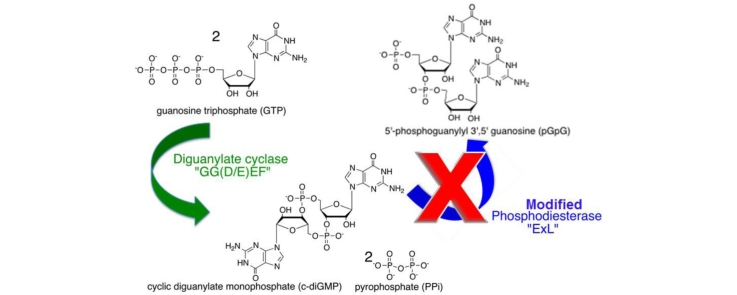
Graphical abstract
Abbrevations: Cyclic-diGMP, bis-3′-5′-cyclic dimeric guanosine monophosphate; GTP, guanosine-5′-triphosphate; pGpG, 5′-phosphoguanylyl-(3′,5′)-guanosine); DGC, diguanylate cyclase; PDE, phosphodiesterase; AvHaCE, Agrobacterium vitis H-NOX associated cyclic-diGMP processing enzyme
Keywords: Cyclic-diGMP, [32P]-cyclic-diGMP, Diguanylate cyclase, Phosphodiesterase, H-NOX associated cyclic-diGMP processing enzyme
Abstract
Cyclic-diGMP is a bacterial messenger that regulates many physiological processes, including many attributed to pathogenicity. Bacteria synthesize cyclic-diGMP from GTP using diguanylate cyclases; its hydrolysis is catalyzed by phosphodiesterases. Here we report the over-expression and purification of a bi-functional diguanylate cyclase-phosphodiesterase from Agrobacterium vitis S4. Using homology modeling and primary structure alignment, we identify several amino acids predicted to participate in the phosphodiesterase reaction. Upon altering selected residues, we obtain variants of the enzyme that efficiently and quantitatively catalyze the synthesis of cyclic-diGMP from GTP without hydrolysis to pGpG. Additionally, we identify a variant that produces cyclic-diGMP while immobilized to NiNTA beads and can catalyze the conversion of [α-32P]-GTP to [32P]-cyclic-diGMP. In short, we characterize a novel cyclic-diGMP processing enzyme and demonstrate its utility for efficient and cost-effective production of cyclic-diGMP, as well as modified cyclic-diGMP molecules, for use as probes in studying the many important biological processes mediated by cyclic-diGMP.
1. Introduction
Cyclic-diGMP [(bis-3′-5′)-cyclic dimeric guanosine monophosphate] is a second messenger ubiquitously produced by bacteria [11], [15], [25] and the eukaryote Dictyostelium discoideum [7]. Since its discovery as a regulator of cellulose biosynthesis in Acetobacter xylinum [45], it has been linked to the regulation of various cellular processes with medical and agricultural implications, including biofilm formation, regulation of virulence factors, pathogenicity, and cell mobility [10], [21], [26], [36], [43], [53]. Because of its role in medically relevant processes, cyclic-diGMP is currently being explored as an anti-infective agent [50], as well as a vaccine adjuvant [6], [22], [38].
The growing interest in understanding the underlying mechanisms by which this molecule regulates these diverse processes has been hampered by the cost associated with obtaining cyclic-diGMP. Because of this, more cost-effective avenues needed to be explored to allow researchers to obtain the nucleotide. Several groups have achieved chemical syntheses of cyclic-diGMP [14], [20], [24], [27], [28], [44], [45]. However, these syntheses require multiple steps that involve protection and deprotection of various functional groups of GTP, which often results in low product yield.
Bacteria synthesize cyclic-diGMP from two molecules of guanosine triphosphate (GTP) using a diguanylate cyclase (DGC) containing a conserved GG(D/E)EF motif [1], [5], [15], [44], [45], [47], [52] or the AGDEF motif [23], [39]. The conserved D/E is proposed to be the active site base responsible for deprotonating the 3′-OH of GTP which facilitates nucleophilic attack of the α-phosphate of the second molecule of GTP thereby producing the cyclized product [5], [26], [57]. Frequently, diguanylate cyclases demonstrate product inhibition. The sequence RxxD has been shown to be required for cyclic-diGMP binding and therefore product inhibition [2], [8], [9], [30], [54]. In bacteria, cyclic-diGMP is hydrolyzed to 5′-phosphoguanylyl-(3′,5′)-guanosine (pGpG) by phosphodiesterases (PDE) containing the signature ExL motif [9], [48], [52], or less commonly the HD-GYP motif [12], [16], [17], [18], [46]. In most cases enzymes containing both GG(D/E)EF and ExL motifs exhibit either diguanylate cyclase or phosphodiesterase activities, but rarely both. However, to date eight bi-functional DGC–PDE enzymes have been identified from various species [13], [19], [31], [33], [34], [35], [49], [55].
Enzymatic synthesis of cyclic-diGMP has been reported by several groups using diguanylate cyclases [9], [29], [40], [51], [58]. In most cases, the enzymes used by these groups contain an RxxD product-inhibition site that limits the amount of cyclic-diGMP produced. One group reported generation a variant of a diguanylate cyclase from the thermophile Thermotoga maritima, encoded by the gene TM1788, in which they changed the RxxD motif to AxxD [40]. However, despite use of this variant, they still observed product inhibition at high concentrations of GTP.
Based on its primary structure, the protein AvHaCE (H-NOX-associated cyclic-diGMP processing enzyme), encoded by the gene Avi_3097 from Agrobacterium vitis strain S4, is predicted to be a bi-functional DGC–PDE. Of particular interest is the fact that the protein lacks the RxxD motif. We hypothesized that due to its lack of the RxxD inhibition site, a variant of AvHaCE without phosphodiesterase activity could efficiently produce large quantities of cyclic-diGMP, and therefore be of great utility to the cyclic-diGMP field. Thus, we sought to investigate whether altering residues thought to be important for phosphodiesterase activity would result in variants of AvHaCE lacking PDE activity while maintaining DGC activity, thus resulting in an improved system for enzymatically producing cyclic-diGMP.
2. Materials and methods
2.1. Materials
5′-GTP was purchased from Promega. [α-32P]-GTP was obtained from PerkinElmer and cyclic di-GMP and pGpG were from Biolog. Restriction endonucleases and Phusion High Fidelity polymerase were purchased from New England Biolabs. PfuTurbo was obtained from Agilent. Qiagen was the source for nickel–nitriloacetic acid (Ni–NTA) agarose. A. vitis S4 cells were a generous gift from Professor Thomas Burr (Cornell University).
2.2. General procedures
Matrix assisted laser desorption ionization (MALDI) analyses were performed at the Stony Brook Proteomics Center or at the Institute of Chemical Biology and Drug Discovery (Stony Brook University) with α-cyano-4-hydroxycinnamic acid as the matrix. High performance liquid chromatography (HPLC) was conducted with a LC-2010A HT liquid chromatography system (Shimadzu). Nucleotides were separated with either a Shim-pack XR-ODS (3 mm × 100 mm) or a Beckman ODS Ultrasphere (4.6 mm × 25 cm) reverse phase C18 column and detection was monitored at 254 nm.
2.3. Construction of wild-type and variant AvHaCE expression vectors
Genomic DNA for cloning A. vitis Avi_3097 was purified from cells of A. vitis S4 by use of the Wizard SV Genomic DNA Purification System by Promega. The gene was cloned from A. vitis genomic DNA by the polymerase chain reaction (PCR) with Pfu Turbo or Phusion as the polymerase. The gene was cloned between the NdeI and XhoI sites of pET20b to generate a C-terminal hexa-histidine tagged protein. QuikChange site directed mutagenesis was used to generate variants of AvHaCE with wild-type Avi_3097 in pET20b plasmid DNA serving as the template for PCR. The Avi_3097 DGC only variant was constructed by introducing a stop codon after the 248th amino acid of the wild-type protein. The presence of the gene and mutations was confirmed at the Stony Brook DNA sequencing facility.
2.4. Expression and purification of hexa-histidine tagged wild-type and variant AvHaCE proteins
Plasmid DNA harboring the gene encoding wild-type or variant AvHaCE was transformed into BL21DE3pLysS competent cells for protein over-expression. Cultures were grown in 2XYT medium (16 g tryptone, 10 g yeast extract, 5 g sodium chloride per liter) supplemented with 100 μg mL−1 ampicillin at 37 °C to an OD600 of ∼1.2. Protein expression was induced by the addition of 10 μM isopropyl β-d-thiogalactopyranoside (IPTG) and was allowed to proceed overnight (∼16 h) at 18 °C after which cells were harvested by centrifugation. Cells were resuspended in Buffer A (50 mM Tris–HCl, pH 7.4, 5 mM β-mercaptoethanol, 10% glycerol, 50 mM arginine, 50 mM glutamic acid, 200 mM sodium chloride, 10 mM imidazole and 500 μM EDTA) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) at room temperature. The suspension was incubated on ice for 20 min and cells were lysed by sonication. Cellular debris was removed by centrifugation at 39,000 × g for one hour at 4 °C. The cleared lysate was applied to a Ni–NTA column equilibrated in Buffer A and the column was washed with 10 column volumes of Buffer A followed by washing with 10 column volumes of buffer containing 50 mM Tris–HCl, pH 7.4, 5 mM β-mercaptoethanol, 10% glycerol, 50 mM arginine, 50 mM glutamic acid, 200 mM sodium chloride, 100 mM imidazole, and 500 μM EDTA. The protein was eluted from the column with buffer containing 50 mM Tris–HCl, pH 7.4, 5 mM β-mercaptoethanol, 10% glycerol, 50 mM arginine, 50 mM glutamic acid, 200 mM sodium chloride, 250 mM imidazole, and 500 μM EDTA. Fractions containing pure protein, as judged by SDS-PAGE analysis, were pooled and dialyzed overnight against 50 mM Tris–HCl, pH 7.4, 5 mM β-mercaptoethanol, 10% glycerol, 50 mM arginine, 50 mM glutamic acid, 200 mM sodium chloride, and 500 μM EDTA and were then stored at −80 °C.
2.5. Protein concentration determination
Protein concentrations were determined by the method of Bradford with bovine serum albumin (BSA) as standard [3]. Protein purity was assessed by SDS-PAGE with a 12.5% gel as described by Laemmli [32].
2.6. Enzyme activity assay
Assays to determine product formation were carried out in 50 mM Tris–HCl, pH 7.5, containing 5 mM MgCl2 and 100 μM GTP. Reactions were initiated by the addition of 1 μM wild-type or variant protein to the assay mixture and were allowed to incubate overnight at room temperature. Reactions were terminated by heating the samples at 95 °C for 5 min. Precipitated proteins were removed by centrifugation after which the supernatant was filtered through a 0.22 μm membrane and analyzed by HPLC with a Shim-pack XR-ODS column. The column was equilibrated with 95% Solvent A (0.1 M triethyl ammonium acetate (TEAA), pH 6.1) −5% Solvent B (70% Solvent A −30% acetonitrile) at a flow rate of 0.1 mL min−1. Nucleotides were eluted with the following gradient: 5–10% Solvent B over 10 min, 10–15% Solvent B over 5 min, 15–20% Solvent B over 5 min, 20–25% Solvent B over 5 min, 25–50% Solvent B over 5 min, 50–5% Solvent B over 5 min and maintained at 5% Solvent B for 5 min. Authentic GTP, cyclic di-GMP, and pGpG were used as standards to determine retention times of the nucleotides. The activity of each enzyme, wild-type or variant, was confirmed a minimum of three times.
2.7. Homology modeling of AvHaCE phosphodiesterase domain
A structural model of the phosphodiesterase domain (residues 249-502) of AvHaCE was generated by use of Modeller 9.9 (http://www.salilab.org/modeller/) with the phosphodiesterase domain (residues 1-250) of YkuI (pdb 2W27) from Bacillus subtilis serving as the template. These residues were chosen for AvHaCE based on its domain prediction with PROSITE (http://prosite.expasy.org/).
2.8. Larger scale synthesis of cyclic-diGMP
Cyclic-diGMP was produced enzymatically with the AVL variant of AvHaCE. GTP (5 μmoles; 2.62 mg) was incubated with 9.9 nmole of enzyme at 30 °C in a 2 mL reaction containing 50 mM Tris–HCl, pH 7.5 and 5 mM MgCl2. Aliquots (100 μL) were removed at various times and reactions were terminated by heating, as described above. Samples were filtered, diluted, and analyzed by HPLC with a Beckman Ultrasphere ODS C18 column with a gradient composed of Solvent A (100 mM KH2PO4, pH 6.1) and Solvent B (70 mM KH2PO4, pH 6.1, and 30% acetonitrile). The column was equilibrated with 10% Solvent B at a flow rate of 0.65 mL min−1. Nucleotides were eluted with the following gradient: 10% Solvent B over 13 min, 10–100% Solvent B over 1 min, hold at 100% Solvent B for 2 min, 100–10% Solvent B over 1 min, and maintained at 10% Solvent B for 3 min.
2.9. Synthesis of cyclic-diGMP with the immobilized AvHaCE AAL variant
Ni–NTA resin, equilibrated with 50 mM Tris–HCl, pH 7.5, was saturated with purified AAL variant of AvHaCE. The resin was washed extensively with 50 mM Tris–HCl, pH 7.5 to remove any AvHaCE not bound to the resin. A reaction mixture containing 500 μM GTP, 5 mM MgCl2 and 50 mM Tris–HCl, pH 7.5 was added to the resin after which the resin was incubated at room temperature with slight agitation. At various time points the reaction sample was centrifuged at 700 × g for 5 min. Aliquots (200 μL) of the supernatant were removed, desalted with a C18 ZipTip and analyzed by MALDI.
2.10. Enzymatic synthesis of radiolabeled cyclic-diGMP
Radiolabeled cyclic-diGMP was synthesized with 2.5 μM purified AAL variant of AvHaCE incubated with 3 mM MgCl2, 300 μM GTP, 100 μCi [α-32P]-GTP and 50 mM Tris–HCl, pH 7.5, at room temperature for 90 min. After 15 min, 60 min, and 90 min, 120 μL of the reaction mixture was removed and the reaction was terminated by heating, as described above. Samples were then centrifuged at 13,000 × g for 1 min to remove precipitates. Supernates were analyzed by HPLC, with a BIOSCAN isotope detector, on a DEAE column (Altex Spherogel-TSK DEAE-5PW; 10 μm, 7.5 mm × 75 mm). The column was developed with a gradient of water (Solvent A) and triethylammonium bicarbonate (1 M TEAB; pH 8; Solvent B) at a flow rate of 1 mL min−1; TEAB is prepared by bubbling CO2 through a solution of 1 M triethylamine until pH 8 is achieved. Nucleotides were eluted with the following gradient: 20% Solvent B over 10 min, 10–40% Solvent B over 25 min, hold at 40% Solvent B for 5 min, 40–95% Solvent B over 5 min, hold at 95% Solvent B for 5 min, 95–20% Solvent B over 5 min and then maintained at 20% Solvent B for 5 min.
3. Results and Discussion
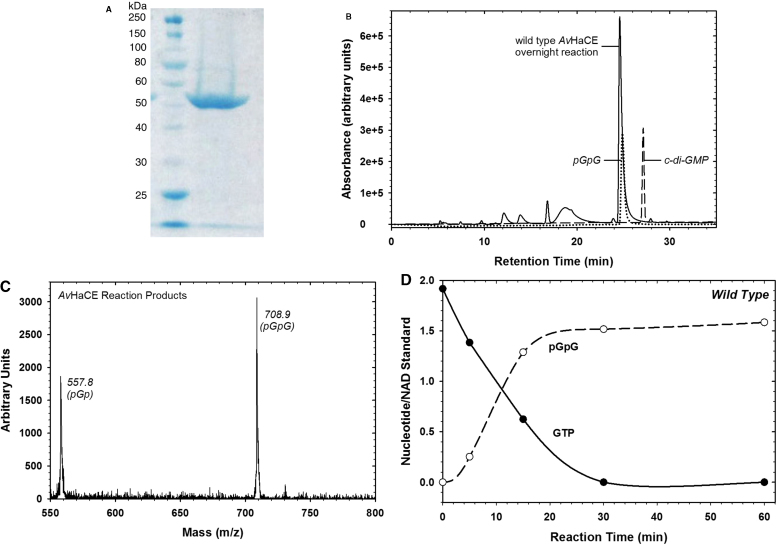
3.1. Avi_3097 encodes a bi-functional diguanylate cyclase-phosphodiesterase
The growing interest in understanding processes regulated by cyclic-diGMP requires one to have access to large quantities of the compound for both in vitro studies as well as for use as a probe in cellular studies. However, obtaining sufficient amounts of pure cyclic-diGMP has presented researchers with a substantial challenge due to the high cost of commercially available cyclic-diGMP and the complexities of its chemical synthesis [14], [20], [24], [27], [28], [44], [45]. Based on the primary structure of its encoded protein, Avi_3097 is predicted to encode a bi-functional diguanylate cyclase-phosphodiesterase. The encoded protein contains both the signature GG(D/E)EF and ExL motifs as well as a conserved loop 6 motif that is proposed to be required for phosphodiesterase activity [41], [56]. To determine whether or not the protein encoded by Avi_3097 contains both diguanylate cyclase and phosphodiesterase activities, we cloned the gene and purified the resulting protein. Upon purification of the protein, we observed a single band with a mass of ∼56 kDa by SDS-PAGE, consistent with the predicted molecular mass of the protein based on its primary structure (Fig. 1A). To confirm that this protein encodes a bi-functional diguanylate cyclase-phosphodiesterase, we incubated the purified wild-type enzyme with GTP. After an overnight incubation, the predominant product eluted from reverse-phase HPLC with a retention time consistent with that of a pGpG standard (Fig. 1B). The eluted peak was collected, analyzed by MALDI, and was confirmed to be pGpG (Fig. 1C; expected molecular mass = 708.5 g/mol). This result suggests that the protein encoded by Avi_3097 is, in fact, a bi-functional diguanylate cyclase-phosphodiesterase and will be referred to as AvHaCE. Fig. 1D illustrates the time-dependent formation of pGpG from 100 μM GTP with 1 μM wild-type AvHaCE. These data indicate the reaction is essentially complete after 1 h under these experimental conditions, with essentially complete conversion of substrate GTP to the ultimate product pGpG.
Fig. 1.
Characterization of wild-type AvHaCE. (A) Coomassie blue stained SDS-PAGE gel (12.5%) of purified AvHaCE. Lane 1, molecular mass standards; lane 2, AvHaCE after purification by immobilized metal affinity chromatography with Ni–NTA as the matrix. (B) HPLC analyses of wild-type AvHaCE reaction product and guanine nucleotide standards, as indicated. (C) MALDI analysis of the HPLC purified AvHaCE reaction product. The species with a molecular mass of 558 corresponds to pGp. (D) Time-course for pGpG production from GTP with wild-type AvHaCE. To correct for variations in injection volumes, pGpG and GTP peak areas were normalized to peak areas of a co-injected NAD standard.
3.2. Identification of residues important for phosphodiesterase activity
Enzymatic synthesis of cyclic-diGMP is a very attractive and cost effective means of producing large amounts of cyclic-diGMP. The price of commercially available cyclic-diGMP is approximately $150 per mg (e.g., from Sigma–Aldrich for orders of more than 250 mg) whereas GTP can be purchased for less than $1 per mg. We therefore sought to identify variants of AvHaCE that maintain efficient cyclic-diGMP synthesis, but lack phosphodiesterase activity.
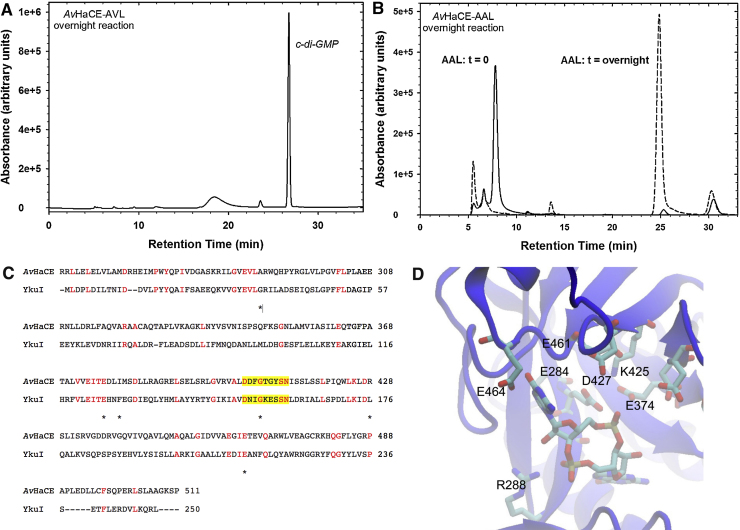
Based on homology with previously characterized cyclic-diGMP phosphodiesterases [37], [41], [42], we predicted that the ExL motif of AvHaCE (EVL in AvHaCE) would be essential for its phosphodiesterase activity. It has been shown that alteration of the conserved Glu within the requisite ExL motif of a homologous enzyme, residue 284 in AvHaCE, results in loss of phosphodiesterase activity [35], [37], [42]. Therefore, we generated the Glu284Ala variant, AVL, of AvHaCE. Prolonged incubation of the AVL variant with GTP resulted in the accumulation of cyclic-diGMP (Fig. 2A), but no pGpG was generated, confirming that this residue is essential for phosphodiesterase activity. Most phosphodiesterases contain an Ala in the “x” position of the ExL motif, therefore we also generated a variant in which both the Glu284 and Val285 were changed to Ala the AAL variant. Unsurprisingly, as we observed with the AVL variant, the AAL variant completely lacked phosphodiesterase activity (Fig. 2B).
Fig. 2.
Residues important for phosphodiesterase activity. (A) HPLC analysis of the reaction products of the AVL variant of AvHaCE after incubation overnight. (B) HPLC analysis of the reaction products of the AAL variant of AvHaCE before and after incubation overnight. The HPLC analysis of the nucleotide mixture immediately after adding AvHaCE is also illustrated, indicting the retention time of GTP. (C) Primary structure alignment of AvHaCE and the phosphodiesterase YkuI from B. subtilis (BSU14090). Conserved residues are highlighted in red. Residues shown to be important for phosphodiesterase activity of RocR from P. aeruginosa[42] are indicated by the asterisk. The yellow highlight indicates the position of loop D. (D) Homology model of residues 249-502, corresponding to the phosphodiesterase domain, of AvHaCE. The model was built with YkuI (pdb 2W27) as the template. Select amino acids proposed to be located within the phosphodiesterase active site of AvHaCE, as well as bound cyclic-diGMP, are rendered in stick format. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To identify other amino acids important for phosphodiesterase activity, the primary structure of the putative phosphodiesterase domain of AvHaCE (residues 249–502) was aligned with residues 1-250 of the phosphodiesterase YkuI from B. subtilis (Fig. 2C). The primary structures of these proteins are 20% identical, and many of the residues shown to be important for activity in other phosphodiesterases are conserved [37], [41], [42]. We then generated a homology model of AvHaCE using YkuI from B. subtilis as the template (pdb 2W27) (Fig. 2D). Our model suggests that a number of the conserved residues are located within the modeled phosphodiesterase active site of AvHaCE. We therefore generated several AvHaCE variants, each with a mutated putative active site residue, to determine the essential residues for phosphodiesterase activity in this bi-functional protein from A. vitis.
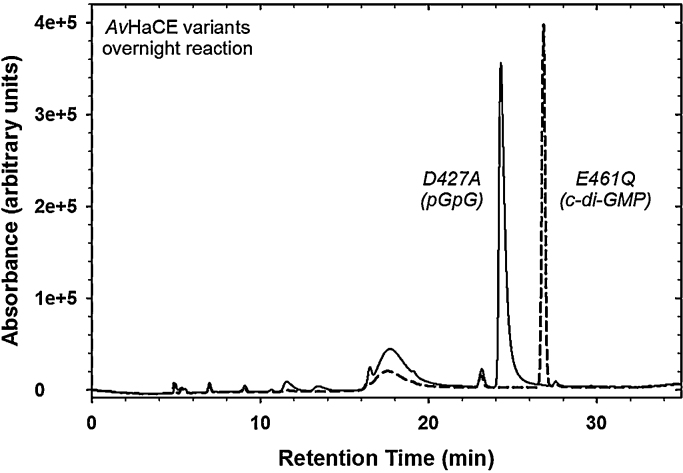
The variants we made, as well as the effect of each mutation on the phosphodiesterase activity of AvHaCE, are summarized in Table 1. Each variant was incubated with GTP overnight and then assessed for product formation by HPLC. In this qualitative assay, accumulation of cyclic-diGMP indicates phosphodiesterase inactivity, while accumulation of pGpG indicates intact phosphodiesterase activity. We found that, as expected, Glu461, which corresponds to the putative active site base [37], [42], [56], is essential for phosphodiesterase activity; alteration of this residue to either Gln or Asp resulted in the accumulation of cyclic-diGMP (Fig. 3). Furthermore, we found that Glu374 and Lys425, which are also located within the modeled active site, also play an essential role in the phosphodiesterase reaction. Mutation of either Arg288, Asp427, or Glu464 to Ala however, was not found to affect phosphodiesterase activity, therefore none of these residues are essential for activity in AvHaCE. At present, the mechanism of PDE phosphodiesterases is not well understood, but presumably the requirement for each of these residues is related to a role in the phosphodiesterase mechanism (e.g., catalysis, substrate binding, structural integrity). Interestingly, a truncated variant of AvHaCE (AvHaCE 1-248) that lacks the phosphodiesterase domain still functions as a diguanylate cyclase (Table 1). This indicates that the presence of the phosphodiesterase domain is not essential for diguanylate cyclase activity in the bi-functional enzyme.
Table 1.
Ability of select AvHaCE variants to produce cyclic-diGMP and/or pGpG.
| Variant | cyclic-diGMP accumulation | pGpG accumulation |
|---|---|---|
| E284A | + | – |
| E284A/V285A | + | – |
| R288A | – | + |
| E374A | + | – |
| K425A | + | – |
| D427A | – | + |
| E461Q | + | – |
| E464A | + | + |
| Cyclase onlya | + | – |
Contains residues 1-248 of AvHaCE.
Fig. 3.
HPLC analyses of reaction products of AvHaCE variant E461Q and D427A, as indicated. Reactions were overnight at room temperature as described in the text.
3.3. The AvHaCE AVL variant efficiently produces cyclic-diGMP
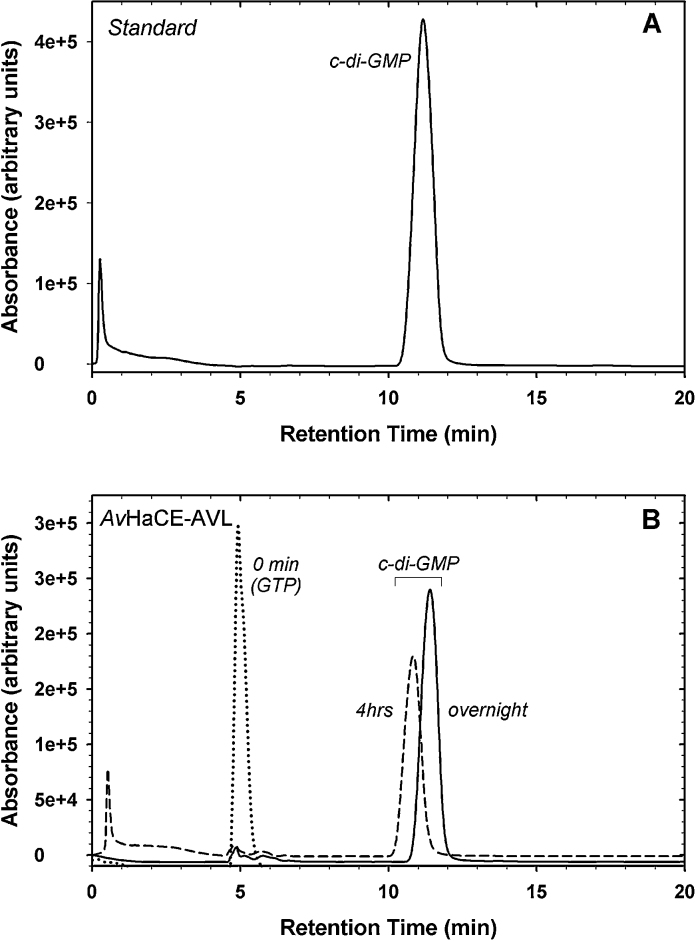
To determine if these variants of AvHaCE lacking phosphodiesterase activity could be used to obtain large amounts of cyclic-diGMP, we increased by 20-fold the scale of the enzyme reaction with the AVL AvHaCE variant. Under our conditions we observed 76% conversion of GTP to cyclic-diGMP within four hours (yielding an average conversion rate of ∼7.5 nmol min−1; Fig. 4B) and 96% conversion after an overnight incubation of AVL with GTP. This reaction produced 2.4 μmol of cyclic-diGMP from 5 μmol of GTP with 9.9 nmol of enzyme.
Fig. 4.
Enzymatic synthesis of cyclic-diGMP with the AvHaCE AVL variant. (A) HPLC analysis of cyclic-diGMP standard. (B) HPLC analysis of reaction products at 0 min (dotted line), after 4 h incubation (dashed line), and overnight incubation (solid line) at room temperature. The species observed at 0 min is GTP.
Therefore, AvHaCE variants can efficiently produce cyclic-diGMP, making this enzymatic synthesis a very attractive and cost effective means of producing large amounts of cyclic-diGMP. Although the enzymatic synthesis of cyclic-diGMP has been reported by several groups [9], [29], [40], [51], [58], our system offers clear advantages over other reported diguanylate cyclases. With these other enzymatic syntheses, it has not been possible to produce large amounts of cyclic-diGMP because most diguanylate cyclases are regulated by product inhibition. For example, Spehr et al. reported the large-scale enzymatic synthesis of cyclic-diGMP from ATP and GMP [51]. However, this system required the coupling enzymes guanosine monophosphate kinase (GMPK) and nucleoside diphosphate kinase (NDK) in addition to a modified diguanylate cyclase from Caulobacter crescentus, and the overall yield of the reaction was less than 50% [51]. Zähringer et al. successfully synthesized cyclic-diGMP using the diguanylate cyclase YdeH from Escherichia coli. However, the reaction required 30 mg of protein to produce 75 mg of cyclic-diGMP and cyclic-diGMP production was limited to 250 μM [58]. As mentioned in the introduction, alteration of the regulatory RxxD product inhibition site of the diguanylate cyclase from T. maritima showed increased cyclic-diGMP production in comparison with the wild-type enzyme. Despite this, however, product inhibition was still observed at concentrations of GTP greater than 0.8 mM [40]. Recently, this variant was engineered to contain the protein's hydrophobic domain and the resulting enzyme was purified from inclusion bodies [29]. This modified T. maritima protein displayed improved enzymatic activity, producing 500 μM of cyclic-diGMP with 2.5 μM of protein. Here, we report using 5 μM of AvHace to produce 1.2 mM of cyclic-diGMP.
3.4. Immobilized AvHaCE AAL variant retains enzymatic activity
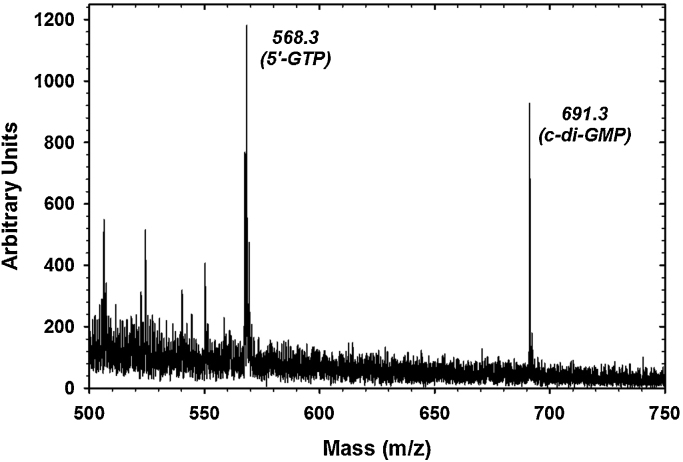
To explore an even more optimal method for the synthesis of large quantities of cyclic-diGMP with variants of AvHaCE, we immobilized the AAL variant of AvHaCE on a matrix, in this case Ni-NTA resin. A 30 min incubation of GTP and MgCl2 with the AAL-variant bound to Ni–NTA beads resulted in the production of cyclic-diGMP. Fig. 5 depicts the mass spectrum we obtained from analysis of the solutes in the flow-through from this column. The major peak with a mass of 691 corresponds to the molecular mass of cyclic-diGMP [M + H]+. The other major peak observed with a molecular mass of 568 corresponds to GTP [M + 2Na]+. It is likely that with optimization of reaction and flow conditions, near quantitative conversion of GTP to cyclic-diGMP could be achieved. With this proof-of-principle, this immobilized enzyme could be used as a reaction column through which GTP could continuously flow to produce correspondingly large quantities of cyclic-diGMP rapidly. To our knowledge, this is the first report of the synthesis of cyclic-diGMP by use of an immobilized enzyme.
Fig. 5.
MALDI analysis of cyclic-diGMP synthesized by immobilized AvHaCE AAL variant.
3.5. AAL variant of AvHaCE can produce radiolabeled cyclic-diGMP
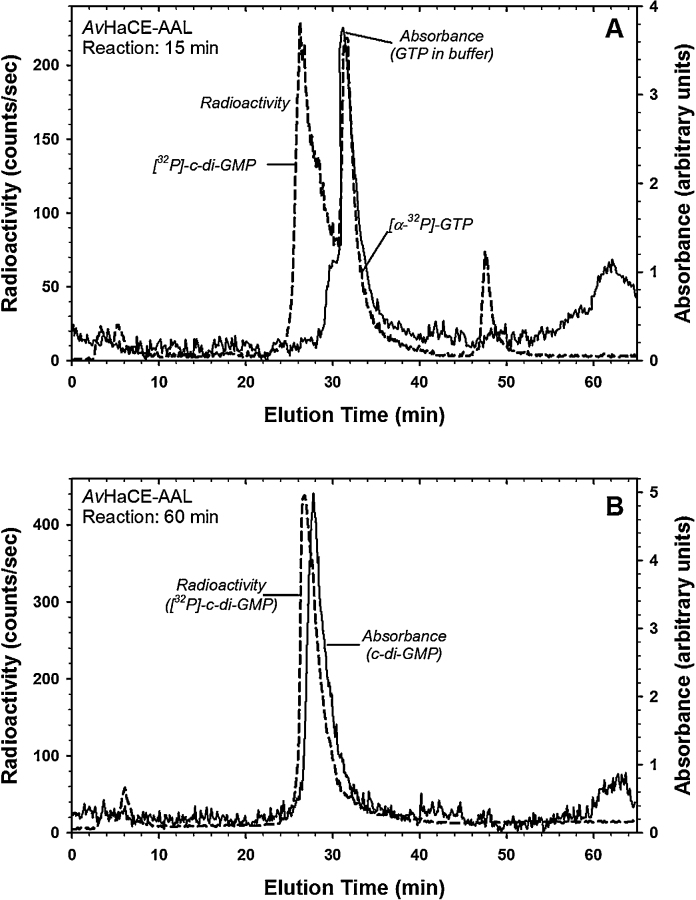
Finally, in order to demonstrate the versatility of our enzymatic cyclic-diGMP production system, the ability of AAL AvHaCE to produce [32P]-cyclic-diGMP from [α-32P]-GTP was explored. The enzymatic reaction was monitored as a radioactive trace from anion-exchange chromatography, comparing the peaks to a UV trace of nucleotide standards (Fig. 6). Production of [32P]-cyclic-diGMP is apparent after a 15 min incubation at room temperature (Fig. 6A). After 60 min of incubation, essentially all of the radioactivity co-elutes with cyclic-diGMP, suggesting 100% conversion from GTP to cyclic-diGMP under our assay conditions (Fig. 6B). Due to the high turnover efficiency of this enzyme, the product obtained is essentially pure. Tritium- or 14C-labeled GTP would be similarly converted to the correspondingly labeled cyclic-di-GMP. Radiolabeled cyclic-diGMP has previously found application in the study of cyclic-diGMP binding/processing proteins [4], [2], [8], [9], [30], [54]. With the simple methods described here, radioactively labeled cyclic-diGMP can be easily prepared, thereby facilitating its use as a probe in studies of cyclic-diGMP signaling in biology.
Fig. 6.
HPLC analysis of radiolabeled cyclic-diGMP synthesis by the AvHaCE AAL variant. Both radioactivity and UV–vis absorbance were monitored, as indicated. A slight shift in the detection of radioactivity and absorbance occurs because of the delay that occurs as eluted material flows first through one detector then the other. (A) Reaction products after 15 min incubation at room temperature. (B) Reaction products after 60 min incubation at room temperature.
4. Conclusions
Generation of a homology model of the PDE domain of AvHaCE allowed us to visualize a putative active site of the enzyme. Guided by this homology model, we used site-directed mutagenesis to generate variants of the enzyme that lack phosphodiesterase activity, therefore resulting in the accumulation of cyclic-diGMP. We showed further that our AVL and AAL variants are capable of producing cyclic-diGMP from GTP, quantitatively, in large amounts, and with only a small amount of enzyme. We anticipate this would be observed for all of our variants that lack phosphodiesterase activity. In addition to these features, our enzymes are extremely attractive for large-scale cyclic-diGMP synthesis because AvHaCE lacks the RxxD motif for product inhibition, which limits the amount of cyclic-diGMP that can be produced by other diguanylate cyclases. We observed no product inhibition under our experimental conditions. Furthermore, our variants are active while immobilized to a solid resin and retain enzymatic activity after several months of storage, making them ideal for long-term storage and repeated cyclic-diGMP syntheses. Lastly, we show that our variants can be used to synthesize radiolabeled cyclic-diGMP from radiolabeled GTP, increasing the versatility and applicability of the cyclic-diGMP production system we describe here.
Author contribution
NMN, DPA, RAJ and EMB designed the research. NMN and DPA carried out the experiments. NMN, RAJ and EMB wrote the paper.
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank Dr. Vadim Patsalo for help with generating the homology model. This work was supported by the Office of Naval Research, grant N00014-10-1-0099 to E.M.B.
Footnotes
Available online 5 May 2015
Contributor Information
Natasha M. Nesbitt, Email: natasha.nesbitt@stonybrook.edu.
Dhruv P. Arora, Email: dhruvparora@gmail.com.
Roger A. Johnson, Email: roger.johnson@stonybrook.edu.
Elizabeth M. Boon, Email: elizabeth.boon@stonybrook.edu.
References
- 1.Ausmees N., Mayer R., Weinhouse H., Volman G., Amikam D., Benziman M., Lindberg M. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 2001;204:163–167. doi: 10.1111/j.1574-6968.2001.tb10880.x. [DOI] [PubMed] [Google Scholar]
- 2.Bordeleau E., Fortier L.C., Malouin F., Burrus V. c-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 2011;7:e1002039. doi: 10.1371/journal.pgen.1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Burdette D.L., Monroe K.M., Sotelo-Troha K., Iwig J.S., Eckert B., Hyodo M., Hayakawa Y., Vance R.E. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan C., Paul R., Samoray D., Amiot N.C., Giese B., Jenal U., Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W., Kuolee R., Yan H. The potential of 3′,5′-cyclic diguanylic acid (c-di-GMP) as an effective vaccine adjuvant. Vaccine. 2010;28:3080–3085. doi: 10.1016/j.vaccine.2010.02.081. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z.H., Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488:680–683. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christen B., Christen M., Paul R., Schmid F., Folcher M., Jenoe P., Meuwly M., Jenal U. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 2006;281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 9.Christen M., Christen B., Folcher M., Schauerte A., Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 10.Cotter P.A., Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 11.D'Argenio D.A., Miller S.I. Cyclic di-GMP as a bacterial second messenger. Microbiology. 2004;150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- 12.Dow J.M., Fouhy Y., Lucey J.F., Ryan R.P. The HD-GYP domain, cyclic di-GMP signaling, and bacterial virulence to plants. Mol. Plant Microbe Interact. 2006;19:1378–1384. doi: 10.1094/MPMI-19-1378. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira R.B., Antunes L.C., Greenberg E.P., McCarter L.L. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 2008;190:851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaffney B.L., Veliath E., Zhao J., Jones R.A. One-flask syntheses of c-di-GMP and the [Rp, Rp] and [Rp, Sp] thiophosphate analogs. Org. Lett. 2010;12:3269–3271. doi: 10.1021/ol101236b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galperin M.Y. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 2004;6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galperin M.Y. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galperin M.Y., Koonin E.V. Functional genomics and enzyme evolution. Homologous and analogous enzymes encoded in microbial genomes. Genetica. 1999;106:159–170. doi: 10.1023/a:1003705601428. [DOI] [PubMed] [Google Scholar]
- 18.Galperin M.Y., Nikolskaya A.N., Koonin E.V. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 19.Gupta K., Kumar P., Chatterji D. Identification, activity and disulfide connectivity of C-di-GMP regulating proteins in Mycobacterium tuberculosis. PloS One. 2010;5:e15072. doi: 10.1371/journal.pone.0015072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayakawa Y., Nagata R., Hirata A., Hyodo M., Kawai R. A facile synthesis of cyclic bis(3′,5′) diguanylic acid. Tetrahedron. 2003;59:6465–6471. [Google Scholar]
- 21.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 22.Hu D.L., Narita K., Hyodo M., Hayakawa Y., Nakane A., Karaolis D.K. c-di-GMP as a vaccine adjuvant enhances protection against systemic methicillin-resistant Staphylococcus aureus (MRSA) infection. Vaccine. 2009;27:4867–4873. doi: 10.1016/j.vaccine.2009.04.053. [DOI] [PubMed] [Google Scholar]
- 23.Hunter J.L., Severin G.B., Koestler B.J., Waters C.M. The Vibrio cholerae diguanylate cyclase VCA0965 has an AGDEF active site and synthesizes cyclic di-GMP. BMC Microbiol. 2014;14:22. doi: 10.1186/1471-2180-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hyodo M., Hayakawa Y. An improved method for synthesizing cyclic bis(3′-5′) diguanylic acid (c-di-GMP) Bull. Chem. Soc. Jpn. 2004;77:2089–2093. [Google Scholar]
- 25.Jenal U. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 2004;7:185–191. doi: 10.1016/j.mib.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Jenal U., Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Ann. Rev. Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 27.Kawai R., Nagata R., Hirata A., Hayakawa Y. A new synthetic approach to cyclic bis(3′ → 5′) diguanylic acid. Nucleic Acids Res. Suppl. 2003:103–104. doi: 10.1093/nass/3.1.103. [DOI] [PubMed] [Google Scholar]
- 28.Kiburu I., Shurer A., Yan L., Sintim H.O. A simple solid-phase synthesis of the ubiquitous bacterial signaling molecule, c-di-GMP and analogues. Mol. Biosyst. 2008;4:518–520. doi: 10.1039/b719423d. [DOI] [PubMed] [Google Scholar]
- 29.Korovashkina A.S., Rymko A.N., Kvach S.V., Zinchenko A. Enzymatic synthesis of c-di-GMP using inclusion bodies of Thermotoga maritima full-length diguanylate cyclase. J. Biotechnol. 2012;164:276–280. doi: 10.1016/j.jbiotec.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Kuchma S.L., Brothers K.M., Merritt J.H., Liberati N.T., Ausubel F.M., O’Toole G.A. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 2007;189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar M., Chatterji D. Cyclic di-GMP: a second messenger required for long-term survival, but not for biofilm formation, in Mycobacterium smegmatis. Microbiology. 2008;154:2942–2955. doi: 10.1099/mic.0.2008/017806-0. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Levet-Paulo M., Lazzaroni J.C., Gilbert C., Atlan D., Doublet P., Vianney A. The atypical two-component sensor kinase Lpl0330 from Legionella pneumophila controls the bifunctional diguanylate cyclase-phosphodiesterase Lpl0329 to modulate bis-(3′-5′)-cyclic dimeric GMP synthesis. J. Biol. Chem. 2011;286:31136–31144. doi: 10.1074/jbc.M111.231340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y., Heine S., Entian M., Sauer K., Frankenberg-Dinkel N. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J. Bacteriol. 2013;195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu N., Pak T., Boon E.M. Characterization of a diguanylate cyclase from Shewanella woodyi with cyclase and phosphodiesterase activities. Mol. Biosyst. 2010;6:1561–1564. doi: 10.1039/c002246b. [DOI] [PubMed] [Google Scholar]
- 36.Mills E., Pultz I.S., Kulasekara H.D., Miller S.I. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell Microbiol. 2011;13:1122–1129. doi: 10.1111/j.1462-5822.2011.01619.x. [DOI] [PubMed] [Google Scholar]
- 37.Minasov G., Padavattan S., Shuvalova L., Brunzelle J.S., Miller D.J., Basle A., Massa C., Collart F.R., Schirmer T., Anderson W.F. Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J. Biol. Chem. 2009;284:13174–13184. doi: 10.1074/jbc.M808221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyabe H., Hyodo M., Nakamura T., Sato Y., Hayakawa Y., Harashima H. A new adjuvant delivery system ‘cyclic di-GMP/YSK05 liposome’ for cancer immunotherapy. J. Control. Release. 2014;184:20–27. doi: 10.1016/j.jconrel.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Oliveira M.C., Teixeira R.D., Andrade M.O., Pinheiro G.M., Ramos C.H., Farah C.S. Cooperative substrate binding by a diguanylate cyclase. J. Mol. Biol. 2014;427:415–432. doi: 10.1016/j.jmb.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 40.Rao F., Pasunooti S., Ng Y., Zhuo W., Lim L., Liu A.W., Liang Z.X. Enzymatic synthesis of c-di-GMP using a thermophilic diguanylate cyclase. Anal. Biochem. 2009;389:138–142. doi: 10.1016/j.ab.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 41.Rao F., Qi Y., Chong H.S., Kotaka M., Li B., Li J., Lescar J., Tang K., Liang Z.X. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J. Bacteriol. 2009;191:4722–4731. doi: 10.1128/JB.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao F., Yang Y., Qi Y., Liang Z.X. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 2008;190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romling U. Cyclic di-GMP, an established secondary messenger still speeding up. Environ. Microbiol. 2011;14:1817–1829. doi: 10.1111/j.1462-2920.2011.02617.x. [DOI] [PubMed] [Google Scholar]
- 44.Ross, Mayer P., Weinhouse R., Amikam H., Huggirat D., Benziman Y., de Vroom M., Fidder E., de Paus A., P, Sliedregt L.A. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 1990;265:18933–18943. [PubMed] [Google Scholar]
- 45.Ross P., Weinhouse H., Aloni Y., Michaeli D., Weinberger-Ohana P., Mayer R., Braun S., de Vroom E., van der Marel G.A., van Boom J.H., Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 46.Ryan R.P., Fouhy Y., Lucey J.F., Crossman L.C., Spiro S., He Y.W., Zhang L.H., Heeb S., Camara M., Williams P., Dow J.M. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Ryjenkov D.A., Tarutina M., Moskvin O.V., Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt A.J., Ryjenkov D.A., Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma I.M., Prakash S., Dhanaraman T., Chatterji D. Characterization of a dual-active enzyme, DcpA, involved in cyclic diguanosine monophosphate turnover in Mycobacterium smegmatis. Microbiology. 2014;160:2304–2318. doi: 10.1099/mic.0.080200-0. [DOI] [PubMed] [Google Scholar]
- 50.Sintim H.O., Smith J.A., Wang J., Nakayama S., Yan L. Paradigm shift in discovering next-generation anti-infective agents: targeting quorum sensing, c-di-GMP signaling and biofilm formation in bacteria with small molecules. Future Med. Chem. 2010;2:1005–1035. doi: 10.4155/fmc.10.185. [DOI] [PubMed] [Google Scholar]
- 51.Spehr V., Warrass R., Hocherl K., Ilg T. Large-scale production of the immunomodulator c-di-GMP from GMP and ATP by an enzymatic cascade. Appl. Biochem. Biotechnol. 2011;165:761–775. doi: 10.1007/s12010-011-9294-z. [DOI] [PubMed] [Google Scholar]
- 52.Tal R., Wong H.C., Calhoon R., Gelfand D., Fear A.L., Volman G., Mayer R., Ross P., Amikam D., Weinhouse H., Cohen A., Sapir S., Ohana P., Benziman M. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 1998;180:4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamayo R., Pratt J.T., Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Ann. Rev. Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamayo R., Schild S., Pratt J.T., Camilli A. Role of cyclic Di-GMP during el tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic Di-GMP phosphodiesterase CdpA. Infect. Immun. 2008;76:1617–1627. doi: 10.1128/IAI.01337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarutina M., Ryjenkov D.A., Gomelsky M. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 2006;281:34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 56.Tchigvintsev A., Xu X., Singer A., Chang C., Brown G., Proudfoot M., Cui H., Flick R., Anderson W.F., Joachimiak A., Galperin M.Y., Savchenko A., Yakunin A.F. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 2010;402:524–538. doi: 10.1016/j.jmb.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wassmann P., Chan C., Paul R., Beck A., Heerklotz H., Jenal U., Schirmer T. Structure of BeF3- -modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure. 2007;15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 58.Zähringer F., Massa C., Schirmer T. Efficient enzymatic production of the bacterial second messenger c-di-GMP by the diguanylate cyclase YdeH from E. coli. Appl. Biochem. Biotechnol. 2011;163:71–79. doi: 10.1007/s12010-010-9017-x. [DOI] [PubMed] [Google Scholar]