Abstract
Background
G-quadruplexes (G4s) are four-stranded nucleic acid structures that form in G-rich sequences. Nucleolin (NCL) is a cellular protein reported for its functions upon G4 recognition, such as induction of neurodegenerative diseases, tumor and virus mechanisms activation. We here aimed at defining NCL/G4 binding determinants.
Methods
Electrophoresis mobility shift assay was used to detect NCL/G4 binding; circular dichroism to assess G4 folding, topology and stability; dimethylsulfate footprinting to detect G bases involved in G4 folding.
Results
The purified full-length human NCL was initially tested on telomeric G4 target sequences to allow for modulation of loop, conformation, length, G-tract number, stability. G4s in promoter regions with more complex sequences were next employed. We found that NCL binding to G4s heavily relies on G4 loop length, independently of the conformation and oligonucleotide/loop sequence. Low stability G4s are preferred. When alternative G4 conformations are possible, those with longer loops are preferred upon binding to NCL, even if G-tracts need to be spared from G4 folding.
Conclusions
Our data provide insight into how G4s and the associated proteins may control the ON/OFF molecular switch to several pathological processes, including neurodegeneration, tumor and virus activation. Understanding these regulatory determinants is the first step towards the development of targeted therapies.
General significance
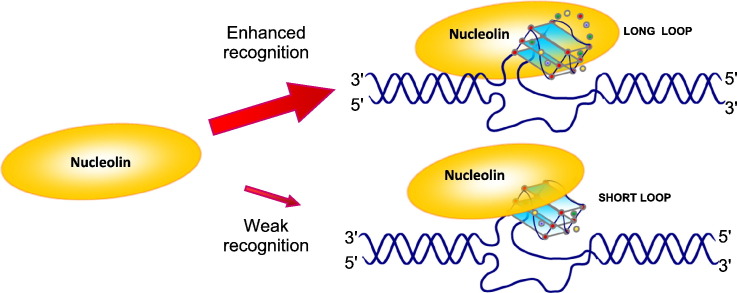
The indication that NCL binding preferentially stimulates and induces folding of G4s containing long loops suggests NCL ability to modify the overall structure and steric hindrance of the involved nucleic acid regions. This protein-induced modification of the G4 structure may represent a cellular mechanosensor mechanism to molecular signaling and disease pathogenesis. This article is part of a Special Issue entitled "G-quadruplex" Guest Editor: Dr. Concetta Giancola and Dr. Daniela Montesarchio.
Abbreviations: G-quadruplexes, G4s; nucleolin, NCL; circular dichroism, CD; electrophoresis mobility shift assay, EMSA; temperature of melting, Tm; dimethylsulfate, DMS; polyehtylene glycol, PEG
Keywords: G-quadruplexes, Nucleolin, Loops, Conformation, Nucleic acids, Protein-nucleic acid binding
Graphical abstract
Highlights
-
•
Nucleolin (NCL) binding to G-quadruplex (G4) nucleic acids is proportional to loop length.
-
•
One/two long loops in G4s enhance NCL binding.
-
•
NCL favors G4 folding with long loops.
-
•
G4 loop length is a stronger driver of NCL binding vs conformation or sequence.
-
•
NCL modification of nucleic acid regions may be a mechanosensor mechanism to disease.
1. Introduction
G-quadruplexes (G4s) are unique four-stranded nucleic acid structures that may form in guanine-rich sequences [1]. Four Gs bind via Hoogsteen-type hydrogen bonds to yield G-quartets that in turn stack on top of each other to form the G4. The presence of K+ cations specifically supports G4 formation and stability. G4s are highly polymorphic, in terms of strand stoichiometry (forming both inter- and intramolecular structures), strand orientation/topology and loop sequence, length and position [2]. G4s have been shown in humans and other eukaryotes [3], [4], in prokaryotes [5], [6], [7], [8] and in viruses [9], [10]. From a functional point of view, in humans G4s play key regulatory and pathological roles, including transcriptional regulation of gene promoters and enhancers, translation, chromatin epigenetic regulation, DNA recombination [11], [12], [13]. Human G4 DNA motifs have been reported to be associated with recombination prone regions [14] and to show mutational patterns that preserved the potential to form G4 DNA structures [15]. G4 misregulation has been associated with a number of relevant human diseases, such as the amyotrophic lateral sclerosis [16], Alzheimer [17] and fragile X syndrome [18], in which expansion of G4-forming regions has been reported. In viruses, G4s have been proposed both as virus cycle modulators and antiviral targets [10]. In the human immunodeficiency virus-1 (HIV-1), the etiologic agent of the acquired immune deficiency syndrome (AIDS), functionally significant G4s in the Nef coding region [19] and in the unique long terminal repeat (LTR) promoter [20], [21], [22] have been identified. These studies have shown that G4 folding at the LTR promoter decreases viral transcription with an effect that is augmented by G4 ligands [23], [24]. G4s have also been reported in the herpes simplex virus 1 [25], Epstein–Barr virus [26], [27], SARS coronavirus [28], human papilloma, hepatitis C, Zika and Ebola virus genomes [29], [30], [31], [32].
The biological effects of G4s are often modulated by the interaction with proteins [33]: the shelterin complex proteins are involved in telomere homeostasis [34], and other G4 interacting proteins either unfold or stabilize the G4 conformation [35], [36]. Mutations in some of these G4-interacting proteins have been linked to genetic diseases, such as the Bloom's and Werner's syndromes, Fanconi anemia [37], [38], and cancer [39].
Among G4 interacting proteins, nucleolin (NCL) is the protein most often reported for its biological functions upon G4 recognition. It is widely believed that NCL plays a chaperone role by helping the correct folding of complex nucleic acids structures. Indeed, NCL has been shown to display a marked preference for both endogenous and exogenous G-rich sequences that can fold into G4 [40]. It has been reported that binding of NCL to the endogenous (GGGGCC)n hexanucleotide repeat expansion (HRE) in C9orf72 is responsible for the initiation of molecular cascades that lead to neurodegenerative diseases [16]. At the promoter level, binding of NCL to G4 structures augments the basal effect of the folded conformation [41], [42], [43], [44]. For example, in the case of HIV-1, it induces potent inhibition of viral transcription by stabilization of the LTR promoter [45]. NCL is mainly present in the nucleolus, but it can be found also in cell membranes and, upon stress stimuli, in the nucleoplasm and cytoplasm to some extent [46], [47]. NCL has been initially described as a RNA binding protein, but recent evidence indicates the relevance, and in some case the preference, of its binding to DNA structures [45]. Because of NCL role in viral mechanisms, such as HIV cell attachment/entry and budding [48], [49], [50] and virus replication and transcription [51], [52], [53], [54], [55], [56], NCL inhibitors have been developed [57], [58], [59]. Interestingly, one of these is a G4-forming oligonucleotide, which displays interesting HIV activity [60], [61].
Despite the involvement of NCL/G4 complex in several human diseases, a deep understanding of the G4 structural features that drive binding to NCL is lacking. A certain degree of selectivity towards nucleic acids has been reported [41], [45]. For instance, in the case of the HIV-1 LTR promoter, where three G4s can form (LTR-II, LTR-III and LTR-IV), NCL binds with high affinity to LTR-III and with intermediate affinity to LTR-II, whereas LTR-IV is not recognized [45].
To understand the structural requirements that stimulate NCL binding to G4s, we tested NCL binding towards different sets of G4 oligonucleotides, considering loop length, topology, sequence, number of G-runs, stability. We found that loop length was always the prevalent determinant of NCL binding to G4s, independently of the conformation and oligonucleotide/loop sequence. These findings both allow a deeper understanding of NCL/G4-regulated physiological and pathological mechanisms, and the possibility to develop new target-specific drugs.
2. Materials and methods
2.1. Oligonucleotides and proteins
Desalted oligonucleotides were purchased from Sigma-Aldrich (Milan, Italy). Oligonucleotides were denatured for 5 min at 95 °C and gradually cooled to room temperature to achieve proper G4 folding. The human recombinant nucleolin (NCL) is from Origene Technologies, Rockville, USA.
2.2. Circular dichroism (CD)
CD spectroscopy was used to probe G4 folding and conformation of the tested DNA oligonucleotides. Oligonucleotides were diluted to a final concentration of 4 μM in lithium cacodylate buffer (10 mM, pH 7.4), KCl or NaCl (100 mM) and 20% of PEG200 where indicated. After heat denaturation samples were folded at room temperature overnight. CD spectra were recorded on a Chirascan-Plus (applied Photophysics, Leatherhead, UK) equipped with a Peltier temperature controller using a quartz cell of 5 mm optical path length and an instrument scanning speed of 50 nm/min. The temperature was set at 20 °C and the spectra were recorded over a wavelength range of 230–320 nm. For Tm determination, spectra were recorded over a temperature range of 20–95 °C, with temperature increase of 5 °C/min. Tm values were calculated according to the van't Hoff equation, applied for a two-state transition from a folded to unfolded state. Acquired spectra were baseline corrected for signal contribution due to the buffer and the observed ellipticities converted to mean residue ellipticity (θ) = deg × cm2 × dmol− 1 (mol ellip).
2.3. Electrophoresis Mobility Shift Assay (EMSA)
EMSA was used to detect G4-NCL interaction and the stoichiometry of G4 folding. Oligonucleotides were 5′-end labeled with [γ-32P]ATP by T4 polynucleotide kinase after 30 min incubation at 37 °C. After DNA precipitation, labeled species were resuspended in lithium cacodylate buffer (10 mM, pH 7.4, KCl 100 mM or NaCl 100 mM and 20% (w/v) PEG200 where indicated), heat denatured and folded at room temperature.
For the determination of G4 stoichiometry samples were run in 20% non-denaturing polyacrylamide gels for ~ 24 h at 60 V. Gels were fresh impressed and visualized by phosphorimaging (Typhoon FLA 9000, GE Healthcare Europe, Milan, Italy).
For NCL binding to DNA, reactions were performed in binding buffer (Tris–HCl 20 mM pH 8, KCl 30 mM, MgCl2 1.5 nM, DTT 1 mM, glycerol 8%, protease inhibitor cocktail – Sigma Aldrich, Milan, Italy – 1%, NaF 5 mM, Na3VO4 1 mM), labeled oligonucleotides (40 nM), human recombinant NCL (300 ng) and incubated for 1 h at 37 °C. Binding reactions were supplemented with 20% (w/v) PEG200 where indicated. To load the same amount of DNA in each reaction, counts per second emitted by the radiolabeled oligonucleotides were measured by a Geiger counter (Mini 900 Scintillation Monitor, Thermo Scientific™, Monza, Italy). Samples were loaded on 8% non-denaturing polyacrylamide gels for ~ 15 h at 40 V. Gels were dried using a gel dryer (Bio Rad Laboratpries, Milan, Italy) and visualized by phosphorimaging (Typhoon FLA 9000, GE Healthcare Europe, Milan, Italy). Quantification of bound DNA molecules was performed by ImageQuant TL Software (GE Healthcare Europe, Milan, Italy). Each gel was performed twice to confirm quantification results; only results consistent throughout the repeated gels were retained and reported.
2.4. Dimethylsulfate footprinting (DMS)
The DNA substrate of interest was gel-purified before use, 5′-end-labeled with [γ-32P]ATP by T4 polynucleotide kinase, purified using MicroSpin G-25 columns (GE Healthcare Europe, Milan, Italy), resuspended in lithium cacodylate buffer 10 mM pH 7.4 with or without KCl 100 mM, heat denatured and folded. Sample solutions were then treated with dimethylsulfate (DMS, 0.5% in ethanol) for 5 min and stopped by addition of 10% glycerol and β-mercaptoethanol. Samples were loaded onto a 16% native polyacrylamide gel and run until the desired resolution was obtained. DNA bands were localized via autoradiography, excised and eluted overnight. The supernatants were recovered, ethanol-precipitated and treated with piperidine 1 M for 30 min at 90 °C. Samples were dried in a speed-vac, washed with water, dried again, and resuspended in formamide gel loading buffer. Reaction products were analyzed on 20% denaturing polyacrylamide gels, visualized by phosphorimaging analysis, and quantified by ImageQuant TL software (GE Healthcare Europe, Milan, Italy).
3. Results
3.1. Nucleolin binding to G-quadruplexes directly correlates with the number of G-tracts
To investigate the determinants required for efficient NCL binding, we initially tested the human telomeric sequence, formed by GGGTTA repeats, and a simplified sequence formed by GGGTTT repeats, which is found in telomeres of ciliates of the genus Paramecium [62]. The interaction of NCL and its four RNA binding domains (RBD) and the Arg-Gly-Gly (RGG) domain to telomeric G4s has been previously reported [41], [63], [64]. Telomeric sequences are rather simple compared to the G4 sequences in oncogene and HIV-1 promoters and were thus chosen to allow assessment of oligonucleotide length, loops, repeats, conformation and sequence contribution to NCL binding. NCL/oligonucleotide binding was tested by electrophoresis mobility shift assays (EMSA) with 32P-labelled nucleic acids and the purified full-length human NCL protein [45].
We initially tested telomeric sequences ((GGGTTA)n and (GGGTTT)n) with increasing number of G-tracts, i.e. n = 4, 6 and 8 (Table 1 ).
Table 1.
Sequences of oligonucleotides used in this study.
| Name | Length (nts) | Sequence 5′-3′’ |
|---|---|---|
| (GGGTTA)4 | 21 | GGGTTAGGGTTAGGGTTAGGG |
| (GGGTTA)6 | 36 | GGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTA |
| (GGGTTA)8 | 54 | TCGAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAC |
| (GGGTTT)4 | 21 | GGGTTTGGGTTTGGGTTTGGG |
| (GGGTTT)6 | 36 | GGGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTTT |
| (GGGTTT)8 | 48 | GGGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTTT |
| (GGGTTT)5 | 27 | GGGTTTGGGTTTGGGTTTGGGTTTGGG |
| n5M1 | 27 | TTTTTTGGGTTTGGGTTTGGGTTTGGG |
| n5M2 | 27 | GGGTTTTTTTTTGGGTTTGGGTTTGGG |
| n5M3 | 27 | GGGTTTGGGTTTTTTTTTGGGTTTGGG |
| n5M4 | 27 | GGGTTTGGGTTTGGGTTTTTTTTTGGG |
| n5M5 | 27 | GGGTTTGGGTTTGGGTTTGGGTTTTTT |
| M43 | 43 | TTTTTGGAGTCGTGTCGCGTGTCGAGCGTGTGTAGTGGTTTTT |
| M30 | 30 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTT |
| M29i | 29 | AAAACAGACAGCTCACTGCGCTCACAAAA |
| M29ii | 29 | TTTTGTGAGCGCGGTGAGGTGTGTGTTTT |
| M26 | 26 | CCTCCTCCTCCTTCTCCTCCTCCTCC |
| M21 | 21 | TTTTTTTTTTTTTTTTTTTTT |
| M14 | 14 | CGGCATGCGTGGCT |
| 111 | 15 | GGGTGGGTGGGTGGG |
| 131 | 17 | GGGTGGGTTTGGGTGGG |
| 161 | 20 | GGGTGGGTTTTTTGGGTGGG |
| 313 | 19 | GGGTTTGGGTGGGTTTGGG |
| 333 | 21 | GGGTTTGGGTTTGGGTTTGGG |
| 363 | 24 | GGGTTTGGGTTTTTTGGGTTTGGG |
| 616 | 25 | GGGTTTTTTGGGTGGGTTTTTTGGG |
| 636 | 27 | GGGTTTTTTGGGTTTGGGTTTTTTGGG |
| 666 | 30 | GGGTTTTTTGGGTTTTTTGGGTTTTTTGGG |
| T111T | 17 | TGGGTGGGTGGGTGGGT |
| T131T | 19 | TGGGTGGGTTTGGGTGGGT |
| T161T | 22 | TGGGTGGGTTTTTTGGGTGGGT |
| c-kit1 | 21 | AGGGAGGGCGCTGGGAGGAGG |
| bcl-2 | 39 | AGGGGCGGGCGCGGGAGGAAGGGGGCGGGAGCGGGGCTG |
| c-myc | 27 | TGGGGAGGGTGGGGAGGGTGGGGAAGG |
| hTert 1–4 | 20 | AGGGGAGGGGCTGGGAGGGC |
| hTert 5–12 | 46 | AGGGGGCTGGGCCGGGGACCCGGGAGGGGTCGGGACGGGGCGGGGT |
| hTert 1–12 | 70 | AGGGGAGGGGCTGGGAGGGCCCGGAGGGGGCTGGGCCGGGGACCCGGGAGGGGTCGGGACGGGGCGGGGT |
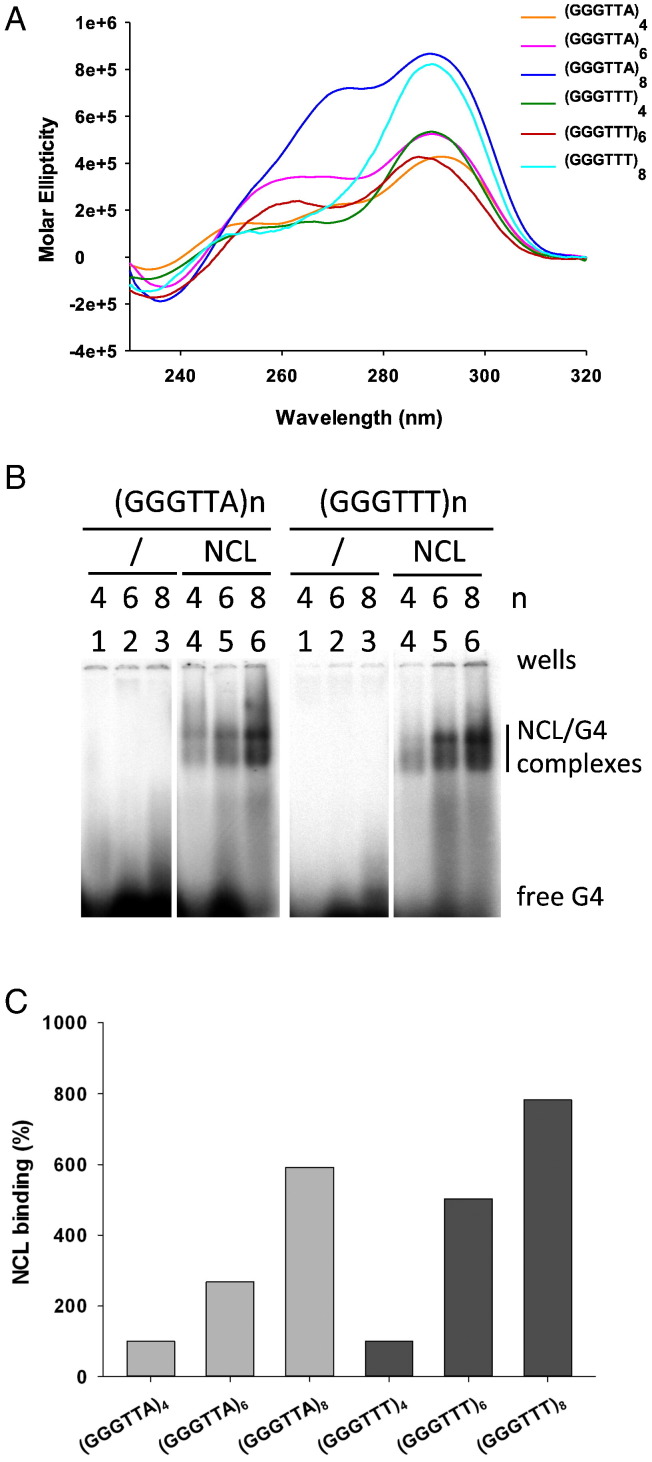
All sequences folded into mixed type G4s as assessed by circular dichroism (CD) analysis: in fact, all oligonucleotides displayed positive peaks around 290 nm and 260 nm, which are signatures of hybrid/mixed type G4 topologies (Fig. 1A). NCL binding to the G4 oligonucleotides was visualized as two bands running slower than the free unbound DNA. The presence of the two bands has been previously demonstrated to be due to the N-terminus self-cleaving activity of NCL [45]: thus the two bands were ascribed to the binding of the full-length (upper) and the N-terminus-minus (lower) NCL protein to the G4s (Fig. 1B). In these conditions, NCL binding was directly proportional to the length of the telomeric sequences and the difference in loop sequence between human and ciliate G4s did not modify this trend (Fig. 1C).
Fig. 1.
NCL binding to G4s with increasing number of G-tracts. (A) CD spectra of telomeric sequences (GGGTTA)n and (GGGTTT)n, where n = 4, 6 and 8. (B) EMSA analysis of NCL binding to telomeric sequences (GGGTTA)n - lane 4, 5, 6 left - and (GGGTTT)n - lane 4, 5, 6 right -. Lanes 1–4 are control reactions without NCL. (C) Quantification of NCL/G4 complex bands obtained in the EMSA shown in panel (B). Binding of lanes 4–6 left and 4–6 right was normalized for (GGGTTA)4 and (GGGTTT)4 NCL/G4 band intensity, respectively.
3.2. Long loops favor NCL binding to G4s
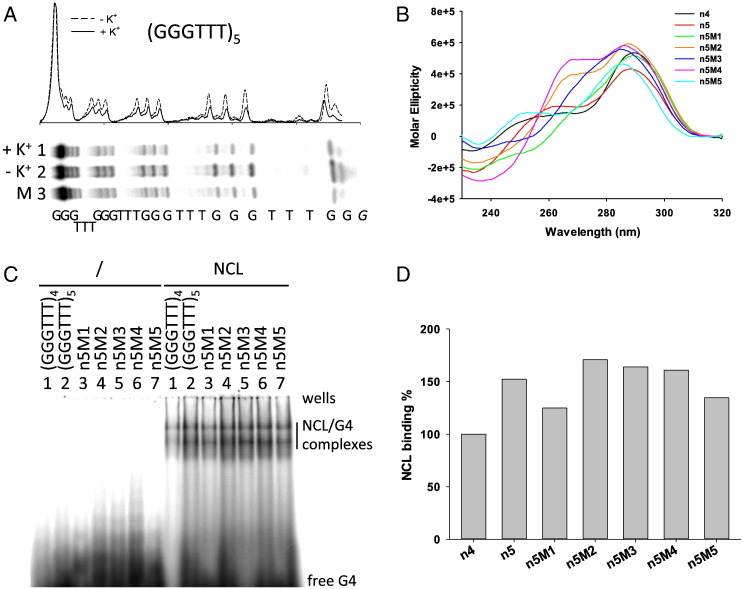
While the sequences with 4 and 8 G-tracts can fold into 1 and 2 stacked G4s, respectively, sequences with 5 and 6 G-tracts can fold into one G4 structure leaving one or two spare G-tracts. To test whether there was a preferred involvement of G-tracts in these conformationally dynamic sequences, oligonucleotide (GGGTTT)5 (Table 1) was folded in the presence and absence of K+ and treated with DMS to highlight G bases protected from DMS-induced methylation when in the G4 folded state. The analysis showed that all Gs were equally protected during folding (Fig. 2A) indicating the equilibrium of multiple G4 structures that involved alternative G-tracts. Similar results have been previously reported [65]. In particular, in the case of the (GGGTTT)5 oligonucleotide, 5 alternative G4 structures, which comprise G-tracts 1-2-3-4, 2-3-4-5, 1-3-4-5, 1-2-4-5, 1-2-3-5, may be envisaged. To test NCL binding to each of these sequences, mutant oligonucleotides where each G-tract was replaced by Ts were employed (Table 1). The (GGGTTT)4 oligonucleotide was used as a control since it displays the same sequence and number of G-tracts as the mutant oligonucleotides, but lacks extra T bases in the loops or at the 5′- or 3′-end (Table 1). All mutant sequences were shown to maintain the G4 folding by CD analysis: oligonucleotides displayed mainly a hybrid/mixed type topology with different levels of parallel/antiparallel contributions (Fig. 2B). The stability of the mutant sequences forming internal long loops was slightly lower than that of the wild-type sequence, as gathered by T melting (Tm) values measured by CD thermal unfolding (Table 2 ).
Fig. 2.
Analysis of NCL binding to alternative G4 structures formed by the (GGGTTT)5 telomeric sequence (A) DMS protection analysis of (GGGTTT)5 folded in the presence or absence of K+. M is a marker lane obtained with the Maxam and Gilbert sequencing protocol. Oligonucleotide sequence is shown below the gel image. The densitogram above the gel shows quantification of cleaved bands intensity. (B) CD spectra of (GGGTTT)5 and mutant derivatives where each G-tract was systematically replaced by Ts. (C) EMSA analysis of NCL binding to (GGGTTT)4, (GGGTTT)5 and mutant oligonucleotides. Oligonucleotide identity is shown above each lane in the gel image. (D) Quantification of NCL/G4 complex bands obtained in the EMSA in panel (C). n4 and n5 stand for (GGGTTT)4 and (GGGTTT)5, respectively.
Table 2.
Stability (Tm values) of oligonucleotides of the (GGGTTT)5 series with G-tracts replaced with Ts. Tm were measured by CD thermal unfolding and reported at the positive CD peaks, signatures of the parallel (265 nm) and antiparallel (285 nm) G4 conformation contribution.
| Oligonucleotide | Tm (265 nm) (°C) | Tm (285 nm) (°C) |
|---|---|---|
| (GGGTTT)4 | 66.6 ± 0.4 | 66.1 ± 0.4 |
| (GGGTTT)5 | 65.5 ± 1.1 | 64.5 ± 0.5 |
| n5M1 | 58.0 ± 0.8 | 55.7 ± 0.5 |
| n5M2 | 53.4 ± 0.5 | 54.5 ± 0.2 |
| n5M3 | 56.7 ± 0.3 | 56.6 ± 0.1 |
| n5M4 | 52.0 ± 0.3 | 53.2 ± 0.1 |
| n5M5 | / | 60.4 ± 0.4 |
EMSA analysis of NCL binding to the mutant sequences showed mild preferential binding towards oligonucleotides where internal G-tracts were mutated (Fig. 2C and D). Binding to sequences with external Ts was consistently lower, but higher than that to the (GGGTTT)4 control. The (GGGTTT)5 oligonucleotide displayed an average binding to all other mutant sequences. These data indicate that NCL preferentially binds G4s with at least one loop longer than 3 nts (e.g. 9 nts, compare n5M2, n5M3 and n5M4 binding, lanes 4–6 NCL, to n5M1 and n5M5, lanes 3 and 7 NCL, Fig. 2C) and with free nts at the 5′ or 3′ termini (compare n5M1, lane 3 NCL, and n5M5, lane 7 NCL, to (GGGTTT)4, lane 1 NCL, Fig. 2C).
3.3. More than two long or extremely short loops destabilize NCL interaction with G4s
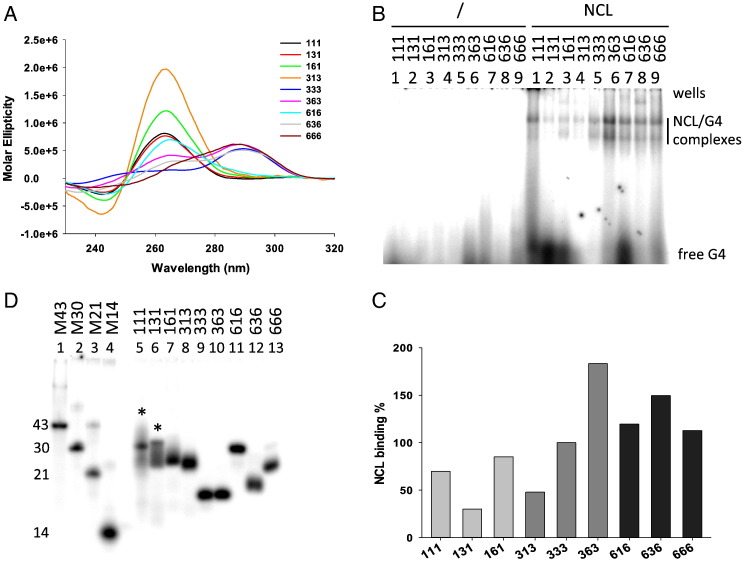
To explore the impact of loop length on NCL binding to G4s, three series of oligonucleotides with different loop-length combinations were tested: in each series, the two lateral loops were kept constant (1, 3 or 6 nts in each series) and the central loop was modified (1, 3 or 6 nts) (Table 1). All oligonucleotides folded into G4: in this case some sequences displayed a neat parallel topology (111, 131, 161, 313, 616, Fig. 3A), while other a mixed/hybrid type one (333, 363, 636, 666, Fig. 3A), in keeping with the notion that G4s with 1-nt loops are constrained to the parallel conformation [66], [67]. In general, G4s with shorter loops were more stable than those with longer loops (Table 3 ).
Fig. 3.
NCL binding to G4s with systematic elongation of loop length. (A) CD spectra of oligonucleotides with two constant lateral (1, 3 or 6 nts) and variable central loops (1, 3 or 6 nts). (B) EMSA analysis of NCL binding to each G4 with different loop-length combination. Oligonucleotides identity is shown above each lane. (C) Quantification of NCL/G4 complex bands obtained in the EMSA in panel (B). (D) Native gel electrophoresis of G4-folded oligonucleotides. Oligonucleotides identity is shown above each lane. Lanes 1–4 are marker oligonucleotides of known length and unable to fold into secondary structures. The symbol * indicates oligonucleotides showing two bands with different migration rate.
Table 3.
Stability (Tm values) of oligonucleotides of the (GGGTTT)4 series with variable-length loops. Tm were measured by CD thermal unfolding and reported at the positive CD peaks, signatures of the parallel (265 nm) and antiparallel (285 nm) G4 conformation contribution.
| Oligonucleotide | Tm (265 nm) (°C) | Tm (285 nm) (°C) |
|---|---|---|
| 111 | > 90 | / |
| 131 | > 90 | / |
| 161 | 72.1 ± 0.3 | / |
| 313 | 66.1 ± 0.4 | / |
| 333 | 66.6 ± 0.4 | 66.1 ± 0.4 |
| 363 | 63.0 ± 0.2 | 64.0 ± 0.2 |
| 616 | 52.7 ± 0.1 | / |
| 636 | 52.9 ± 0.3 | 53.0 ± 0.1 |
| 666 | 45.3 ± 0.3 | 44.4 ± 0.1 |
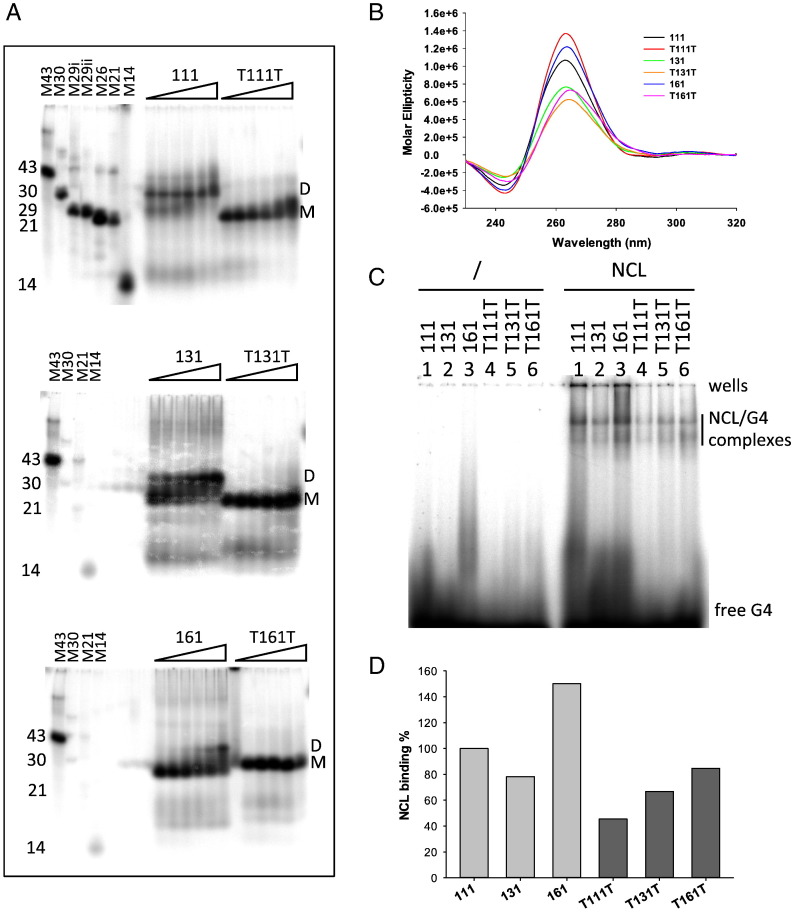
EMSA binding analysis showed a general increase in NCL binding upon loop extension. In particular, considering the series where the lateral loops were kept constant, an increase in binding was observed with the increasing length of the central loop; in the series where the central loop was kept constant, an increased binding was observed upon the increasing length of the lateral loops (Fig. 3B and C). Two notable exceptions were found: when all loops were 1 nt- or 6-nt long, NCL binding was higher or lower, respectively, than expected by the above-described trend (oligonucleotides 111 and 666 in Fig. 3B and C).
The observed behavior was apparently independent of the G4 conformation: oligonucleotides adopted both mixed and parallel-like G4 conformations (Fig. 3A), while no straightforward correlation with the intensity of NCL binding could be found (Fig. 3B and C). However, when testing G4 folding in EMSA, we observed one main band for all oligonucleotides, except oligonucleotides 111 and 131, which displayed two bands running at different migration rates (Fig. 3D). To test the molecularity of the upper running band, each labeled oligonucleotide was added with increasing amounts of the same unlabeled nucleic acid. As shown in Fig. 4A, the amount of the upper running band increased with increasing amounts of the unlabeled oligonucleotide, indicating the dimeric nature of the upper band. It has been reported that the presence of one T base at the end of a G4 forming oligonucleotide induces a preferential monomeric G4 [68], [69]. Indeed, when one T base was added at each end of the 111, 131 and 161 oligonucleotides (T111T, T131T, T161T, Table 1), the oligonucleotides maintained the G4 conformation (Fig. 4B) and only the monomeric faster running band was obtained (Fig. 4C). These T-added oligonucleotides bound NCL to a lower extent than those without T, probably due to steric hindrance, however, the binding trend was restored: the increase in the amount of NCL/DNA complex paralleled the lengthening of the loops (Fig. 4D).
Fig. 4.
Analysis of monomeric and dimeric G4 folding of relevant oligonucleotides. (A) Native gel electrophoresis at increasing oligonucleotide concentration (20, 45, 120, 415, 1600, 6200 nM). Samples identity is indicated above the gel image. Marker oligonucleotides with known length and unable to fold into secondary structures are loaded. (B) CD spectra of G4s showing dimeric folding and their corresponding monomolecular form when one T base was added at each of the two ends. (C) EMSA analysis of NCL binding. Oligonucleotides identity is shown above each lane. (D) Quantification of NCL/G4 complex bands obtained in the EMSA in panel (C).
The above data indicate that in G4-forming sequences formed by GGG tracts and T loops, NCL binding is in general favored by the presence of one or two long loops (i.e. 6 nt), whereas three long loops are less favored. The presence of one or two 1-nt long loops in the absence of at least one 6-nt loops disfavors NCL binding.
3.4. NCL binding to G4 oligonucleotides is independent of the G4 conformation
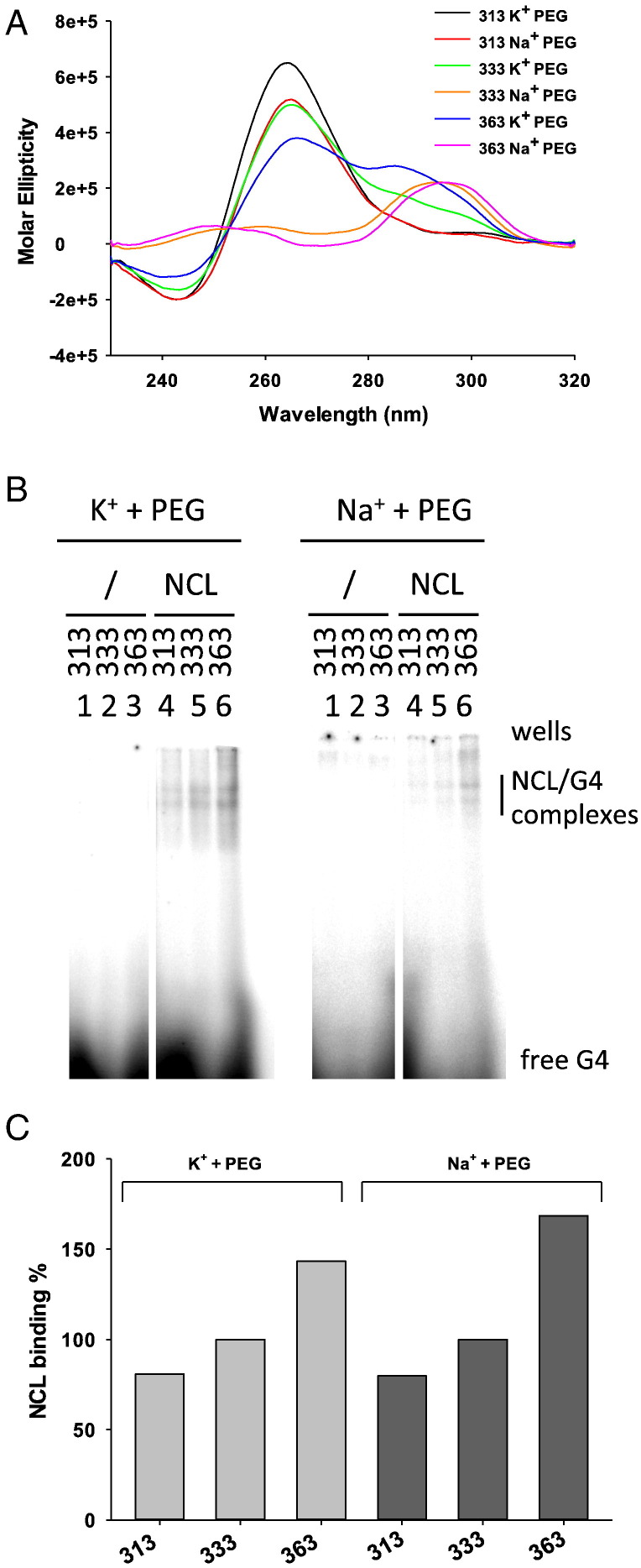
It has been reported that oligonucleotide G4 folding depends on the presence of the monovalent cation in solution. For instance, the human telomeric sequence adopts a hybrid conformation in K+ solution [70], while it is antiparallel in a Na+ solution [71]. Moreover, the presence in solution of cosolvents, such as polyethylene glycol (PEG), during G4 folding is commonly used to perturb G4 topology [72]. The human telomeric G4 DNA in the presence of PEG is reported to adopt a parallel G4 conformation, which is not the predominant form in dilute solutions [73], [74], [75], [76]. To investigate the effect of G4 conformation on NCL binding, we folded oligonucleotides previously tested in KCl alone (Fig. 3B), in KCl and NaCl in the presence of PEG 20% (w/v). In these conditions we observed in most cases a clear-cut conformational change (Fig. 5A): for instance, oligonucleotide 333 was mixed with a prevalent antiparallel conformation in K+, while it shifted to a prevalent parallel topology in K+ + PEG (compare Figs. 3A and 5A). The binding of NCL was assessed on these oligonucleotides folded in PEG (Fig. 5B). We observed the maintenance of the increased NCL binding to longer loop sequences trend (Fig. 5C). These data indicate that binding of NCL to telomeric G4 oligonucleotides is in general independent of the conformation and that loop length is the major determinant of NCL binding efficiency to telomeric G4s.
Fig. 5.
Analysis of NCL binding towards oligonucleotides with variable G4s conformation. (A) CD spectra of 313, 333, 363 oligonucleotides folded in the presence of 20% (w/v) PEG200 and K+ or Na+. (B) EMSA analysis of NCL binding to 313, 333, 363 oligonucleotides folded in the presence of 20% (w/v) PEG200 and K+ or Na+ as indicated above the gel image. (C) Quantification of NCL/G4 complex bands obtained in the EMSA in panel (B). For the K+ or Na+ experiment series, binding was normalized to the intensity of the 333 NCL/G4 complex band in the corresponding salt condition.
3.5. Sequence dependence: NCL binding to oncogenes promoter G4s
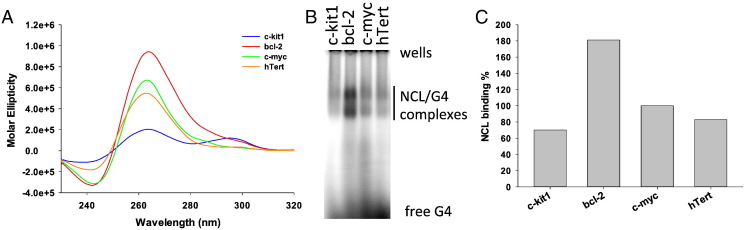
To verify if NCL binding determinants observed in telomeric (GGGTTT)n G4s were maintained in G4s with more complex sequences, we tested NCL binding towards four oncogene promoters i.e. c-myc, c-kit1, bcl-2 and hTert 1–4 [77], [78], [79], [80]. These were shown to form stable G4s whose conformations have been characterized by deep structural analysis. All these oncogene promoter G4s display loops with different length, composition and conformation (Table 1 and Fig. 6A). NCL bound preferentially to bcl-2, followed by c-myc; hTert 1–4 and c-kit1 displayed similar and lower NCL binding (Fig. 6B and C). Among these G4s, bcl-2 is the only structure exhibiting a long loop (7 nts) and two shorter loops (3 and 1 nts), whereas c-myc, c-kit1 and hTert 1–4 they all have loops ≤ 4 nts (Table 1).
Fig. 6.
NCL binding to human oncogene promoter G4s. (A) CD spectra of G4s formed in the c-kit1, bcl-2, c-myc and hTert 1–4 promoters. (B) EMSA analysis of NCL binding to oncogene promoter G4s. (C) Quantification of NCL/G4 complexes obtained in the EMSA in panel (B).
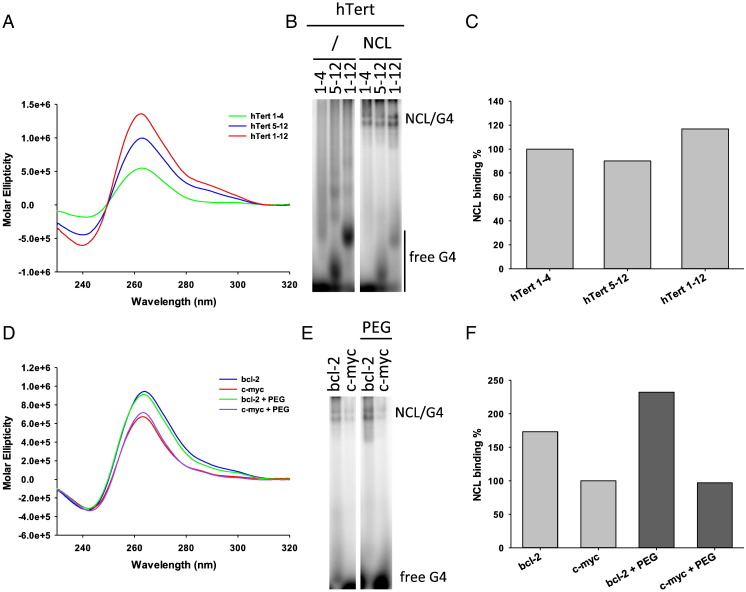
It has very recently been shown that the hTert full-length sequence is composed of 12 G-tracts that fold in two adjacent G4s: our tested hTert 1–4 comprises the first four G-tracts; the second G4, spanning G-tracts 5–12, displays a peculiar structure where a 26 nt-long loop forms an extended stem-hairpin [81]. We wished to test NCL binding to the two separate hTert G4s (hTert 1–4 and hTert 5–12) and to the full-length sequence (hTert 1–12). All three hTert G4s have similar CD spectra with a major peak at 260 nm and a shoulder at 295 nm, with molar ellipticity values at 260 nm increasing with the number of G-tracts in the G4-forming oligonucleotides (Fig. 7A). NCL binding to hTert 1–4 and hTert 5–12 was similar, indicating that the stem-hairpin suppresses the enhancing binding effect of the long loop (Fig. 7B and C). Binding of NCL to hTert 1–12 was slightly enhanced (Fig. 7B and C), retracing the behavior observed in the (GGGTTA)n and (GGGTTT)n oligonucleotides, where NCL preferentially bound to oligonucleotides with longer loops when multiple G-tracts were available for folding (Fig. 1, Fig. 2). It is thus possible that also in the case of hTert 1–12 NCL induces folding of a less stable but favorite long-looped G4. In alternative, the 7 nt spacer between the two G4 s (hTert 1–4 and hTert 5–12) is recognized as a long loop and therefore stimulates NCL binding. We next tested bcl-2 and c-myc in the presence of PEG 20% (w/v) to rule out an effect of PEG in NCL binding. Both oligonucleotides in K+ are folded in a parallel G4 conformation, which is not modified in the presence of PEG (Fig. 7D). NCL maintained a similar trend and degree of binding to these G4s in the absence/presence of PEG, confirming that PEG did not significantly alter NCL binding (Fig. 7E).
Fig. 7.
NCL binding to G4s in special conditions. (A) CD spectra of G4s formed by hTert 1–4, hTert 5–12 and hTert 1–12 oligonucleotides. Numbers indicate G4-tracts. (B) EMSA analysis of NCL binding to hTert G4s. (C) Quantification of NCL/G4 complexes obtained in the EMSA in panel (B). (D) CD spectra of G4s formed by c-myc and bcl-2 in the presence/absence of PEG 20% (w/v). (E) EMSA analysis of NCL binding to c-myc and bcl-2 G4s in the presence/absence of PEG 20% (w/v). (F) Quantification of NCL/G4 complexes obtained in the EMSA in panel (E).
These data are in prefect agreement with the NCL binding trend observed in telomeric (GGGTTT)n G4s. In addition, they also fit with NCL binding to HIV-1 LTR promoter G4s, previously reported and obtained by both EMSA and FRET analysis [45]. In that case, as well, the best bound G4 LTR-III displayed 1, 11, 3 nt-long loops, followed by LTR-II with 8, 11, 1 nt-long loops; in contrast LTR-IV, which presented 1, 4, 1 nt-long loops, was bound to a negligible extent [21], [45].
These data indicate that the results collected on (GGGTTT)n G4s apply in general to G4s with random sequences and that, again, the loop length is the major determinant in driving NCL binding to G4s.
4. Discussion
Despite the fact that NCL binding to G4 nucleic acids has been proposed to be responsible for human neurodegenerative diseases [16], pathological molecular mechanisms in cells [41], [42], [43], [44] and viruses [48], [49], [50], [51], [52], [53], [54], [55], [56], a deep characterization of the G4 features that induce NCL binding have been attempted only by Gonzalez et al. [41]. This group, who performed pioneer studies on NCL binding to the oncogene promoter c-myc G4, reported a preferential binding of NCL to parallel G4s with short loops. In contrast, our present work indicates that long loops in the G4 structure are preferred and this behavior is independent of the G4 conformation or loop/oligonucleotide sequence. These apparent differences may be explained by several reasons: 1) the use of a recombinant NCL purified in bacteria [41] vs the full-length human NCL purified from human cells (this work): the recombinant protein included all RBDs and C-terminal domain of the natural human protein but lacked the N-terminal domain which in turn was fused to the Escherichia coli maltose-binding protein; even if the nucleic acid binding activity was maintained in these conditions, the specific recognition of G4s, and in particular of their steric features, may be severely distorted by the introduction of a different N-terminal domain. Indeed, the N-terminal domain of NCL is a long unstructured (300 amino acids) domain that, besides its reported interaction with other proteins, may support the binding specificity of the RBD and RGG domains that directly make contact with the nucleic acid. 2) It has been demonstrated that short loops tend to constrain G4 structures into a parallel conformation, regardless of base composition [66], [67], therefore the suggested preferential binding to short-looped G4s in the parallel conformation may derive from the analysis of two dependent variables. 3) In our present work we performed a systematic analysis of loop sequence and oligonucleotide conformation working on a simplified (GGGTTT)n sequence. We unambiguously showed that NCL prefers G4s with at least one long loop (≥ 3 nts), while three short loops (1 nt) or three long loops (6 nts) are disfavored. The analysis on the simplified sequences allowed us to modify the conformation of the oligonucleotides working in different salt (K+ or Na+) and crowding conditions (PEG 20% w/v) [68], [72], [75] to show that NCL binding is in general independent of the oligonucleotide conformation and that loop lengths is the driving force that enhances NCL binding to G4s. By extending the results from the telomeric sequences to cellular oncogene and HIV-1 promoter G4s, we were able to show that loop length is the major determinant in the enhancement of NCL binding to G4s even in more variable and complex sequences. This observation is in line with the reported recognition of G4s by the cellular protein EWS: EWS shares with NCL the RGG domain which mediates recognition of G4s [82] and binds to G4s independently of base composition in long loops [64].
Finally, our results may shed some light on NCL recognition of repeats of G4 forming regions, such as the hexanucleotide repeat involved in the neurodegenerative diseases amyotrophic lateral sclerosis and frontotemporal dementia [16] and telomeric repeats [83]. We (Fig. 2A present work) and others [65] have shown that regions containing multiple G-tracts can fold into several conformations involving the alternative use of different G-tracts. We here indicate that NCL binding stimulates folding of these regions to G4s containing long loops (i.e. some G-tracts may be included to the loop moiety) therefore modifying the overall 3D structure and steric hindrance of these nucleic acid repeated sequences. Interestingly, indeed, NCL preferentially stabilized G4s that were less intrinsically stable, likely because of the possibility in this condition to fold them in the best protein-fitting conformation, i.e. G4s with longer and loose loops: this conformational modification may represent a marker to molecular signaling and disease pathogenesis. Our data perfectly fit with the very recent demonstration that protein binding to G4 and iMotif structures modify complex stability and the overall nucleic acid conformation to provide a mechanosensor-regulated cellular mechanism [84].
5. Conclusions
Our work has unambiguously shown that loop length of G4 structures is a fundamental feature recognized by NCL to promote its binding to G4-folded nucleic acids. Understanding the determinants that regulate NCL/G4 binding is the first step towards the development of therapies that specifically alter this molecular switch that triggers cellular signaling, such as initiation of transcription in the HIV-1 promoter and hence viral production, virus infection, tumor formation, maintenance and metastasis, and neurodegeneration.
Transparency Document
Transparency document.
Acknowledgments
Funding: This work was supported by the Bill & Melinda Gates Foundation, Seattle, WA [GCE grant numbers OPP1035881, OPP1097238]; the European Research Council [ERC Consolidator grant 615879]; and the Italian Ministry of University and Research [FIRB-Ideas RBID082ATK].
Footnotes
This article is part of a Special Issue entitled "G-quadruplex" Guest Editor: Dr. Concetta Giancola and Dr. Daniela Montesarchio.
The Transparency document associated with this article can be found, in online version.
References
- 1.Lipps H.J., Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Sen D., Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 3.Verma A., Halder K., Halder R., Yadav V.K., Rawal P., Thakur R.K., Mohd F., Sharma A., Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 4.Hershman S.G., Chen Q., Lee J.Y., Kozak M.L., Yue P., Wang L.S., Johnson F.B. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaume N., Pathak R., Yadav V.K., Kota S., Misra H.S., Gautam H.K., Chowdhury S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013;41:76–89. doi: 10.1093/nar/gks1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawal P., Kummarasetti V.B., Ravindran J., Kumar N., Halder K., Sharma R., Mukerji M., Das S.K., Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thakur R.S., Desingu A., Basavaraju S., Subramanya S., Rao D.N., Nagaraju G. Mycobacterium tuberculosis DinG is a structure-specific helicase that unwinds G4 DNA: implications for targeting G4 DNA as a novel therapeutic approach. J. Biol. Chem. 2014;289:25112–25136. doi: 10.1074/jbc.M114.563569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wieland M., Hartig J.S. Investigation of mRNA quadruplex formation in Escherichia coli. Nat. Protoc. 2009;4:1632–1640. doi: 10.1038/nprot.2009.111. [DOI] [PubMed] [Google Scholar]
- 9.Harris L.M., Merrick C.J. G-quadruplexes in pathogens: a common route to virulence control? PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metifiot M., Amrane S., Litvak S., Andreola M.L. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 2014;42:12352–12366. doi: 10.1093/nar/gku999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maizels N. G4-associated human diseases. EMBO Rep. 2015;16:910–922. doi: 10.15252/embr.201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou B., Liu C., Geng Y., Zhu G. Topology of a G-quadruplex DNA formed by C9orf72 hexanucleotide repeats associated with ALS and FTD. Sci. Rep. 2015;5:16673. doi: 10.1038/srep16673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mani P., Yadav V.K., Das S.K., Chowdhury S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakken S., Rognes T., Hovig E. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009;37:5749–5756. doi: 10.1093/nar/gkp590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R., Rothstein J.D., Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisette J.F., Montagna D.R., Mihailescu M.R., Wolfe M.S. A G-rich element forms a G-quadruplex and regulates BACE1 mRNA alternative splicing. J. Neurochem. 2012;121:763–773. doi: 10.1111/j.1471-4159.2012.07680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fry M., Loeb L.A. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. U. S. A. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perrone R., Nadai M., Poe J.A., Frasson I., Palumbo M., Palu G., Smithgall T.E., Richter S.N. Formation of a unique cluster of G-quadruplex structures in the HIV-1 Nef coding region: implications for antiviral activity. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amrane S., Kerkour A., Bedrat A., Vialet B., Andreola M.L., Mergny J.L. Topology of a DNA G-quadruplex structure formed in the HIV-1 promoter: a potential target for anti-HIV drug development. J. Am. Chem. Soc. 2014;136:5249–5252. doi: 10.1021/ja501500c. [DOI] [PubMed] [Google Scholar]
- 21.Perrone R., Nadai M., Frasson I., Poe J.A., Butovskaya E., Smithgall T.E., Palumbo M., Palu G., Richter S.N. A dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J. Med. Chem. 2013;56:6521–6530. doi: 10.1021/jm400914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piekna-Przybylska D., Sullivan M.A., Sharma G., Bambara R.A. U3 region in the HIV-1 genome adopts a G-quadruplex structure in its RNA and DNA sequence. Biochemistry. 2014;53:2581–2593. doi: 10.1021/bi4016692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perrone R., Butovskaya E., Daelemans D., Palu G., Pannecouque C., Richter S.N. Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 2014;69:3248–3258. doi: 10.1093/jac/dku280. [DOI] [PubMed] [Google Scholar]
- 24.Perrone R., Doria F., Butovskaya E., Frasson I., Botti S., Scalabrin M., Lago S., Grande V., Nadai M., Freccero M., Richter S.N. Synthesis, binding and antiviral properties of potent core-extended naphthalene diimides targeting the HIV-1 long terminal repeat promoter G-quadruplexes. J. Med. Chem. 2015;58:9639–9652. doi: 10.1021/acs.jmedchem.5b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Artusi S., Nadai M., Perrone R., Biasolo M.A., Palu G., Flamand L., Calistri A., Richter S.N. The herpes simplex virus-1 genome contains multiple clusters of repeated G-quadruplex: implications for the antiviral activity of a G-quadruplex ligand. Antivir. Res. 2015;118:123–131. doi: 10.1016/j.antiviral.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murat P., Zhong J., Lekieffre L., Cowieson N.P., Clancy J.L., Preiss T., Balasubramanian S., Khanna R., Tellam J. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014;10:358–364. doi: 10.1038/nchembio.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norseen J., Johnson F.B., Lieberman P.M. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 2009;83:10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tluckova K., Marusic M., Tothova P., Bauer L., Sket P., Plavec J., Viglasky V. Human papillomavirus G-quadruplexes. Biochemistry. 2013;52:7207–7216. doi: 10.1021/bi400897g. [DOI] [PubMed] [Google Scholar]
- 30.Wang S.R., Min Y.Q., Wang J.Q., Liu C.X., Fu B.S., Wu F., Wu L.Y., Qiao Z.X., Song Y.Y., Xu G.H., Wu Z.G., Huang G., Peng N.F., Huang R., Mao W.X., Peng S., Chen Y.Q., Zhu Y., Tian T., Zhang X.L., Zhou X. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti-hepatitis C target. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1501535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleming A.M., Ding Y., Alenko A., Burrows C.J. Zika virus genomic RNA possesses conserved G-quadruplexes characteristic of the Flaviviridae family. ACS Infect. Dis. 2016;2:674–681. doi: 10.1021/acsinfecdis.6b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S.R., Zhang Q.Y., Wang J.Q., Ge X.Y., Song Y.Y., Wang Y.F., Li X.D., Fu B.S., Xu G.H., Shu B., Gong P., Zhang B., Tian T., Zhou X. Chemical targeting of a G-quadruplex RNA in the ebola virus L gene. Cell Chem. Biol. 2016;23:1113–1122. doi: 10.1016/j.chembiol.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Sissi C., Gatto B., Palumbo M. The evolving world of protein-G-quadruplex recognition: a medicinal chemist's perspective. Biochimie. 2011;93:1219–1230. doi: 10.1016/j.biochi.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palm W., de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 35.Cogoi S., Paramasivam M., Membrino A., Yokoyama K.K., Xodo L.E. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010;285:22003–22016. doi: 10.1074/jbc.M110.101923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cogoi S., Shchekotikhin A.E., Xodo L.E. HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res. 2014;42:8379–8388. doi: 10.1093/nar/gku574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohaghegh P., Karow J.K., Brosh R.M., Jr., Bohr V.A., Hickson I.D. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seal S., Thompson D., Renwick A., Elliott A., Kelly P., Barfoot R., Chagtai T., Jayatilake H., Ahmed M., Spanova K., North B., McGuffog L., Evans D.G., Eccles D., C. Breast Cancer Susceptibility, Easton D.F., Stratton M.R., Rahman N. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat. Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 39.Paramasivam M., Membrino A., Cogoi S., Fukuda H., Nakagama H., Xodo L.E. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009;37:2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Girvan A.C., Teng Y., Casson L.K., Thomas S.D., Juliger S., Ball M.W., Klein J.B., Pierce W.M., Jr., Barve S.S., Bates P.J. AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006;5:1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez V., Guo K., Hurley L., Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez V., Hurley L.H. The C-terminus of nucleolin promotes the formation of the c-MYC G-quadruplex and inhibits c-MYC promoter activity. Biochemistry. 2010;49:9706–9714. doi: 10.1021/bi100509s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uribe D.J., Guo K., Shin Y.J., Sun D. Heterogeneous nuclear ribonucleoprotein K and nucleolin as transcriptional activators of the vascular endothelial growth factor promoter through interaction with secondary DNA structures. Biochemistry. 2011;50:3796–3806. doi: 10.1021/bi101633b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei P.C., Wang Z.F., Lo W.T., Su M.I., Shew J.Y., Chang T.C., Lee W.H. A cis-element with mixed G-quadruplex structure of NPGPx promoter is essential for nucleolin-mediated transactivation on non-targeting siRNA stress. Nucleic Acids Res. 2013;41:1533–1543. doi: 10.1093/nar/gks1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tosoni E., Frasson I., Scalabrin M., Perrone R., Butovskaya E., Nadai M., Palu G., Fabris D., Richter S.N. Nucleolin stabilizes G-quadruplex structures folded by the LTR promoter and silences HIV-1 viral transcription. Nucleic Acids Res. 2015;43:8884–8897. doi: 10.1093/nar/gkv897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daniely Y., Dimitrova D.D., Borowiec J.A. Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol. Cell. Biol. 2002;22:6014–6022. doi: 10.1128/MCB.22.16.6014-6022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storck S., Shukla M., Dimitrov S., Bouvet P. Functions of the histone chaperone nucleolin in diseases. Subcell. Biochem. 2007;41:125–144. doi: 10.1007/1-4020-5466-1_7. [DOI] [PubMed] [Google Scholar]
- 48.Callebaut C., Blanco J., Benkirane N., Krust B., Jacotot E., Guichard G., Seddiki N., Svab J., Dam E., Muller S., Briand J.P., Hovanessian A.G. Identification of V3 loop-binding proteins as potential receptors implicated in the binding of HIV particles to CD4(+) cells. J. Biol. Chem. 1998;273:21988–21997. doi: 10.1074/jbc.273.34.21988. [DOI] [PubMed] [Google Scholar]
- 49.Gao W., Li M., Zhang J. Tandem immunoprecipitation approach to identify HIV-1 Gag associated host factors. J. Virol. Methods. 2014;203:116–119. doi: 10.1016/j.jviromet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 50.Ueno T., Tokunaga K., Sawa H., Maeda M., Chiba J., Kojima A., Hasegawa H., Shoya Y., Sata T., Kurata T., Takahashi H. Nucleolin and the packaging signal, psi, promote the budding of human immunodeficiency virus type-1 (HIV-1) Microbiol. Immunol. 2004;48:111–118. doi: 10.1111/j.1348-0421.2004.tb03496.x. [DOI] [PubMed] [Google Scholar]
- 51.Balinsky C.A., Schmeisser H., Ganesan S., Singh K., Pierson T.C., Zoon K.C. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J. Virol. 2013;87:13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bender B.J., Coen D.M., Strang B.L. Dynamic and nucleolin-dependent localization of human cytomegalovirus UL84 to the periphery of viral replication compartments and nucleoli. J. Virol. 2014;88:11738–11747. doi: 10.1128/JVI.01889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y.L., Liu C.D., Cheng C.P., Zhao B., Hsu H.J., Shen C.L., Chiu S.J., Kieff E., Peng C.W. Nucleolin is important for Epstein-Barr virus nuclear antigen 1-mediated episome binding, maintenance, and transcription. Proc. Natl. Acad. Sci. U. S. A. 2014;111:243–248. doi: 10.1073/pnas.1321800111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strang B.L., Boulant S., Kirchhausen T., Coen D.M. Host cell nucleolin is required to maintain the architecture of human cytomegalovirus replication compartments. MBio. 2012;3 doi: 10.1128/mBio.00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su P.Y., Wang Y.F., Huang S.W., Lo Y.C., Wang Y.H., Wu S.R., Shieh D.B., Chen S.H., Wang J.R., Lai M., Chang C.F. Cell surface nucleolin facilitates enterovirus 71 binding and infection. J. Virol. 2015;89:4527–4538. doi: 10.1128/JVI.03498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tayyari F., Marchant D., Moraes T.J., Duan W., Mastrangelo P., Hegele R.G. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat. Med. 2011;17:1132–1135. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
- 57.Hovanessian A.G. Midkine, a cytokine that inhibits HIV infection by binding to the cell surface expressed nucleolin. Cell Res. 2006;16:174–181. doi: 10.1038/sj.cr.7310024. [DOI] [PubMed] [Google Scholar]
- 58.Nisole S., Said E.A., Mische C., Prevost M.C., Krust B., Bouvet P., Bianco A., Briand J.P., Hovanessian A.G. The anti-HIV pentameric pseudopeptide HB-19 binds the C-terminal end of nucleolin and prevents anchorage of virus particles in the plasma membrane of target cells. J. Biol. Chem. 2002;277:20877–20886. doi: 10.1074/jbc.M110024200. [DOI] [PubMed] [Google Scholar]
- 59.Said E.A., Courty J., Svab J., Delbe J., Krust B., Hovanessian A.G. Pleiotrophin inhibits HIV infection by binding the cell surface-expressed nucleolin. FEBS J. 2005;272:4646–4659. doi: 10.1111/j.1742-4658.2005.04870.x. [DOI] [PubMed] [Google Scholar]
- 60.Metifiot M., Amrane S., Mergny J.L., Andreola M.L. Anticancer molecule AS1411 exhibits low nanomolar antiviral activity against HIV-1. Biochimie. 2015;118:173–175. doi: 10.1016/j.biochi.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 61.Perrone R., Butovskaya E., Lago S., Garzino-Demo A., Pannecouque C., Palu G., Richter S.N. The G-quadruplex-forming aptamer AS1411 potently inhibits HIV-1 attachment to the host cell. Int. J. Antimicrob. Agents. 2016;47:311–316. doi: 10.1016/j.ijantimicag.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCormick-Graham M., Romero D.P. A single telomerase RNA is sufficient for the synthesis of variable telomeric DNA repeats in ciliates of the genus Paramecium. Mol. Cell. Biol. 1996;16:1871–1879. doi: 10.1128/mcb.16.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J. Structure of the human telomere in K + solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takahama K., Sugimoto C., Arai S., Kurokawa R., Oyoshi T. Loop lengths of G-quadruplex structures affect the G-quadruplex DNA binding selectivity of the RGG motif in Ewing's sarcoma. Biochemistry. 2011;50:5369–5378. doi: 10.1021/bi2003857. [DOI] [PubMed] [Google Scholar]
- 65.Dexheimer T.S., Sun D., Hurley L.H. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bugaut A., Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tippana R., Xiao W., Myong S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014;42:8106–8114. doi: 10.1093/nar/gku464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Largy E., Marchand A., Amrane S., Gabelica V., Mergny J.L. Quadruplex turncoats: cation-dependent folding and stability of quadruplex-DNA double switches. J. Am. Chem. Soc. 2016;138:2780–2792. doi: 10.1021/jacs.5b13130. [DOI] [PubMed] [Google Scholar]
- 69.Smargiasso N., Rosu F., Hsia W., Colson P., Baker E.S., Bowers M.T., De Pauw E., Gabelica V. G-quadruplex DNA assemblies: loop length, cation identity, and multimer formation. J. Am. Chem. Soc. 2008;130:10208–10216. doi: 10.1021/ja801535e. [DOI] [PubMed] [Google Scholar]
- 70.Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 72.Buscaglia R., Miller M.C., Dean W.L., Gray R.D., Lane A.N., Trent J.O., Chaires J.B. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res. 2013;41:7934–7946. doi: 10.1093/nar/gkt440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heddi B., Phan A.T. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 74.Xue Y., Kan Z.Y., Wang Q., Yao Y., Liu J., Hao Y.H., Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K + solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 75.Boncina M., Vesnaver G., Chaires J.B., Lah J. Unraveling the thermodynamics of the folding and interconversion of human telomere G-quadruplexes. Angew. Chem. 2016;55:10340–10344. doi: 10.1002/anie.201605350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miyoshi D., Nakao A., Sugimoto N. Molecular crowding regulates the structural switch of the DNA G-quadruplex. Biochemistry. 2002;41:15017–15024. doi: 10.1021/bi020412f. [DOI] [PubMed] [Google Scholar]
- 77.Dai J., Chen D., Jones R.A., Hurley L.H., Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lim K.W., Lacroix L., Yue D.J., Lim J.K., Lim J.M., Phan A.T. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010;132:12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- 79.Rankin S., Reszka A.P., Huppert J., Zloh M., Parkinson G.N., Todd A.K., Ladame S., Balasubramanian S., Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kang H.J., Cui Y., Yin H., Scheid A., Hendricks W.P., Schmidt J., Sekulic A., Kong D., Trent J.M., Gokhale V., Mao H., Hurley L.H. A pharmacological chaperone molecule induces cancer cell death by restoring tertiary DNA structures in mutant hTERT promoters. J. Am. Chem. Soc. 2016 doi: 10.1021/jacs.6b07598. [DOI] [PubMed] [Google Scholar]
- 82.Thandapani P., O'Connor T.R., Bailey T.L., Richard S. Defining the RGG/RG motif. Mol. Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 83.Ishikawa F., Matunis M.J., Dreyfuss G., Cech T.R. Nuclear proteins that bind the pre-mRNA 3' splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sutherland C., Cui Y., Mao H., Hurley L.H. A mechanosensor mechanism controls the G-quadruplex/i-motif molecular switch in the MYC promoter NHE III1. J. Am. Chem. Soc. 2016 doi: 10.1021/jacs.6b09196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.