Figure 3 is superimposed on the PSI trimer structures (red); the inset on the left shows the resulting arrangement of chlorophyll and carotenoid pigments; the dashed line inset on the right shows the protrusions of the PsaC-D-E subunits out of the membrane plane (red, surface representation), which can be seen as three elongated topological features on each PSI trimer.
The arrangement of PSI trimers in Figure 3 shows that PSI complexes are more closely packed within the membrane bilayer than is apparent from their lateral spacing, as judged by the distances between surface protrusions arising from the PsaC-D-E subunits (Figures 2B to 2D). This “tip of the iceberg” effect masks the true packing below the membrane surface. Analysis of the PSI trimers in Figure 3 also reveals a degree of long-range order, expressed in terms of a strong spatial correlation between the positions (Supplemental Figure 7A) of neighboring trimers within a cutoff distance of 25 nm; a moderate orientational correlation is also present for the planar symmetry axes of the trimers (Supplemental Figure 7B). The relative positions of neighboring trimers show a distribution with a mean value of 1.3 nm from the grid average across the entire membrane (with the boundary trimers excluded for analysis as shown in Supplemental Figure 7C). The six clusters visible in Supplemental Figure 7A correspond to the hexagonal packing pattern inherent in the AFM topograph, suggesting a natural packing order of PSI trimers (Figure 3, left inset). We note that some areas of the AFM topograph in Figures 1A and 2B show PSI trimers that are less well packed than the example in the inset (left) in Figure 3 and that other interactions between trimers are also found within such patches. Also, the occasional presence of monomeric and dimeric PSI complexes, as found in Figure 5B, will lead to variations in the packing within PSI-only regions.
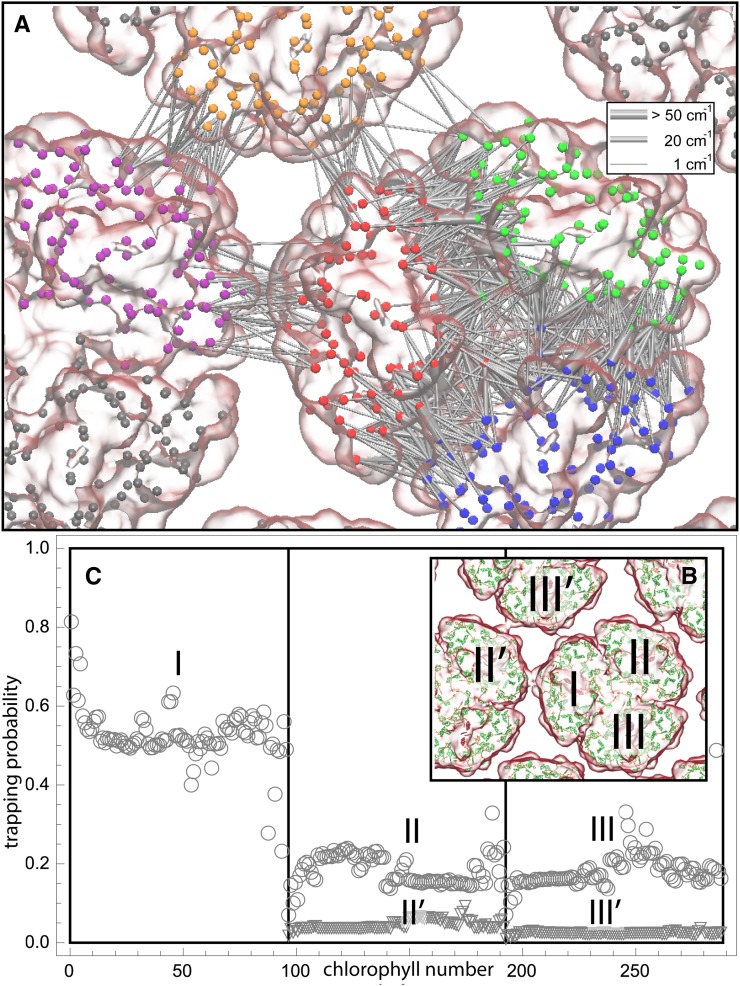
We examined the effect of the trimer packing pattern (Figures 1 and 3) on the excitonic connectivity of the pigment network, in terms of the excitation transfer probabilities across neighboring PSI monomers, computed in an effective Hamiltonian formalism following Sener et al. (2004) and Damjanović et al. (2002). Due to the approximate symmetry of the trimer array (Supplemental Figure 7), a system comprising immediate neighbors of a given monomer is considered: These are the five PSI monomers as shown in Figure 4. The PSI monomers are labeled as I, II, and III, which together comprise a PSI trimer corresponding to PDB:1JB0, and II' and III', which are the immediate neighbors of monomer I in the nearby trimers corresponding to the symmetry copies of the monomers II and III, respectively. The corresponding effective Hamiltonian 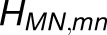 is
is
| (1) |
where 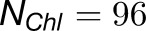 is the number of chlorophylls in one monomer (Sener et al., 2004; Damjanović et al., 2002; Sener et al., 2002) . Figure 4A illustrates the couplings
is the number of chlorophylls in one monomer (Sener et al., 2004; Damjanović et al., 2002; Sener et al., 2002) . Figure 4A illustrates the couplings 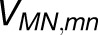 across the five monomers, I, II, III, II', and III', as labeled in Figure 4B.
across the five monomers, I, II, III, II', and III', as labeled in Figure 4B.
Figure 4.
Excitonic Connectivity between PSI Complexes within and across Trimers Corresponding to the Hexagonal Pattern in Figure 3.
Shown in (A) are the couplings HMN,mn between pairs of pigments m,n of PSI monomers M,N, respectively, for M≠N; intramonomer couplings are not shown for clarity. The connections have cross sections proportional to the coupling |HMN,mn|; couplings below 1/cm are not shown. Due to the approximate symmetry displayed by the PSI grid (Supplemental Figure 7), only the couplings of immediate neighbors of one PSI monomer (red in [A], I in [B]) are considered; two of the neighbors are in the same trimer (green in [A], II in [B]; blue in [A], III in [B]) and two are corresponding symmetry-copies in neighboring trimers (purple in [A], II' in [B]; orange in [A], III' in [B]). Excitation sharing between monomers is shown in (C) in terms of the trapping probability 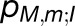 of excitation at RCI as a function of initial site across the PSI monomers M = I, II, III, II', III' as indicated in (B). Chlorophyll number m corresponds to the order given in PDB:1JB0. As suggested by the connections in (A), excitation sharing within the monomers of the same trimer is more prominent than across trimers.
of excitation at RCI as a function of initial site across the PSI monomers M = I, II, III, II', III' as indicated in (B). Chlorophyll number m corresponds to the order given in PDB:1JB0. As suggested by the connections in (A), excitation sharing within the monomers of the same trimer is more prominent than across trimers.
The excitonic connectivity of the pigment array in Figure 4 is described by the quantum yield q, namely, the probability of an excitation to be trapped by a reaction center (RC), and the cross-monomer trapping probability 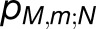 , namely, the probability for an excitation initiated at chlorophyll M,m to be absorbed at RCN. (See Methods for definitions of q and
, namely, the probability for an excitation initiated at chlorophyll M,m to be absorbed at RCN. (See Methods for definitions of q and 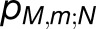 in terms of
in terms of 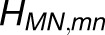 .) The quantum yield of the array, q = 0.97, is almost indistinguishable from the value computed earlier for a single PSI trimer (Sener et al., 2004). The cross-monomer trapping probabilities
.) The quantum yield of the array, q = 0.97, is almost indistinguishable from the value computed earlier for a single PSI trimer (Sener et al., 2004). The cross-monomer trapping probabilities 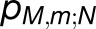 describe the level of excitation sharing between neighboring monomers and are plotted in Figure 4C for the reference monomer
describe the level of excitation sharing between neighboring monomers and are plotted in Figure 4C for the reference monomer 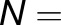 I. The cross-trapping probabilities averaged over each monomer
I. The cross-trapping probabilities averaged over each monomer
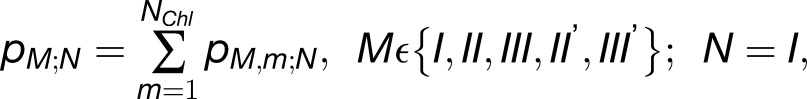 |
(2) |
are 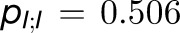 ,
, 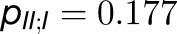 ,
, 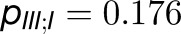 ,
, 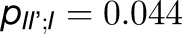 , and
, and 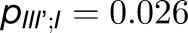 . Thus, the total cross-monomer trapping within a given trimer (I,II,III),
. Thus, the total cross-monomer trapping within a given trimer (I,II,III), 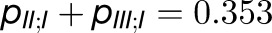 , is 5 times greater than cross-monomer trapping between monomers belonging to adjacent trimers (I,II',III'),
, is 5 times greater than cross-monomer trapping between monomers belonging to adjacent trimers (I,II',III'), 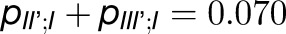 . Excitation sharing between trimers as represented by the cross-trapping probabilities,
. Excitation sharing between trimers as represented by the cross-trapping probabilities, 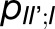 and
and 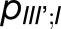 , is not negligible; however, the packing pattern of trimers (Figure 3) does not provide a significant improvement of quantum yield (Supplemental Figure 7E) over that of an isolated trimer (Sener et al., 2002) due to the short trapping time of PSI. Since the quantum yields for the observed trimer array (Figure 3) and for isolated trimers are nearly identical, the primary role of the trimer packing pattern appears to maximize the packing efficiency of PSI complexes in the membrane rather than to facilitate intertrimer energy transfer.
, is not negligible; however, the packing pattern of trimers (Figure 3) does not provide a significant improvement of quantum yield (Supplemental Figure 7E) over that of an isolated trimer (Sener et al., 2002) due to the short trapping time of PSI. Since the quantum yields for the observed trimer array (Figure 3) and for isolated trimers are nearly identical, the primary role of the trimer packing pattern appears to maximize the packing efficiency of PSI complexes in the membrane rather than to facilitate intertrimer energy transfer.
T. elongatus Thylakoids with PSI Interspersed among Other Complexes
AFM topographs of a second membrane patch from T. elongatus showed trimeric complexes that match those in Figures 1A and 1D but with a seemingly random distribution throughout the membrane (Figures 5A and 5B). Other examples are shown in Supplemental Figure 5. The heights of 88 complexes were measured giving an average height of 11.5 ± 0.7 nm (Supplemental Figure 8), much higher than the 9.3 nm expected for a PSI-only membrane (Figure 2G, left). Although most PSI complexes were trimeric, as outlined by the black triangles in Figure 5B, several monomeric and putative dimeric complexes were also present, proposed to be PSI based on the match of their heights to the trimeric PSI complexes, which were 11.9 ± 0.5 nm (n = 29) and 11.4 ± 0.9 nm (n = 42), respectively, from the mica surface. These figures are comparable with h3 in Figure 5 for PSI trimers, which is 11.5 ± 0.7 nm. Adjacent areas in the membrane patch were between 9.0 and 10.5 nm from the mica substrate, which tallies with the overall heights of PSII and cytb6f complexes measured from their respective crystallographic structures (Figure 5D); the lack of protruding features on the cytoplasmic face of each of these complexes is also consistent with the relatively featureless zones in the AFM topographs in Figure 5B, which are exaggerated by the color representation used in Figure 5A to show that there is some topology in these areas of the membrane. The lumenal subunits of PSII and the cytb6f complex protrude from the surface of the lipid bilayer (Figure 5D); the consequence of this pronounced topology is to elevate the thylakoid membrane from the mica surface, as depicted in Figure 5D. The height of the PSI complexes above the cytoplasmic surface of the membrane, h2 in Figure 5C, is 3.2 ± 0.6 nm. This height is consistent with the measurement made for the PSI protrusions in the membrane patch in Figures 1 and 2.
The area of membrane in Figure 5B not occupied by PSI is available for housing PSII and cytb6f complexes, so we determined the PSI:PSII:cytb6f stoichiometry for our membrane preparation, which was 1.0 PSI trimer:0.38 PSII dimer:0.31 cytb6f dimer. We note that at the level of AFM, each membrane is different from the next, so it is not straightforward to compare individual membranes with estimates of the abundance of complexes obtained from bulk measurements that sum all the PSI-only and PSI+PSII membrane regions. Nonetheless, given the known structures and sizes of these complexes, and taking into account the presence of other membrane proteins of unknown identity and size, the area of the minimal topology non-PSI membrane regions is sufficient to accommodate the expected amounts of PSII and cytb6f.
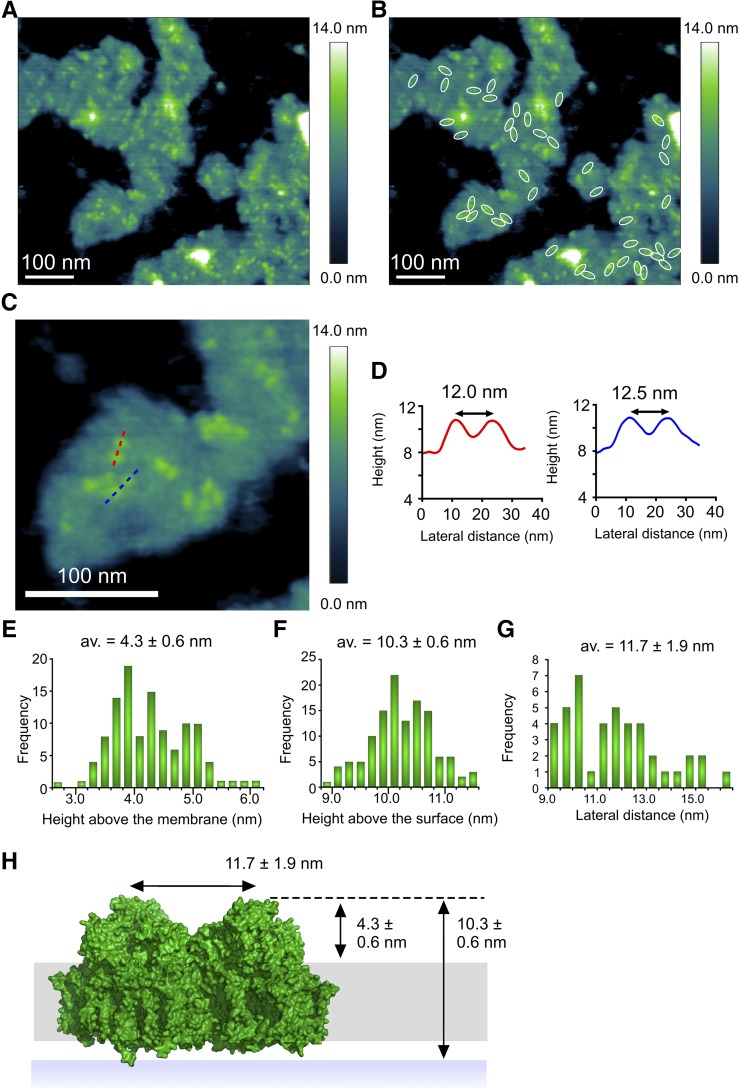
Cyanobacterial membranes attach to the mica substrate, leaving the cytoplasmic face exposed and available for imaging by AFM. A different method for immobilizing digitonin-treated thylakoids was found, similar to that used for Supplemental Figure 4, whereby the membranes were dried onto the mica under a constant stream of nitrogen and washed with imaging buffer (see Methods), making it possible to find membranes attached in the reverse orientation. The distribution of PSII complexes in this membrane topograph mirrors the distribution of PSI in Figure 5; now the weak topographic features for the lumenal face of PSI are scarcely visible, but protruding water-oxidizing complexes make it possible to identify PSII. At the low resolution of the AFM topograph in Figure 6, the PSII dimer appears as a pair of 4.3 ± 0.6 nm protrusions separated by 11.7 ± 1.9 nm (Figure 6D), consistent with previous AFM studies (Sznee et al., 2011; Johnson et al., 2014) and the structure of the water oxidizing component of the PSII core complex (Ferreira et al., 2004; Loll et al., 2005; Umena et al., 2011; Suga et al., 2015).
Figure 6.
AFM of Putative PSII Complexes in a Thylakoid Membrane Patch from T. elongatus.
(A) AFM of membrane patches from T. elongatus showing dimeric protein complexes.
(B) Forty-three dimeric complexes have been outlined in white.
(C) Zoomed-in view in which dimeric complexes can be seen. Red and blue dashed lines show the locations of the sections shown in (D).
(D) Sections of dimeric complexes in (C) showing that the spacing between the constituent monomers is ∼12 nm.
(E) The heights of 113 protein complexes above the surface of the lipid bilayer were measured; the average height is 4.3 ± 0.6 nm, which is consistent with the crystal structure of PSII PDB:3WU2 (Umena et al., 2011).
(F) The heights of 129 complexes were measured from the mica surface giving an average height of 10.3 ± 0.6 nm above the mica.
(G) The distance between the two halves of the dimeric complexes was measured giving an average separation of 11.7 ± 1.9 nm.
(H) Diagram showing the height of the PSII complex (green) from the surface of the mica (blue band), lipid bilayer (gray band), and the distance between the water-oxidizing complexes of the two monomers in the dimeric PSII complex. The dashed black line shows the position of the apex of the PSII dimer on the lumenal face of the thylakoid membrane.
Lateral Segregation of PSI in Thylakoids from Synechococcus sp PCC 7002 and Synechocystis sp PCC 6803
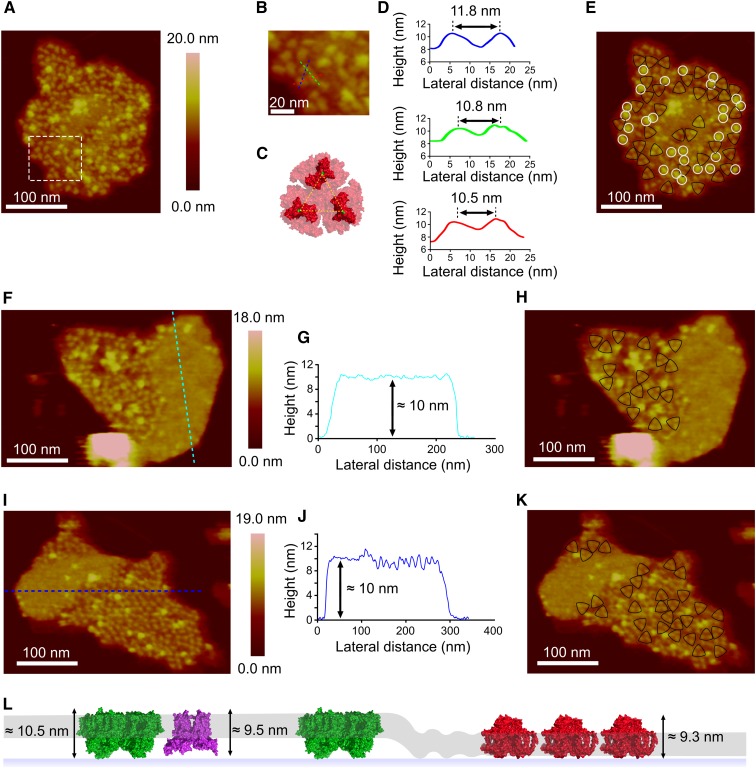
To explore the notion that PSI arrays are a general feature of cyanobacterial thylakoids, membranes from the cyanobacteria Synechocystis and Synechococcus 7002 were isolated and fractionated in the same way as for T. elongatus to produce membrane patches that were amenable to AFM analysis. We recorded topographs for a number of membranes from Synechocystis (Figures 7A, 7F, and 7I), and we observed regions consisting largely of trimeric complexes (outlined in Figures 7E, 7H, and 7K); the dimensions of these topographic features were comparable to the PSI complexes imaged in T. elongatus membranes (Figures 7B to 7D) and were therefore assigned as PSI. While it was possible to image PSI-enriched regions in membrane patches from Synechocystis (Figures 7A, 7F, and 7I), the PSI complexes were not hexagonally packed as was seen in T. elongatus membrane patches (Figure 1A). Instead, they had an apparently random distribution. PSI complexes were also observed in both monomeric and dimeric configurations (complexes not outlined in Figures 7E, 7H, and 7K).
Figure 7.
AFM of PSI in Thylakoids from Synechocystis sp PCC 6803.
(A) Topograph of a PSI-rich region.
(B) Zoomed image from the inset outlined in (A) with the positions of three height profiles shown (red, blue, and green dashed lines), taken across the approximate positions of maximum height.
(C) Structure of the cytoplasmic face of the PSI complex taken from PDB:1JB0 for the T. elongatus PSI complex.
(D) Height profiles corresponding to the lines in (B), with lateral distances shown. The dashed yellow lines are drawn between proline 29 of the PsaC subunit on each monomer, shown in green, and represent a distance of 9.1 nm.
(E) The topograph in (A), with PSI trimers outlined in black and other topographic features with an average height of 13.0 ± 0.6 nm above the mica and 6.0 ± 0.8 nm above the lipid bilayer outlined in white.
(F) and (I) PSI arrays bordering membrane regions with minimal topology.
(G) and (J) Sections across these featureless regions corresponding to the dashed lines in (F) and (I).
(H) and (K) the topographs in (F) and (I)with the PSI trimers outlined in black.
(L) Diagram depicting a model for the lateral segregation of PSI (red) from PSII (green) and cytb6f (purple) complexes in the lipid bilayer (gray band).
Another feature of the Synechocystis thylakoid membranes was the presence of a second protein complex (highlighted in Figures 7E with white circles and numbered in Supplemental Figure 9). This complex had an average height of 13.0 ± 0.6 nm above the mica and 6.0 ± 0.8 nm above the lipid bilayer, too large to be PSI (measured as 9.3 ± 0.4 nm and 3.2 ± 0.3 nm above the mica and lipid bilayer, respectively, from the membrane patch in Figure 1A). These dimensions are consistent with the cyanobacterial NDH-1 complex (Folea et al., 2008b), which has been shown to be present in cyanobacterial thylakoid membranes (Ogawa, 1992; Zhang et al., 2004; Xu et al., 2008).
It was also possible to image trimeric protein complexes in membrane patches from Synechococcus 7002, which were assigned as PSI (Supplemental Figure 10). AFM topographs show a closely packed array of ∼46 PSI trimers; the array is contiguous with another region of the thylakoid membrane that houses no PSI complexes but does contain protein, as shown by the ∼10-nm height above the mica (Supplemental Figure 10). This value is significantly higher than the 5.0 ± 0.2 nm measured for the empty lipid bilayer in Supplemental Figure 6, indicating that this area is occupied by protein complexes with minimal topology on the cytoplasmic face of the membrane.
The presence of PSI arrays bordering membrane regions with minimal topology was also a feature in Synechocystis membranes; Figures 7F and 7I show membrane patches where densely packed PSI complexes are contiguous with featureless areas ∼10 nm above the mica support (Figures 7G and 7J), significantly higher than the 5.0 ± 0.2 nm expected for lipid-only regions of membrane. From the height data and the lack of distinct topology, it is possible these regions represent the location of PSII and the cytb6f complex, both of which would elevate the membrane to the observed height by virtue of their large protrusions on the underside (lumenal face) of the immobilized thylakoid membrane; neither complex has significant topology on the upper side (cytoplasmic face) of the membrane. A model for the segregation of PSI from PSII and cytb6f complexes is proposed in Figure 7L. This is in contrast to membrane patches from T. elongatus where large areas of featureless membranes are not observed; rather, PSI complexes can be seen interspersed among complexes with minimal topology on the cytoplasmic side of the membrane, again proposed to be PSII and cytb6f complexes (Figure 5B).
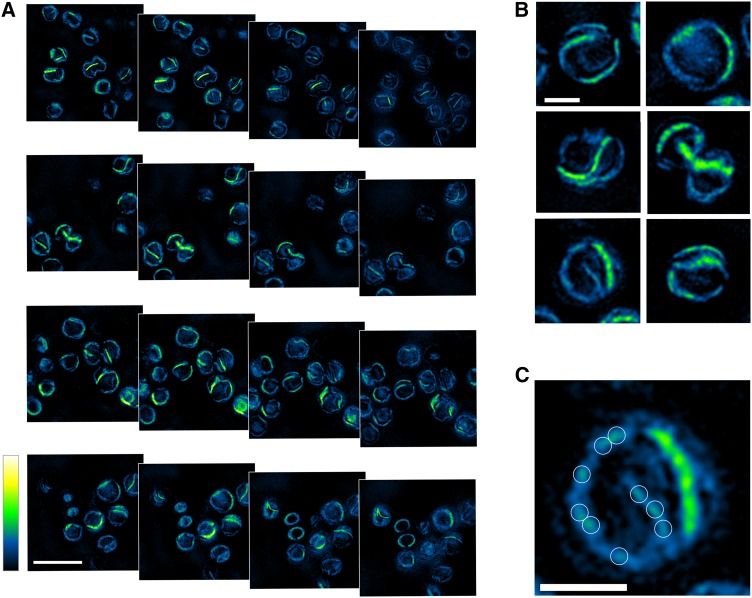
HCFM and 3D-SIM of PSI-YFP Complexes in Synechocystis sp PCC 6803 Cells
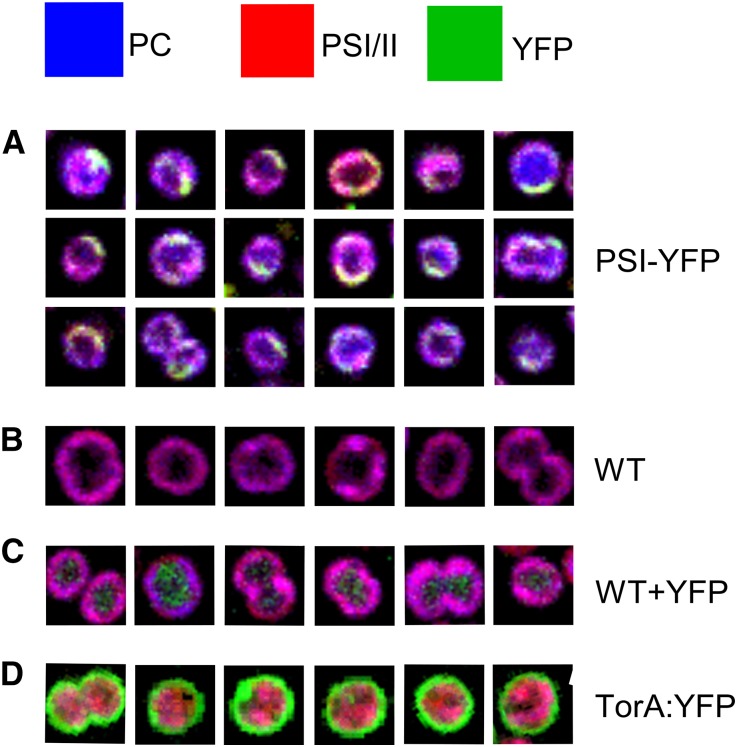
To visualize thylakoid domains in vivo, we used two forms of optical microscopy to image PSI, which had been labeled with the SYFP2 variant of the yellow fluorescent protein (YFP). This strain of Synechocystis bears a fluorescent tag on the cytoplasmic face of the PSI complex, attached to the C terminus of the PsaF subunit. HCFM (Vermaas et al., 2008) was used to image the in vivo distribution of PSI-YFP complexes; this form of microscopy combines spatial and spectral information within every pixel in the image, and the overlapping spectra for exogenous fluorescent protein and endogenous phycobilisome and photosystem components are resolved computationally by multivariate curve resolution (MCR). This approach has already been used to image the spatial distribution of pigments in Synechocystis (Vermaas et al., 2008; Collins et al., 2012; Liberton et al., 2013a). Here, we used spectra for phycobilisome and photosystem and YFP components and employed controls in which non-thylakoid cell compartments in Synechocystis were labeled, namely, “free” YFP in the cytoplasm and YFP exported to the periplasm using the Twin-arginine translocase (Spence et al., 2003). Figure 8A shows the cellular distribution of phycocyanin, chlorophyll in PSI/PSII, and YFP based on spectra representing the pure fluorescent components. The PSI-YFP cells showed a heterogeneous distribution of components, with the yellow/green signal confined mainly to the periphery of the cells. Distinct green and yellow membrane patches are observable corresponding to areas of PSI-YFP and PSI-YFP with phycocyanin, respectively. These patches are interspersed with areas rich in magenta signal that arises from superposition of phycocyanin (blue) and PSI/PSII (red). This superposition is clearly seen in the wild-type control (Figure 8B), indicating that at the diffraction-limited spatial resolution of HCFM images (250 nm lateral, 600 nm axial), the combination of phycobilisome and chlorophyll signals produces a fairly homogeneous image of thylakoid membranes. Comparison of Figures 8A and 8B shows that substantial zones of PSI-YFP are found in thylakoid membranes. Figure 8C clearly shows YFP expressed uniformly throughout the cytoplasm, and Figure 8D shows that the YFP is efficiently targeted to the periplasm by the TorA signal peptide. Overall, Figure 8 shows that the cellular organization of PSI-YFP in thylakoid membranes differs from the distribution of YFP targeted to other cellular compartments and that in some cells the area of YFP-labeled PSI covers a quarter of the cell circumference, implying distances of 1 μm or more and significant areas of the Synechocystis thylakoid membrane allocated to PSI-enriched regions.
Figure 8.
HCFM of PSI-YFP Complexes in Synechocystis sp PCC 6803 Cells.
(A) Cellular distribution of phycocyanin, PSI/PSII chlorophyll, and YFP based on spectra representing the pure fluorescent components and color coded as shown at the top of the figure. Green and yellow membrane patches correspond to areas of PSI-YFP and PSI-YFP with phycocyanin, respectively. The magenta signal arises from superposition of phycocyanin (blue) and PSI/PSII (red).
(B) Wild-type control showing only the magenta phycocyanin plus PSI/PSII signal.
(C) Control showing YFP expressed uniformly throughout the cytoplasm.
(D) Control for the periplasmic compartment, in which YFP is efficiently targeted to the periplasm by the TorA signal peptide. Box sizes = 3 × 3 µm.
Cells of the PSI-YFP strain of Synechocystis were imaged using 3D-SIM (Gustafsson et al., 2008), which excites the sample with a variety of precisely known illumination patterns, allowing reconstruction of a high-resolution optical image. The benefit of 3D-SIM is a 2-fold improvement compared with diffraction-limited imaging, and for three-dimensional volumetric imaging there is an 8-fold improvement; typical resolutions are ∼120 nm (xy) and ∼320 nm (z). The data set comprises 19 sequential images, each of which corresponds to 125 nm in the axial plane. The images were recorded using a 528/48-nm filter for YFP, which excludes phycobilisome and chlorophyll fluorescence. Figure 9A shows four fields of images of PSI-YFP cells, each one with four consecutive, but nonsequential optical slices through each cell. In these images, maximal YFP fluorescence is displayed in green/yellow with lower levels of YFP fluorescence in blue, as shown in the vertical scale bar. These images show that PSI-YFP is the dominant complex in some thylakoid regions, which can be extensive in some Synechocystis cells, less so in others. As the field of view moves through each cell, PSI-YFP membrane regions fade and others come into view. We did not perform complete three-dimensional reconstructions and quantifications of the PSI-YFP membrane areas within each cell, as this is beyond the scope of this study. However, inspection of single slices through six cells in Figure 9B shows that there are PSI-YFP-enriched membrane regions of 1 μm or more in length, consistent with the HCFM images in Figure 8, and that PSI-only regions occupy sizeable areas of the Synechocystis thylakoid membrane. Other noteworthy features include much smaller, faint zones of YFP fluorescence (Figure 9C), likely to come from areas of the thylakoid where PSI and PSII are intermixed, as also shown in the AFM images in Figures 5 to 7. Finally, these 3D-SIM images show that PSI-YFP is present in all areas of the thylakoid membrane; low concentrations of PSI are indicated by the blue signal, which traces the outlines of paired thylakoids as they traverse the cell and also shows points where they appear to converge (Figure 9B, bottom left panel). These features have also been seen in tomographic reconstructions from negative stain electron microscopy (van de Meene et al., 2006).
Figure 9.
3D-SIM of PSI-YFP Complexes in Synechocystis Cells.
(A) Four fields of images of PSI-YFP cells, each one with four consecutive, but nonsequential, optical slices through each cell. YFP fluorescence is displayed on the Green Fire Blue scale in ImageJ, with highest levels of YFP green/yellow and lower levels in blue (bar = 4 µm).
(B) Zoomed images to show PSI-YFP membranes in more detail (bar = 1 µm).
(C) Image showing smaller, faint zones of PSI-YFP fluorescence outlined in white (bar = 1 µm).
DISCUSSION
The AFM topographs in Figures 1, 2, 5, 6, and 7 and Supplemental Figures 5, 8, 9, and 10 show the molecular arrangements of PSI-enriched and mixed PSI/PSII thylakoid membrane regions from T. elongatus, Synechococcus sp PCC 7002, and Synechocystis sp PCC 6803. In intact thylakoid membranes within the cell, such PSI-only and PSI/PSII zones are expected to be contiguous, as seen in Figure 5 and Supplemental Figures 5, 8, 9, and 10. This is in line with previous evidence supporting some degree of spatial heterogeneity in cyanobacterial thylakoids (Vermaas et al., 2008; Sherman et al., 1994; Agarwal et al., 2010). The identification of PSI-rich membrane regions could have been anticipated, since PSI is the major photosystem in T. elongatus (PSI:PSII = 3.94); nevertheless, the existence of PSI-only membrane domains showing long-range order, and with no other membrane proteins such as Complex I present, was unexpected as no such arrangement has been seen previously despite extensive analyses of cyanobacterial thylakoid membranes by freeze-fracture and negative stain electron microscopy (Olive et al., 1986, 1997; Mörschel and Schatz, 1987; Vernotte et al., 1990; Westerman et al., 1994; Folea et al., 2008a). The close packing of PSI trimers seen in the AFM topographs in Figures 1 and 2 leaves no room for other complexes such as cytb6f or ATP synthase and further work, for example, using fluorescent labeling and microscopy of whole cells, will have to be performed to confirm the exclusion of other complexes from such PSI arrays in vivo. We found PSI clusters in membranes purified without the aid of digitonin (Supplemental Figure 4), albeit not in an ordered array in the single example we were able to examine; treatment with 0.1% (w/w) digitonin maintains the overall PSI:PSII ratio, and it appears to separate PSI-enriched membrane domains from other regions of the thylakoid, rendering them more amenable to high-resolution AFM imaging, as in Figure 1A. The AFM topographs in Figure 7 and Supplemental Figure 10 indicate a similar PSI organization in Synechocystis and Synechococcus 7002, so the partitioning of thylakoids into PSI-enriched domains could be widespread in cyanobacteria; surveys of thylakoids from a wide variety of cyanobacteria are required to examine this point further.
Visualization of PSI domains by AFM required removal of phycobilisomes from membranes, but HCFM and 3D-SIM identify PSI domains in whole cells, which do contain phycobilisomes, showing that our AFM topographs reflect the in vivo molecular organization of PSI. We have shown by AFM that PSI domains can be found in three different cyanobacteria, T. elongatus, Synechococcus 7002, and Synechocystis, and have recorded HCFM and 3D-SIM images of intact Synechocystis cells that establish PSI domains within the context of the complete thylakoid system. The 3D-SIM images show that extensive PSI arrays occupy a significant proportion of the thylakoid system in some cells, and other, much smaller, PSI regions are interspersed throughout the thylakoid membrane system. Consistent with these mixed regions, the AFM images in Figures 5 to 7 show membranes where both PSI and PSII complexes reside. We cannot quantify the thylakoid membrane area occupied by PSI-only domains, but it is possible to make a rough estimate; we use a number of 1.5 × 107 chlorophyll molecules/cell (Keren et al., 2004), of which ∼85% is accounted for by PSI and the rest by PSII. Thus, there are ∼47,000 PSI trimers and 34,000 PSII dimers per cell. Using the membrane packing density of PSI from Figure 2A, we calculate that PSI trimers can occupy up to 19 μm2 of thylakoid area in a Synechocystis cell. Similarly, using an estimated packing density for PSII of 4000 dimers per μm2, these complexes occupy ∼8.6 μm2 of thylakoid area. Given the small size of a Synechocystis cell, ∼1 μm diameter, multiple layers of paired thylakoids are required to provide the internal membrane area necessary for accommodating PSI and PSII, as shown by tomographic reconstructions of Synechocystis (van de Meene et al., 2006). Inspection of the cells visualized by 3D-SIM in Figure 9B shows that generally there are between one and three PSI-enriched thylakoid domains per cell, and their total area likely accounts for only a few μm2 of the Synechocystis thylakoid membrane. Thus, PSI-enriched domains are significant, but much of the PSI is probably situated in much smaller zones (Figure 9C), alongside PSII.
Our study uniquely combines optical microscopy on the cellular scale and AFM on the molecular scale, and the availability of the PSI structure provides an opportunity to construct membrane models at a high level of detail. We have shown previously that such models of purple bacterial photosynthetic membranes (Sener et al., 2007; Chandler et al., 2014) provide insights into the factors that limit the conversion of solar energy to ATP. The inherent resolution of such models is still determined by the underlying AFM data and does not represent an atomic resolution structure prediction. However, the assignment of atomic coordinates according to the supramolecular arrangement permits the computational determination of primary energy conversion properties such as excitation transfer kinetics, rate-limiting diffusion processes, or the rate kinetics of ATP synthesis (Cartron et al., 2014; Sener et al., 2016). In this work, the PSI structure (Jordan et al., 2001) and AFM membrane topographs were used to build a structural model of a PSI-only membrane, which reveals the intramembrane packing of these complexes. The structural model reveals long-range order across the array corresponding to strong correlations between PSI trimer locations and, to a lesser extent, between trimer orientations, suggesting a repeated ordered array of trimers as an approximate building block of the PSI-rich domains. The small degree of exciton sharing between PSI trimers (Figure 4C) confers little advantage in light-harvesting efficiency when compared with unconnected PSI trimers (Supplemental Figure 7E), although the arrangement revealed by our AFM topographs achieves a high packing density of PSI complexes in some regions of the thylakoid membrane, possibly to accommodate the excess of PSI complexes in relation to PSII.
Some AFM topographs of T. elongatus membranes show that PSI trimers can intermingle with other complexes (Figure 5); we exploited the precision of the height data in the AFM images, which are typically accurate to 0.1 nm and are not dependent on the “sharpness” of the AFM probe. These analyses showed that the neighbors of PSI are likely to be PSII, as depicted in Figure 5D. The pronounced topology of the PSII water-oxidizing complex on the lumenal side of the membrane aids its identification in AFM topographs of plant thylakoids (Sznee et al., 2011; Johnson et al., 2014; Kirchhoff et al., 2008; Phuthong et al., 2015), but in our measurements, this side of the membrane adheres to the mica support and is not available for imaging. Figure 6 shows that, using a different method to immobilize membranes, PSII complexes can be imaged at low resolution, and they likely sit alongside PSI complexes. The mixed PSI/PSII T. elongatus membranes identified in Figures 5A and 5B, and those from Synechocystis (Figure 7) and Synechococcus 7002 (Supplemental Figure 10) could be the origin of the megacomplex consisting of PSI, PSII, and a phycobilisome (Liu et al., 2013); the PSI-PSII contact zones shown in this work could provide sites for docking phycobilisomes, which are then proposed to donate excitation energy to either one of these photosystems (Liu et al., 2013). The regions with PSI and PSII in close proximity in Figures 5A and 5B also create the conditions for “spillover” of excitations from PSII to PSI (Biggins and Bruce, 1989). State transitions in cyanobacteria involve responses to fluctuations in the quantity and quality of incident light and the redistribution of energy harvested by phycobilisomes to mainly PSI or PSII (van Thor et al., 1998). The PSI/PSII regions identified in Figures 5A, 5B, and 7 and Supplemental Figure 10 represent a functionally flexible array of traps where small adjustments in phycobilisome-photosystem interactions, either through local conformational changes in the phycobilisome or in underlying membrane proteins (Biggins et al., 1984) or through movement of phycobilisomes on the membrane surface (Joshua and Mullineaux, 2004), alter the destination for energy harvested by the phycobilisome. Whether the PSI-only zones in Figure 1A are also visited by phycobilisomes as part of the state transition mechanism is not known. We note that there are distinct yellow and green patches in the whole-cell HCFM images in Figure 8A, indicating that there are at least some PSI-enriched zones without phycobilisomes. There are some monomeric and dimeric topographic features in Figure 5B that we tentatively assign as PSI on the basis of their heights above the membrane bilayer and the mica surface. Monomeric PSI complexes are routinely resolved by separation of cyanobacterial membranes on CN-PAGE and, as clarified recently, this complex is not an artifact of PSI trimer disassembly but it is synthesized independently with fewer de novo chlorophyll molecules required than for trimers (Kopečná et al., 2012). On the other hand, almost no dimeric PSI complexes are detected by native gels (Kopečná et al., 2012); therefore, an association of two monomeric PSI is expected to be rather weak and transient, or unstable.
PSII complexes were identified in Figure 6, and the heights of their membrane-extrinsic features are consistent with intact water-oxidizing complexes, as seen in previous studies (Sznee et al., 2011; Johnson et al., 2014). The AFM height data do not positively identify cytb6f complexes in the membranes with random organization of PSI and PSII (Figures 6A and 6B). The ratio of PSII to cytb6f complexes in cyanobacteria has previously been measured to be between 1.08 and 1.38, depending on growth conditions (Fujita and Murakami, 1987), so they are likely present in the topographs, but there is only 0.4 nm difference in the height of PSII and cytb6f complexes on the lumenal surface of the thylakoid and the low resolution in the topograph in Figure 6A is insufficient for positive identification of cytb6f complexes. Johnson et al. (2014) overcame this difficulty by using a form of peak-force quantitative nanomechanical mapping in which AFM probes were functionalized with oxidized plastocyanin, which transiently interacts with the reduced cytb6f complexes encountered during the scan over the membrane surface. This work showed that cytb6f complexes were distributed throughout the PSII-enriched regions of the granal lamellae from spinach (Spinacia oleracea); a similar approach could be used to discover the membrane distribution of cytb6f complexes in cyanobacterial thylakoids. In plants, such a colocalization of PSII and cytb6f complexes allows fast exchange of plastoquinol/plastoquinone molecules between the two complexes enabling efficient electron transport from PSII to cytb6f (Johnson et al., 2014). Although we cannot demonstrate the location of cytb6f complexes in these mixed PSI/PSII membrane regions, we can exclude the possibility that cytb6f, or indeed any other complex, are present in the PSI-only membrane imaged in Figures 1 and 2 and modeled in Figure 3. We also note that we did not see ATP synthase complexes so they are absent from the membranes we isolated, or they were damaged during the isolation procedure or during imaging. However, we tentatively assign the complexes in Synechocystis thylakoid membranes (Figure 7E, white circles) to the cyanobacterial NDH-1 complex; the proximity of Complex 1 and PSI has been demonstrated recently in Arabidopsis using single particle reconstruction (Yadav et al., 2017).
There are some parallels between the presence of PSI-enriched regions in cyanobacterial thylakoids and the segregation of PSI into stromal lamellae in higher plants. Linear electron flow in plants requires that plastocyanin diffuses from granal cytb6f complexes to PSI in stromal lamellae over distances of tens or even hundreds of nanometers (Johnson et al., 2014; Kirchhoff et al., 2004). This significant physical separation of granal and stromal lamellae, and obstruction of plastocyanin diffusion at the membrane surface by protein protrusions such as the PSII water-oxidizing complex, are proposed to impede the reduction of stromal PSI (Kirchhoff et al., 2004). In cyanobacteria, PSI turnover in PSI-enriched membrane domains requires that reduced plastocyanin or cytochrome c6 molecules, generated by distal cytb6f complexes in the mixed membrane zones, diffuse to their docking sites on the lumenal side of the PSI array. Given that cyanobacterial thylakoids are closely appressed, and enclose a lumen of 5 to 9 nm (van de Meene et al., 2006; Liberton et al., 2013b), plastocyanin migration from cytb6f complexes and reduction of the PSI in large arrays might be impeded, as in plants. The cytochrome c6 to PSI ratio for T. elongatus, for example, has been determined as 1.7 (Nguyen et al., 2017), so ensuring that they are oxidized by one of the PSI complexes in an array could be important. Having reached a PSI array, reduced plastocyanin or cytochrome c6 molecules will find many options for docking to an oxidized PSI within a confined area of a few hundred nm2, optimizing the supply of oxidized plastocyanin or cytochrome c6 for turnover at cytb6f complexes. The product of PSI turnover is reduced ferredoxin, like plastocyanin a small, rapidly diffusible electron carrier; PSI arrays act as a central point of supply of reduced ferredoxin for cyclic electron transport, pigment biosynthesis, nitrate, nitrite, and sulfite metabolism, and CO2 fixation (Lea-Smith et al., 2015).
In conclusion, we used AFM to image the supramolecular organization of PSI complexes, building on the crystallographic work that determined the three-dimensional structure of PSI (Jordan et al., 2001). The molar excess of PSI over PSII results in a heterogeneity in thylakoid composition (Liu, 2016), so there are areas of the thylakoid where the two complexes are interspersed and others where PSI trimers are arranged into regular arrays, which are clearly visualized by high-resolution optical microscopy. A detailed structural membrane model shows that intertrimer energy migration is possible. A more complete model of these processes, and a detailed understanding of the light-harvesting and electron transport processes of photosynthesis in cyanobacteria, await further identification of PSII and cytb6f complexes, and then of other major components such as the ATP synthase and Complex I.
METHODS
Growth of Cells and Strain Generation
Thermosynechococcus elongatus BP-1 was grown as a 20-liter culture in medium D (Castenholz, 1969), bubbled with 5% (v/v) CO2-enriched air under a constant white light intensity of 250 μmol photons/m2/s at 57°C. Synechococcus sp PCC 7002 and Synechocystis sp PCC 6803 were grown as 1-liter cultures in A+ (Sakamoto and Bryant, 1998) and BG-11 (Rippka et al., 1979) media, respectively. Both were grown under a constant white light intensity of 60 μmol photons/m2/s at 30°C in a rotary shaker at 150 rpm. For growth on plates, BG-11 was supplemented with 10 mM N-Tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid, 5 mM glucose, 1.5% (w/v) agar, 0.3% (w/v) sodium thiosulfate, and appropriate antibiotics (for PSI-YFP, 34 µg/mL chloramphenicol, and for YFP/TorA-YFP, 40 µg/mL kanamycin).
To generate the strains of Synechocystis sp PCC 6803 expressing YFP or a TorA-YFP fusion, genes encoding the respective proteins were cloned into the NdeI/BglII sites of the pPD-FLAG vector (Hollingshead et al., 2012), replacing the 3xFLAG tag coding sequence. When the resulting plasmids were introduced into Synechocystis (as described in Hollingshead et al., 2012), the inserted gene was integrated in place of psbAII (slr1311) by double homologous recombination. Transformants were selected on BG-11 plates containing 5 µg/mL kanamycin, and genome copies were segregated by plating with sequential doubling of the kanamycin concentration up to 40 µg/mL. Successful strain generation was confirmed by PCR.
Preparation of Crude Thylakoid Membranes
Crude membranes were prepared in the presence of 1.2 M betaine as described (Boussac et al., 2004). Cells were grown and harvested in the log phase at a final OD685 ≈ 1.6 to 2.0, pelleted by centrifugation, and washed with buffer 1 (40 mM MES-NaOH, pH 6.5, 15 mM MgCl2, 15 mM CaCl2, 1.2 M betaine, and 10% [v/v] glycerol). After adjusting the chlorophyll a concentration to 1.5 mg/mL and adding 0.2% (w/v) BSA, 50 μg/mL DNase I, 1 mM amino-caproic acid, and 1 mM benzamidine, cells were disrupted via two passes in a French press at 6000 p.s.i. at 4°C. All subsequent steps were performed under dim light conditions at 4°C. Unbroken cells were removed by centrifugation for 5 min at 4°C and 1000g (JA14 rotor; Beckman Coulter) and the crude membranes pelleted and washed three times with buffer 1 by centrifugation for 20 min at 4°C and 149,000g (45Ti rotor; Beckman Coulter). Crude membranes were finally resuspended in buffer 2 (20 mM MES-NaOH, pH 6.5, 10 mM MgCl2, 20 mM CaCl2, and 25% [v/v] glycerol). Membranes were then either used for digitonin treatment (see below) or frozen in liquid nitrogen and stored at a chlorophyll a concentration of 2 to 3 mg/mL at –80°C.
Digitonin Treatment of Membranes
For the preparation of membranes for imaging by AFM, crude membrane samples were loaded onto a 11.5-mL continuous sucrose gradient made up from equal volumes of 30% (w/w) and 50% (w/w) sucrose that contained 0.1% (w/w) digitonin made up in a buffer containing 25 mM KH2PO4/K2HPO4 (pH 7.4), 100 mM NaCl, and 10 mM MgCl2. The gradient was made in a SW41 centrifuge tube which was centrifuged at 40,000 rpm in a SW41 rotor for 2 h at 4°C. The membrane bands were then harvested via the use of a peristaltic pump. For assessing the effect of digitonin on the PSI:PSII ratio of membrane samples, membranes were prepared in the same way with the exception that the sucrose gradient contained 1.0% (w/w) digitonin.
Tween 20 Treatment of Membranes
Membranes were prepared in the same way as for the digitonin treatment of membranes with the exception that the sucrose gradient contained 0.1% (w/w) Tween 20.
AFM
Instrumentation
A Multimode VIII AFM with Nanoscope 8.0 controller (Bruker Nano Surfaces Business) was used to image biological samples.
Sample Preparation
Typically, 5 μL of sample at a chlorophyll concentration of 50 to 100 µg/mL was pipetted onto freshly cleaved mica followed by 45 μL of adsorption buffer [10 mM (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), pH 7.5, 150 mM KCl, and 25 mm MgCl2]. The sample was then either incubated for 1 to 2 h to allow membrane patches to adsorb to the mica surface or was dried under a constant stream of nitrogen and washed with imaging buffer (10 mM HEPES, pH 7.5, 100 mM KCl, and 10 mm MgCl2). The sample was mounted onto the piezoelectric scanner for imaging.
Sample Imaging
Samples were imaged in peak force quantitative nanomechanical mapping mode under liquid with a peak force frequency of 2 kHz unless otherwise stated. When imaging under liquid, the standard fluid cell was used to house and control the AFM probe. Once the probe had been inserted into the fluid cell, the reservoirs were filled with standard imaging buffer and mounted on top of the sample where the laser was aligned with the probe. The peak force amplitude used during imaging was 10 nm, and images were taken at 256 × 256 or 512 × 512 pixel arrays. The peak force set point was varied between 50 and 1000 pN, and the scan rate was selected to be between 0.5 and 1.2 Hz. The probes used during imaging were SNL probes (Bruker), and all image processing was done in NanoScope Analysis v1.40r1 or Gwyddion v2.47 software packages.
CN-PAGE
CN-PAGE was conducted using a 4 to 16% gradient gel according to the protocol described (Wittig et al., 2007). Gels were poured using 16 × 20-cm plates separated with 10-mm spacers. One hundred microliters of sample was loaded per lane with a concentration of 0.1 to 0.2 mg/mL of chlorophyll. Gels were run at 4°C.
Electroelution of Protein Complexes from Polyacrylamide Gels
Bands containing protein complexes were cut out of the CN-PAGE gel and sectioned into pieces with the approximate dimensions of 2 × 2 mm. The gel pieces were placed in the electro-eluter (Bio-Rad model 422 Electro-Eluter), which was run at a constant current of 10 mA. The elution buffer contained 500 mM Tricine, 150 mM Bis-Tris/HCl (pH 7.0), 0.5% (w/v) sodium deoxycholate, and 0.02% (w/v) n-dodecyl-β-maltoside detergent.
Cytochrome b6f Concentration Calculation
Absorbance spectra of isolated membranes from T. elongatus in oxidized and reduced states (dithionite was used as the reductant) were taken, and the concentration of cytb6f was calculated from the difference spectrum using an extinction coefficient of 14 mM−1 cm−1 for the difference in absorption between 563 and 570 nm (Cramer and Whitmarsh, 1977).
Absorbance Spectroscopy
Absorbance spectra of whole cells and membrane suspensions were taken at room temperature using a Cary 50 spectrophotometer between the wavelengths of 250 and 800 nm. The Cary WinUV software package was used to display and analyze spectra.
Structural Model of PSI Arrays
The structural model does not constitute a prediction of coordinates at atomic resolution as its inherent resolution is still determined by AFM data. The structural model instead constitutes a representative structure assignment to permit computations requiring atomic coordinates such as excitation transfer kinetics. The model of the array shown in Figure 3 was built by fitting the protrusion profile of the PSI trimer to the AFM topograph. For this purpose, the peaks of the PSI trimer, PDB:1JB0, in Figure 1D are mapped on the density in Figure 1A, thus determining the position and orientation of 96 trimers. The resulting planar structural model was visualized using VMD (Humphrey et al., 1996) and refined by removing steric clashes through manual adjustment of PSI placements (Supplemental Figure 11). Steric clashes do not indicate real, physiological steric clashes in the membrane, but in the initial assignment of trimer positions only. These apparent steric clashes (similar issues also occurred initially when modeling the bacterial chromatophore vesicle; Cartron et al., 2014) arise because (1) the AFM resolution is coarser than the atomic resolution; (2) local fluctuations in image quality can be lower than what the overall resolution indicates; and (3) the image recognition applied to the placement of the protein complexes is imperfect.
Excitation Transfer Kinetics and Excitonic Connectivity between PSI Monomers
Excitation transfer kinetics for the PSI array shown in Figure 4 follows the formalism in Sener et al. (2004), described below for completeness. The effective Hamiltonian, 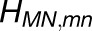 , is stated in Equation 1. The site energies
, is stated in Equation 1. The site energies 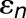 were taken from Damjanović et al. (2002). The intramonomer couplings between chlorophylls
were taken from Damjanović et al. (2002). The intramonomer couplings between chlorophylls 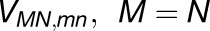 were taken from the full Coulomb computations reported by Cramer and Whitmarsh (1977). The intermonomer couplings,
were taken from the full Coulomb computations reported by Cramer and Whitmarsh (1977). The intermonomer couplings, 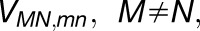 were computed using the transition-dipole approximation according to Sener et al. (2004). Excitation transfer kinetics in the PSI arrays can be described using Förster theory as an approximation (Sener et al., 2002, 2004; Croce and van Amerongen, 2013; Yang et al., 2003; van Stokkum et al., 2013).
were computed using the transition-dipole approximation according to Sener et al. (2004). Excitation transfer kinetics in the PSI arrays can be described using Förster theory as an approximation (Sener et al., 2002, 2004; Croce and van Amerongen, 2013; Yang et al., 2003; van Stokkum et al., 2013).
The transfer rate 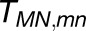 between chlorophylls (M,m) and (N,n) is given according to Förster theory (Van Amerongen et al., 2000)
between chlorophylls (M,m) and (N,n) is given according to Förster theory (Van Amerongen et al., 2000)
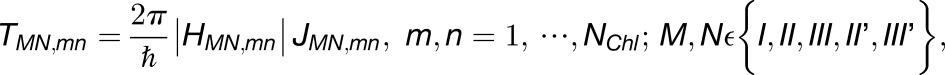 |
where 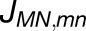 are the spectral overlap integrals defined by Sener et al. (2004). Accordingly, the time evolution of the
are the spectral overlap integrals defined by Sener et al. (2004). Accordingly, the time evolution of the  dimensional state vector
dimensional state vector 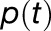 , where
, where 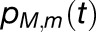 is the probability that chlorophyll (M,m) is excited at time t, is governed by
is the probability that chlorophyll (M,m) is excited at time t, is governed by 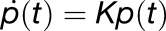 , where the matrix K is (Sener et al., 2004).
, where the matrix K is (Sener et al., 2004).
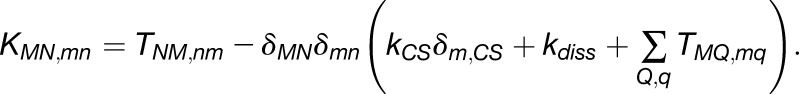 |
Here, 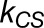 and
and 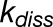 are the rates for charge separation at a RC and dissipation, respectively (van Grondelle et al., 1994);
are the rates for charge separation at a RC and dissipation, respectively (van Grondelle et al., 1994); 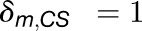 when m is a charge separation site and 0 otherwise. The quantum yield q for an initial state
when m is a charge separation site and 0 otherwise. The quantum yield q for an initial state 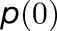 (Sener et al., 2004) is given by
(Sener et al., 2004) is given by
whereas the cross-monomer trapping probability 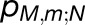 for an excitation initiated at chlorophyll (M,m) to be absorbed at RCN, describing the excitonic connectivity between monomers, is given by
for an excitation initiated at chlorophyll (M,m) to be absorbed at RCN, describing the excitonic connectivity between monomers, is given by
HCFM
Spectral images of Synechocystis cells were obtained on a custom hyperspectral confocal fluorescence microscope (Sinclair et al., 2006). A 488-nm laser excitation (Coherent) was focused onto the cells using a 60× oil immersion objective (Nikon Plan Apochromat, NA 1.4). Spectral data for each voxel of the image were dispersed using a prism spectrometer (500–800 nm) and collected with an electron-multiplied CCD camera (Andor Technologies). During imaging, a combination of laser and stage scanning provided diffraction limited voxels with a spatial resolution of 240 × 240 × 600 nm and an integration time of 240 µs/spectrum. Laser power at the sample was ∼700 nW. For all imaging, 5 μL of cultured cells suspended in BG-11 media was placed upon BG-11 containing, agar-coated slides, spread to a thin layer, covered with a No. 1.5 cover glass, and sealed with nail polish. Samples were immediately imaged and were not kept under the cover slip for more than 1 h. A minimum of 100 cells was imaged for each Synechocystis strain.
HCFM images were preprocessed and underlying component spectra were extracted using multivariate curve resolution as described elsewhere (Jones et al., 2012; Timlin et al., 2015). In brief, HCFM images of all the Synechocystis strains were compiled into a composite image set. Principal component analysis of the preprocessed data set indicated five underlying spectral factors were present. A five-factor MCR model was developed, consisting of a flat baseline to describe detector offset and four pigment spectra: YFP, phycocyanin, allophycocyanin, and chlorophyll. This MCR model described ∼90% of the spectral variance, and no significant spectral residuals were observed. Concentration maps for each of the spectral components identified by MCR were predicted using classical least squares.
3D-SIM
PSI-YFP Synechocystis cells in PBS (pH 7.4) were incubated on a piranha-cleaned, poly-l-lysine-coated cover slip for 30 min before being washed with PBS and mounted with VectaShield (Vector Laboratories). Samples were imaged in the Wolfson Light Microscopy Facility at the University of Sheffield, on a DeltaVision OMX V4 microscope (GE Healthcare) with the Blaze 3D-SIM module equipped with a 60× 1.42 NA oil objective. Illumination was performed with a 488-nm laser line for YFP excitation, and light was collected through a 528/48-nm band-pass emission filter. Axial sectioning was achieved with a step size of 125 nm, and the resultant image stacks were reconstructed with SoftWoRx 6.0 software (GE Healthcare).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: PsaA (Slr1834), BAA17437; PsaB (Slr1835), BAA17438; PsaC (Ssl0563), BAA10036; PsaD (Slr0737), BAA16688; PsaE (Ssr2831), BAA18383; PsaF (Sll0819), BAA18108; PsaI (Smr0004), BAA18774; PsaJ (Sml0008), BAA18107; PsaK (Sll0629), BAA18671; PsaL (Slr1655), BAA18773; PsaM (Smr0005), BAA17005; and PsaX (Tsr0813), NP_681602.
Supplemental Data
Supplemental Figure 1. Protein complexes of the oxygenic photosynthetic electron transport chain in cyanobacterial thylakoid membrane.
Supplemental Figure 2. Purification of membrane patches on sucrose gradients.
Supplemental Figure 3. CN-PAGE of solubilized Thermosynechococcus elongatus thylakoid membranes from digitonin gradients.
Supplemental Figure 4. Low-resolution AFM topograph of a thylakoid membrane patch prepared from a digitonin-free gradient.
Supplemental Figure 5. Gallery of membrane patches prepared from Thermosynechococcus elongatus.
Supplemental Figure 6. AFM of Thermosynechococcus elongatus membrane patches devoid of protein.
Supplemental Figure 7. Long-range order of PSI trimers and the role of excitation sharing.
Supplemental Figure 8. AFM of mixed PSI and PSII membrane patches showing the height measurements for both complexes.
Supplemental Figure 9. AFM of Synechocystis sp PCC 6803 thylakoid membranes.
Supplemental Figure 10. PSI array in a thylakoid membrane from Synechococcus sp PCC 7002.
Supplemental Figure 11. Refinement of the structural model.
Supplementary Material
Acknowledgments
This work was supported as part of the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award DE-SC 0001035. PARC’s role was to fund C.M.-C., M.C.B.-D., and J.A.T. and to provide partial support for C.N.H. and K.S. C.N.H. also acknowledges financial support from Advanced Award 338895 from the European Research Council. C.N.H. and A.H. acknowledge funding from the Biotechnology and Biological Sciences Research Council (award number BB/M000265/1). K.S. acknowledges support from National Science Foundation Grants MCB1157615 and PHY0822613, and National Institutes of Health Grants 9P41GM104601 and 5R01GM098243-02. S.F.H.B. was supported by a University of Sheffield 2022 Futures Studentship. The SIM imaging was performed at the University of Sheffield Wolfson Light Microscopy Facility and was partly funded by MRC Grant MR/K015753/1. We are grateful for the PSI-YFP strain provided by Roman Sobotka, Institute of Microbiology, Třeboň, Czech Republic. We acknowledge Michael Sinclair for the use and maintenance of the hyperspectral confocal fluorescence microscope as well as Howland Jones and Stephen Anthony for the previous development of the multivariate curve resolution software. Sandia National Laboratories is a multi-mission laboratory managed and operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the U.S. Department of Energy’s National Nuclear Security Administration under Contract DE-AC04-94AL85000.
AUTHOR CONTRIBUTIONS
C.M.-C., M.S., K.S., J.B., J.A.T., and C.N.H. designed the research. C.M.-C., S.F.H.B., A.H., K.M., M.C.B.-D., J.A.T., and M.S. performed the research. C.N.H., C.M.-C., M.S., and K.S. wrote the article.
Glossary
- AFM
atomic force microscopy
- HCFM
hyperspectral confocal fluorescence microscopy
- 3D-SIM
three-dimensional structured illumination microscopy
- CN-PAGE
Clear Native-PAGE
- MCR
multivariate curve resolution
- RC
reaction center
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Agarwal R., Matros A., Melzer M., Mock H.-P., Sainis J.K. (2010). Heterogeneity in thylakoid membrane proteome of Synechocystis 6803. J. Proteomics 73: 976–991. [DOI] [PubMed] [Google Scholar]
- Biggins J., Bruce D. (1989). Regulation of excitation energy transfer in organisms containing phycobilins. Photosynth. Res. 20: 1–34. [DOI] [PubMed] [Google Scholar]
- Biggins J., Campbell C.L., Bruce D. (1984). Mechanism of the light state transition in photosynthesis. II. Analysis of phosphorylated polypeptides in the red alga, Porphyridium cruentum. Biochim. Biophys. Acta 767: 138–144. [Google Scholar]
- Boussac A., Rappaport F., Carrier P., Verbavatz J.-M., Gobin R., Kirilovsky D., Rutherford A.W., Sugiura M. (2004). Biosynthetic Ca2+/Sr2+ exchange in the photosystem II oxygen-evolving enzyme of Thermosynechococcus elongatus. J. Biol. Chem. 279: 22809–22819. [DOI] [PubMed] [Google Scholar]
- Bryant D.A., ed (2006). The Molecular Biology of Cyanobacteria, Vol. 1. (Dordrecht, The Netherlands: Springer Science & Business Media). [Google Scholar]
- Cartron M.L., Olsen J.D., Sener M., Jackson P.J., Brindley A.A., Qian P., Dickman M.J., Leggett G.J., Schulten K., Hunter C.N. (2014). Integration of energy and electron transfer processes in the photosynthetic membrane of Rhodobacter sphaeroides. Biochim. Biophys. Acta 1837: 1769–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castenholz R.W. (1969). Thermophilic blue-green algae and the thermal environment. Bacteriol. Rev. 33: 476–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler D.E., Strümpfer J., Sener M., Scheuring S., Schulten K. (2014). Light harvesting by lamellar chromatophores in Rhodospirillum photometricum. Biophys. J. 106: 2503–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis V.P., Chitnis P.R. (1993). PsaL subunit is required for the formation of photosystem I trimers in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 336: 330–334. [DOI] [PubMed] [Google Scholar]
- Collins A.M., Liberton M., Jones H.D., Garcia O.F., Pakrasi H.B., Timlin J.A. (2012). Photosynthetic pigment localization and thylakoid membrane morphology are altered in Synechocystis 6803 phycobilisome mutants. Plant Physiol. 158: 1600–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer W.A., Whitmarsh J. (1977). Photosynthetic cytochromes. Annu. Rev. Plant Physiol. 28: 133–172. [Google Scholar]
- Croce R., van Amerongen H. (2013). Light-harvesting in photosystem I. Photosynth. Res. 116: 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damjanović A., Vaswani H.M., Fromme P., Fleming G.R. (2002). Chlorophyll excitations in photosystem I of Synechococcus elongatus. J. Phys. Chem. B 106: 10251–10262. [Google Scholar]
- Farah J., Rappaport F., Choquet Y., Joliot P., Rochaix J.D. (1995). Isolation of a psaF-deficient mutant of Chlamydomonas reinhardtii: efficient interaction of plastocyanin with the photosystem I reaction center is mediated by the PsaF subunit. EMBO J. 14: 4976–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira K.N., Iverson T.M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838. [DOI] [PubMed] [Google Scholar]
- Folea I.M., Zhang P., Aro E.-M., Boekema E.J. (2008a). Domain organization of photosystem II in membranes of the cyanobacterium Synechocystis PCC6803 investigated by electron microscopy. FEBS Lett. 582: 1749–1754. [DOI] [PubMed] [Google Scholar]
- Folea I.M., Zhang P., Nowaczyk M.M., Ogawa T., Aro E.-M., Boekema E.J. (2008b). Single particle analysis of thylakoid proteins from Thermosynechococcus elongatus and Synechocystis 6803: localization of the CupA subunit of NDH-1. FEBS Lett. 582: 249–254. [DOI] [PubMed] [Google Scholar]
- Fotiadis D., Müller D.J., Tsiotis G., Hasler L., Tittmann P., Mini T., Jenö P., Gross H., Engel A. (1998). Surface analysis of the photosystem I complex by electron and atomic force microscopy. J. Mol. Biol. 283: 83–94. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Murakami A. (1987). Regulation of electron transport composition in cyanobacterial photosynthetic system: stoichiometry among photosystem I and II complexes and their light-harvesting antennae and cytochrome b6/f complex. Plant Cell Physiol. 28: 1547–1553. [Google Scholar]
- Gustafsson M.G., Shao L., Carlton P.M., Wang C.J., Golubovskaya I.N., Cande W.Z., Agard D.A., Sedat J.W. (2008). Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94: 4957–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead S., Kopecná J., Jackson P.J., Canniffe D.P., Davison P.A., Dickman M.J., Sobotka R., Hunter C.N. (2012). Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J. Biol. Chem. 287: 27823–27833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14: 33–38, 27–28. [DOI] [PubMed] [Google Scholar]
- Johnson M.P., Vasilev C., Olsen J.D., Hunter C.N. (2014). Nanodomains of cytochrome b6f and photosystem II complexes in spinach grana thylakoid membranes. Plant Cell 26: 3051–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H.D., Haaland D.M., Sinclair M.B., Melgaard D.K., Collins A.M., Timlin J.A. (2012). Preprocessing strategies to improve MCR analyses of hyperspectral images. Chemom. Intell. Lab. Syst. 117: 149–158. [Google Scholar]
- Jordan P., Fromme P., Witt H.T., Klukas O., Saenger W., Krauss N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411: 909–917. [DOI] [PubMed] [Google Scholar]
- Joshua S., Mullineaux C.W. (2004). Phycobilisome diffusion is required for light-state transitions in cyanobacteria. Plant Physiol. 135: 2112–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetyan N.V., Holzwarth A.R., Rögner M. (1999). The photosystem I trimer of cyanobacteria: molecular organization, excitation dynamics and physiological significance. FEBS Lett. 460: 395–400. [DOI] [PubMed] [Google Scholar]
- Keren N., Aurora R., Pakrasi H.B. (2004). Critical roles of bacterioferritins in iron storage and proliferation of cyanobacteria. Plant Physiol. 135: 1666–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H., Lenhert S., Büchel C., Chi L., Nield J. (2008). Probing the organization of photosystem II in photosynthetic membranes by atomic force microscopy. Biochemistry 47: 431–440. [DOI] [PubMed] [Google Scholar]
- Kirchhoff H., Schöttler M.A., Maurer J., Weis E. (2004). Plastocyanin redox kinetics in spinach chloroplasts: evidence for disequilibrium in the high potential chain. Biochim. Biophys. Acta 1659: 63–72. [DOI] [PubMed] [Google Scholar]
- Kopečná J., Komenda J., Bucinská L., Sobotka R. (2012). Long-term acclimation of the cyanobacterium Synechocystis sp. PCC 6803 to high light is accompanied by an enhanced production of chlorophyll that is preferentially channeled to trimeric photosystem I. Plant Physiol. 160: 2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea-Smith D.J., Bombelli P., Vasudevan R., Howe C.J. (2015). Photosynthetic, respiratory and extracellular electron transport pathways in cyanobacteria. Biochim. Biophys. Acta 1857: 247–255. [DOI] [PubMed]
- Liberton M., Collins A.M., Page L.E., O’Dell W.B., O’Neill H., Urban V.S., Timlin J.A., Pakrasi H.B. (2013a). Probing the consequences of antenna modification in cyanobacteria. Photosynth. Res. 118: 17–24. [DOI] [PubMed] [Google Scholar]
- Liberton M., Page L.E., O’Dell W.B., O’Neill H., Mamontov E., Urban V.S., Pakrasi H.B. (2013b). Organization and flexibility of cyanobacterial thylakoid membranes examined by neutron scattering. J. Biol. Chem. 288: 3632–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhang H., Niedzwiedzki D.M., Prado M., He G., Gross M.L., Blankenship R.E. (2013). Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science 342: 1104–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.-N. (2016). Distribution and dynamics of electron transport complexes in cyanobacterial thylakoid membranes. Biochim. Biophys. Acta 1857: 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005). Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II. Nature 438: 1040–1044. [DOI] [PubMed] [Google Scholar]
- Mörschel E., Schatz G.H. (1987). Correlation of photosystem-II complexes with exoplasmatic freeze-fracture particles of thylakoids of the cyanobacterium Synechococcus sp. Planta 172: 145–154. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U., Chauvat F. (1996). Gene transfer and manipulation in the thermophilic cyanobacterium Synechococcus elongatus. Mol. Gen. Genet. 252: 93–100. [DOI] [PubMed] [Google Scholar]
- Naithani S., Hou J.-M., Chitnis P.R. (2000). Targeted inactivation of the psaK1, psaK2 and psaM genes encoding subunits of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth. Res. 63: 225–236. [DOI] [PubMed] [Google Scholar]
- Nguyen K., Vaughn M., Frymier P., Bruce B.D. (2017). In vitro kinetics of P700(+) reduction of Thermosynechococcus elongatus trimeric photosystem I complexes by recombinant cytochrome c6 using a Joliot-type LED spectrophotometer. Photosynth. Res. 131: 79–91. [DOI] [PubMed] [Google Scholar]
- Ogawa T. (1992). Identification and characterization of the ictA/ndhL gene product essential to inorganic carbon transport of Synechocystis PCC6803. Plant Physiol. 99: 1604–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive J., Ajlani G., Astier C., Recouvreur M., Vernotte C. (1997). Ultrastructure and light adaptation of phycobilisome mutants of Synechocystis PCC 6803. Biochim. Biophys. Acta 1319: 275–282. [Google Scholar]
- Olive J., M’bina I., Vernotte C., Astier C., Wollman F.A. (1986). Randomization of the EF particles in thylakoid membranes of Synechocystis 6714 upon transition from state I to state II. FEBS Lett. 208: 308–312. [Google Scholar]
- Phuthong W., Huang Z., Wittkopp T.M., Sznee K., Heinnickel M.L., Dekker J.P., Frese R.N., Prinz F.B., Grossman A.R. (2015). The use of contact mode atomic force microscopy in aqueous medium for structural analysis of spinach photosynthetic complexes. Plant Physiol. 169: 1318–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111: 1–61. [Google Scholar]
- Sakamoto T., Bryant D.A. (1998). Growth at low temperature causes nitrogen limitation in the cyanobacterium Synechococcus sp. PCC 7002. Arch. Microbiol. 169: 10–19. [DOI] [PubMed] [Google Scholar]
- Schluchter W.M., Shen G., Zhao J., Bryant D.A. (1996). Characterization of psaI and psaL mutants of Synechococcus sp. strain PCC 7002: a new model for state transitions in cyanobacteria. Photochem. Photobiol. 64: 53–66. [DOI] [PubMed] [Google Scholar]
- Sener M.K., Park S., Lu D., Damjanovic A., Ritz T., Fromme P., Schulten K. (2004). Excitation migration in trimeric cyanobacterial photosystem I. J. Chem. Phys. 120: 11183–11195. [DOI] [PubMed] [Google Scholar]
- Sener M.K., Lu D., Ritz T., Park S., Fromme P., Schulten K. (2002). Robustness and optimality of light harvesting in cyanobacterial photosystem I. J. Phys. Chem. B 106: 7948–7960. [Google Scholar]
- Sener M.K., Olsen J.D., Hunter C.N., Schulten K. (2007). Atomic-level structural and functional model of a bacterial photosynthetic membrane vesicle. Proc. Natl. Acad. Sci. USA 104: 15723–15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sener M., Strümpfer J., Singharoy A., Hunter C.N., Schulten K. (2016). Overall energy conversion efficiency of a photosynthetic vesicle. eLife 5: e09541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sétif P., Fischer N., Lagoutte B., Bottin H., Rochaix J.-D. (2002). The ferredoxin docking site of photosystem I. Biochim. Biophys. Acta 1555: 204–209. [DOI] [PubMed] [Google Scholar]
- Sherman D.M., Troyan T.A., Sherman L.A. (1994). Localization of membrane proteins in the cyanobacterium Synechococcus sp. PCC7942 (radial asymmetry in the photosynthetic complexes). Plant Physiol. 106: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair M.B., Haaland D.M., Timlin J.A., Jones H.D. (2006). Hyperspectral confocal microscope. Appl. Opt. 45: 6283–6291. [DOI] [PubMed] [Google Scholar]
- Spence E., Sarcina M., Ray N., Møller S.G., Mullineaux C.W., Robinson C. (2003). Membrane-specific targeting of green fluorescent protein by the Tat pathway in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 48: 1481–1489. [DOI] [PubMed] [Google Scholar]
- Suga M., Akita F., Hirata K., Ueno G., Murakami H., Nakajima Y., Shimizu T., Yamashita K., Yamamoto M., Ago H., Shen J.-R. (2015). Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 517: 99–103. [DOI] [PubMed] [Google Scholar]
- Sznee K., Dekker J.P., Dame R.T., van Roon H., Wuite G.J.L., Frese R.N. (2011). Jumping mode atomic force microscopy on grana membranes from spinach. J. Biol. Chem. 286: 39164–39171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Thor J.J., Mullineaux C.W., Matthijs H.C.P., Hellingwerf K.J. (1998). Light harvesting and state transitions in cyanobacteria. Bot. Acta 111: 430–443. [Google Scholar]
- Timlin J.A., Haaland D.M., Jones H.D. (2015). Experimental and Data Analytical Approaches to Automating Multivariate Curve Resolution in the Analysis of Hyperspectral Images (No. SAND2015-11147B). (Albuquerque, NM: Sandia National Laboratories). [Google Scholar]
- Ting C.S., Rocap G., King J., Chisholm S.W. (2002). Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10: 134–142. [DOI] [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J.-R., Kamiya N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60. [DOI] [PubMed] [Google Scholar]
- Van Amerongen H., Valkunas L., van Grondelle R. (2000). Photosynthetic Excitons. (Singapore: World Scientific; ). [Google Scholar]
- van de Meene A.M., Hohmann-Marriott M.F., Vermaas W.F., Roberson R.W. (2006). The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184: 259–270. [DOI] [PubMed] [Google Scholar]
- van Grondelle R., Dekker J.P., Gillbro T., Sundström V. (1994). Energy transfer and trapping in photosynthesis. Biochim. Biophys. Acta 1187: 1–65. [Google Scholar]
- van Stokkum I.H., Desquilbet T.E., van der Weij-de Wit C.D., Snellenburg J.J., van Grondelle R., Thomas J.-C., Dekker J.P., Robert B. (2013). Energy transfer and trapping in red-chlorophyll-free photosystem I from Synechococcus WH 7803. J. Phys. Chem. B 117: 11176–11183. [DOI] [PubMed] [Google Scholar]
- Vermaas W.F., Timlin J.A., Jones H.D., Sinclair M.B., Nieman L.T., Hamad S.W., Melgaard D.K., Haaland D.M. (2008). In vivo hyperspectral confocal fluorescence imaging to determine pigment localization and distribution in cyanobacterial cells. Proc. Natl. Acad. Sci. USA 105: 4050–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernotte C., Astier C., Olive J. (1990). State 1-state 2 adaptation in the cyanobacteria Synechocystis PCC 6714 wild type and Synechocystis PCC 6803 wild type and phycocyanin-less mutant. Photosynth. Res. 26: 203–212. [DOI] [PubMed] [Google Scholar]
- Westerman M., Ernst A., Brass S., Böger P., Wehrmeyer W. (1994). Ultrastructure of cell wall and photosynthetic apparatus of the phycobilisome-less Synechocystis sp. strain BO 8402 and phycobilisome-containing derivative strain BO 9201. Arch. Microbiol. 162: 222–232. [Google Scholar]
- Wittig I., Karas M., Schägger H. (2007). High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics 6: 1215–1225. [DOI] [PubMed] [Google Scholar]
- Xu Q., Yu L., Chitnis V.P., Chitnis P.R. (1994). Function and organization of photosystem I in a cyanobacterial mutant strain that lacks PsaF and PsaJ subunits. J. Biol. Chem. 269: 3205–3211. [PubMed] [Google Scholar]
- Xu M., Ogawa T., Pakrasi H.B., Mi H. (2008). Identification and localization of the CupB protein involved in constitutive CO2 uptake in the cyanobacterium, Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 49: 994–997. [DOI] [PubMed] [Google Scholar]
- Xu W., Tang H., Wang Y., Chitnis P.R. (2001). Proteins of the cyanobacterial photosystem I. Biochim. Biophys. Acta 1507: 32–40. [DOI] [PubMed] [Google Scholar]
- Yadav K.N., Semchonok D.A., Nosek L., Kouřil R., Fucile G., Boekema E.J., Eichacker L.A. (2017). Supercomplexes of plant photosystem I with cytochrome b6f, light-harvesting complex II and NDH. Biochim. Biophys. Acta 1858: 12–20. [DOI] [PubMed] [Google Scholar]
- Yang M., Damjanović A., Vaswani H.M., Fleming G.R. (2003). Energy transfer in photosystem I of cyanobacteria Synechococcus elongatus: model study with structure-based semi-empirical Hamiltonian and experimental spectral density. Biophys. J. 85: 140–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Battchikova N., Jansen T., Appel J., Ogawa T., Aro E.-M. (2004). Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16: 3326–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.