Highlights
-
•
A DNA-sensor was developed to monitor the hybridization of complementary ssDNAs.
-
•
The signal strength was proportional to the length of short oligonucleotides.
-
•
Elevated temperature was observed to reduce non-specific interactions.
-
•
The study proposes sandwich hybridization approach to improve signal strength.
Keywords: Sandwich hybridization, DNA-sensor, Polytyramine
Abstract
A capacitive DNA-sensor model system was used to monitor the capture of complementary single-stranded DNAs. The sensor chip consisted of a gold electrode, which was carefully insulated with a polytyramine layer and covalently tagged with 25-mer oligo-C. As low as 10−11 moles per liter of target oligo-G could be detected by injecting 250 μL of sample. Elevated temperature was used to reduce non-specific hybridization. Less than 10% of non-target 25-mer oligo-T interacted nonspecifically with the oligo-C probes when hybridization process was performed at 50 °C. Studying the relationship of length of the analyte to the signal strength, the output from the capacitive DNA-sensor increased to almost the double; from 50 to 88-nF cm−2, when a 25-mer oligo-G was used instead of a 15-mer. By sandwich hybridization at room temperature, it was possible to further increase the signal, from 78-nF cm−2 for the target 50-mer oligo-G alone, to 114-nF cm−2.
1. Introduction
The development of sensitive, selective and real-time sensors for monitoring DNA in biological samples is very important. Determination of specific DNA-sequences in clinical or food samples can result in the detection and identification of certain infectious organisms [1]. Various DNA-sensors with labeled probes have been reported; where the use of radioisotope-labeled (125I or 132P) DNA-probes have been reported frequently [2], [3], [4]. However, apart from high sensitivities, the use of isotope-labeled reagents is restricted because of the potential danger of radioactivity. Therefore, new strategies have been introduced for labeling of DNA such as use of avidin–biotin [5], ferrocenium [6], chemiluminescent agent [7], [8], fluorescent dye [9], and various metal nanoparticles [10] such as gold-nanoparticles [11]. Assays based on labeled reagents are among the most sensitive reported, but in general they are costly, complex and time-consuming.
Alternatively, various DNA-sensors with label-free probes have been developed. Among these are piezoelectric [12], acoustic [13], optical [14] and electrochemical transduction [1]. In particular, electrochemical DNA sensors are robust, cheap and allow fast detection. They make use of single-stranded DNA probes that are attached to the surface of the sensing devices which have the potential to allow rapid and quantitative monitoring of label-free hybridization. Electrochemical detection of DNA hybridization usually involves changes in electrochemical parameters such as; capacitance [15], impedance [16] and electrochemical quartz crystal microbalance measurements [17] at fixed potential or detecting complementary target, using both direct electrochemical oxidation of guanine and redox of the electroactive indicator methylene blue [17], [18], [19]. The above listed electrochemical DNA-sensors that use label-free probes are cost effective alternatives adopted for real-time monitoring, however with serious drawbacks; low selectivity and low sensitivity [15], [17].
This paper describes the use of a capacitive DNA-sensor application, where a surface-bound label-free oligonucleotide probes captures a target complementary DNA-sequence and real time measurement is performed. Nevertheless, the application of elevated temperature to reduce non-specific hybridization (interaction of non-complementary oligos) in order to increase the selectivity, the influence of oligo length to the signal strength, and application of sandwich hybridization approach in order to amplify the signal strength of the long DNA molecules are reported.
2. Materials and methods
2.1. Materials
All single stranded oligonucleotides were obtained from Eurofins MWG Operon (Ebersberg, Germany): 25-mer oligonucleotides-C (oligo-C); 15-, 25- and 50-mer oligonucleotides-G (oligo-G); and 25-mer oligonucleotides-T (oligo-T). Absolute ethanol and sodium hydroxide (NaOH) were obtained from VWR International (Leuven, Belgium). Tyramine, N-hydroxysuccinimide (NHS), N-(3-dimethylaminopropyl) N-ethylcarbodiimide hydrochloride (EDC), ethanesulfonic acid (MES), and 1-dodecane thiol were obtained from Sigma–Aldrich (Steinheim, Germany). All other chemicals used were of analytical grade. All buffers and regeneration solutions were prepared with double distilled water from a Milli-Q system (Millipore, Massachusetts, USA). All solutions were filtered through a membrane (pore size 0.22 μm) and degassed prior to use.
2.2. Methods
2.2.1. Electrode surface modification
A gold electrode (99.9% purity, custom-made, ϕ = 3 mm) with a surface area of 0.07 cm2 was used as a working electrode. Prior to the modification with oligonucleotides, the gold electrode was polished with alumina slurry with a particle size of 0.1 μm (Struers, Ballerup, Denmark) and cleaned through sonication in distilled water and subsequently in absolute ethanol, for 15 min in each solvent. It was then washed with distilled water and dried with pure nitrogen gas [20], followed by plasma cleaning, PDC-3XG (Harrick, New York, USA) for 20 min, and after that coated by the electropolymerization of tyramine on the electrode surface [21]. The coated electrode was rinsed with distilled water to remove any loosely bound polymer and it was finally carefully dried with pure nitrogen gas prior to immobilizaton.
To immobilize nucleotide-probes on the modified electrode, 20 μL of a mixture that contained 20 μM 25-mer oligo-C, 9.9 μM NHS and 1.65 μM EDC in 40 mM MES buffer, pH 6.5 was added on the surface of the electrode. The reaction was left at room temperature for 30 min, while covered to prevent evaporation. The electrode was then transferred to 4 °C and kept there for 24 h. Prior to use, the sensor electrode was washed with 10 mM potassium phosphate buffer pH 7.2, and distilled water before being dried by pure nitrogen gas. The modified electrode was then immersed into 10 mM 1-dodecane thiol for 20 min in order to provide insulation and to block any pin holes. Cyclic voltammetry, CV/Auto-lab (Utrecht, Netherlands) was used to monitor the results of insulation and immobilization processes [21].
2.2.2. Experimental set up
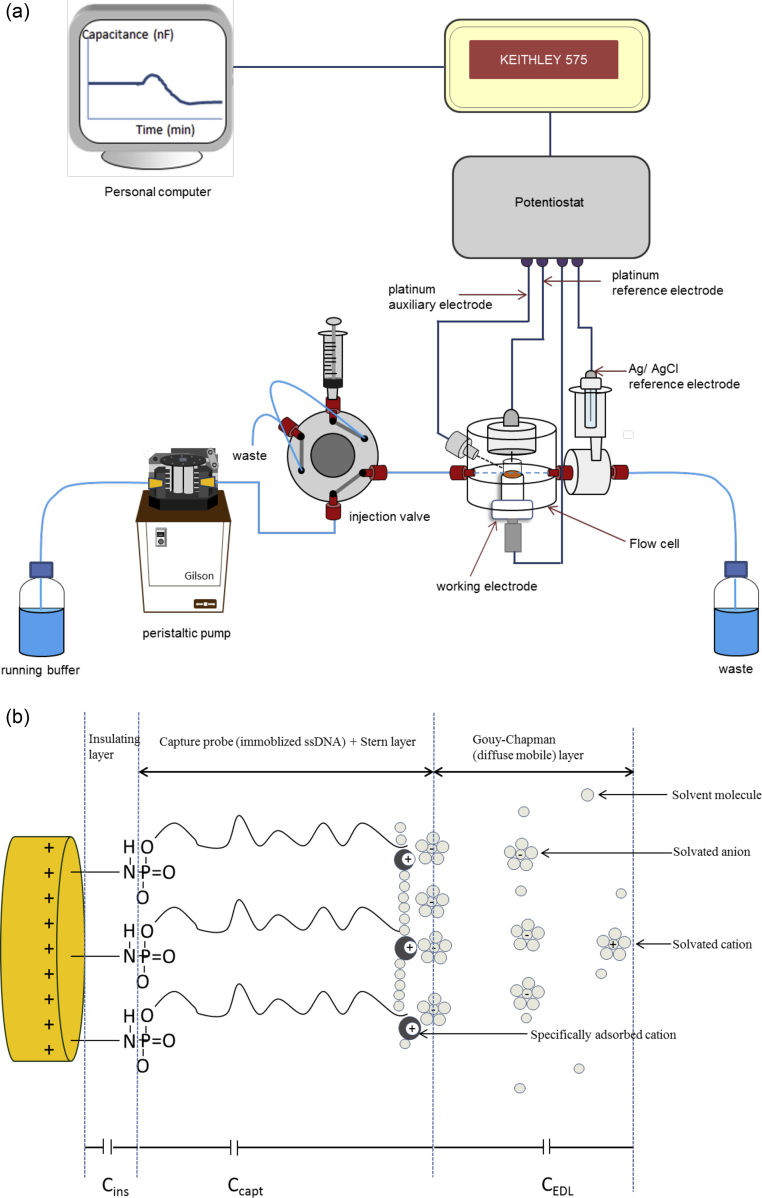
All experiments were performed in a conventional four-electrode flow cell with a dead volume of 10 μL, using a data acquisition unit (Keithley Instruments, Cleveland, OH, USA) and a potentiostat interfaced with a personal computer (Fig. 1a). Details of the experimental set-up of the four-electrode flow cell injection capacitive sensor system were described previously [22]. A modified electrode, using 25-mer oligo-C probe immobilized on the surface, was placed into the flow cell and then equilibrated with running buffer (10 mM potassium phosphate buffer pH 7.2) at flow rate of 100 μL/min until a stable base line was obtained, followed by injecting 250 μL of a sample in the same buffer. NaOH (50 mM) was applied for intermediate regeneration after hybridization step [23] in order to break the binding between oligo-C probe and an analyte (oligo-G), and hence, to facilitate additional measurements. All the measurements made in this study were performed in triplicates, either at room temperature (23 °C, RT) or at elevated temperatures. For the studies that involve the use of elevated temperatures, a column-thermostat, Jetstream 2 (Vienna, Austria) was used.
Fig. 1.
(a) Experimental set up of the flow injection capacitive DNA-sensor system. (b) Schematic of the electrical double layer structure of a capacitive DNA-sensor electrode with the built-up of capacitors in series.
In principle, when a bare electrode surface is subjected to the electrolyte solution, an electrical double layer which consists of adsorbed fixed layer (Stern layer) and a diffuse mobile layer (Gouy–Chapman diffuse layer) is formed at the electrode surface/electrolyte solution interface. The interface between electrode surface and the electrolyte solution (the electric double layer) behaves like a capacitor; i.e., it is capable of storing electric charge [24]. The electrical double layer capacitance could be described by Eq. (1).
| (1) |
where, is the capacitance of the electrical double layer, is the capacitance of Stern (adsorption layer) layer and is the capacitance of the Gouy–Chapman (diffuse mobile) layer.
However, in typical capacitive DNA-sensor, the capacitance at the electrode surface/electrolyte solution interface is built up of several capacitors in series [25]; (i) the capacitance of insulating layer (polytyramine layer), Cins (ii) the capacitance of capture probe (immobilized DNA), Ccapt and any contribution from adsorbed fixed layer (iii) and the capacitance of diffuse mobile layer, Cdl as depicted in Fig. 1b. Therefore total capacitance (CTot) at electrode surface/electrolyte solution interface could be described by Eq. (2).
| (2) |
When the analyte hybridizes on capture probe, consequently this increases the thickness and the length of the capture probe layer. The displacement of the diffuse mobile layer created during the potentiostatic pulse will cause a decrease in total capacitance, which is strictly proportional to the analyte concentration. The surface should be designed so that, the capacitance of the insulating layer, Cins is high as possible that allows the capacitance from the binding of analyte to be detected. This change in capacitance due to binding of analyte was used for detection.
2.2.3. Capacitance measurement
A positive potential pulse of 50 mV was applied each sixty second at the modified electrode (working electrode), which gives a current response signal. The current was sampled and the total capacitance was obtained by taking the logarithm of Eq. (3)
| (3) |
where, i(t) is the current in the open circuit (RC model) as a function of time, u is the applied pulse potential, Rs is the dynamic resistance of capture probe layer, CTot is the total capacitance measured between the gold electrode surface and the electrolyte solution interface, and t is the time elapsed after the potentiostatic step was applied. The technique is described in detail elsewhere [22].
Hybridization of single stranded DNA (ssDNA) on the capture probe caused CTot to decrease. Then, the capacitance change, ΔC, could be determined as a difference between the two base lines, before and after injection of the sample. A baseline was considered stable when a standard deviation of an average of the last five measuring points of a registered total capacitance is <1 nF. The necessity to evaluate an average of five capacitance values was previously mathematically proved [26]. However, standard deviation of <1 nF was introduced based on previous observations (data not shown) that the signal for the lowest concentration (10−12 M) of the target analyte tested in this study, was clearly observed when the standard deviation of the 5 average points of the baseline before injection of the analyte was <1 nF.
2.2.4. DNA-hybridization on the electrode surface at different temperatures
Hybridization of target DNA was initially performed at RT. Oligo-G probes of different lengths (15-, 25- and 50-mer) were injected into the system at different concentrations, i.e. 10−8, 10−9, 10−10 and 10−11 M. The result in capacitance change of each oligo-probe length was registered and evaluated.
In the analytical step using DNA-sensors, higher temperatures are often needed in order to improve the selectivity of the sensor. However, it is necessary to know the influence of the temperature on the electrode modified surface in order to understand whether a measured capacitance is caused by changes to temperature or by any other event on the electrode surface. For this reason, the behavior of the modified electrode surface with respect to capacitance changes was initially studied at RT, 30, 40, 50, and 60 °C, and kept for 30 min at each temperature.
Specific and non-specific hybridizations at RT, 30, 40, 50, and 60 °C were also studied by applying target DNA, 10−8 M of 25-mer oligo-G on the modified electrode surface. Later, the same concentration of non-specific DNA, 25-mer oligo-T was also applied under identical conditions and the results were compared to each other. This study offers a predictable optimum temperature that discriminates non-specific hybridization without significantly affecting the specific hybridization.
2.2.5. Sandwich hybridization
Sandwich hybridization was performed at RT by injecting 50-mer oligo-G at different concentrations (10−8, 10−9, 10−10 and 10−11 M). Once a stable base line was observed, the same concentration of 25-mer oligo-C was injected. These results were compared with those obtained from injection of the 50-mer oligo-G, alone.
3. Results and discussion
3.1. Cyclic voltammetry
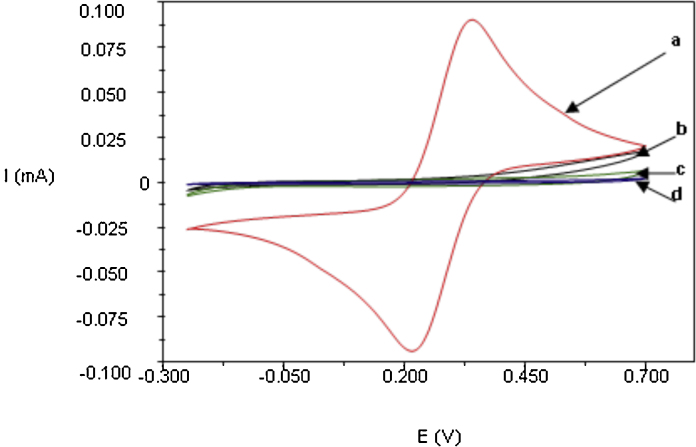
The electrochemical behavior of the electrode was studied after each modification step (Fig. 2) by oxidizing and reducing a redox couple on the bare gold electrode surface. After electropolymerization of tyramine on the electrode surface, the redox peak was decreased markedly. The deposited polytyramine, besides of providing free amino groups for covalent binding to the phosphate group of oligonucleotides by forming phosphoramide bond [27], it also provides an insulating property on the electrode surface.
Fig. 2.
Cyclic voltammograms recorded in 10 mM K3[Fe(CN)6] in 0.1 M KCl. The potential was swept in the range between −250 and 700 mV (vs Ag/AgCl) with a sweep rate of 100 mV s−1: (a) bare gold electrode (red); (b) gold electrode modified with polytyramine layer (black); (c) the same as (b) after immobilization of 25-mer oligo-C (green); (d) as in (c) after treatment with 1-dodecanethiol (blue).(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The oligo-C probe coupled to the polytyramine layer also contributed to the insulating behavior of the polytyramine layer. Therefore, a further decrease of redox peak was observed after subsequent immobilization of oligo-C. However, after treatment with 1-dodecanethiol the cyclic voltammograms showed complete blockage of redox reaction. The electrode surface was assumed to be completely covered so that the all influence from pin holes were considered negligible based on, that makes the electrode/solution interface to be described by resistor–capacitor in series (RC) model (Eq. (2)) above. Otherwise the capacitance would be in parallel with resistor (R(RC) model), resulting in a decrease in sensitivity due to leakage of current.
3.2. Analytical characterization
3.2.1. Capacitance change due to hybridization of target DNA
The value of registered capacitance depends on the dielectric and insulating features at the working electrode and solution interface. Fig. 3 shows the basic features of the registered capacitance; before injection of analyte, Cbeforeanalyte; after injection of analyte, Cafteranalyte; and after regeneration, Cafterregeneration.
Fig. 3.
Time course graph showing the capacitance; (i) before injection of analyte (Cbeforeanalyte) (ii) after injection of analyte (Cafteranalyte) and (iii) after regeneration (Cafterregeneration). Inset shows how to determine the capacitance change upon injection of analyte.
Upon injection of oligo-G, the hybridization with immobilized oligo-C on the electrode surface took place that resulted into a decrease in capacitance. The observed little increase in capacitance immediately after injection of oligo-G might be due to an increase in negative charge density as the polyanion DNA-probes approach the electrode. A similar argument has been applied for 5′-end biotin modified oligo-C–avidin–polyaniline films system where a decrease in electron transfer resistance was observed during hybridization [16], [18]. However, once the base-pairing between oligo-G and oligo-C took place, water and electrolyte ions (diffuse mobile layer) were displaced. The diffuse mobile layer contains high abundance of negatively charged ions that outweighed the polyanion on the DNA surface. The capacitance change was then dominated by the displacement of the diffuse mobile layer away from the electrode surface as a result of an increase in thickness and length of the capture probe layer; hence decrease in capacitance was registered [15]. Regeneration of the modified electrode surface by injecting 50 mM NaOH was used to distrupt the hydrogen bonds between the paired DNA strands (oligo-C and oligo-G) without damaging the oligo-C (capture probe). The capacitance was then returned to the original base line ready for additional measurements. Fig. 3 inset, shows how the capacitance change upon injection of analyte change was determined.
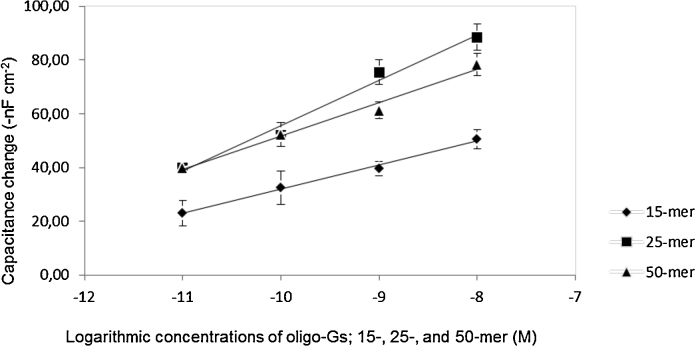
The capacitive change was proportional to the applied concentrations of the oligo-Gs, (15-, 25- and 50-mer) as depicted in Fig. 4. Applying higher number of oligo-G molecules, could lead to displacement of more number of electrolyte ions (the diffuse mobile layer) further away from the electrode surface, therefore a larger decrease in total capacitance was registered [28].
Fig. 4.
Capacitance change with an increased concentration of 15-, 25- and 50-mer oligo-G.
Nevertheless, the magnitude of registered capacitance change was also found to some extent to be dependent on the length of applied oligo-G. For instance, applying 25-mer oligo-G at electrode modified surface resulted in a capacitance shift which was approximately twice as high as that caused by a 15-mer oligo-G (Table 1). However, there was no significant difference for the capacitance change, when the same concentration of 25- and 50-mer oligo-Gs was applied on the surface. In theory, the effect of 50-mer oligo-G was expected to be twice of that 25-mer oligo-G and three times of that 15-mer oligo-G; this is because the longer DNA molecule hybridizes on the surface, the longer the capture probe layer it becomes, then the further distance the diffuse mobile layer is displaced, which would lead to larger decrease in total capacitance.
Table 1.
Calculated capacitance change upon applying of oligo-G of different lengths on the electrode surface coupled with 25-mer oligo-C (capture probe).
| Oligo-G length (-mer) | Capacitance change: average ± SD (-nF cm−2) |
||
|---|---|---|---|
| at 10−8 M | at 10−9 M | at 10−11 M | |
| 15 | 51 ± 4 | 40 ± 3 | 23 ± 5 |
| 25 | 89 ± 5 | 76 ± 5 | 40 ± 2 |
| 50 | 78 ± 4 | 61 ± 3 | 40 ± 2 |
On the contrary, the bending behavior of the long molecules, like DNA, could be the explanation of the observed results for 50-mer oligo-G. The long DNA molecules exhibit intrinsic bending behavior due to various factors, such as van der Waals force and aromatic–aromatic (π–π) interaction between the bases of the same DNA molecule. Nonetheless, Kelly et al. (1998) reported that, when an electrode surface is positively charged (by applying a positive potential pulse), the intrinsic negatively charged DNA is pulled towards the electrode and hence adopts a tilted orientation [29]. Since, the longer the DNA molecule the more negative charges it has, a 50-mer would be pulled towards the electrode surface even more (more tilted) than 25-mer oligo-G, resulted into less signal strength than expected. However, by introducing a sandwich hybridization approach, it was possible to increase the signal strength of the 50-mer oligo-G. The results of this approach are described in Section 3.2.3.
3.2.2. The effect of elevated temperature on hybridization of oligonucleotides on the sensor surface
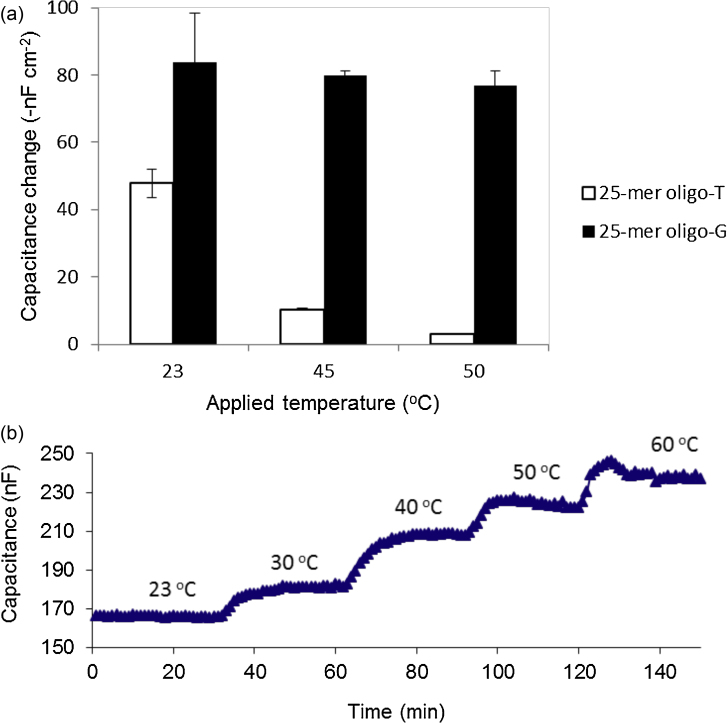
Initially, the behavior of the modified electrode surface with reference to capacitance change at different temperatures was studied (Fig. 5a). It was observed that the capacitance increased with increasing temperature. It may be suggested that, with increasing temperature, the mobility of ions in the diffuse mobile layer increases too, resulting into an increase in electrical conductivity of the electrolyte. The latter leads to an increase in the dielectric “constant” of the medium [30], hence, resulted into an increase in registered capacitance. But also, the increase in temperature could lead to reorientation of the oligo-C (capture probe) on the electrode surface from its initial tilted orientation [29], but also, became less dense which then allows the electrolyte ions to reach closer to the electrode surface and hence, a further increase in capacitance is observed.
Fig. 5.
(a) Capacitance change of modified electrode surface at different temperatures: 23, 30, 40, 50 and 60 °C. (b) Capacitance change from an electrode with immobilized 25-mer oligo-C, for injections of 10−8 M complementary (oligo-G) and non-complementary (oligo-T), at different temperatures.
The modified electrode surface seems to withstand temperatures up to 50 °C; however at 60 °C, the baseline became unstable. Observations indicated that the accumulation of released gas bubbles on the electrode surface was the probable cause of the baseline instability at higher temperatures. Therefore, it was concluded that, the maximum suitable temperature for the present experimental set-up was 50 °C. Since the hybridization of DNA is often carried out at even lower temperature, this temperature range is sufficient for most application of the DNA sensors.
The capacitance change, ΔC, due to non-specific hybridization, 25-mer oligo-T was found to decrease drastically; from 48 to 3 nF cm−2 as the temperature increased from RT to 50 °C, respectively (Fig. 5b). However, there was no significant decrease in target hybridization (25-mer oligo-G) capacitance change with respect to the increase in temperature. The capacitance changes at RT compared to 50 °C, were 84 and 77 nF cm−2, respectively.
The hybridization between the non-target (non-complementary) oligonucleotide with the capture probe could be explained by the different weak interactions such as aromatic–aromatic (π–π) interaction and van der Waals forces. The non-specific interaction could have been more efficiently reduced at 50 °C if small amounts of formamide had been added in the running buffer, without affecting the target DNA. Formamide helps to reduce the thermal stability of double stranded nucleic acid [31], [32]. However, our results suggest that, working at high temperature up 50 °C, could efficiently reduce non-specific hybridization by more than 90% without significantly altering the specific interaction. Carrarra et al. [28] studied the efficiency of the hybridization reaction at elevated temperature (80 °C), and they found that 80% of the target DNA molecules reacted with probes forming double helix while less than 10% of the non-target DNA molecules reacted with probes forming double helix. As the temperature increases, the kinetic energy increases which causes increasing molecular motion and thereby breaking the weak interactions and hence, reducing non-specific DNA hybridization.
There must be a trade-off between raising the temperature to eliminate non-specific binding and the temperature effect on the specific binding. This is an aspect that needs to be kept under control. However, it does not seem to be a problem at temperatures below 50 °C as were used in this study.
3.2.3. Sandwich hybridization
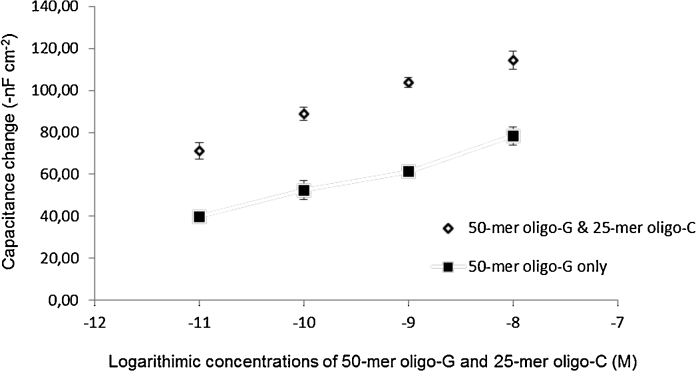
Hybridization of 50-mer oligo-G with immobilized 25-mer oligo-C on the electrode surface was initially performed. Subsequently, another 25-mer oligo-C was injected to the system at the same concentration as that of oligo-G. This resulted in a higher capacitive response as compared to response from hybridization of 50-mer oligo-G alone to the sensor surface (Fig. 6). In this study, the 50-mer oligo-G was expected to be long enough to give the intrinsic bending behavior, but also to experience higher attraction force towards the electrode surface than others (25- and 15-mer). For example, the signal from the 50-mer oligo-G at concentration of 10−8 M was lower than expected, 78-nF cm−2, but after subsequent injection of the same concentration of the shorter 25-mer oligo-C, the hybridization of partial bent oligo-G with oligo-C occurs, resulting in further increase of capacitance change to 114-nF cm−2. The subsequent injected short complementary oligonucleotide hybridized with bases from a partially bent long oligonucleotide molecule, and resulted in an amplification of the signal, which has indicated that the diffuse mobile layer was even further displaced from the surface of the gold electrode due to hybridization of DNA molecules. Increasing in signal strength could lead to an increase in sensitivity of an analytical device too. However, in some cases, signal strength is somewhat not very important when improving sensitivity of an analytical device; because the signal can be very big but the detection limit cannot be very good due to poor signal to noise level.
Fig. 6.
Sandwich hybridization 50-mer oligo-G followed by 25-mer oligo-C (♦) compared with 50-mer oligo-G alone (■).
4. Conclusions
The application of polymer chemistry (polytyramine) for insulation of a gold electrode surface and immobilization of oligo-nucleotides to that surface is a simple and repeatable method for DNA based sensors. This work has demonstrated that the capacitance change, ΔC, is proportional to the concentration of and the length of the hybridized oligo-G for the developed system. However, longer DNA molecules have to be treated differently. This was solved by using sandwich hybridization, which increased the amplitude of the signal. Non-specific hybridization was handled by elevating the temperature up to 50 °C, resulting in a tenfold decrease of the signal compared to RT. In order to adopt the developed model system for diagnostic and research purposes, it is necessary to use real bacteria/virus DNA samples. This will be the future work, to further optimize selectivity and sensitivity for the developed model system.
Acknowledgements
This work was funded by World Bank supported project titled “Capacity Building in Science, Technology and Higher education” (STHEP) which is being implemented at University of Dar es salaam, Tanzania. The support from the Swedish Research Council is gratefully acknowledged.
Footnotes
Available online 12 June 2014
References
- 1.Basselet P., Wegrzyn G., Enfors S.O., Gabig-Ciminska M. Sample processing for DNA chip array-based analysis of enterohemorrhagic Escherichia coli (EHEC) Microb. Cell Fact. 2008;7:29. doi: 10.1186/1475-2859-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake T.A., Hindler J.A., Berlin O.G.W., Bruckner D.A. Rapid identification of mycobacterium-avium complex in culture using DNA probes. J. Clin. Microbiol. 1987;25:1442–1445. doi: 10.1128/jcm.25.8.1442-1445.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakamoto H., Lemaire O., Merdinoglu D., Guesdon J.L. Comparison of enzyme-linked immunosorbent-assay (ELISA) with dot hybridization using P-32 acetylaminofluorene or 2-acetylaminoflurene (AFF)-labeled cDNA probes for the detection and characterization of beet nectrotic yellow vein virus. Mol. Cell. Probe. 1989;3:159–166. doi: 10.1016/0890-8508(89)90026-1. [DOI] [PubMed] [Google Scholar]
- 4.Wetherall B.L., McDonald P.J., Johnson A.M. Detection of campylobacter-pylori DNA by hybridization with non-radioactive probes in comparison with a P-32-labeled probe. J. Med. Microbiol. 1988;26:257–263. doi: 10.1099/00222615-26-4-257. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Elsholz B., Enfors S.O., Gabig-Ciminska M. Confirmative electric DNA array-based test for food poisoning Bacillus cereus. J. Microbiol. Method. 2007;70:55–64. doi: 10.1016/j.mimet.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Teles R., Rodrigues F., Prazeres F., Lima-Filho D.M., Luiz J. Electrochemical detection of a dengue-related oligonucleotide sequence using ferrocenium as a hybridization indicator. Sensors. 2007;7:2510–2518. doi: 10.3390/s7112510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X., Zhang X.-E., Chai Y.Q., Hu W.-P., Zhang Z.-P., Zhang X.-M., Cass E.A.G. DNA optical sensor: a rapid method for the detection of DNA hybridization. Biosens. Bioelectron. 1998;13:451–458. doi: 10.1016/s0956-5663(97)00095-x. [DOI] [PubMed] [Google Scholar]
- 8.Xu D.-K., Ma L.-R., Liu Y.-Q., Jiang Z.-H., Liu Z.-H. Development of chemiluminescent biosensing of nucleic acids based on oligonucleotide-immobilized gold surfaces. Analyst. 1999;124:533–536. [Google Scholar]
- 9.Livache T., Fouque B., Roget A., Marchand J., Bidan G., Téoule R., Mathis G. Polypyrrole DNA chip on a silicon device: example of hepatitis C virus genotyping. Anal. Biochem. 1998;255:188–194. doi: 10.1006/abio.1997.2462. [DOI] [PubMed] [Google Scholar]
- 10.Fritzsche W., Taton T.A. Metal nanoparticles as labels for heterogeneous, chip-based DNA detection. Nanotechnology. 2003;14:63–73. doi: 10.1088/0957-4484/14/12/R01. [DOI] [PubMed] [Google Scholar]
- 11.Sauthier M.L., Carroll R.L., Gorman C.B., Franzen S. Nanoparticle layers assembled through DNA hybridization: characterization and optimization. Langmuir. 2002;18:1825–1830. [Google Scholar]
- 12.Tombelli S., Minunni M., Mascini M. Piezoelectric biosensors: strategies for coupling nucleic acids to piezoelectric devices. Methods. 2005;37:48–56. doi: 10.1016/j.ymeth.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H., Tan H., Wang R., Wei W., Yao S. Immobilization of DNA on silver surface of bulk acoustic wave sensor and its application to the study of UV-C damage. Anal. Chim. Acta. 1998;374:31–38. [Google Scholar]
- 14.Francia G.D., Ferrara V.L., Manzo S., Chiavarini S. Towards a label-free optical porous silicon DNA sensor. Biosens. Bioelectron. 2005;21:661–665. doi: 10.1016/j.bios.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Berggren C., Staelhandske P., Brundell J., Johansson G. A feasibility study of a capacitive biosensor for direct detection of DNA hybridization. Electroanalysis. 1999;11:156–160. [Google Scholar]
- 16.Dharuman V., Grunwald T., Nebling E., Albers J., Blohm L., Hintsche R. Label-free impedance detection of oligonucleotide hybridisation on interdigitated ultramicroelectrodes using elctrochemical redox probes. Biosens. Bioelectron. 2005;21:645–654. doi: 10.1016/j.bios.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 17.Tenreiro A., Cordas C.M., Abrantes L.M. Oligonucleotide immobilisation on polytyramine-modified electrodes suitable for electrochemical DNA biosensors. Portugalie Electrochim. Acta. 2003;21:361–370. [Google Scholar]
- 18.Arora K., Prabhakar N., Chand S., Malhotra B.D. Ultrasensitive DNA hybridization biosensor based on polyaniline. Biosens. Bioelectron. 2007;23:613–662. doi: 10.1016/j.bios.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Mascini M. Affinity electrochemical biosensors for pollution control. Pure Appl. Chem. 2001;73:23–30. [Google Scholar]
- 20.Limbut W., Kanatharana P., Mattiasson B., Asawatreratanakul P., Thavarungkul P. A comparative study of capacitive immunosensors based on self-assembled monolayers formed from thiourea, thioctic acid, and 3-mercaptopropionic acid. Biosens. Bioelectron. 2006;22:233–240. doi: 10.1016/j.bios.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 21.Teeparuksapun K., Hedström M., Wong E.Y., Tang S., Hewlett I.K., Mattiasson B. Ultrasensitive detection of HIV-1 p24 antigen using nanofunctionalized surfaces in a capacitive immunosensor. Anal. Chem. 2010;82:8406–8411. doi: 10.1021/ac102144a. [DOI] [PubMed] [Google Scholar]
- 22.Limbut W., Hedström M., Thavarungkul P., Kanatharana P., Mattiasson B. Capacitive biosensor for detection of endotoxin. Anal. Bioanal. Chem. 2007;389:517–525. doi: 10.1007/s00216-007-1443-4. [DOI] [PubMed] [Google Scholar]
- 23.Ananthanawat C., Vilaivan T., Mekboonsonglarp W., Hoven V.P. Thiolated pyrrolidinyl peptide nucleic acids for the detection of DNA hybridization using surface plasmon resonance. Biosens. Bioelectron. 2009;24:3544–3549. doi: 10.1016/j.bios.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Bard A.J., Faulkner L.R. second ed. John Wiley & Sons; New York: 2001. Electrochemical Methods:Fundamentals Applications. [Google Scholar]
- 25.Berggren C., Bjarnason B., Johansson G. Capacitive biosensors. Electroanalysis. 2001;13:3. doi: 10.1016/s0956-5663(98)00058-x. [DOI] [PubMed] [Google Scholar]
- 26.Ramanaviciene A., Ramanavicius A. Pulsed amperometric detection of DNA with an ssDNA/polypyrrole-modified electrode. Anal. Bioanal. Chem. 2004;379:287–293. doi: 10.1007/s00216-004-2573-6. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y.-D., Pang D.-W., Hu S., Wang Z.-L., Cheng J.-K., Dai H.-P. DNA-modified electrodes; part 4: optimization of covalent immobilization of DNA on self-assembled monolayers. Talanta. 1999;49:751–756. doi: 10.1016/s0039-9140(99)00078-8. [DOI] [PubMed] [Google Scholar]
- 28.Carraraa S., Gürkaynakb F.K., Guiduccic C., Stagnic C., Beninic L., Leblebicib Y., Bruno S., Michelib G. Interface layering phenomena in capacitance detection of DNA with biochips. Sens. Transducers. 2007;76:969–977. [Google Scholar]
- 29.Kelly S.O., Barton J.K., Jackson N.M., McPherson L.D., Potter A.B., Spain E.M., Allen M.J., Hill M.G. Orienting DNA helices on gold using applied electric fields. Langmuir. 1998;14:6781–6784. [Google Scholar]
- 30.Hong J., Yoon D.S., Park M., Choi J., Kim T.S., Im G., Kim S., Pak Y.E., No K. A dielectric biosensor using the capacitance change with ac frequency integrated on glass substrates. Jpn. J. Appl. Phys. 2004;43:5639–5645. [Google Scholar]
- 31.McConaughy B.L., Laird C.D., McCarthy B.J. Nucleic acid reassociation in formamide. Biochemistry. 1969;8:3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- 32.Blake R.D., Delcourt S.G. Thermodynamic effects of formamide on DNA stability. Nucleic Acid Res. 1996;24:2095–2103. doi: 10.1093/nar/24.11.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]