Highlights
-
•
Human Kv2.1 was expressed in the inner membrane of E. coli using prokaryotic codon.
-
•
Bacterial spheroplasts large enough for the automated patch clamp were prepared by microfluidic chips.
-
•
Kv2.1 current was recorded from the giant spheroplast by the automated patch clamp.
-
•
E. coli spheroplasts were used for dose–response assay of potassium channel inhibitors.
-
•
Our system will become the simple and sensitive drug assay method anyone can use.
Keywords: Ion channel, Human Kv2.1, Escherichia coli, Giant spheroplast, Automated patch clamp, TEA (tetraethylammonium), 4-AP (4-aminopyridine), Drug screening
Abstract
Kv2.1, the voltage-gated ion channel, is ubiquitously expressed in variety of tissues and dysfunction of this ion channel is responsible for multiple diseases. Electrophysiological properties of ion channels are so far characterized with eukaryotic cells using the manual patch clamp which requires skilful operators and expensive equipments. In this research, we created a simple and sensitive drug screen method using bacterial giant spheroplasts and the automated patch clamp which does not require special skills. We expressed a eukaryotic voltage-gated ion channel Kv2.1 in Escherichia coli using prokaryotic codon, and prepared giant spheroplasts large enough for the patch clamp. Human Kv2.1 currents were successfully recorded from giant spheroplasts with the automated system, and Kv2.1-expressed E. coli spheroplasts could steadily reacted to the dose–response assay with TEA and 4-AP. Collectively, our results indicate for the first time that the bacterial giant spheroplast can be applied for practical pharmaceutical assay using the automated patch clamp.
1. Introduction
Ion channels play essential roles in wide range of biological phenomena including neural transduction and muscle contraction [9]. Voltage-gated K+ channels are ubiquitously expressed in various organs including brain, heart, kidney, skeletal muscle, retina and pancreas, and Kv2.1 is responsible for regulating neurotransmitter release, heart rate, muscle contraction, insulin secretion, and cell volume [21], [25], [17], [11], which means many diseases are related to the misconduct of this ion channel, making Kv2.1 valuable pharmaceutical target.
Patch clamp, developed by [18], is the most sensitive technique for ion channel analysis and has been the gold standard to characterize electrophysiological properties of drug candidates [3], [10], [24]. Patch clamp, so far, refers to the manual patch clamp whose samples are typically tissue slices, Xenopus oocytes, yeasts, cell lines and the lipid bilayer which are relatively easy to pick manually under visual aids. Conventional manual patch clamp requires skilful operators and expensive equipments and that is the bottleneck of the high throughput screening of pharmaceutical compounds [3]. In order to resolve this situation, several automated patch clamp systems have been developed in the past decade [24]. Automated patch clamp systems, such as Port-a-Patch from Nanion has made electrophysiological observation accessible for anyone thanks to the automated system and cost effectiveness [10], [24]. With this novel system, even if the operator was a patch clamp beginner, high quality data can be obtained without fuss.
Escherichia coli, a Gram-negative bacterium, is the most extensively characterized organism genetically and biochemically. Many gene manipulation methods are already established to handle E. coli and many mutants are available. Despite differences in physiological background and the post-translational modification mechanism, multiple sequence alignment of eukaryotic and prokaryotic voltage-gated channels indicates charged residues within transmembrane helices are strikingly conserved [19], and prokaryotic ion channels have almost the same physical properties with eukaryotic channels and are comparable to eukaryotic channels [16], [20]. The bacterial membrane, therefore, can serve as a valid model for studying channel structure [2]. Until now, E. coli has been used only to produce foreign recombinant proteins, yet the E. coli expression system can be also comparable to eukaryotic expression systems like Xenopus oocytes for studying heterologous ion channels. The use of E. coli has several technical advantages over existing eukaryotic expression systems. In case of expressing prokaryotic channels, their functions can be better reproduced in E. coli than using eukaryotic system. In addition, since genetic code is mutual, the channel expression and its electrophysiological characterization can be seamlessly accomplished by only one construct; we do not need to prepare mRNA in eukaryotic code for Xenopus oocyte injection nor purify membrane proteins for reconstitution in the lipid bilayer for patch clamp, saving tremendous time and effort [13]. Furthermore, E. coli can be used not only to express prokaryotic proteins but also to express eukaryotic proteins. Although E. coli does not possess the glycosylation machinery and the expression of eukaryotic membrane proteins often turns out to be toxic to the bacterium, E. coli can still be a feasible alternative to eukaryotic system. Precise analysis of the target membrane protein is possible if knockout strains were prepared to silence genes which interfere with the exogenous target protein. Besides, if membrane proteins were expressed in eukaryotic cells, they can be sorted into organelles, not into the outermost plasma membrane; however, if E. coli was selected for the expression system, membrane proteins are assured to be expressed in the inner membrane, which is convenient for the patch clamp [23]. Consequently, E. coli can be a realistic alternative to eukaryotic systems for studying ion channels. The major obstacle which makes the use of E. coli difficult for patch clamp is that bacterial cells are usually too small for patch clamp electrodes, and not so many cases have been reported at this time. Ion channels of E. coli have been studied with the patch clamp since its development by Matrinac et al. [14], who made gigantic bacteria called “snakes” using antiseptics and produced giant spheroplasts large enough for the patch clamp by enzymatic digestion of the elongated bacteria.
The combination of giant bacterial spheroplasts and the easy-to-operate automated patch clamp system will prove extremely powerful for primary drug candidate screening through characterization of biophysical properties of membrane proteins. The purpose of this project was to develop a simple and sensitive pharmaceutical assay method using the bacterial giant spheroplast and the automated patch clamp system. This paper describes the process in which human Kv2.1 was expressed in E. coli cells which are then digested to produce giant spheroplasts suitable for the automated patch clamp; Kv2.1 currents were measured with the automated patch clamps system and time-course recordings of the dose–response assay were demonstrated by the administration and wash-out of TEA (tetraethylammonium) and 4-AP (4-aminopyridine).
2. Materials and methods
2.1. The E. coli strain and chemicals
Overnight Express C43 (DE3) SOLOs chemically competent cells (Lucigen, Wisconsin, USA) were used for spheroplast preparation. LB broth (Sigma, Missouri, USA) and Terrific Broth (Fluka, Missouri, USA) were prepared by manufacturers’ instruction. Ready-Lyse Lysozyme (Epicenter, Chicago, USA) and OmniCleave Endonuclease (Epicentre) were diluted to 1/5 with attached substrate diluents for easier handling. d(+)-Glucose, sucrose, 1 M Tris–HCl, pH 8.0, ampicillin, cephalexine, IPTG and TEA were purchased from Wako (Osaka, Japan). Ampicillin, cephalexine and IPTG were prepared in 50 mg/ml, 10 mg/ml and 1 M, respectively. 4-AP and NFA (niflumic acid) were purchased from Sigma (Missouri, USA).
2.2. Expression vector and transformation
The artificial gene of human Kv2.1 was synthesized using prokaryotic codon (Operon Biotechnology, Tokyo, Japan) and inserted into pET-23a vector (Novagen, Massachusetts, USA) at BamHI site with In-Fusion HD Cloning Kit (Clontech, California, USA) according to the manufacturer’s instruction. Then, Stellar competent cells (Clontech) were transformed and seeded on LB/Amp plate and the insert was confirmed with colony check PCR. The colony with right insert was cultured in 100 ml LB/Amp for plasmid extraction by Plasmid Midi Kit (QIAGEN, Hilden, Germany) using low-copy plasmid midiprep protocol. Overnight Express C43 (DE3) SOLOs chemically competent cells were then transformed with the plasmid and plated on LB/Amp plates.
2.3. Western blotting
Single colony of Kv2.1-expressed C43 was inoculated with 100 ml Overnight Express Instant TB Medium (Novagen) and incubated at 37 °C for 4 h and 30 °C for overnight with shaking of 200 rpm. C43 with plain pET vector was used as a control. The culture was spun at 3000 × g for 5 min to collect the pellet. Membrane protein fractions were extracted from the C43 pellet with ReadyPrep Protein Extraction Kit (Bio-Rad Laboratories, California, USA) according to the manufacturer’s instruction. Denatured samples were separated by SDS-PAGE using SuperSep Ace 7.5% gel (Wako) at 200 V for 1 h. Proteins were transferred onto 0.2 μm PVDF membrane (Bio-Rad Laboratories) with Trans-Blot Turbo Blotting System (Bio-Rad Laboratories), and 0.5% Block Ace (DS Pharma Biomedical, Osaka, Japan) was used for blocking overnight. The blotted membrane was exposed to primary (His-probe (G-18) Rabbit polyclonal IgG, Santa Cruz Biotechnology, Texas, USA) and secondary (Anti-Rabbit IgG heavy and light chain HRP conjugated, Bethyl Laboratories, Texas, USA) antibody using SNAP i.d. (Merck Millipore). The membrane was incubated with ImmunoStar LD (Wako) at room temperature for 1 min and bands were detected by C-DiGit Blot Scanner (LI-COR Biosciences, Nebraska, USA).
2.4. Spheroplast preparation
E. coli spheroplasts were prepared by enzymatic digestion described in these literatures [1], [5], [8], [14]. In short, single colony of E. coli was inoculated in 2 ml LB medium and cultured at 37 °C with the shaking of 220 rpm overnight. 200 μl of this pre-culture was diluted into 20 ml TB medium with 20 μl of 50 mg/ml ampicillin and cultured for 70 min. When the O.D.600 reached 0.12, cephalexin, a septation inhibitor, was added to the final concentration of 60 μg/ml, and the elongation of bacterial filaments, referred as “snakes” were continued for 2.5 h. At the end of the culture, 1 mM IPTG was added and incubated at 37 °C with shaking for 20 min for Kv2.1 induction.
2 ml culture was then dispensed into 2 ml micro tubes and “snakes” were harvested by centrifugation of 1000 × g for 3 min. The pellet was resuspended in 500 μl of 1 M glucose by gentle tapping. The bacterial cell wall was digested by successively adding 30 μl 1 M Tris–HCl, pH 8.0, 2 μl (5760 U) diluted lysozyme, 2 μl (80 U) diluted endonuclease, and 6 μl 125 mM EDTA, pH 8.0. The tube was mixed every time the reagents were added by inverting for several times, and incubated at room temperature for 10 min. 100 μl of stop solution (10 mM Tris–HCl, pH 8.0, 0.7 M sucrose, 20 mM MgCl2) was added to end digestion and spheroplast suspension was stored at −80 °C and used within 2 weeks from the preparation.
On the day of electrophysiological recording, a tube of frozen spheroplasts was thawed on ice and filtered with 40 μm and 20 μm nylon mesh (Kyosin Riko, Tokyo, Japan) to remove large debris. Filtered spheroplast suspension was then diluted to 10 times by adding 1 M glucose with 100 mM Tris–HCl, pH 8.0. Diluted sample was loaded in a 15 ml syringe and sorted by particle size using a spiral sorter chip (microfluidic ChipShop, Jena, Germany) and a syringe pump (YMC Keyboard Chemistry, Kyoto, Japan) at a flow rate of 0.5 ml/min. Spheroplasts sorted at outlet #4 of the microfluidic chip were spun down at 700 × g for 3 min and resuspended in 50 μl 1 M glucose with 100 mM Tris–HCl, pH 8.0.
2.5. Electrophysiology
Whole-cell patch clamp recording of the E. coli spheroplast was performed by the Port-a-Patch® system (Nanion, München, Germany) using NPC-1 borosilicate glass chips with a resistance of 10–15 MΩ. All experiments were conducted at room temperature (22–27 °C). The Nanion’s standard internal solution was composed of 50 mM KCl, 10 mM NaCl, 60 mM KF, 20 mM EGTA, 10 mM Hepes, KOH, pH 7.2. For the external solution, 400 mM sucrose was supplemented to the Nanion’s standard external solution which contained 140 mM NaCl, 4 mM KCl, 1 mM MgCl2, 2 mM (experimental) or 5 mM (sealing) CaCl2, 5 mM glucose, 10 mM Hepes, NaOH, pH 7.4. 10 μM NFA was supplemented to experimental solutions to block chloride current contamination. For observation of Kv2.1 inhibition, 100 mM and 300 mM TEA and 1 mM and 10 mM 4-AP were added to the experimental solution.
First, the external solution with 5 mM Ca2+ was used for the patch formation on the glass chip aperture. 5 μl of the spheroplast suspension was applied to the well. Once the spheroplast achieved the whole-cell recording configuration, 5 mM Ca2+ solution was replaced with 2 mM Ca2+ external solution using the external perfusion module, leaving for at least 2 min until the membrane became stable. The membrane usually got stable after 3–10 min after membrane break-in in 2 mM Ca2+ external solution. Patches were rejected if current levels were unstable during the recording, and if currents following replacement with 2 mM Ca2+ external solution did not recover to the baseline values, suggesting that rundown of the patch had occurred.
K+ channel currents were recorded with the EPC-10 USB Patch Clamp Amplifier (HEKA Electronik Dr. Schulze GmbH, Germany), and data acquisition and analysis were performed with PatchMaster (HEKA Electronik). A voltage step protocol with 20 mV increments was performed between −60 mV to +60 mV for 500 ms duration with 15 s intervals from a holding potential of −80 mV to observe Kv2.1 and endogenous K+ channel activity.
For observation of Kv2.1 current inhibition using TEA and 4-AP, control currents were first measured in 2 mM Ca2+ external solution for 4 min at 12 points when the membrane became stable. Then 2 mM Ca2+ external solution supplemented with TEA or 4-AP was applied into the well using the external perfusion system and the currents were measured for 4 min at 12 points. After one set of the recording was finished, the inhibitor was immediately washed out with 2 mM Ca2+ external solution. A time-course pharmacology protocol was used to monitor Kv2.1 currents by giving +40 mV pulses for 500 ms duration from a holding potential of −80 mV with 20 ms intervals. Current recording was initiated 3–10 min after the membrane break-in, and continued up to 60 min as long as the membrane was stable.
2.6. Data analysis
To analyze Kv2.1 current modulation with TEA and 4-AP, first and last points of each measuring section were omitted and mid 10 points were used for calculation. Currents were first normalized to the average of 10 wash-out current magnitudes prior to inhibitor administration. Extreme values were excluded with Smirnov–Grubbs test. Statistical significance was determined by Tukey–Kramer test. P values of <0.05 were considered significant, and results are indicated as means ± S.E.M.
3. Results
3.1. Spheroplast morphology
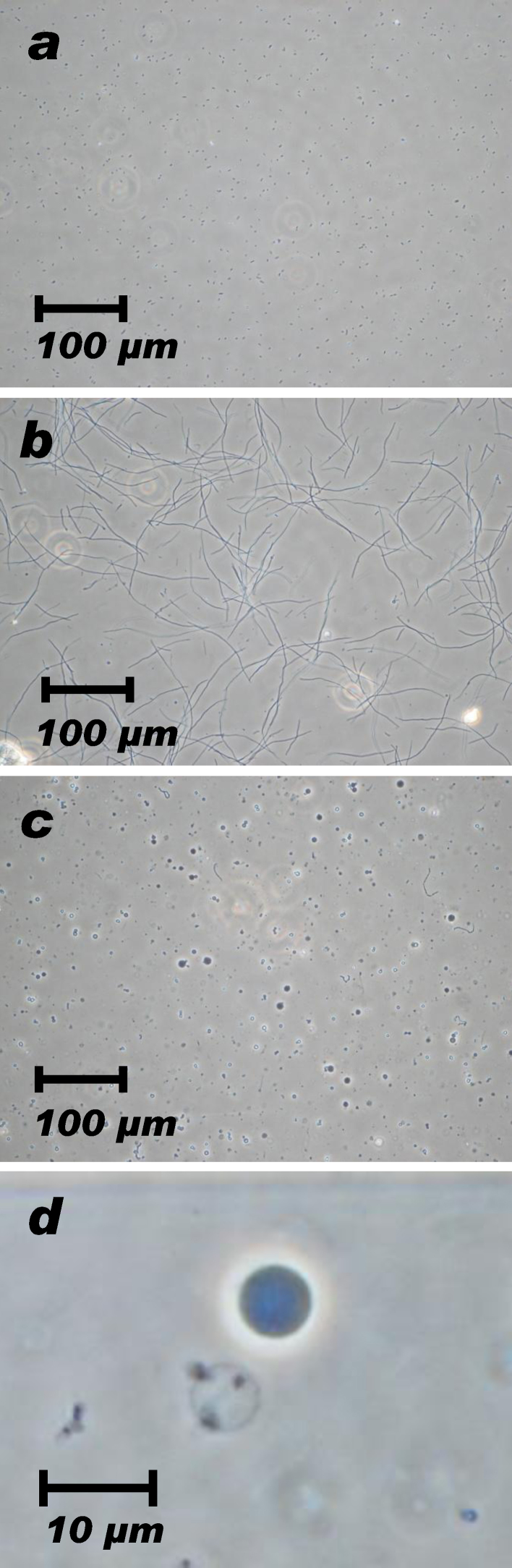
During the culture with 60 μg/ml cephalexin, bacterial “snakes” elongated to 100–150 μm (Fig. 1a–b) and their digestion with enzymes produced spheroplasts ranging Φ 1–7 μm (Fig. 1c). Enzymatic digestion of bacterial snakes produced 4 types of spheroplasts: large (Φ 4–7 μm) and gray, medium (Φ 2–4 μm) and gray, small (less than 2 μm) and shiny, and shady and transparent ones referred as “ghosts”.
Fig. 1.
E. coli giant spheroplast preparation.
(a) C43 in the original culture. (b) C43 cultured for 3 h in presence of 60 mg/ml cephalexin. Bacteria were elongated to become “snakes” ranging 100–150 μm. (c) Bacterial “snakes” were digested with 5760 U lysozyme and 80 U DNase to produce giant spheroplasts ranging 1–7 μm in diameter. (d) A giant spheroplast suitable for the patch clamp recording looks dark-colored (upper right); a shady and fragile “ghost” spheroplast was located on lower left of the dark-colored spheroplast.
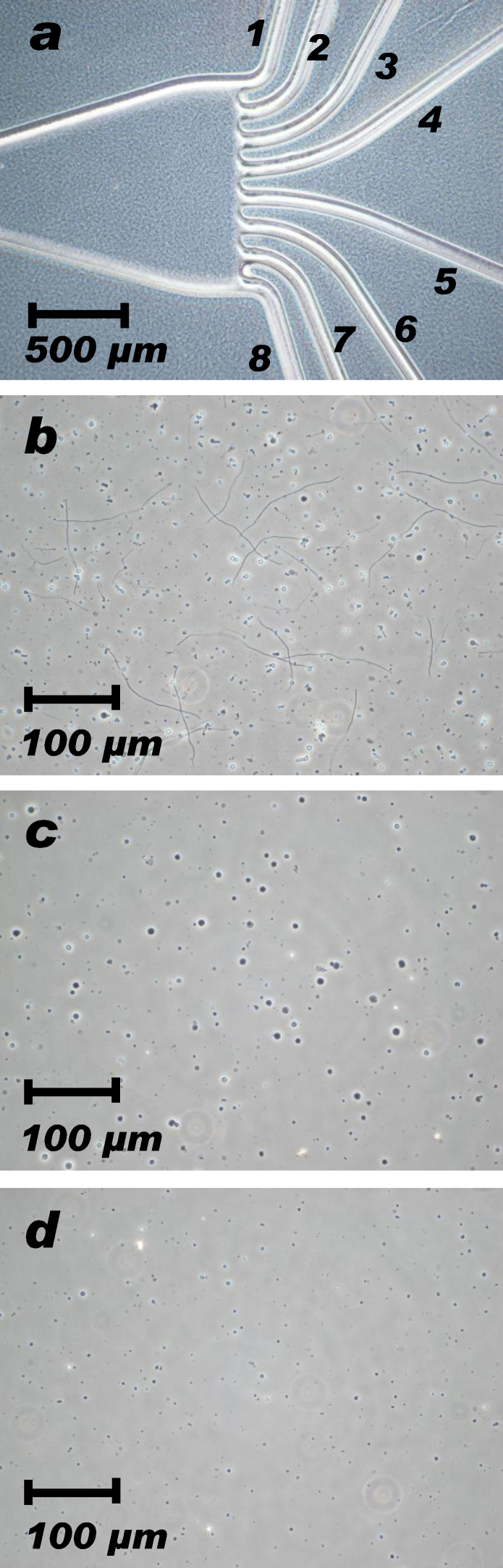
3.2. Spheroplast sorting by particle size
Large spheroplasts between Φ 4 and 7 μm were successfully separated by microfluidic chips (Fig. 4a) and the selected spheroplasts were condensed at 700 × g centrifugation (Fig. 4c). Majority of smaller spheroplasts and debris were sorted into other outlets of the chip (Fig. 4d); however, some population of small spheroplasts could not be completely excluded from the population of large spheroplasts. Besides, microfluidic chips did not discriminate the condition of the spheroplast membrane; therefore, ghost spheroplasts were also sorted into the selected fraction as long as the particle size was between Φ 4 and 7 μm. Although some small spheroplasts and fragile ghost spheroplasts could not be perfectly removed from the selected fraction, 90% of acquired patches reached giga-seal within 2 min from the sample application into the well, and 70% of the acquired patches were in sufficient conditions for electrophysiological recording.
Fig. 4.
Spheroplast separation by particle size.
Spheroplast suspension was sorted by particle size using spiral sorter chips. (a) Outlet ports of a spiral sorter chip. (b) Untreated spheroplast sample. Many undigested “snakes” and debris were present and large spheroplasts were sparsely distributed. (c) Large spheroplasts between Φ 4 and 7 μm were sorted to the outlet #4. (d) Middle-sized spheroplasts between Φ 2 and 4 μm were sorted to the outlet #3. Even smaller spheroplasts and debris were sorted to the outlet #1 and #2.
3.3. Kv2.1 current recording
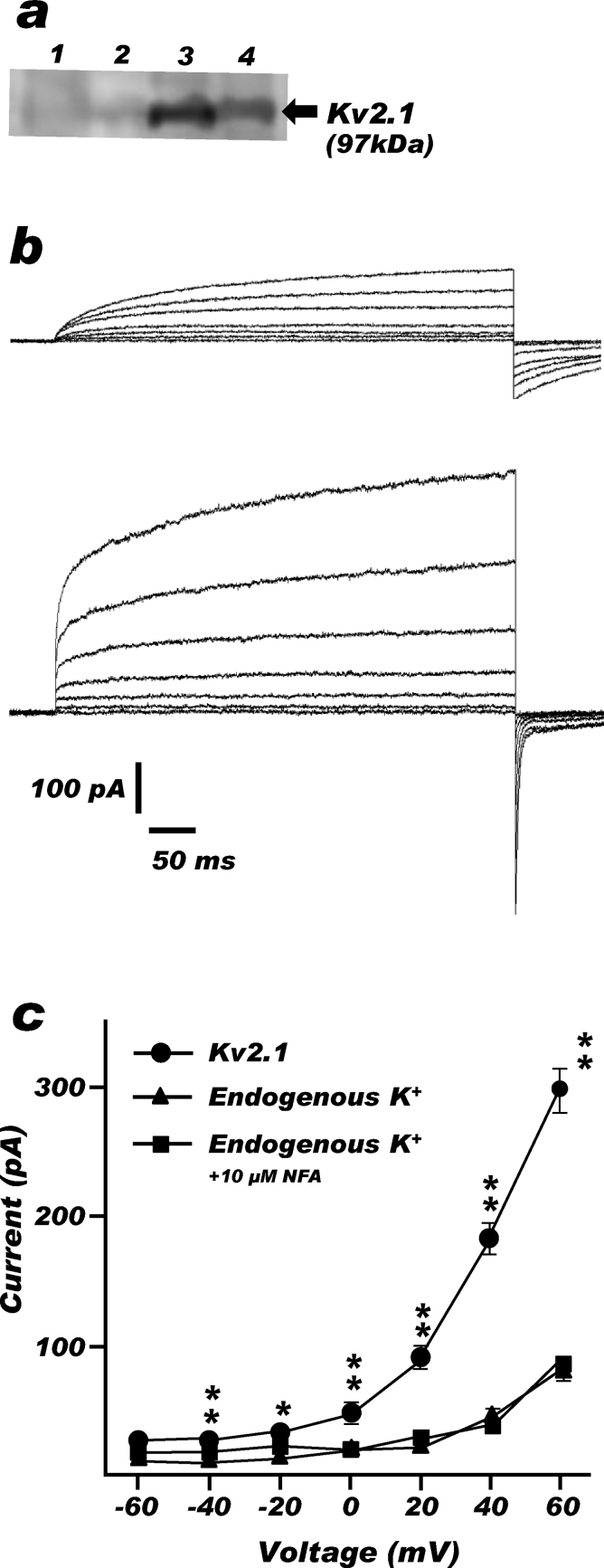
Human Kv2.1 was expressed in C43 and the expression was confirmed with the western blotting (Fig. 2a). Human Kv2.1 currents were successfully monitored with the Port-a-Patch automated patch clamp system and the representative traces of Kv2.1 and endogenous K+ channels evoked by the voltage-step protocol between −60 mV and +60 mV were shown in Fig. 2b. As summarized in Fig. 2c, the average current from C43 spheroplasts expressing human Kv2.1 was significantly higher than C43 spheroplasts transformed with control vector at voltage steps over −40 mV (P < 0.05). 10 μM NFA was added to one group of native K+ and Kv2.1 measurements to avoid chloride current contamination; still native K+ current magnitudes were not altered significantly at all voltage steps by the presence of NFA in the system (P < 0.05).
Fig. 2.
Human Kv2.1 expressed in C43.
(a) Kv2.1 bands, estimated to be 97 kDa, were detected in Kv2.1-expressed C43 (lane 3) and BL21 (lane 4) by western blotting; no bands were detected from C43 (lane 1) and BL21 (lane 2) with control vector. BL21 was used for expression check only. (b) Representative traces showing outward K+ currents triggered by a series of pulses (from −60 mV to +60 mV, 500 ms) from a holding potential of −80 mV in plain C43 spheroplasts (upper) and spheroplasts expressing human Kv2.1 channel (lower). (c) IV-relationship summary of (b). Results are indicated as mean ± S.E.M. of 18 measurement for native K+ channel with no NFA, 17 for native K+ channel with 10 μM NFA and 12 for Kv2.1 with 10 μM NFA, respectively. *, significantly higher (P < 0.05) than native K+ channels.
3.4. Kv2.1 current inhibition with TEA and 4-AP
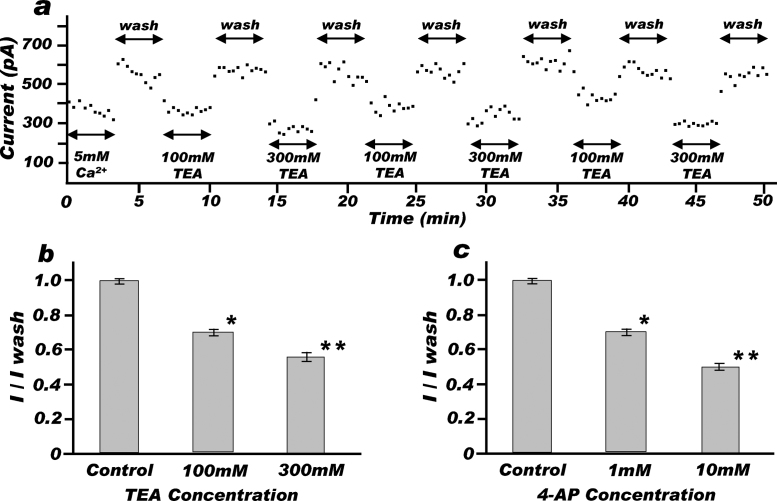
Human Kv2.1-expressed C43 giant spheroplasts steadily responded to the repeated administration of inhibitors and wash solution. Once a strong seal was achieved, the spheroplast was stable for 30 min for most of times, and even lasted for 60 min. Fig. 3a shows a representative time-course recording of Kv2.1 current inhibition with TEA administration and the current recovery with wash-out. The Kv2.1 current blocked by TEA always returned to the original control level as soon as the well was flushed with 2 mM Ca2+ external solution. In addition, Kv2.1-expressed giant spheroplasts reacted to TEA with dose-dependent manner. As shown in Fig. 3b, 100 mM and 300 mM TEA significantly reduced Kv2.1 current magnitudes (0.706 ± 0.009 and 0.569 ± 0.009, respectively; P < 0.05) to the control. Furthermore, Kv2.1 current inhibition was observed using another K+ channel inhibitor, 4-AP. Kv2.1 currents returned to the control level with wash-out and were suppressed dose-dependent manner, like observed in TEA. Fig. 3c shows that 1 mM and 10 mM 4-AP significantly reduced Kv2.1 current magnitudes (0.667 ± 0.023 and 0.481 ± 0.019, respectively; P < 0.05) to the control.
Fig. 3.
Kv2.1 inhibition with TEA and 4-AP.
(a) Representative time-course recording of Kv2.1 currents. This spheroplast had been stable for almost 1 h, enduring repeated drug administration and wash with 100 mM or 300 mM TEA and 2 mM Ca2+ solution. (b) The inhibition of Kv2.1 currents with TEA was observed by giving +40 mV pulses (500 ms) with 20 s intervals from a holding potential of −80 mV. Current recording was initiated 10 min after the membrane break-in, and lasted 30–60 min while the membrane kept stability. Current magnitudes were normalized to the wash current magnitude immediately before the TEA addition. Results are mean ± S.E.M. for 60 sweeps from 6 separate experiments. *, significantly lower (P < 0.05) than 2 mM Ca2+ control; **, significantly lower (P < 0.05) than 2 mM Ca2+ control and 100 mM TEA. (c) The inhibition of Kv2.1 currents with 4-AP was observed in the same way with TEA. Results are mean ± S.E.M. for 10 sweeps from 2 separate experiments. 10 μM NFA was added to avoid chloride channel current. *, significantly lower (P < 0.05) than 2 mM Ca2+ control; **, significantly lower (P < 0.05) than 2 mM Ca2+ control and 1 mM 4-AP.
4. Discussion
4.1. The density and the uniformity of the sample count for the automated patch clamp
In order to use bacterial spheroplasts for automated patch clamp system, the spheroplast suspension needed to be dense and uniform. In our preliminary experiment with spheroplast sample without size selection, only 20% of all acquired patches reached giga-seal, whose success coincided with the percentage of large and gray spheroplasts present in whole spheroplast suspension. According to the past reports, ideal whole-cell patches could be obtained with large and gray spheroplasts, and no stable results were obtained with “ghost” spheroplasts which were leaky and fragile, and with small and shiny spheroplasts which were simply hard to catch with the electrode [1], [8]. So, we isolated large spheroplast population ranging Φ 4–7 μm with microfluidic chips which were capable of sorting particles by inertial force and condensed the selected fraction with centrifugation. In result, the automated system can now capture spheroplasts within 10 s from the sample application to the well and 90% reached giga-seal in 2 min; 70% of all obtained patches were good enough for electrophysiological recording. When spheroplast sample was condensed by simple centrifugation without size selection, Port-a-Patch caught spheroplasts within 10 s from the sample application but only 20% of the acquired patches were good for recording; when untreated spheroplast suspension was used for Port-a-Patch, the apparatus needed 2 min, or often failed, to capture a spheroplast. The automated patch captured spheroplasts efficiently when concentration of the spheroplast suspension was over 1 × 107 spheroplasts/ml, which was in the range reported by [4] for cells (5 × 105–5 × 107 cells/ml). The microfluidic sorter chip not only accelerated the experimental processes but also saved time; if ultracentrifugation had been used for separating spheroplasts of desired size, it would have taken more than 2 h, while pumping the sample through a sorter chip took only 11 min and 700G spin-down with an ordinary microcentrifuge took 3 min.
4.2. The patch clamp detects subtle membrane protein expression
The automated patch clamp system stably obtained sensible Kv2.1 current readings from the bacterial giant spheroplast. Sparsely-expressed ion channels in the membrane are hard to detect with immunological assays. As shown in the result, Kv2.1 expression was barely observed with western blotting after the tedious membrane protein extraction by ultracentrifugation and the detection with the latest scanner, while the patch clamp could sharply detect such a subtle but important signal in the bacterial membrane. The patch clamp clearly reveals the presence of weakly-expressed membrane proteins which could be otherwise overlooked so far, giving detailed biophysical properties.
4.3. Stable drug assays were achieved by bacterial spheroplasts
Time-course observation of Kv2.1 channel was finely achieved with the E. coli spheroplast using the external perfusion unit. Kv2.1 currents sharply responded to TEA and 4-AP administration and rapid wash and the dose–response relationship could be observed stably for 30 min in average.
4.4. More advantages of bacterial spheroplasts
As stated in the introduction, the bacterial giant spheroplast has many technical benefits over eukaryotic system and liposome reconstitution. In addition to these advantages, the bacterial spheroplast can be a better option when the target protein is ligand-gated ion channel because rapid exchange of bath solution is necessary for dose–response assays, and rapid wash-out is difficult to accomplish with the planner lipid bilayer patch. Moreover, the orientation of the membrane protein polarity is correctly arranged in the natural bacterial membrane while the orientation of the protein insertion cannot be controlled in the artificial lipid bilayer [13]. Again, the most important advantage of all is that the direct use of bacterial spheroplasts can skip painstaking steps needed to set up the eukaryotic system for electrophysiological analysis. Now we are refining a novel technique named PERRIS (intra periplasm secretion and selection) method, a kind of in vitro directed evolution in which a target protein and the interacting peptide are expressed in E. coli inner membrane and periplasmic space, respectively, to search and optimize interacting peptides [12]. In this process, tremendous number of recombinant E. coli would be produced and we needed a fast and inexpensive way of primary screening. It is true that fancy multi-channel automated patch clamp gears are available today and using eukaryotic cells is much better for screening; however, many laboratories in the world are still using E. coli for genetic research and many of them would not be able to afford such expensive equipments. Relatively affordable single channel patch clamp and the direct use of bacterial spheroplasts may help researchers in small labs not to give up electrophysiological studies simply because of the budget problem.
4.5. Many ion channels are awaiting to be screened as pharmaceutical targets.
Today membrane proteins occupy the large number of promising pharmaceutical targets; of 6650 predicted potential drug targets, G-protein-coupled receptors and ion channels comprise of 30% and 15%, respectively [22]. Ion channel modulators are attracting interests these years due to their pharmacological values which can treat various diseases such as hypertension, long QT syndrome, diabetes, epilepsy, schizophrenia, depression and pain [6], [7], [15]. Our novel method, the combination of the bacterial spheroplast and the automated patch clamp system, will be able to evaluate the interaction of these membrane proteins and drug candidates with great details without annoying manipulation. Furthermore, with the introduction of the multi-channel automated patch clamp and the spheroplast fractionation, this screening method will become even more powerful and expedite the screening process in future.
5. Conclusion
The eukaryotic voltage-gated ion channel Kv2.1 was expressed in E. coli cells and giant spheroplasts large enough for the patch clamp were prepared. Human Kv2.1 currents could be successfully recorded from the giant spheroplast with the automated planner patch clamp which did not require any special skills for handling. Kv2.1-expressed E. coli spheroplasts could be stably used for the dose-response assay with time-course recording using TEA and 4-AP, making it the first application of the bacterial giant spheroplast for practical pharmaceutical assay using the automated patch clamp system.
Acknowledgements
We are grateful to skillful personnel from Nanion Technologies GmbH: Mr. Atsushi Ohtsuki nicely taught me overall instructions concerning the Port-a-Patch system and electrophysiology basics, and Ms. Maria Barthmes gave me kind advices for spheroplast preparation. This work was partly supported by JSPS KAKENHI Grant-in-Aid for Scientific Research (C); number 25350984, and Business Development Task Force from AIST. The authors of this paper declare that there are no conflicts of interest.
Footnotes
Available online 5 May 2015
References
- 1.Berrier C., Besnard M., Ajouz B., Coulombe A., Ghazi A. Multiple mechanosensitive ion channels from Escherichia coli, activated at different thresholds of applied pressure. J. Membr. Biol. 1996;151:175–185. doi: 10.1007/s002329900068. [DOI] [PubMed] [Google Scholar]
- 2.Blount P., Sukharev S.I., Moe P.C., Schroeder M.J., Guy H.R., Kung C. Membrane topology and multimeric structure of a mechanosensitive channel protein of Escherichia coli. EMBO J. 1996;15(18):4798–4805. [PMC free article] [PubMed] [Google Scholar]
- 3.Brüggemann A., Farre C., Haarmann C., Haythornthwaite A., Kreir M., Stoelzle S., George M., Fertig N. Planar patch clamp: advances in electrophysiology. Methods Mol. Biol. 2008;491:165–176. doi: 10.1007/978-1-59745-526-8_13. [DOI] [PubMed] [Google Scholar]
- 4.Brueggemann A., George M., Klau M., Beckler M., Steindl J., Behrends J.C., Fertig N. Ion channel drug discovery and research: the automated Nano-Patch-Clamp technology. Curr. Drug Discov. Technol. 2004;1(1):91–96. doi: 10.2174/1570163043484833. [DOI] [PubMed] [Google Scholar]
- 5.Buechner M., Delcour A.H., Martinac B., Adler J., Kung C. Ion channel activities in the Escherichia coli outer membrane. Biochim. Biophys. Acta. 1990;1024(1):111–121. doi: 10.1016/0005-2736(90)90214-9. [DOI] [PubMed] [Google Scholar]
- 6.Camerino D.C., Tricarico D., Desaphy J.F. Ion channel pharmacology. Neurotherapeutics. 2007;4(2):184–198. doi: 10.1016/j.nurt.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Cannon S.C. Physiologic principles underlying ion channelopathies. Neurotherapeutics. 2007;4(2):174–183. doi: 10.1016/j.nurt.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Cui C., Smith D.O., Adler J. Characterization of mechanosensitive channels in Escherichia coli cytoplasmic membrane by whole-cell patch clamp recording. J. Membrane Biol. 1995;144:31–42. doi: 10.1007/BF00238414. [DOI] [PubMed] [Google Scholar]
- 9.Dworakowska B., Dołowy K. Ion channels-related diseases. Acta Biochim. Pol. 2000;47(3):685–703. [PubMed] [Google Scholar]
- 10.Farre C., Haythornthwaite A., Haarmann C., Stoelzle S., Kreir M., George M., Brüggemann A., Fertig N. Port-a-Patch and Patchliner: high fidelity electrophysiology for secondary screening and safety pharmacology. Comb. Chem. High Throughput Screen. 2009;12(1):24–37. doi: 10.2174/138620709787047966. [DOI] [PubMed] [Google Scholar]
- 11.Hölter P., Kunst S., Wolloscheck T., Kelleher D.K., Sticht C., Wolfrum U., Spessert R. The retinal clock drives the expression of Kcnv2, a channel essential for visual function and cone survival. Invest Ophthalmol. Vis. Sci. 2012;53(11):6947–6954. doi: 10.1167/iovs.12-10234. [DOI] [PubMed] [Google Scholar]
- 12.Kubo T., Ono S., Kimura T., Kobayashi S., Kondo T., Fukuda E., Haga T., Kameyama K. Random peptide library based on spider neurotoxin, and utilization of the library in in vitro evolution derected to GPCR ligands. Toxicon. 2012;60:113. [Google Scholar]
- 13.Kuo M.M., Saimi Y., Kung C., Choe S. Patch clamp and phenotypic analyses of a prokaryotic cyclic nucleotide-gated K+ channel using Escherichia coli as a host. J. Biol. Chem. 2007;282(33):24294–24301. doi: 10.1074/jbc.M703618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinac B., Buechner M., Delcour A.H., Adler J., Kung C. Pressure-sensitive ion channel in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2297–2301. doi: 10.1073/pnas.84.8.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathie A. Ion channels as novel therapeutic targets in the treatment of pain. J. Pharm. Pharmacol. 2010;62(9):1089–1095. doi: 10.1111/j.2042-7158.2010.01131.x. [DOI] [PubMed] [Google Scholar]
- 16.Milkman R. An Escherichia coli homologue of eukaryotic potassium channel proteins. Proc. Natl. Acad. Sci. U. S. A. 1994;91(9):3510–3514. doi: 10.1073/pnas.91.9.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misonou H., Mohapatra D.P., Trimmer J.S. Kv2.1: a voltage-gated K+ channel critical to dynamic control of neuronal excitability. Neurotoxicology. 2005;26(5):743–752. doi: 10.1016/j.neuro.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Neher E., Sakmann B. Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature. 1976;260(5554):799–802. doi: 10.1038/260799a0. [DOI] [PubMed] [Google Scholar]
- 19.Santos J.S., Lundby A., Zazueta C., Montal M. Molecular template for a voltage sensor in a novel K+ channel. I. Identification and functional characterization of KvLm, a voltage-gated K+ channel from Listeria monocytogenes. J. Gen. Physiol. 2006;128(3):283–292. doi: 10.1085/jgp.200609572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schrempf H., Schmidt O., Kümmerlen R., Hinnah S., Müller D., Betzler M., Steinkamp T., Wagner R. A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. EMBO J. 1995;14(21):5170–5178. doi: 10.1002/j.1460-2075.1995.tb00201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shieh C.C., Coghlan M., Sullivan J.P., Gopalakrishnan M. Potassium channels: molecular defects, diseases, and therapeutic opportunities. Pharmacol. Rev. 2000;52(4):557–594. [PubMed] [Google Scholar]
- 22.Terstappen G.C., Reggiani A. In silico research in drug discovery. Trends Pharmacol. Sci. 2001;22(1):23–26. doi: 10.1016/s0165-6147(00)01584-4. [DOI] [PubMed] [Google Scholar]
- 23.Uozumi N. Escherichia coli as an expression system for K(+) transport systems from plants. Am. J. Physiol. Cell Physiol. 2001;281(3) doi: 10.1152/ajpcell.2001.281.3.C733. C733-9. [DOI] [PubMed] [Google Scholar]
- 24.Yajuan X., Xin L., Zhiyuan L. A comparison of the performance and application differences between manual and automatedpatch-clamp techniques. Curr. Chem. Genomics. 2012;6:87–92. doi: 10.2174/1875397301206010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida M., Nakata M., Yamato S., Dezaki K., Sugawara H., Ishikawa S.E., Kawakami M., Yada T., Kakei M. Voltage-dependent metabolic regulation of Kv2.1 channels in pancreatic beta-cells. Biochem. Biophys. Res. Commun. 2010;396(2):304–309. doi: 10.1016/j.bbrc.2010.04.088. [DOI] [PubMed] [Google Scholar]