Abstract
Background: Despite the certain contribution of metabolic and haemodynamic factors in diabetic nephropathy (DN), many lines of evidence highlight the role of immunologic and inflammatory mechanisms. To elucidate the contribution of the immune system in the development of DN, we explored the contribution of gene variants (polymorphisms) in relevant pathophysiologic pathways.
Methods: We selected six major pathways related to immune response from the Kyoto Encyclopaedia of Genes and Genomes database and thereafter we traced all available genetic association studies (GASs) involving gene variants in these pathways from PubMed and HuGE Navigator. Finally, we used meta-analytic methods for synthesizing the results of the GASs.
Results: One hundred three GASs were retrieved that included 443 variants from 75 genes. Of those variants, 138 were meta-analysed and 61 produced significant results; seven variants were investigated in single GASs and showed significant association. Variants in CCL2, CCR5, IL6, IL8, EPO, IL1A, IL1B, IL100, IL1RN, GHRL, MMP9, TGFB1, VEGFA, MMP3, MMP12, IL12RB1, PRKCE, TNF and TNFRSF19 genes were associated with an increased risk of DN.
Conclusions: There is evidence that variants related with immunologic response affect the course of DN. However, the present results should be interpreted with caution since the current number of available GASs is limited.
Keywords: association, diabetic nephropathy, inflammation, meta-analysis, pathway
Introduction
Diabetic nephropathy (DN) is a major microvascular complication in the course of both type 1 (T1DM) and type 2 diabetes mellitus (T2DM). It is the leading cause of end-stage renal disease (ESRD) in the Western world and is the primary cause of morbidity and mortality in diabetic patients [1, 2], accounting for 25–40% of incident patients [3]. DN, characterized by high blood pressure, proteinuria, a progressive decline in renal function and an increased risk of cardiovascular disease, is becoming more and more prevalent to the extent that it has reached epidemic proportions. Worldwide, ∼347 million people have diabetes and this number is expected to increase to 430 million by 2030 [4].
Despite extensive study of the cellular mechanisms of DN and the benefits of current therapeutics, alarming proportions of diabetic patients are still reaching ESRD. Wide variation is attributed to both race and gender, with Caucasians and males having the lowest rates compared with African Americans and females, respectively. Reduction of overall rates remains an unfulfilled target, although it has been achieved by some subgroups, including Caucasians, females and patients ≤44 years of age (http://www.usrds.org/2014/view/v2_02.aspx). Therefore, reducing the occurrence of DN remains a challenge for nephrologists and there is an urgent need for the investigation of new molecular targets.
DN is believed to be a multifactorial disease that results from the effects of glucose-dependent and genetic factors, blood pressure, vasoactive hormones such as angiotensin II and other environmental factors, which combine in a complicated synergistic way [5]. Familial clustering of DN, as well as racial variation, indicates the importance of genetic factors in the development of DN, regardless of the type of diabetes [6–10].
Despite the known contribution of genetic determinants, DN is caused by a complex interplay of metabolic and haemodynamic factors. Although DN is considered to be a non-immune disease, many lines of evidence indicate a principal role for immunological and inflammatory mechanisms in its occurrence and progression [11, 12].
Inflammation constitutes a physiological response to injury and infection and is necessary for tissue healing. The hallmark symptom of DN, proteinuria, is not only evidence of underlying glomerular injury, but also contributes to further tubular and interstitial damage due to the abnormal filtration of proteins and the activation of cellular responses to renal injury itself [13, 14]. In vitro studies using proximal tubular cells have shown that the plasma protein load triggers the synthesis of endothelin-1, monocyte chemoattractant protein-1 and RANTES (Regulated on Activation, Normal T Cell Expressed and Secreted), which attract monocytes/macrophages and T lymphocytes [15–17], as well as albumin-upregulated tubular gene expression and the production of interleukin 8 (IL-8) [18]. Cytokine secretion by tubular epithelial cells is upregulated by the presence of high molecular weight proteins and cytokines in the glomerular filtrate. Intersitial inflammation and fibrosis may also occur as a result of alterations in blood flow and cytokine secretion by tubular epithelial cells [19]. In addition, the synthesis of proinflammatory cytokines occurs in several types of renal cells (glomerular, endothelial, tubular and mesangial), as well as in monocytes, macrophages and T cells [20]. Accumulating evidence also suggests that individuals who progress to diabetes display features of inflammation years before the onset of the disease [21], and population-based studies indicate that inflammatory parameters are strong predictors of the development of diabetes [22].
The crucial role of inflammatory mechanisms in the development of DN is reflected by the anti-inflammatory actions of current therapeutics. Blockade of the renin–angiotensin–aldosterone system (RAAS) provides anti-inflammatory actions that are potentially relevant to the therapeutic approach [23–25]. Utimura et al. [26] demonstrated that mycophenolate mofetil prevents the development of albuminuria and glomerular injury in experimental DN, which is unrelated to effects on glomerular haemodynamics or the improvement of metabolic control. Other studies have reported the effect of the chimeric anti-tumour necrosis factor α(TNF-α) antibody infliximab and the agent pentoxifylline as having significant anti-inflammatory properties, indicating the importance of TNF-α modulation [27]. Inflammatory cells (mainly macrophages) are also seen in the glomeruli and interstitium of patients with DN, indicating a contribution of inflammation in the development of DN [28]. Thus the key features of inflammation are seen in DN, although to a mild extent compared with classic inflammatory diseases [29].
Materials and methods
Identification and eligibility of relevant studies
In an effort to elucidate the contribution of the immune system in the development and progression of DN, we conducted a candidate pathway analysis focusing on genes involved in six pathways, as classified by the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database (Box 1). The meanings of the acronyms of statistically significant variants are shown in Supplementary data, Table S1. In the meta-analysis, we included up-to-date genetic association studies (GASs) concerning genes in the aforementioned KEGG pathways, and a few others with either strong affinity regarding their function or their relatedness to the other genes (Supplementary data, Table S2).
Box 1. Pathways involved in immune responses in KEGG regarding diabetic nephropathy
| Cytokine–cytokine receptor interaction |
| BMP2, BMP4, BMP7, CCL2, CCL5, CCR5, EPO, GHR, IL10, IL1B, IL1R1, IL4, IL6, IL6R, IL8, LTA, TGFB1, TGFBR1, TGFBR2, TNF, TNFRSF1A, TNFRSF1B, VEGA |
| Antigen processing and presentation |
| HLA-A, HLA-B, HLA-DQA1, HLA-DQB1, HLA-DRB1, HSPA1A, HSPA1B, HSPA1L, LTA |
| Natural killer cell-mediated cytotoxicity |
| BID, CASP3, HLA-A, HLA-B, ICAM1, NFAT5, PRKCB, TNF |
| T cell receptor signalling pathway |
| CTLA4, IL10, IL4, MALT1, NFAT5, NFKB1, TNF |
| B cell receptor signalling pathway |
| GSK3B, MALT1, NFAT5, NFKB1, PRKCB |
| Leucocyte transendothelial migration |
| GSK3B, MALT1, NFAT5, NFKB1, PRKCB |
| Other |
| GHRL, IL1RN, CCR2, ICAM4, CASP12, NOX4, NOX3, PRKCE, PRKCSH, MMP1, MMP3, MMP7, MMP8, MMP10, MMP11, MMP12, MMP13, MMP14, MMP15, MMP16, MMP17, MMP19, MMP20, MMP24, MMP25, MMP26, MMP27, MMP28, TNFRSF9, TNFRSF19 |
The included studies were published in English and were recorded in HuGE (human genome epidemiology) Phenopedia (last update of database on 1 July 2015) regarding the disease term ‘diabetic nephropathies’. We also retrieved articles from genome-wide association studies (GWASs) in HuGE Pub Lit (published literature database in human genome epidemiology) and the National Human Genome Research Institute (NHGRI) Catalogue of Published Genome-Wide Association Studies (http://www.genome.gov/gwastudies/). Meta-analyses of the included genes were also screened. We searched PubMed with search terms such as ‘diabetic nephropathy’ AND ‘association’ AND (‘gene symbol’ OR ‘gene name’) (accessed on 1 July 2015) and the abstracts of the selected articles were screened to assess their eligibility. Finally, references in the eligible articles were retrieved to identify articles not indexed in PubMed or HuGE Navigator.
The inclusion criteria that studies had to meet were (i) they involved cases with persistent micro-/macroalbuminuria with or without diabetic retinopathy; (ii) they involved diseased controls with diabetes and normoalbuminuria or normal renal function, and/or healthy controls; (iii) they provided full genotypic data either via genotype counts or allele frequencies, with articles presenting results after merging genotypes being excluded and (iv) they included human subjects. The diabetes could be either T1DM or T2DM.
Studies investigating disease progression, severity, phenotype modification, response to treatment or survival were excluded. Case reports, editorials, reviews and non-English articles were also excluded, as well as studies with other study designs, such as family-based studies. The eligibility of the articles was assessed independently by two investigators (M.T. and E.Z.), the results were compared and any disagreements were resolved by reaching consensus.
Data extraction
The following information was extracted from each article: first author, year of publication, ethnicity, PubMed unique identifier, type of diabetes, country and phenotype. For cases and controls, we recorded their number, duration of diabetes, the selection criteria and the implementation of matching criteria. With regard to genotypic data, we extracted (if available) the full genotype counts or allele frequencies.
Data synthesis and analysis
The association between genotype distribution and DN was examined using the generalized linear odds ratio (ORG) [30, 31]. The ORG expresses the probability of a subject being more diseased relative to the probability of being less diseased, given that diseased subjects have a higher mutational load. Explicitly, the ORG shows how many cases/healthy control pairs exist in the study, for which the cases have a larger mutational load relative to the number of pairs for which the healthy controls have a larger mutational load; alternatively, ORG indicates whether the mutational load of a variant is implicated in disease susceptibility [30, 31].
ORG was calculated for each variant with available genotypic counts. For the variants with available allele frequencies, the examined model was the allele contrast. The threshold for meta-analysis was the presence of two studies. The pooled OR was estimated using the DerSimonian and Laird [32] random effects model. The associations are presented with ORs, either generalized for genotypic data or pooled for allelic data, with corresponding 95% confidence intervals (CIs). We tested for between-study heterogeneity with Cochran’s Q statistic (considered statistically significant at P < 0.10) and assessed its extent with the I2 statistic, which is independent of the number of studies in the meta-analysis and takes values between 0 and 100%, with higher values denoting a greater degree of heterogeneity [33, 34]. ORG was calculated using a software for implementing the generalized odds ratio methodology for the analysis and meta-analysis of GAS (ORGGASMA) (http://biomath.med.uth.gr) [31].
For each study, we examined whether controls confronted with Hardy–Weinberg equilibrium (HWE) predicted genotypes using Fisher’s exact test. For studies providing only allele counts, we relied on the authors’ assessment of deviations from HWE. We also tested for small-study effects with the Egger test [35].
Results
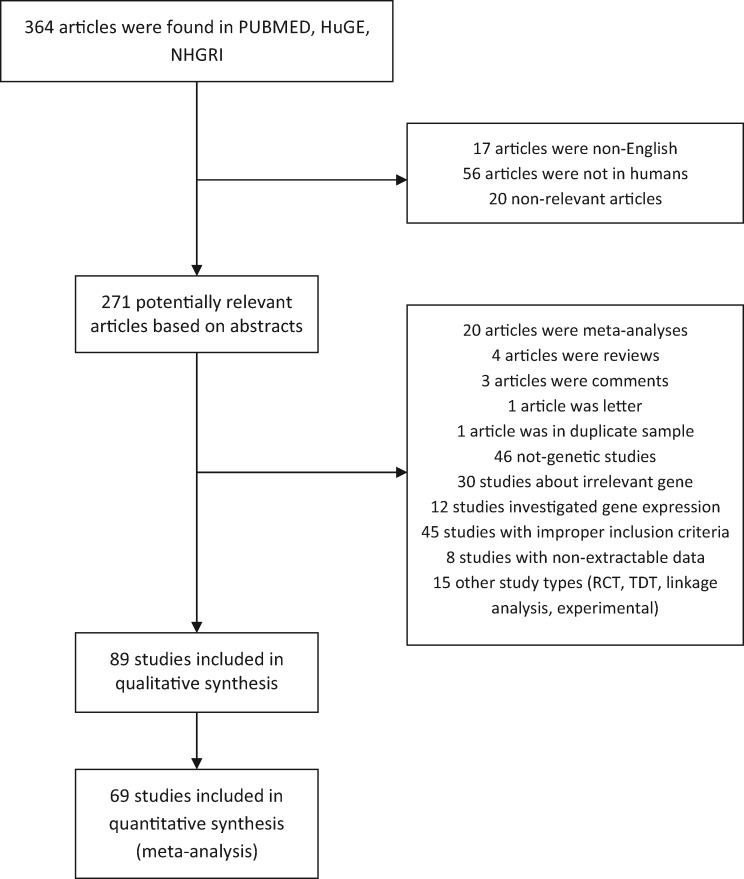
The literature review identified 365 titles. We independently read the abstracts of the articles and their references to assess their eligibility for the meta-analysis, as well as the published meta-analyses for the relevant genes. The results were compared and disagreements were resolved by consensus. The full articles for the remaining studies were evaluated for compliance with the inclusion criteria. When an article provided data for different populations, then each population was considered as a different study. Data from 89 articles representing 103 studies were included in the qualitative synthesis and 68 articles in the quantitative synthesis. Figure 1 presents a flow chart of retrieved articles, as well as excluded articles with and the reasons for exclusion. Overall, 75 candidate genes from 6 candidate pathways and 443 polymorphisms harboured in these genes were investigated in 103 gene–disease association studies that were identified in the field of DN. The characteristics of each study and their references are shown in Supplementary data, Table S3. The studies involved 12 148 healthy controls, 26 215 diseased controls and 28 520 cases. The studies were published between 1996 and 2015.
Fig. 1.
Flow chart showing how studies were selected for meta-analysis.
Study characteristics and association results
Studies were conducted in various populations of different racial descent: 54 studies involved Caucasians, 37 studies recruited Asians (of which 22 included East Asians), 8 studies involved Africans and 4 studies were conducted in ethnically mixed populations. The mean age of the cases in the individual studies was 54.5 years, while the mean ages of the diseased and healthy controls were 53.4 and 51.6 years, respectively. The mean diabetes duration of cases was 19.9 years and of diseased controls was 18.6 years. The distribution of genotypes in the control group departed from HWE in 25 studies, while for 19 studies there were not enough data to test the compliance with HWE.
In the analyses in which ORG was calculated, statistically significant results were revealed for the variants of interleukin 1 beta (IL1B), heat shock protein family A (Hsp70) member 1A (HSPA1A) (two variants), heat shock protein family A (Hsp70) member 1B (HSPA1B), BH3 interacting domain death agonist (BID), protein kinase C substrate 80K-H (PRKCSH), tumor necrosis factor (TNF) and marginally for protein kinase C beta (PRKCB), in cases versus diseased controls. In analyses with three groups (healthy controls, diseased controls, cases), statistically significant associations were reported for C-C motif chemokine receptor 5 (CCR5), intercellular adhesion molecule 1 (ICAM1), interleukin 6 (IL6), heat shock protein family A (Hsp70) member 1A (HSPA1A) (two variants), heat shock protein family A (Hsp70) member 1B (HSPA1B), interleukin 1 beta (IL1B), cytochrome b-245 alpha chain (CYBA) and interleukin 1 alpha (IL1A). In cases versus healthy controls, statistically significant associations were reported for ICAM1, HSPA1A (two variants), HSPA1B, IL1B, CYBA and ghrelin and obestatin prepropeptide (GHRL), and marginally for TNF and IL1A.
In the allele contrast model of cases versus diseased controls, significant associations were observed for glycogen synthase kinase 3 beta (GSK3B) (two variants), IL1B, interleukin 6 receptor (IL6R) (two variants), MALT1 paracaspase (MALT1), nuclear factor of activated T-cells 5 (NFAT5) (two variants), transforming growth factor beta 2 (TGFB2), TNF, TGFB1, matrix metallopeptidase 1 (MMP1) (three variants), MMP2, MMP20 (two variants), MMP17, MMP16 (seven variants), MMP10, MMP8, NADPH oxidase 1 (NOX1) and MMP9, whereas in the allele contrast model of cases versus healthy controls, significant associations were observed for IL6R, MALT1 and major histocompatibility complex, class II, DQ alpha 1 (HLA-DQA1).
Meta-analysis results
In total, 138 variants were investigated in two or more studies and their results were subject to meta-analysis. Table 1 shows the statistically significant results of meta-analyses exploring the presence of association between the relevant genetic variants and DN based on genotype counts. The statistically significant results of meta-analyses exploring the presence of association between the genetic variants and DN based on allele counts are presented in Supplementary data, Table S4. However, for completeness, all the results are presented in Supplementary Tables S5–S9 regardless of evidence of statistical significance. The meaning of the acronyms of statistically significant variants are shown in Supplementary data, Table S1. Figures 2–4 are forest plot representations of the genetic variants significantly associated with DN. The references of the included studies per single- nucleotide polymorphism are presented in Supplementary data, Tables S10–S13.
Table 1.
Statistically significant results from meta-analyses based on genotype counts
| Diseased controls versus cases | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Variant | RS | Studies (n) | Cases/controls (n) | RE ORG (95% CI) | I2 (%) | PQ | PE |
| CCL2 | Insertion/deletion | rs3917887 | 2 | 336/347 | 2.08 (1.61–2.68) | 0.00 | 0.32 | NA |
| CCR5 | −59029 A>G | rs1799987 | 8 | 2125/2127 | 0.69 (0.53–0.91 | 83.21 | <0.01 | 0.04 |
| All in HWE | 6 | 1789/1780 | 0.79 (0.60–1.03) | 80.27 | <0.01 | 0.07 | ||
| IL6 | −634 C>G | rs1800796 | 2 | 580/332 | 1.30 (0.86–1.96) | 39.75 | 0.20 | NA |
| All in HWE | 1 | 292/162 | 1.56 (1.05–2.32) | NA | NA | NA | ||
| IL8 | −251 T>A | rs4073 | 2 | 336/347 | 1.60 (1.23–2.09) | 0.00 | 0.54 | NA |
| All in HWE | 1 | 240/255 | 1.52 (1.11–2.08) | NA | NA | NA | ||
| EPO | G>T | rs1617640 | 3 | 1618/954 | 1.64 (1.43–1.89) | 0.00 | 0.78 | 0.03 |
| IL1B | C-511T | rs16944 | 2 | 268/312 | 1.77 (1.32–2.37) | 0.00 | 0.88 | NA |
| IL10 | C-819T | rs1800871 | 2 | 539/425 | 0.73 (0.58–0.93) | 0.00 | 0.83 | NA |
| All in HWE | 1 | 515/402 | 0.73 (0.58–0.93) | NA | NA | NA | ||
| GHRL | Leu72Met | rs696217 | 2 | 307/297 | 0.44 (0.24–0.81) | 51.73 | 0.15 | NA |
| MMP9 | Arg279Gln | rs17576 | 2 | 336/347 | 1.95 (1.51–2.52) | 0.00 | 0.74 | NA |
| IL1A | C-889T | rs1800587 | 2 | 283/310 | 2.09 (1.49–2.92) | 0.00 | 0.44 | NA |
| Healthy controls versus cases | ||||||||
| CCR5 | −59029 A>G | rs1799987 | 2 | 643/793 | 0.52 (0.41–0.66) | 30.43 | 0.23 | NA |
| IL10 | C-819T | rs1800871 | 2 | 539/773 | 1.35 (1.09–1.68) | 0.00 | 0.93 | NA |
| All in HWE | 1 | 515/748 | 1.35 (1.08–1.68) | NA | NA | NA | ||
| IL1RN | 86 bp VNTR | – | 2 | 40/261 | 3.41 (1.93–6.00) | 0.00 | 0.56 | NA |
| TGFB1 | T869C | rs1800470 | 6 | 814/1450 | 1.30 (0.86–1.96) | 83.64 | <0.01 | 0.18 |
| All in HWE | 4 | 706/1103 | 1.73 (1.46–2.04) | 0.00 | 0.41 | 0.21 | ||
| TGFB1 | −509 (C>T) | rs1800469 | 2 | 600/915 | 1.33 (1.10–1.60) | 0.00 | 0.48 | NA |
| Healthy controls versus diseased controls versus cases | ||||||||
| IL10 | C-819T | rs1800871 | 2 | 539/425/773 | 1.29 (1.11–1.50) | 0.00 | 0.97 | NA |
| All in HWE | 1 | 515/402/748 | 1.29 (1.11–1.50) | NA | NA | NA | ||
| TGFB1 | T869C | rs1800470 | 5 | 770/787/1332 | 1.36 (1.08–1.70) | 73.18 | 0.01 | 0.24 |
| All in HWE | 4 | 1.47 (1.24–1.75) | 45.66 | 0.14 | 0.45 | |||
| TGFB1 | −509 (C>T) | rs1800469 | 2 | 600/612/915 | 1.22 (1.07–1.39) | 0.00 | 0.65 | NA |
NA: not applicable.
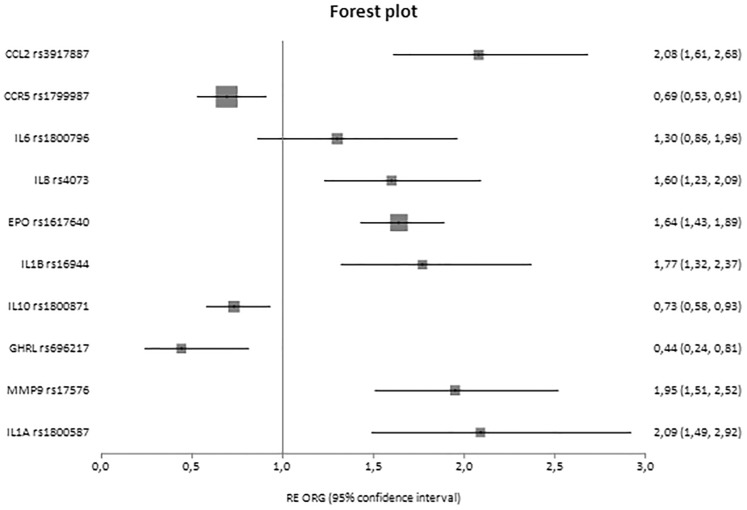
Fig. 3.
Forest plot of healthy controls versus cases displaying only significant results based on genotype counts.
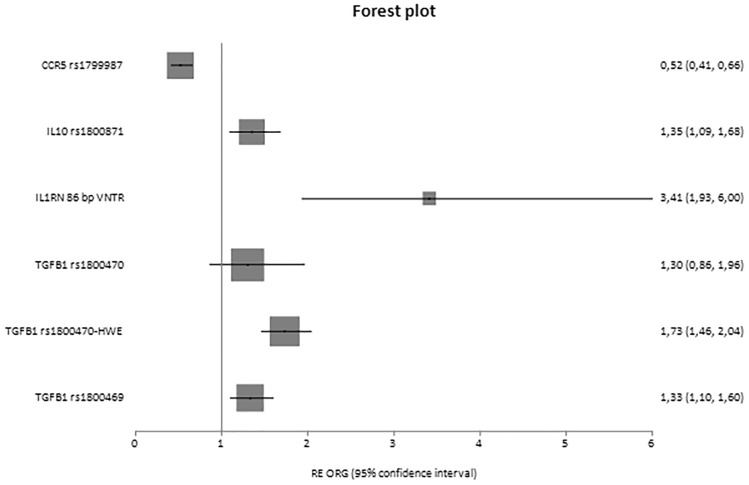
Fig. 2.
Forest plot of diseased controls and cases displaying only significant results based on genotype counts.
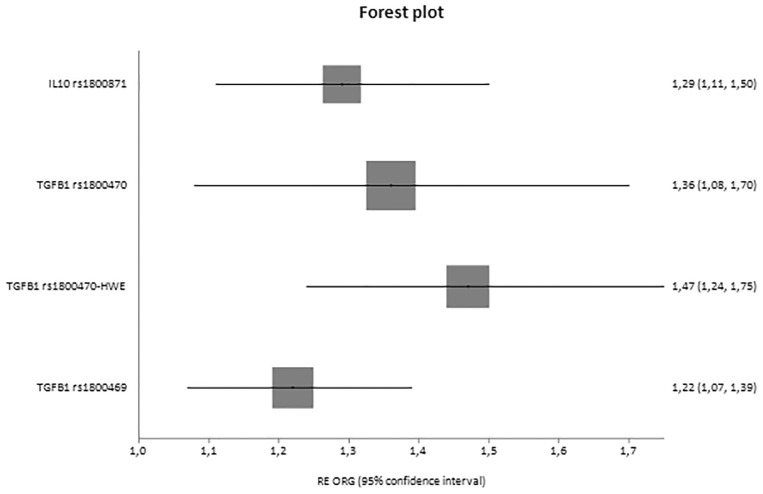
Fig. 4.
Forest plot of healthy controls versus diseased controls versus cases displaying only significant results based on genotype counts.
In analyses based on genotypes in which ORG was calculated, significant results were shown for the variants of C-C motif chemokine ligand 2 (CCL2) rs3917887 [ORG = 2.08 (95% CI 1.61–2.68)], CCR5 rs1799987 [ORG = 0.69 (95% CI 0.53–0.91)], IL6 rs1800796 [ORG = 1.56 (95% CI 1.05–2.32)], IL8 rs4073 [ORG = 1.60 (95% CI 1.23–2.09)], erythropoietin (EPO) rs1617640 [ORG = 1.64 (95% CI 1.43–1.89), IL1B rs16944 [ORG = 1.77 (95% CI 1.32–2.37)], IL10 rs1800871 [ORG = 0.73 (95% CI 0.58–0.93)], GHRL rs696217 [ORG = 0.44 (95% CI 0.24–0.81)] and MMP9 rs17576 [ORG = 1.95 (95% CI 1.51–2.52)], as well as IL1A rs1800587 [ORG = 2.09 (95% CI 1.49–2.92)] in cases versus diseased controls.
In analyses with three groups, significant associations were reported for IL10 rs1800871 [ORG = 1.29 (95% CI 1.11–1.50)] and TGFB1 rs1800470 [ORG = 1.47 (95% CI 1.24–1.75)] and rs1800469 [ORG = 1.22 (95% CI 1.07–1.39)], and in cases versus healthy controls, CCR5 rs1799987 [ORG = 0.52 (95% CI 0.41–0.66)], IL10 rs1800871 [ORG = 1.35 (95% CI 1.09–1.68)], TGFB1 rs1800470 [ORG = 1.73 (95% CI 1.46–2.04)], TGFB1 rs1800469 [ORG = 1.33 (95% CI 1.10–1.60)] and interleukin 1 receptor antagonist (IL1RN) 86 bp VNTR in intron 2 [ORG = 3.41 (95% CI 1.93–6.00)].
Regarding the allele contrast model and the statistically significant results, significant associations were observed for IL1RN, vascular endothelial growth factor A (VEGFA), MMP3 (32 variants) and MMP12 (18 variants) in comparisons using cases versus diseased controls and interleukin 12 receptor subunit beta 1 (IL12RB1), protein kinase C epsilon (PRKCE) and TNFRSF19 in comparisons using cases versus healthy controls (Supplementary data, Table S4).
However, in general these results were based on a small number of studies (2–11) and therefore they should be interpreted with caution.
GWAS
Of the 15 available GWASs for DN, only 2 were included in the meta-analysis. The other studies either mainly referred to irrelevant genes or did not provide extractable data.
Potential bias
The sensitivity analyses excluding studies not conforming to HWE altered the pattern of results for CCR5 (rs1799987) and IL6 (rs1800796) in cases versus diseased controls and for TGFB1 (rs1800470) in cases versus healthy controls. No differential magnitude of effect in large versus small studies was detected for the variants except for those harboured in TGFB1 (rs1800469) in the analysis of diseased controls versus cases and in VEGFB (rs12366035) and IL12A (rs583911, IL12A_6489, rs2243135) in an allele contrast model in cases versus diseased controls. Concerning the heterogeneity across studies, the random effects model was implemented in 33 of 77 meta-analyses with full genotypic data; significant heterogeneity was found.
Discussion
In this systematic review, 443 polymorphisms from 75 candidate genes belonging to 6 candidate pathways were synthesized. Meta-analysis of several individual genetic variants in relation to DN was performed previously; however, this systematic review constitutes the most comprehensive overview to date, assessing all genetic variants that are associated with inflammation and the immune response in DN according to KEGG pathways.
In total, 138 variants were meta-analysed. The key finding of our study was that polymorphisms in CCL2, CCR5, IL6, IL8, EPO, IL1A, IL1B, IL10, IL1RN, GHRL, MMP9, TGFB1 (2 variants), VEGFA, MMP3 (31 variants), MMP12 (18 variants), IL12RB1, PRKCE, TNF (2 variants) and TNFRSF19 gave significant results, 7 of which were results from single studies. These results support the contribution of inflammation and the immune response in the pathogenesis of DN. Functional studies remain to be performed to elucidate the exact roles of these genetic variants.
To the best of our knowledge, our analysis provides the highest level of evidence for a contribution of the immune system and inflammation in DN. However, it is worth noting that Nazir et al. [36] explored the association between 18 genes involved in inflammatory cytokines and angiogenesis pathways with DN, where 11 genetic variants in or near 9 genes revealed statistically significant associations with DN.
Our analysis has several strengths. Regarding phenotype definition, the ideal would have been the inclusion of patients with macroalbuminuria only, but this would have meant the exclusion of many studies. In order to guarantee adequacy regarding phenotype definition, we only included cases where subjects had microalbuminuria that was characterized as persistent, because microalbuminuria that is not persistent can be reversible and this could lead to underestimation of the genetic effects. Furthermore, all of the different combinations of groups were compared in order to make the trait discrimination feasible, to reveal which genes are responsible for DN independent of diabetic predisposition, because any GAS examining the risk of developing DN in either type of diabetes might be readily confounded if the genetic factors under investigation were also predisposing to diabetes. For instance, in the case of IL1RN, the most significant association was detected when healthy controls were compared with cases [OR = 3.41 (95% CI 1.93–6.00)], whereas comparison of diseased controls and cases detected no association [OR = 2.16 (95% CI 1.00–4.65)]; this suggests that this variant does not need the diabetic milieu to exercise its influence and that inflammation could be an inherent condition in DN.
However, our analysis also has some limitations. Publication bias is a concern in all meta-analyses. In this analysis, only studies published in journals were included. Studies with negative results are not usually published, leading to an overestimation of genetic effects. Furthermore, the results should be interpreted with caution since both the number of studies in each meta-analysis and the sample sizes of some studies were small. However, these methodological issues are present in the literature of GASs in DN. Failure to account for haplotypic structure or ethnicity/gender-specific interactions between genetic polymorphisms and environmental factors may have also contributed to the pattern of results observed. This may reflect the divergent genetic background of different ethnic and disease groups. The type of diabetes should also be taken into consideration, as findings of renal biopsy revealed more structural heterogeneity in patients with T2DM. These lines of evidence suggest that the genetic background of DN secondary to T2DM is more complicated compared with DN due to T1DM [37]. Many conditions contribute to this complexity, such as the fact that T2DM occurs more in elderly people than T1DM, is accompanied by other comorbidities and that environmental factors have more time to affect the phenotype. Finally, in the findings of the present synopsis, bias may have been introduced since it solely included variants of the inflammatory pathways and thus the impact of the current (or other additional) variants in different related pathways has not been examined.
The studies included in the present systematic review showed heterogeneity in terms of ethnicity, type of diabetes, study design and sample size that could affect the results. More specifically, some studies selected healthy unrelated populations instead of uncomplicated diabetic patients as control groups, as well as the diabetic controls possibly having diabetic retinopathy in a few studies. This disease was the only one accepted in diseased controls, because in this case the evidence of non-susceptibility in DN is stronger than in the case of completely uncomplicated diabetic patients. The type of diabetes should also be taken into consideration. For some of the meta-analyses, the clinical heterogeneity was accompanied by statistical heterogeneity with an I2 statistic of up to 96%. To account for potential heterogeneity, a random effects model was performed by default. Subgroup analyses according to diabetes type and ethnicity were not performed due to the small number of studies in these meta-analyses.
DN is a complex disease that involves epistatic and gene–environment interactions. Therefore, a single type of genetic study, such as candidate gene association studies, has a reduced likelihood of yielding conclusive inferences. Hypothesis-free studies such as GWASs [38, 39], microarrays, gene expression analyses [40] and whole-genome linkage scans [41, 42] may assist in providing more conclusive evidence regarding the contribution of genetics in DN. This can be achieved by examining the genomic convergence of these different types of studies [39]. Although GWASs represent a superior strategy for unravelling genetic complexity, the findings of candidate gene association studies may be supportive in replicating existing evidence and in revealing genuine genetic effects that could merit prioritization in future studies. Furthermore, GWASs themselves lack replication; therefore, replication of their findings from different investigators and different methodologies is essential for the interpretation of the mass of associations likely to result from GWASs [38, 43, 44].
Conclusion
In summary, the overall meta-analyses showed a significant role for 66 variants in 18 genes associated with advanced DN. However, these positive associations resulted from pooling a small number of studies, therefore these results must be interpreted with caution. In a polygenic complex disorder like DN, association of individual polymorphisms in genes may be small and sometimes non-informative, whereas combinations of specific genotypes may be more relevant. However, few studies have examined multiple alleles simultaneously for determining the risk of DN. Functional studies have yet to establish the role of genes highlighted in the present systematic review. Nonetheless, the practical implications of the hypothesis-generating findings of the current meta-analysis for practicing clinicians are unknown for the time being and yet to be defined. Clearly, DN is not a classic inflammatory disease, although the key features of inflammation are seen to be quite mild compared with classic inflammatory diseases. Furthermore, it should be emphasized that immunosuppressive therapy has been shown to be of no benefit in clinical practice for DM nephropathy, and this is the reason for the clear lack of immunomodulating therapy in DN. Nonetheless, the current therapies exert anti-inflammatory effects; for instance, blockade of the RAAS, which is a treatment for DN, also has a pleiotropic anti-inflammatory effect. Although the findings of the present meta-analysis are not applicable to clinical practice, they contribute to a better understanding of the pathophysiology of DN and therefore may be used to improve current therapy strategies and initiate the development of new treatments targeting inflammation at any stage of the disease.
Supplementary Data
Supplementary data are available online at http://ckj.oxfordjournals.org
Supplementary Material
Conflicts of interest
None declared.
References
- 1. Kato M, Arce L, Natarajan R.. MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol 2009; 4: 1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kanwar YS, Wada J, Sun L. et al. Diabetic nephropathy: mechanisms of renal disease progression. Exp Biol Med 2008; 233: 4–11 [DOI] [PubMed] [Google Scholar]
- 3. Collins AJ, Foley RN, Chavers B. et al. US Renal Data System 2013 Annual Data Report. Am J Kidney Dis 2014; 63: A7. [DOI] [PubMed] [Google Scholar]
- 4. Fernandez-Fernandez B, Ortiz A, Gomez-Guerrero C. et al. Therapeutic approaches to diabetic nephropathy–beyond the RAS. Nat Rev Nephrol 2014; 10: 325–46 [DOI] [PubMed] [Google Scholar]
- 5. Ravid M, Brosh D, Ravid-Safran D. et al. Main risk factors for nephropathy in type 2 diabetes mellitus are plasma cholesterol levels, mean blood pressure, and hyperglycemia. Arch Intern Med 1998; 158: 998–1004 [DOI] [PubMed] [Google Scholar]
- 6. Seaquist ER, Goetz FC, Rich S. et al. Familial clustering of diabetic kidney disease. Evidence for genetic susceptibility to diabetic nephropathy. N Engl J Med 1989; 320: 1161–1165 [DOI] [PubMed] [Google Scholar]
- 7. Cowie CC, Port FK, Wolfe RA. et al. Disparities in incidence of diabetic end-stage renal disease according to race and type of diabetes. N Engl J Med 1989; 321: 1074–1079 [DOI] [PubMed] [Google Scholar]
- 8. Pettitt DJ, Saad MF, Bennett PH. et al. Familial predisposition to renal disease in two generations of Pima Indians with type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia 1990; 33: 438–443 [DOI] [PubMed] [Google Scholar]
- 9. Freedman BI, Tuttle AB, Spray BJ.. Familial predisposition to nephropathy in African-Americans with non-insulin-dependent diabetes mellitus. Am J Kidney Dis 1995; 25: 710–3 [DOI] [PubMed] [Google Scholar]
- 10. Quinn M, Angelico MC, Warram JH. et al. Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia 1996; 39: 940–945 [DOI] [PubMed] [Google Scholar]
- 11. Tuttle KR. Linking metabolism and immunology: diabetic nephropathy is an inflammatory disease. J Am Soc Nephrol 2005; 16: 1537–1538 [DOI] [PubMed] [Google Scholar]
- 12. Mora C, Navarro JF.. Inflammation and diabetic nephropathy. Curr Diab Rep 2006; 6: 463–468 [DOI] [PubMed] [Google Scholar]
- 13. Zandi-Nejad K, Eddy A.. Why is proteinuria an ominous biomarker of progressive kidney disease? Kidney Int 2004; 66: 76–89 [DOI] [PubMed] [Google Scholar]
- 14. Abbate M, Zoja C, Remuzzi G.. How does proteinuria cause progressive renal damage? J Am Soc Nephrol 2006; 17: 2974–2984 [DOI] [PubMed] [Google Scholar]
- 15. Zoja C, Morigi M, Figliuzzi M. et al. Proximal tubular cell synthesis and secretion of endothelin-1 on challenge with albumin and other proteins. Am J Kidney Dis 1995; 26: 934–941 [DOI] [PubMed] [Google Scholar]
- 16. Wang Y, Chen J, Chen L. et al. Induction of monocyte chemoattractant protein-1 in proximal tubule cells by urinary protein. J Am Soc Nephrol 1997; 8: 1537–1545 [DOI] [PubMed] [Google Scholar]
- 17. Zoja C, Donadelli R, Colleoni S. et al. Protein overload stimulates RANTES production by proximal tubular cells depending on NF-kappa B activation. Kidney Int 1998; 53: 1608–1615 [DOI] [PubMed] [Google Scholar]
- 18. Tang S, Leung JCK, Abe K. et al. Albumin stimulates interleukin-8 expression in proximal tubular epithelial cells in vitro and in vivo. J Clin Invest 2003; 111: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rutledge JC, Ng KF, Aung HH. et al. Role of triglyceride-rich lipoproteins in diabetic nephropathy. Nat Rev Nephrol 2010; 6: 361–370 [DOI] [PubMed] [Google Scholar]
- 20. Sugimoto H, Shikata K, Wada J. et al. Advanced glycation end products-cytokine-nitric oxide sequence pathway in the development of diabetic nephropathy: aminoguanidine ameliorates the overexpression of tumour necrosis factor-alpha and inducible nitric oxide synthase in diabetic rat glomeruli. Diabetologia 1999; 42: 878–886 [DOI] [PubMed] [Google Scholar]
- 21. Bloomgarden ZT. Inflammation and insulin resistance. Diabetes Care 2003; 26: 1922–1926 [DOI] [PubMed] [Google Scholar]
- 22. Spranger J, Kroke A, Mohlig M. et al. Inflammatory cytokines and the risk to develop type 2 diabetes: results of the prospective population-based European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Diabetes 2003; 52: 812–817 [DOI] [PubMed] [Google Scholar]
- 23. Dagenais NJ, Jamali F.. Protective effects of angiotensin II interruption: evidence for antiinflammatory actions. Pharmacotherapy 2005; 25: 1213–1229 [DOI] [PubMed] [Google Scholar]
- 24. Dandona P, Dhindsa S, Ghanim H. et al. Angiotensin II and inflammation: the effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockade. J Hum Hypertens 2007; 21: 20–27 [DOI] [PubMed] [Google Scholar]
- 25. Takebayashi K, Matsumoto S, Aso Y. et al. Aldosterone blockade attenuates urinary monocyte chemoattractant protein-1 and oxidative stress in patients with type 2 diabetes complicated by diabetic nephropathy. J Clin Endocrinol Metab 2006; 91: 2214–2217 [DOI] [PubMed] [Google Scholar]
- 26. Utimura R, Fujihara CK, Mattar AL. et al. Mycophenolate mofetil prevents the development of glomerular injury in experimental diabetes. Kidney Int 2003; 63: 209–216 [DOI] [PubMed] [Google Scholar]
- 27. Moriwaki Y, Inokuchi T, Yamamoto A. et al. Effect of TNF-alpha inhibition on urinary albumin excretion in experimental diabetic rats. Acta Diabetol 2007; 44: 215–218 [DOI] [PubMed] [Google Scholar]
- 28. Shikata K, Makino H.. Role of macrophages in the pathogenesis of diabetic nephropathy. Contrib Nephrol 2001; 134: 46–54 [DOI] [PubMed] [Google Scholar]
- 29. Shikata K, Makino H.. Microinflammation in the pathogenesis of diabetic nephropathy. J Diabetes Investig 2013; 4: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zintzaras E. The power of generalized odds ratio in assessing association in genetic studies. J Appl Stat 2012; 39: 2569–2581 [Google Scholar]
- 31. Zintzaras E. The generalized odds ratio as a measure of genetic risk effect in the analysis and meta-analysis of association studies. Stat Appl Genet Mol Biol 2010; 9: article 21 [DOI] [PubMed] [Google Scholar]
- 32. DerSimonian R, Laird N.. Meta-analysis in clinical trials. Control Clin Trials 1986; 7: 177–188 [DOI] [PubMed] [Google Scholar]
- 33. Cochran WG. The combination of estimates from different experiments. Biometrics 1954; 10: 101–129 [Google Scholar]
- 34. Higgins JPT, Thompson SG.. Quantifying heterogeneity in a meta-analysis. Stat Med 2002; 21: 1539–1558 [DOI] [PubMed] [Google Scholar]
- 35. Egger M, Davey Smith G, Schneider M. et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315: 629–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nazir N, Siddiqui K, Al-Qasim S. et al. Meta-analysis of diabetic nephropathy associated genetic variants in inflammation and angiogenesis involved in different biochemical pathways. BMC Med Genet 2014; 15: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fioretto P, Mauer M.. Histopathology of diabetic nephropathy. Semin Nephrol 2007; 27: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zintzaras E, Lau J.. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol 2008; 61: 634–645 [DOI] [PubMed] [Google Scholar]
- 39. Kitsios GD, Zintzaras E.. Genomic convergence of genome-wide investigations for complex traits. Ann Hum Genet 2009; 73: 514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zintzaras E, Ioannidis JP.. Meta-analysis for ranked discovery datasets: theoretical framework and empirical demonstration for microarrays. Comput Biol Chem 2008; 32: 38–46 [DOI] [PubMed] [Google Scholar]
- 41. Zintzaras E, Ioannidis JP.. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol 2005; 28: 123–137 [DOI] [PubMed] [Google Scholar]
- 42. Zintzaras E, Ioannidis JP. . HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics 2005; 21: 3672–3673 [DOI] [PubMed] [Google Scholar]
- 43. Thomas DC. Are we ready for genome-wide association studies? Cancer Epidemiol Biomarkers Prev 2006; 15: 595–598 [DOI] [PubMed] [Google Scholar]
- 44. Storey JD, Tibshirani R.. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 2003; 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.