Highlights
-
•
A residue in active site which affects the optimum pH for catalysis was found.
-
•
A mutant with an extended optimum pH 7–9 was constructed.
-
•
The catalytic mechanism explains the extended optimum pH of the mutant.
Key words: Phenylalanine ammonia-lyase, Rhodotorula glutinis JN-1, Friedel–Crafts-type mechanism, Site-directed mutagenesis, Resolution, The optimum pH
Abstract
Phenylalanine ammonia-lyase (RgPAL) from Rhodotorula glutinis JN-1 stereoselectively catalyzes the conversion of the l-phenylalanine into trans-cinnamic acid and ammonia, and was used in chiral resolution of dl-phenylalanine to produce the d-phenylalanine under acidic condition. However, the optimum pH of RgPAL is 9 and the RgPAL exhibits low catalytic efficiency at acidic side. Therefore, a mutant RgPAL with a lower optimum pH is expected. Based on catalytic mechanism and structure analysis, we constructed a mutant RgPAL-Q137E by site-directed mutagenesis, and found that this mutant had an extended optimum pH 7–9 with activity of 1.8-fold higher than that of the wild type at pH 7. As revealed by Friedel–Crafts-type mechanism of RgPAL, the improvement of the RgPAL-Q137E might be due to the negative charge of Glu137 which could stabilize the intermediate transition states through electrostatic interaction. The RgPAL-Q137E mutant was used to resolve the racemic dl-phenylalanine, and the conversion rate and the eeD value of d-phenylalanine using RgPAL-Q137E at pH 7 were increased by 29% and 48%, and achieved 93% and 86%, respectively. This work provides an effective strategy to shift the optimum pH which is favorable to further applications of RgPAL.
1. Introduction
New asymmetrically biocatalytic methods continue to be developed for the production of enantiomerically pure chiral amino acids which constitute a significant fraction of the chiral building blocks that are required as intermediates for a range of target molecules, including pharmaceuticals and agrochemicals [31], [35]. Traditional biocatalytic methods for the production of chiral chemicals have relied on hydrolytic enzymes, such as lipase [5], acylases [15], [28] and hydantoinases [19], and ammonia lyase and aminomutase [9], [13], [23], [31]. Among these enzymes, the phenylalanine ammonia lyase (PAL) (EC 4.3.1.5) and phenylalanine aminomutase (PAM) have been used for the synthesis of a broad range of arylalanines [9], [21], [29], [31].
The industrial-scale production of PAL mainly utilizes the strains of the Rhodotorula genus [12], [32]. We previously screened strains from soil and identified a Rhodotorula glutinis strain with higher PAL activity, which was denoted JN-1 (CCTCC M2011490). The full-length gene of the phenylalanine ammonia lyase (RgPAL) from R. glutinis JN-1was isolated and successfully expressed in E. coli [38]. The RgPAL is a member of the 4-methylene-imidazol-5-one (MIO)-dependent enzyme family, which includes PAL, histidine ammonia-lyase (HAL) [27], tyrosine ammonia-lyase (TAL) [20], and PAM and tyrosine aminomutases (TAM) [13], [14], [29]. The MIO is a highly electrophilic prosthetic group that is formed post-translationally from a highly conserved Ala–Ser–Gly motif (Fig. 1), which attacks the substrate to facilitate the elimination of ammonia [24]. The RgPAL is shown to region-and-stereo selectively catalyze l-phenylalanine to trans-cinnamic acid and can be used to resolve dl-phenylalanine to produce the d-phenylalanine. The solubility of the trans-cinnamic acid is low at acidic side (about 0.006 g/L in aqueous solution at 25 °C), and the d-phenylalanine could be easily separated from the reaction solution through pH controlling. Therefore, the asymmetric resolution of racemic dl-phenylalanine by PAL is an attractive route and exhibits commercial application prospects. However, the optimum pH of RgPAL is 9 and the RgPAL exhibits low catalytic efficiency at acidic side; the trans-cinnamic acid exhibits high solubility at pH 9 and the accumulated trans-cinnamic acid during the reaction inhibits the catalysis, which presents a significant barrier to RgPAL application. Therefore, a mutant RgPAL with a lower optimum pH is expected.
Fig. 1.
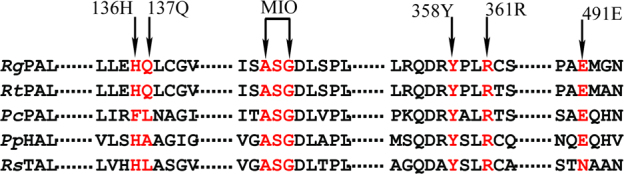
Amino acid sequence alignment of RgPAL with other PALs, HAL, and TAL.
The sequences shown are from Rhodotorula glutinis (RgPAL), R. toruloides (RtPAL), P. crispum (PcPAL), Pseudomonas putida (PpHAL), and Rhodobacter sphaeroides (RsTAL). The MIO prosthetic group, and His136, Gln137, Arg361, Tyr358, and Glu491 are indicated.
The optimum pH of enzymatic activity is often determined by the ionizable amino acids at active site that are involved in catalysis and substrate binding [30], [36]. The key issue is that which ionizable amino acids can be accurately picked out, and the catalytic mechanism and structure analysis can provide useful information in this aspect [37]. The RgPAL acts through the Friedel–Crafts-type mechanism (Fig. S1) [1], [22], [25]. In the reaction, the MIO attack the phenyl ring of the substrate to form carbocation 1 which would stabilize intermediate 2 formed by removal of the substrate’s C-3 hydrogen [1], [3]. Collapse of the system to product occurs with the elimination of NH3 and the release of trans-cinnamic acid from the MIO. Reservations on this mechanism center on the potentially large energy barrier that must be surpassed in forming the carbocation intermediate [3]. Therefore, negative charge around the phenyl ring might be involved in stabilizing the carbocation intermediate through electrostatic interaction and could facilitate the elimination of NH3.
In this work, we aim to shift the optimum pH of RgPAL toward the acidic side. Based on analyses of catalytic mechanism and structure, the His136 and Gln137 residues of RgPAL were found to form a hairpin motif to clamp the phenyl ring of substrate. The RgPAL-Q137E mutant extended the optimum pH to the range of 7–9. The specific activity of RgPAL-Q137E mutant was increased 1.8-fold at pH 7. The effective strategy for improving the catalytic activity and shifting the optimum pH is favorable to further applications of RgPAL.
2. Materials and methods
2.1. Plasmids and strains
The plasmids pMD18-T (Takara, Japan) and pET-28a (+) (Novagen, USA) were used for cloning and expression. The pET-28a-pal that encodes the RgPAL gene from R. glutinis JN-1 (CCTCC M2011490) was constructed in our previous study [38]. The E. coli strains JM109 and BL21 (DE3) (Novagen, USA) were used as a host strains for plasmid amplification and enzyme expression, respectively.
2.2. Site-directed mutagenesis
The mutants were constructed using site-directed mutagenesis. The PCR reaction was conducted using the PrimeSTAR HS DNA polymerase (Takara, Japan) and the pET-28a-pal plasmid as the template DNA. The primers are shown in Supplementary Table S1. The PCR product was digested by DpnI (Takara, Japan) at 37 °C for 1 h. The PCR product was transformed into competent cells of E. coli JM109. After the sequence verified, the extracted plasmid was transformed into E. coli BL21 (DE3) for enzyme expression.
2.3. Expression and purification of RgPAL and its mutants
The wild type and mutant proteins were expressed with N-terminal His-tag using the pET-28a (+) vector. The cells were grown to an OD600 of 0.6, and the enzyme expression was induced using 0.4 mM IPTG. After the cells were shaken at 24 °C for 20 h, the cells were collected by centrifugation (5 min, 4 °C, 10,000 × g), washed twice with 50 mM sodium phosphate buffer (containing 10 mM imidazole, and 150 mM NaCl, pH 7.5) and sonificated on ice at 40% power. After centrifugation, the supernatant was stored at 4 °C. The enzymes were purified by His-tag-purification using an Akta-purifier (GE Healthcare). The proteins were loaded onto a 1 mL HisTrap FF crude column (GE Healthcare), and the column was then washed using the same buffer and 58.3% of the elution-buffer (containing 250 mM imidazole, 150 mM NaCl). After elution, the enzyme was desalted using a HiPrep 26/10 desalting column (GE Healthcare) equilibrated with buffer (50 mM Tris–HCl, pH 8.6). The purity of the sample was detected through SDS-PAGE, and the concentration of enzyme protein was measured by Bradford method [2].
2.4. Computer-aided modeling and docking
The model of RgPAL was created through the submission of the sequence to SWISS-MODEL (http://swissmodel.expasy.org/) using the RtPAL (PDB ID: 1T6J) from R. toruloides with 75% identity as the template. The model was analyzed using the SWISS-MODEL server as described by Bartsch, Donnelly, and Rother [1], [4], [26]. Substrates were docked into the active site using Autodock (version 4), and the substrate binding was analyzed according to the method described by Wang [33] and Bartsch [1].
2.5. Circular dichroism (CD)
The CD spectra were measured on MOS-450/AF-CD-STP-A (Bio-Logic, France) at a protein concentration of 0.1 mg/mL in 50 mM Tris/HCl buffer (pH 8.6) using a 1 cm path-length quartz cuvette. To minimize the signal baseline drift, the spectropolarimeter and xenon lamp were warmed up at least 30 min prior to each experiment. The enzyme data in the 190–240 nm bands were collected, and which the spectrum obtained for a buffer blank was subtracted from these data.
2.6. Activity assay of RgPAL
The assay to determine the kinetic parameters were performed using different concentrations of l-phenylalanine (1–20 mM) (Sigma–Aldrich, Germany). The reactions were initiated by the addition of an appropriate quantity of RgPAL to each reaction system. The reaction was conducted at 40 °C and stopped by addition of 0.5 mL of methanol. The formation of trans-cinnamic acid was measured by HPLC (Hitachi, Japan) at 290 nm, the mobile phase contained 50% methanol. The obtained experimental dependences of the initial catalytic rates on the substrate concentrations were fitted to Michaelis–Menten equation through nonlinear regression analysis using Origin (7.5 versions). One enzyme activity unit was defined as the amount of enzyme that produced 1 μmol trans-cinnamic acid per minute at 40 °C.
The effects of the pH were determined at 40 °C using a series of buffers with various pH values (pH 5.0–7.0, 50 mM sodium acetate buffer; pH 7.0–9.0, 50 mM Tris–HCl buffer; pH 9.0–12.0, 50 mM sodium carbonate buffer).
2.7. The resolution of racemic dl-phenylalanine using RgPAL and RgPAL-Q137E
The chiral resolutions of dl-phenylalanine using RgPAL and RgPAL-Q137E were performed at pH 7 and pH 9, respectively. The experiments were carried out in 500 mL batch conical flasks with lid in a rotating shaker and contained 300 mL of dl-phenylalanine (100 mM) and 250 μg of pure enzyme at 40 °C. The conversion rate of l-phenylalanine and the eeD value of d-phenylalanine were calculated by the following equations:
where the eeD is the enantiomeric excess of d-phenylalanine; the Lphe, in is the initial concentration of l-phenylalanine; the Lphe,out is the residual concentration of l-phenylalanine after resolution; the Dphe is the concentration of d-phenylalanine. The d-phenylalanine and l-phenylalanine are detected through HPLC (Hitachi, Japan) at 205 nm according to the method described by Fukuhara [7]. The mobile phase contained 20% methanol and a complex of optically active L-Pro-Cu(II) (1.5 mM L-Pro and 0.75 mM CuSO4).
2.8. Data analysis
The “mutational effect” was determined by dividing the kcat value of the mutant enzyme by that of the wild type, and the free energy (ΔΔG‡) was calculated from the following equation: ΔΔG‡ = −RTln (mutational effect) as described by Olucha (2011, 2012) [17], [18].
3. Results
3.1. Selection of mutation sites based on homology modeling of the RgPAL structure
The active site typically contains ionizable groups (Arg, Lys, His, Glu, and Asp) that are involved in substrate binding and catalysis and that determine the pH activity profile of an enzyme [30]. To successfully select those residues in the active site, a theoretical model of RgPAL was constructed through homology modeling using RtPAL (PDB ID: 1T6J) as the template. As shown in Fig. 2, all of the residues that were the superimposed with RtPAL showed an RMSD of 0.224 Å (Fig. 2A), and the Ramachandran plot suggests that 94.9%, 3.2%, and 1.9% of the residues in derived model are in acceptable region, marginal region and disallowed region, respectively (Fig. 2B), These finding indicated that the model is reasonable and could be used in further molecular docking simulation. Using the AutoDock global–local evolutionary algorithm, we searched for those sites with the lowest free energy of binding between the ligand and the enzyme. As shown in Fig. 3, the active site cavity of RgPAL was bisected into two regions (Fig. 3A): one binds the amide group adjacent to the aromatic ring and the other binds the carboxyl group of the substrate. The phenyl ring of the substrate is roughly orthogonal to the plane of the MIO, and the methylidene of the MIO points to C2 of the aromatic ring (Fig. 3A and B). In the carboxyl group binding pocket, the Arg361 residue is 3.2 Å from the carboxyl group of the substrate, and this residue might play a role in the binding of the carboxylate moiety of the substrate through a salt bridge. The Tyr358 residue is 2.7 Å from the β-H of substrate, which is close enough to act as the β-H abstracted base (Fig. 3C). The Glu491 residue is the closet residue to the amino group of the substrate (2.8 Å, Fig. 3C) and might accept the amino group of substrate as the enzyme base, which is consistent with the results reported by Bartsch [1]. The Tyr358, Arg361 and Glu491 are highly conserved in PAL (Fig. 1). In the aromatic ring binding pocket, the His136 residue points to the phenyl ring of the substrate. The imidzaole group of His136 is parallel to the phenyl ring and might generate a π–π interaction. Moreover, the imidazole of His136 and the adjacent amide group of Gln137 which points to the phenyl ring within a distance of 4.5 Å, form a hairpin motif to clamp the phenyl ring (Fig. 3B and C).
Fig. 2.
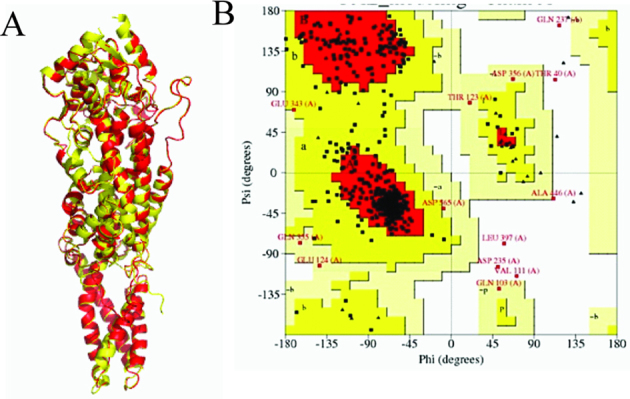
Comparative modeling of RgPAL and RtPAL. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
(A) Superimposed monomer structures of RgPAL and RtPAL. The model of RgPAL (red) was constructed using SWISS-MODEL and was superimposed with that of RtPAL (yellow); (B) Ramachandran plot of RgPAL. The red, yellow, pale-yellow, and grey white areas represent highly reliable, acceptable, marginal, and disallowed regions, respectively.
Fig. 3.
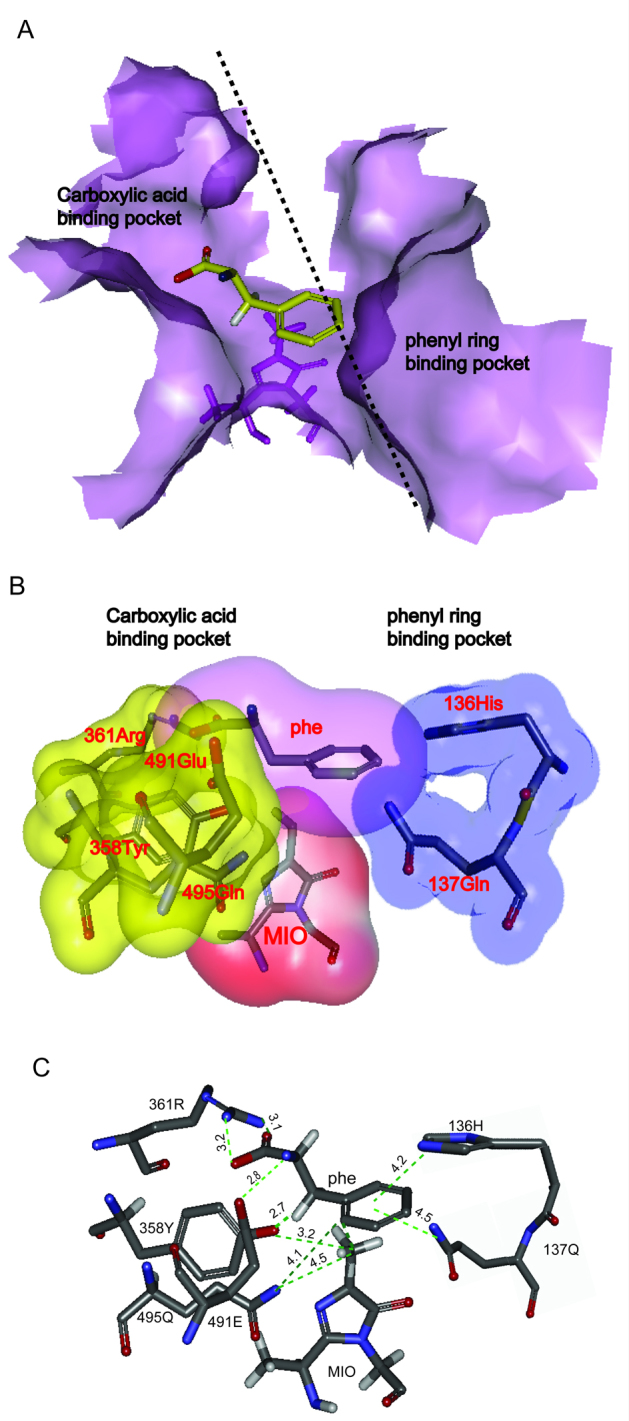
Structural analysis and molecular docking of RgPAL. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
(A) Solvent accessible surface of RgPAL in the active site that binds to the substrate. The front part of the RgPAL tetramer was removed to reveal the internal cavity in the vicinity of the MIO cofactor. The position of a substrate molecule bound to RgPAL is shown in yellow. MIO is labeled (violet). (B) Two regions adjacent to the aromatic ring and the carboxyl group of the substrate in active site. The binding site of the carboxylic acid and the amine group of the substrate are indicated in yellow, and the binding site for the aromatic ring is shown in blue. (C) The relative positions of the residues in the active site. The distances from the substrate to His136, Gln137, Arg361, Tyr358, Glu491, and MIO in the active site are shown. The oxygen, nitrogen, and carbon atoms are shown in red, blue, and gray, respectively.
3.2. Effects of mutations at 136 and 137 sites on enzymatic activity
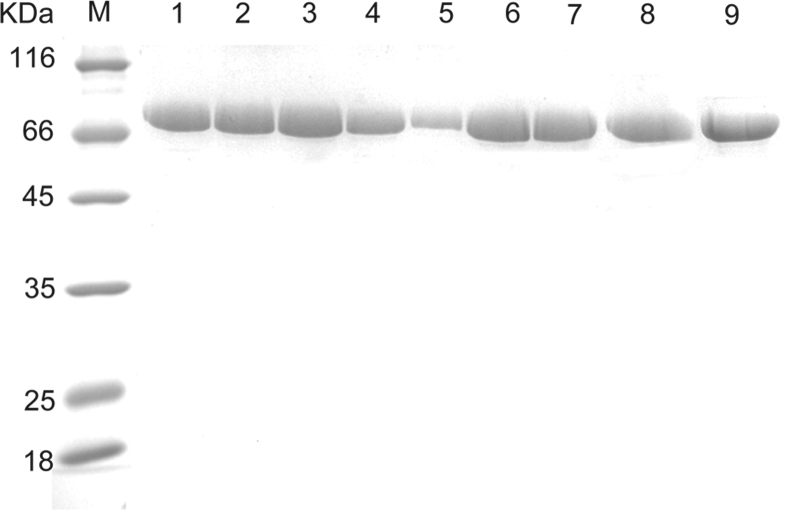
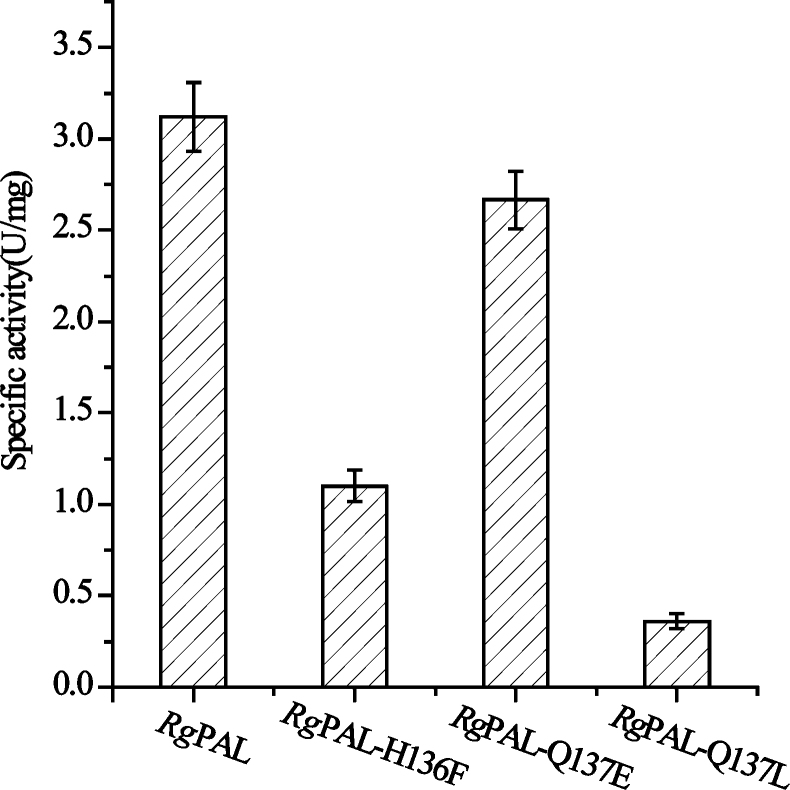
To verify the function of the hairpin, the His136, Gln137 were deleted (RgPAL-Δ136H, RgPAL-Δ137Q) and mutated to negative (RgPAL-H136E, RgPAL-Q137E) and positive charges (RgPAL-H136K, RgPAL-Q137K) as well as uncharged amino acids (RgPAL-H136F, RgPAL-Q137L), respectively. The mutant and wild type RgPAL proteins appeared a single band of about 75 kDa on SDS-PAGE (Fig. 4). The activities of RgPAL-Δ136H and RgPAL-Δ137Q were not detected (data not shown), suggesting that the residues at the two sites were essential for catalysis. The RgPAL-H136K, RgPAL-Q137K and RgPAL-H136E lost the enzymatic activity (data not shown), and the RgPAL-H136F, RgPAL-Q137L sharply decreased the activity (Fig. 5). Compared with those mutants, the activity of RgPAL-Q137E decreased slightly (Fig. 5). These findings are consistent with the Friedel–crafts-type mechanism; the positively charged or uncharged amino acids at active site were disadvantageous to the electrophilic attack on the aromatic ring of the substrate by the MIO.
Fig. 4.
SDS-PAGE analysis of the purified wild-type and mutant proteins.
Lanes: M, molecular marker; 1, wild-type RgPAL; 2, RgPAL-H136F; 3, RgPAL-H136E; 4, RgPAL-H136K; 5, RgPAL-Δ136H; 6, RgPAL-Q137E; 7, RgPAL-Q137L; 8, RgPAL-Q137K; 9, RtPAL-Δ137Q, (Δ) indicated that the residue was deleted.
Fig. 5.
Effects of mutation at 136 and 137 sites on the enzymatic activity.
The reaction was conducted for 30 min at pH 9 and 40 °C, the activity of RgPAL-H136E, RgPAL-H136 K, RgPAL-Δ136H, RgPAL-Q137K, and RtPAL-Δ137Q were not detected (data not shown).
3.3. Effects of pH on the activity of RgPAL and RgPAL-Q137E
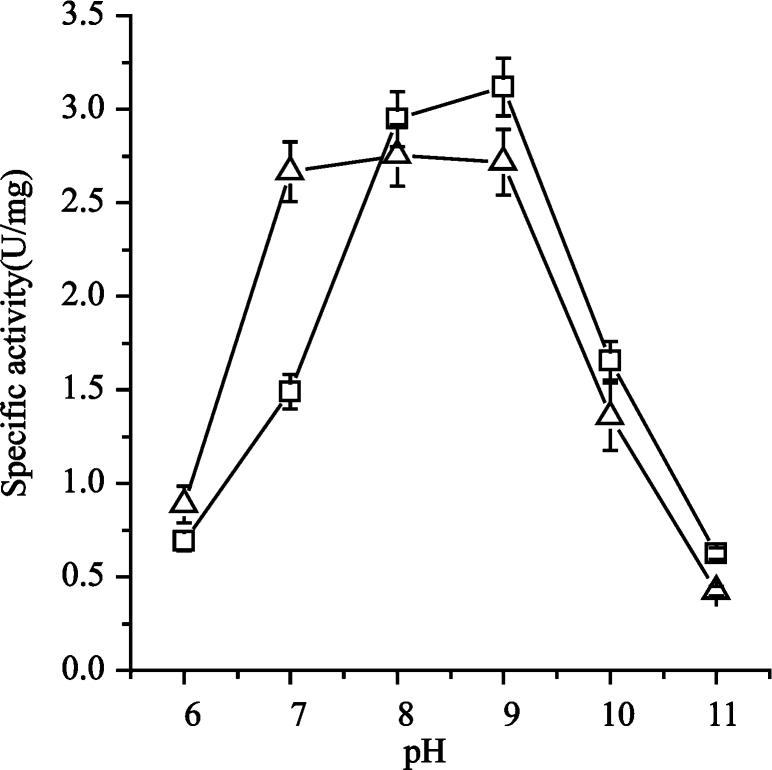
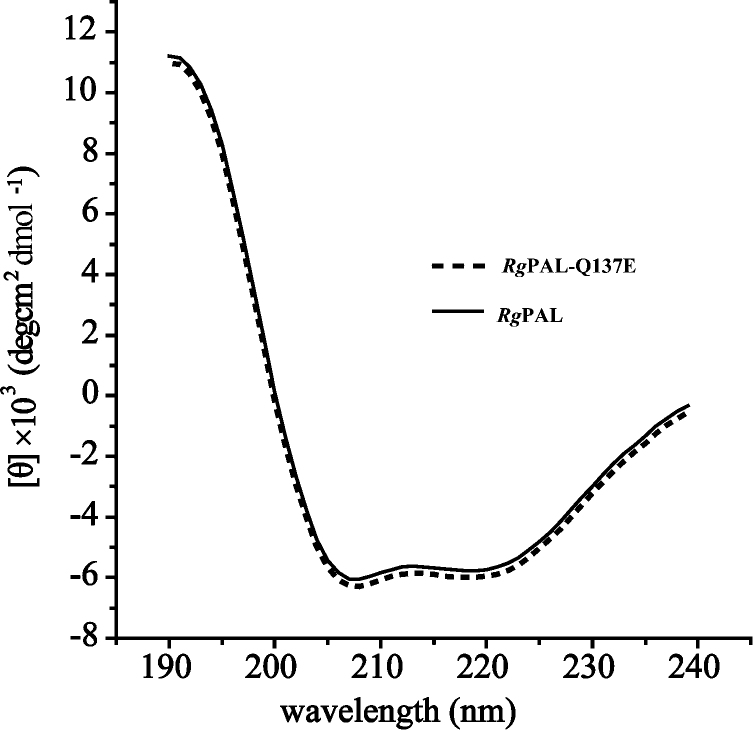
The effects of pH on the catalysis of RgPAL-Q137E were further studied because the mutation at 136 and 137 sites decreased the activity except for RgPAL-Q137E. The activity was determined over the pH range from 7–10 using a buffer system to maintain a constant ionic strength. Interestingly, the optimal pH of RgPAL-Q137E was extended to 7–9, the activity of RgPAL-Q137E at pH 7 (2.7 U/mg) is 1.8-fold higher than that of the wild type (1.5 U/mg) (Fig. 6). The CD spectrum of the mutant was similar to that of the wild type (Fig. 7) indicating that this mutant did not change the secondary structure of RgPAL. These findings suggested that the pH range extension of RgPAL-Q137E might results from the negative charge of Glu137, but not the secondary structure change.
Fig. 6.
Effects of pH on the enzymatic activities of RgPAL and RgPAL-Q137E.
The activity was determined over the pH range from 7–10 using a buffer system maintaining a constant ionic strength (pH 5.0–7.0, 50 mM sodium acetate buffer; pH 7.0–9.0, 50 mM Tris–HCl buffer; pH 9.0–12.0, 50 mM sodium carbonate buffer). (Δ) RgPAL-Q137E; (□) RgPAL.
Fig. 7.
The conformation change of the mutants was checked with circular dichroism (CD).
The CD spectra were measured at a protein concentration of 0.1 mg/mL in 50 mM Tris/HCl buffer (pH 8.6). solid line: RgPAL; dotted line: RgPAL-Q137E.
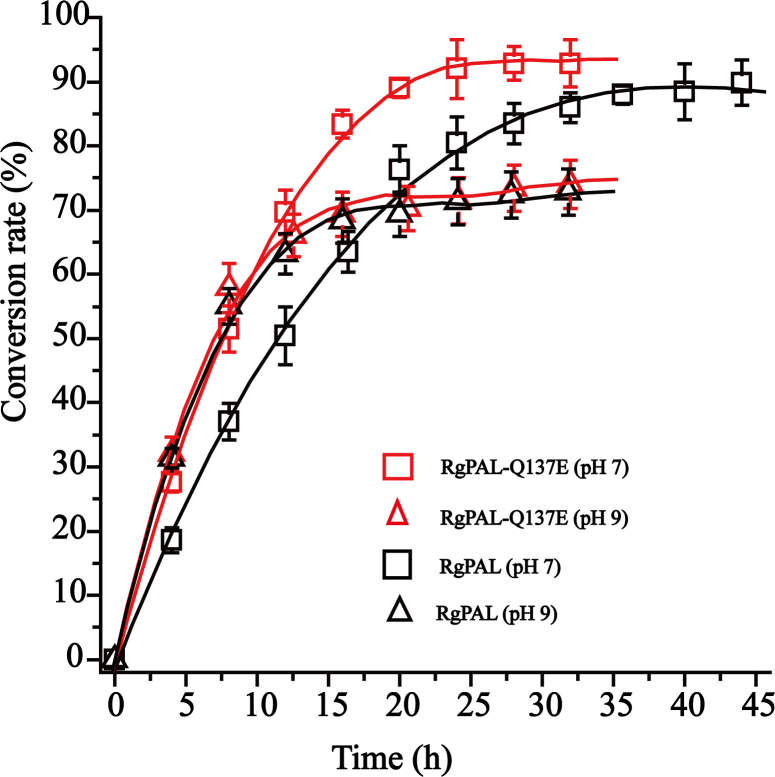
3.4. Asymmetric resolution of racemic dl-phenylalanine
The dl-phenylalanine was resolved using RgPAL and RgPAL-Q137E at pH 7 and pH 9, respectively. As shown in Fig. 8, under the condition of pH 9, about 65% of l-phenylalanine was converted in both reactions after 16 h, and the conversion rates hardly increased after 16 h and the ultimate conversion rate and eeD value were 72% and 58%, respectively. This may be due to the inhibition of the accumulated trans-cinnamic acid. On the other hand, when the reaction was carried out at pH 7, the precipitation of trans-cinnamic acid was observed, and the inhibition effect was obviously relieved. The conversion rate and eeD value using RgPAL-Q137E at pH 7 achieved 93% and 86% within 26 h, respectively, while the RgPAL needed more than 45 h to achieve the same conversion rate at pH 7. These findings indicated that RgPAL-Q137E was benefit for chiral resolution of dl-phenylalanine.
Fig. 8.
The resolution process of dl-phe using RgPAL and RgPAL-Q137E.
The resolution of dl-phenylalanine using RgPAL and RgPAL-Q137E were performed at pH 7 and pH 9, respectively.
4. Discussion
The His136 and Gln137 of RgPAL seemed to form a hairpin motif to clamp the phenyl ring (Fig. 3). The imidazole of His and the amide group of Gln in the hairpin motif contain lone pair electrons, which might increase the electron density of the phenyl ring of the substrate. According to Friedel–Crafts-type mechanism, the phenyl ring of the substrate with higher electron density is vulnerable to the attack by the MIO [3], [22]. Although the His and Phe present a similar structure, and both of His136 and F136 are likely to form π–π interaction with the phenyl ring of substrate (Fig. 3B), the imidazole of His which contains richer electron rather than the phenyl ring of Phe at pH 9, is accessible to enhance the electron density of the phenyl ring of the substrate [1]. Therefore, the activity of RgPAL-H136F was lower than that of RgPAL at pH 9. Moreover, the amino acid at 136 site (His or Phe, Fig. 1) is involved in recognizing the substrate [16], [34], the other mutations at this site would affect substrate binding. As a result, RgPAL-H136E and RgPAL-H136K lost the activity.
The Gln137 is only conserved in RgPAL and RtPAL but not in other PALs, HAL and TAL (Fig. 1). The mutations at this site decreased or lost the activity except the RgPAL-Q137E. The RgPAL-Q137E mutant had an extended optimum pH 7–9 with the activity of about 1.8-fold higher than that of the wild type at pH 7 (Fig. 6). The optimum pH of an enzyme depends on the ionizable amino acids at active site which are involved in catalysis [10], [11] or stabilization of transition state by electrostatic interaction [17], [18] and enzyme substrate binding [6]. The Glu137 might be involved in the stabilization of transition state by electrostatic interaction. According to the Friedel–Crafts-type mechanism, the carbocation intermediate is large energy barrier which must be surpassed [3]. The improvements of RgPAL-Q137E at pH 7 may be due to the negative charge of Glu137 which facilitates stabilization of carbocation intermediate to reduce the energy barrier through electrostatic interaction. The kcat value provides an assessment of the specificity and is directly related to the free energy of activation of the transition state [8], [17], [18]. Therefore, the calculation of mutational effects using kcat provides insights on the contribution of electrostatics. As shown in Table 1, the kcat mutational effect was 0.56 at pH 7, which represents a ΔΔG‡ of 1.56 kcal/mol, and the kcat mutational effect was found to be 0.93 at pH 9, which represents a ΔΔG‡ of 0.19 kcal/mol. Therefore, a negative charge at position 137contributes 1.32 kcal/mol (1.56 kcal/mol − 0.19 kcal/mol) of net free energy toward the electrostatic stabilization of the transition state. In addition, the negative charge of Glu137 is also likely to counteract the adverse effects of the positive charge of His136, which favors a protonated state at pH 7. The pKa value of the imidazole group of His is approximately 5–7, and the His136 tends to exhibit the protonated state with a positive charge at pH 7, which is disadvantageous to the electrophilic attack MIO on the aromatic ring of the substrate by the MIO.
Table 1.
Contribution of the negative charge at 137 site to activity.
| pH | Enzyme | kcat (s−1) × 103 | Mutational effecta | ΔΔG‡ (kJ M−1) |
|---|---|---|---|---|
| 7 | RgPAL | 13.2 ± 0.7 | ||
| RgPAL-Q137E | 23.6 ± 1.9 | 0.56 | 1.51 | |
| 9 | RgPAL | 23.1 ± 1.2 | ||
| RgPAL-Q137E | 24.7 ± 1.4 | 0.93 | 0.19 | |
| 1.32b |
RgPAL kcat value divided by the mutant kcat value.
The ΔΔG‡ at pH 7 subtracted by the ΔΔG‡ at pH 9.
5. Conclusions
RgPAL could be used to resolve dl-phenylalanine to produce optical pure d-phenylalanine, since the pH is one of the most important quality parameters for PAL catalytic reaction to resolve the dl-phenylalanine. In this study, the optimum pH of RgPAL was shifted toward the acidic side through site-directed mutagenesis based on the analysis of catalytic mechanism and structure. The RgPAL-Q137E mutant exhibited a wide pH range from 7 to 9. When this mutant was used to resolve the dl-phenylalanine, the conversion rate and eeD value increased by 29% and 48%, and the ultimate conversion rate and eeD value achieved 93% and 86%, respectively. However, the eeD value and conversion rate using RgPAL-Q137E need to be further improved, such research is currently carrying out in our lab. This work provides an effective strategy to shift the optimum pH for the enzyme application.
Acknowledgments
This work is financially supported by the National High Technology Research and Development Program of China (863 Program, 2011AA100905), the Program for New Century Excellent Talents in University (NCET-10-0461), the High Foreign Experts Project(GDW20123200114), the Key project of Chinese Ministry of Education (311023), the Priority Academic Program Development of Jiangsu Higher Education Institutions, the 111 Project (No. 111-2-06).
Footnotes
Available online 6 June 2014
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.btre.2014.06.001.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Bartsch S., Bornscheuer U.T. A single residue influences the reaction mechanism of ammonia lyases and mutases. Angew. Chem. Int. Ed. Engl. 2009;48(18):3362–3365. doi: 10.1002/anie.200900337. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Calabrese J.C., Jordan D.B., Boodhoo A., Sariaslani S., Vannelli T. Crystal structure of phenylalanine ammonia lyase: multiple helix dipoles implicated in catalysis. Biochemistry. 2004;43(36):11403–11416. doi: 10.1021/bi049053+. [DOI] [PubMed] [Google Scholar]
- 4.Donnelly M., Fedeles F., Wirstam M., Siegbahn P.E., Zimmer M. Computational analysis of the autocatalytic posttranslational cyclization observed in histidine ammonia-lyase: a comparison with green fluorescent protein. J. Am. Chem. Soc. 2001;123(20):4679–4686. doi: 10.1021/ja004009c. [DOI] [PubMed] [Google Scholar]
- 5.Forro E., Fulop F. Recent lipase-catalyzed hydrolytic approaches to pharmacologically important β- and γ-amino acids. Curr. Med. Chem. 2012;19(36):6178–6187. [PubMed] [Google Scholar]
- 6.Fu D., Huang H., Meng K., Wang Y., Luo H., Yang P., Yuan T., Yao B. Improvement of Yersinia frederiksenii phytase performance by a single amino acid substitution. Biotechnol. Bioeng. 2009;103(5):857–864. doi: 10.1002/bit.22315. [DOI] [PubMed] [Google Scholar]
- 7.Fukuhara T.Y.S. Novel ligand-exchange chromatographic resolution of dl-amino acids using nucleotides and coenzymes. J. Chromatogr. Sci. 1990;28(3):114–117. [Google Scholar]
- 8.Ghosh A., Sakaguchi R., Liu C., Vishveshwara S., Hou Y.M. Allosteric communication in cysteinyl tRNA synthetase: a network of direct and indirect readout. J. Biol. Chem. 2011;286(43):37721–37731. doi: 10.1074/jbc.M111.246702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gloge A., Zon J., Kovari A., Poppe L., Retey J. Phenylalanine ammonia-lyase: the use of its broad substrate specificity for mechanistic investigations and biocatalysis–synthesis of l-arylalanines. Chemistry. 2000;6(18):3386–3390. doi: 10.1002/1521-3765(20000915)6:18<3386::aid-chem3386>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 10.Hirata A., Adachi M., Sekine A., Kang Y.N., Utsumi S., Mikami B. Structural and enzymatic analysis of soybean β-amylase mutants with increased pH optimum. J. Biol. Chem. 2004;279(8):7287–7295. doi: 10.1074/jbc.M309411200. [DOI] [PubMed] [Google Scholar]
- 11.Hirata A., Adachi M., Utsumi S., Mikami B. Engineering of the pH optimum of Bacillus cereus β-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type. Biochemistry. 2004;43(39):12523–12531. doi: 10.1021/bi049173h. [DOI] [PubMed] [Google Scholar]
- 12.Jia S.R., Cui J.D., Li Y., Sun A.Y. Production of l-phenylalanine from trans-cinnamic acids by high-level expression of phenylalanine ammonia lyase gene from Rhodosporidium toruloides in Escherichia coli. Biochem. Eng. J. 2008;42(3):193–197. [Google Scholar]
- 13.Klettke K.L., Sanyal S., Mutatu W., Walker K.D. β-styryl- and β-aryl-β-alanine products of phenylalanine aminomutase catalysis. J. Am. Chem. Soc. 2007;129(22):6988–6989. doi: 10.1021/ja071328w. [DOI] [PubMed] [Google Scholar]
- 14.Krug D., Muller R. Discovery of additional members of the tyrosine aminomutase enzyme family and the mutational analysis of CmdF. ChemBioChem. 2009;10(4):741–750. doi: 10.1002/cbic.200800748. [DOI] [PubMed] [Google Scholar]
- 15.Kulkarni R.S., Kalkote U.R. Resolution of dl-Phenylglycine by Penicillin G acylase. Hindustan Antibiot. Bull. 2005;47–48(1–4):41–44. [PubMed] [Google Scholar]
- 16.Louie G.V., Bowman M.E., Moffitt M.C., Baiga T.J., Moore B.S., Noel J.P. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem. Biol. 2006;13(12):1327–1338. doi: 10.1016/j.chembiol.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olucha J., Meneely K.M., Lamb A.L. Modification of residue 42 of the active site loop with a lysine-mimetic side chain rescues isochorismate-pyruvate lyase activity in Pseudomonas aeruginosa PchB. Biochemistry. 2012;51(38):7525–7532. doi: 10.1021/bi300472n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olucha J., Ouellette A.N., Luo Q., Lamb A.L. pH Dependence of catalysis by Pseudomonas aeruginosa isochorismate-pyruvate lyase: implications for transition state stabilization and the role of lysine 42. Biochemistry. 2011;50(33):7198–7207. doi: 10.1021/bi200599j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park J.H., Kim G.J., Kim H.S. Production of d-amino acid using whole cells of recombinant Escherichia coli with separately and coexpressed d-hydantoinase and N-carbamoylase. Biotechnol. Prog. 2000;16(4):564–570. doi: 10.1021/bp0000611. [DOI] [PubMed] [Google Scholar]
- 20.Pilbak S., Farkas O., Poppe L. Mechanism of the tyrosine ammonia lyase reaction-tandem nucleophilic and electrophilic enhancement by a proton transfer. Chemistry. 2012;18(25):7793–7802. doi: 10.1002/chem.201103662. [DOI] [PubMed] [Google Scholar]
- 21.Poppe L., Paizs C., Kovacs K., Irimie F.D., Vertessy B. Preparation of unnatural amino acids with ammonia-lyases and 2,3-aminomutases. Methods Mol. Biol. 2012;794:3–19. doi: 10.1007/978-1-61779-331-8_1. [DOI] [PubMed] [Google Scholar]
- 22.Poppe L., Retey J. Friedel–Crafts-type mechanism for the enzymatic elimination of ammonia from histidine and phenylalanine. Angew. Chem. Int. Ed. Engl. 2005;44(24):3668–3688. doi: 10.1002/anie.200461377. [DOI] [PubMed] [Google Scholar]
- 23.Raj H., Szymanski W., de Villiers J., Rozeboom H.J., Veetil V.P., Reis C.R., de Villiers M., Dekker F.J., de Wildeman S., Quax W.J., Thunnissen A.M., Feringa B.L., Janssen D.B., Poelarends G.J. Engineering methylaspartate ammonia lyase for the asymmetric synthesis of unnatural amino acids. Nat. Chem. 2012;4(6):478–484. doi: 10.1038/nchem.1338. [DOI] [PubMed] [Google Scholar]
- 24.Retey J. Discovery and role of methylidene imidazolone, a highly electrophilic prosthetic group. Biochim. Biophys. Acta. 2003;1647(1–2):179–184. doi: 10.1016/s1570-9639(03)00091-8. [DOI] [PubMed] [Google Scholar]
- 25.Retey J. Enzymatic catalysis by Friedel–Crafts-type reactions. Naturwissenschaften. 1996;83(10):439–447. doi: 10.1007/BF01144012. [DOI] [PubMed] [Google Scholar]
- 26.Rother D., Poppe L., Morlock G., Viergutz S., Retey J. An active site homology model of phenylalanine ammonia-lyase from Petroselinum crispum. Eur. J. Biochem. 2002;269(12):3065–3075. doi: 10.1046/j.1432-1033.2002.02984.x. [DOI] [PubMed] [Google Scholar]
- 27.Rother D., Poppe L., Viergutz S., Langer B., Retey J. Characterization of the active site of histidine ammonia-lyase from Pseudomonas putida. Eur. J. Biochem. 2001;268(23):6011–6019. doi: 10.1046/j.0014-2956.2001.02298.x. [DOI] [PubMed] [Google Scholar]
- 28.Schutt H., Schmidt-Kastner G., Arens A., Preiss M. Preparation of optically active d-arylglycines for use as side chains for semisynthetic penicillins and cephalosporins using immobilized subtilisins in two-phase systems. Biotechnol. Bioeng. 1985;27(4):420–433. doi: 10.1002/bit.260270406. [DOI] [PubMed] [Google Scholar]
- 29.Strom S., Wanninayake U., Ratnayake N.D., Walker K.D., Geiger J.H. Insights into the mechanistic pathway of the Pantoea agglomerans phenylalanine aminomutase. Angew. Chem. Int. Ed. Engl. 2012;51(12):2898–2902. doi: 10.1002/anie.201108525. [DOI] [PubMed] [Google Scholar]
- 30.Tomschy A., Brugger R., Lehmann M., Svendsen A., Vogel K., Kostrewa D., Lassen S.F., Burger D., Kronenberger A., van Loon A.P., Pasamontes L., Wyss M. Engineering of phytase for improved activity at low pH. Appl. Environ. Microbiol. 2002;68(4):1907–1913. doi: 10.1128/AEM.68.4.1907-1913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner N.J. Ammonia lyases and aminomutases as biocatalysts for the synthesis of α-amino and β-amino acids. Curr. Opin. Chem. Biol. 2011;15(2):234–240. doi: 10.1016/j.cbpa.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Wall M.J., Quinn A.J., D'Cunha G.B. Manganese (Mn2+)-dependent storage stabilization of Rhodotorula glutinis phenylalanine ammonia-lyase activity. J. Agric. Food Chem. 2008;56(3):894–902. doi: 10.1021/jf072614u. [DOI] [PubMed] [Google Scholar]
- 33.Wang L., Gamez A., Archer H., Abola E.E., Sarkissian C.N., Fitzpatrick P., Wendt D., Zhang Y., Vellard M., Bliesath J., Bell S.M., Lemontt J.F., Scriver C.R., Stevens R.C. Structural and biochemical characterization of the therapeutic Anabaena variabilis phenylalanine ammonia lyase. J. Mol. Biol. 2008;380(4):623–635. doi: 10.1016/j.jmb.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watts K.T., Mijts B.N., Lee P.C., Manning A.J., Dannert Schmidt-, C Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem. Biol. 2006;13(12):1317–1326. doi: 10.1016/j.chembiol.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Wohlgemuth R. Asymmetric biocatalysis with microbial enzymes and cells. Curr. Opin. Microbiol. 2010;13(3):283–292. doi: 10.1016/j.mib.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Yang F., Zhang S., Tang W., Zhao Z.K. Identification of the orotidine-5'-monophosphate decarboxylase gene of the oleaginous yeast Rhodosporidium toruloides. Yeast. 2008;25(9):623–630. doi: 10.1002/yea.1607. [DOI] [PubMed] [Google Scholar]
- 37.Yang H.Q., Liu L., Shin H.D., Chen R.R., Li J., Du G., Chen J. Structure-based engineering of histidine residues in the catalytic domain of α-amylase from Bacillus subtilis for improved protein stability and catalytic efficiency under acidic conditions. J. Biotechnol. 2013;164(1):59–66. doi: 10.1016/j.jbiotec.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Zhu L., Cui W., Fang Y., Liu Y., Gao X., Zhou Z. Cloning, expression and characterization of phenylalanine ammonia-lyase from Rhodotorula glutinis. Biotechnol. Lett. 2013;35(5):751–756. doi: 10.1007/s10529-013-1140-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.