Abstract
The present study emphasizes the biosurfactant mediated anthracene degradation by a marine alkaliphile Bacillus licheniformis (MTCC 5514). The isolate, MTCC 5514 degraded >95% of 300 ppm anthracene in an aqueous medium within 22 days and the degradation percentage reduced significantly when the concentration of anthracene increased to above 500 ppm. Naphthalene, naphthalene 2-methyl, phthalic acid and benzene acetic acid are the products of degradation identified based on thin layer chromatography, high performance liquid chromatography, gas chromatography and mass analyses. It has been observed that the degradation is initiated by the biosurfactant of the isolate for solubilization through micellation and then the alkali pH and intra/extra cellular degradative enzymes accomplish the degradation process. Encoding of genes responsible for biosurfactant production (licA3) as well as catabolic reactions (C23O) made with suitable primers designed. The study concludes in situ production of biosurfactant mediates the degradation of anthracene by B. licheniformis.
Keywords: Anthracene; Marine Bacillus sp; Biosurfactants; Catechol 2,3 dioxygenase; Degradation
1. Introduction
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous pollutants generated from various anthropogenic activities [20]. These compounds are grouped under hazardous aromatic compounds, having two or more fused benzene rings arranged in such a way [25] and insoluble in water [13] and persistence nature. These compounds have been identified as toxic, carcinogenic and some compounds demonstrated teratogenic effects [23].
The insoluble and persistence nature of PAHs are the major limitations on the removal or remediation from the soil or aqueous media. The old as well as newer treatment technologies employed in the removal of contaminants are ineffective in the case of PAHs [32]. Though chemical oxidants able to cleave the fused rings and the formation of hydroxylated or oxygenated metabolites needs immediate attention. The only option available is the use of microorganisms. Among the microorganisms, some of the microbial species have the capacity to tolerate PAHs and some species even try to metabolize. But the time taken to catabolism and the transformations of catabolized products were the major drawbacks realized. It has been understood that less than 25 numbers of bacterial species exhibited the degradation of PAHs [31] and the screening and identification of potential species need intensive research.
The degradative capacity of the demonstrated bacterial species was through the dissolution and the genes responsible for the catabolism. The surface-active agent produced by these organisms mediates the dissolution. These surface-active agents interact with the insoluble compounds by reducing the interfacial tension and make them available to the microbes [11]. The role of surface-active agents for the degradation of PAHs is in reports [16]. Furthermore, it has been realized that compared to the terrestrial species, microorganisms of marine origin displayed the higher percentage of production of surface-active agents [18]. Since, the marine source is the ultimate contaminated site, the micro flora of marine source may have the inbuilt capacity to remediate the contaminants at the fastest rate and have robustness in solubilizing as well as degrading the PAHs [22]. It is challenging to have terrestrial microbes with complete robustness, and most of the organisms require an external addition of surface active agents as reported [18]. The present study reveals the potency of marine bacterial isolate in the degradation of the selected PAHs, namely anthracene.
Anthracene, together with other polycyclic aromatic hydrocarbons (PAHs), is a persistent and toxic soil contaminant [14]. Anthracene is sparingly soluble in water, highly resistant to nucleophilic attack and hence, recalcitrant to biodegradation [12] and accumulate easily in the ecosystem. In powdered form it causes irritation to the eyes, nose or lungs and is a probable inducer of tumors [8]. Once anthracene enters the body, it appears to target the skin, stomach, intestines and the lymphatic system. It may even cause burning, itching and edema. Due to its low solubility, most of the researchers attempt to remove anthracene in soil/sediment. Only very few studies are there on the biological removal of anthracene from aqueous media. Microbial degradation of anthracene is an inexpensive way of removing/remediating anthracene from soil and water. Microbial remediation removes or immobilizes the pollutants and reducing the toxicity with a very low environmental impact. A variety of bacterial species have been isolated to utilize anthracene as the sole source of carbon and energy [24]. Considerable attention has been paid on the metabolic pathways and genetics of degradation of low molecular mass PAHs, such as naphthalene, phenanthrene and anthracene, by Gram −ve bacteria, particularly, the genus, Pseudomonas and Sphingomonas [5]. However, less attention has been intended on the degradation of PAHs by Gram +ve bacteria, Bacillus species.
Bacillus species are the versatile microbial species studied extensively for the production of secondary metabolites and surface active agents, such as surfactin, fengycin, lichenysin, iturin, pumilacidin and bacillomycin [30]. Since the available literatures suggested that the ring opening followed by further cleavage of PAHs takes place at pH above neutral, Bacillus species with the said robustness (biosurfactant production as well as growth at alkali pH) have been the choice to study the degradation of PAHs.
Thus, the present study exemplifies the biosurfactant mediated anthracene degradation efficacy of marine bacterial species in an aqueous medium. In brief, the study explores degradation of anthracene and finger printing of the degradative products using TLC, HPLC and GC–MS analyses. Further, the study extended to identify the genes responsible for the biosurfactant production and said degradation, elucidation of degradation pathway and the schematic representation on the degradation process.
2. Materials and methods
2.1. Chemicals
Anthracene (99% purity) was purchased from HiMedia. Bacteriological media, chemicals, silica gel coated TLC plates and solvents were purchased from Hi-Media and Sisco Research Laboratory (SRL), Mumbai, India.
2.2. Isolate MTCC 5514
Isolate MTCC 5514 was initially screened from marine samples, characterized and identified according to the standard protocol and procedures and deposited in Microbial Type Culture Collection (MTCC), Chandigarh, India and used for the study. The 16S rRNA gene sequence was submitted to NCBI with the accession number HM145910.
2.3. Biodegradation studies: experimental setup
To the pre-sterilized medium (Zobell Marine Broth, (HiMedia)), anthracene at 100–1000 ppm concentrations were supplemented aseptically and inoculated with the 1 × 105 cells/mL of MTCC 5514, incubated at 37 °C under shaking condition (200 rpm) for the period of 10, 16 and 22 days.
2.4. Growth and pH analysis
Growth of the marine isolate MTCC 5514 in the presence of anthracene at varying concentrations, viz., 0, 100, 300, 500, 750 and 1000 ppm was observed by measuring the optical density of the culture broth at 600 nm at 24 h intervals using UV–visible spectrophotometer (UV-2450, Shimadzu, Japan). The pH of the growth medium measured at 24 h intervals till 22 days using Elico pH meter, model CL 54.
2.5. Surfactant activity measurements
The surfactant property of the extracellular medium during the growth of the isolate was qualitatively measured by drop collapse test and quantitatively by plate method using GBX-3S tensiometer (DM) at room temperature [3]. Both synthetic (SDS, Tween 20, Triton X 100 (at 1% concentration)) and commercially available surfactant (Lecithin (at 10% concentration)) were used for comparison.
2.6. Instrumental analysis for the identification of the degraded products
2.6.1. Thin layer chromatography (TLC) analysis
Thin layer chromatography was used as a primary tool to identify the degraded products. Followed by removal of the samples, the cell free supernatant was mixed with ethyl acetate and the ethyl acetate fraction was separated and subjected to TLC analysis using chloroform:ethyl acetate:acetic acid (5:5:0.1) (v/v) as a solvent system and exposed to 2% Gibbs reagent after drying.
2.6.2. High performance liquid chromatography (HPLC) analysis
Followed by the extraction with ethyl acetate, the samples were filtered through 0.2 mm syringe filter and analyzed in a high performance liquid chromatography. HPLC (SHIMADZU, SPD-10 A VP) with the silicon C18 column was used to separate and analyze PAHs under isocratic condition (solvent – acetonitrile:water (80:20) (v/v) detection wavelength – 254 nm). The flow rate of the mobile phase (acetonitrile) was maintained at 0.5 mL/min. The samples (20 μL) were injected to HPLC analyzer for the analysis of PAHs. Based on the retention time, the fractions were collected and further subjected to analysis.
2.6.3. Gas chromatography–mass spectrometry (GC–MS) analysis
A Hewlett-Packard 689 gas chromatography equipped with 5973 mass spectrometer with HP-5MS (30 m × 0.25 mm I D × 0.25 μm) fused silica capillary column was used for the analysis. The column temperature program was set at 100 °C hold for 1 min, 15 °C/min to 160 °C and 5 °C/min to 300 °C hold for 7 min. The GC injector was held isothermally at 280 °C with a splitless period of 3 min. Helium was used as the carrier gas, at a flow rate of 1 mL/min by using electronic pressure control. The GC–MS interface temperature was maintained at 280 °C. The MS was operated in electron impact (EI) ionization mode with electron energy of 70 eV and the scan to determine appropriate masses for selected ion monitoring ranged from 50 to 500 amu (atom to mass unit). Standards from Sigma Aldrich were used for the PAH (anthracene) and their metabolites. GC–MS library search was used to confirm the metabolites without standards.
2.7. PCR amplification and sequencing of 16S rRNA
Genomic DNA (gDNA) of MTCC 5514 was extracted from using DNeasy Blood & Tissue Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer’s protocol for Gram +ve bacteria. The 16S rRNA was PCR amplified using the universal primers 8F: 5′-AGAGTTTGATCCTGGCTCAG-3′ and 1492R: 5′-GGCTACCTTGTTACGACTT-3′ as described by Turner et al. [29]. Homology of the 16S rRNA sequence was compared with sequences available in databases using Blast from the National Center for Biotechnology Information [2] and the Ribosomal Database Project [7]. Alignment of obtained 16S rRNA sequence and sequences from the databases, were all trimmed to the same length using CLUSTAL Omega algorithm [26]. The sequence details were already submitted to NCBI with the wide accession no. HM145910.
2.8. Encoding of genes responsible for surface-active agent production and degradative enzyme
The genes encoding the biosurfactant (licA3) and catechol 2,3 dioxygenase (C23O) of the chosen organism was studied and the details were summarized in the following paragraph.
2.8.1. PCR primer design
The primers for both, surfactant (licA3) and catechol 2,3 dioxygenase (C23O) genes were designed from earlier reports [6], [27] and were synthesized at Eurofins Genomics India Pvt. Ltd. A portion of surfactant gene 0.26 kb (licA3) gene was pulled out from the genomic DNA using F: 5′- CAA AAG CGC ATC ATA CCA CGT TGA G - 3′ and R: 5′-AGC GGC ACA TAT TGA TGC GGT TC - 3′ primers, with 2.5 U of Taq DNA polymerase in a 25 μL reaction mixture, consisting of 100 ng of genomic DNA, 20 pmol of each primer, 200 μM dNTPs and 1X Taq buffer with 2 mM MgCl2. PCR was conducted using the following temperature profile: initial denaturation at 93 °C for 2 min, then 38 cycles of 35 s at 93 °C, 35 s at 48 °C, and 45 s at 72 °C; and finally an extension reaction of 5 min at 72 °C.
Likewise a portion of catechol gene 1.27 kb (C23O) was pulled out using F: 5′- ATG AGC AAC AAA TAC GAA TT- 3′ and R: 5′- TCA AAC GGT CAA TCT GAT AT- 3′ primers, with 1.5 U of Taq DNA polymerase in a 25 μL reaction mixture, consisting of 100 ng of genomic DNA, 20 pmol of each primer, 200 μM dNTPs and 1X Taq buffer with 1.5 mM MgCl2. PCR was conducted using the following temperature profile: initial denaturation at 93 °C for 2 min, then 30 cycles of 1 min at 93 °C, 35 s at 45 °C, and 1.5 min at 72 °C; and finally an extension reaction of 5 min at 72 °C.
PCR products were analyzed by electrophoresis on 1% agarose TAE gels. The expected DNA bands of 0.26/1.27 kb were excised from gel and purified using the Gel Extraction Kit (Sigma–Aldrich, USA) as per the manufacturer’s protocol. Sequencing reactions were carried out with a Big Dye Terminator cycle sequencing kit by using ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA, USA).
3. Results
3.1. Characterization and identification of the isolate MTCC 5514
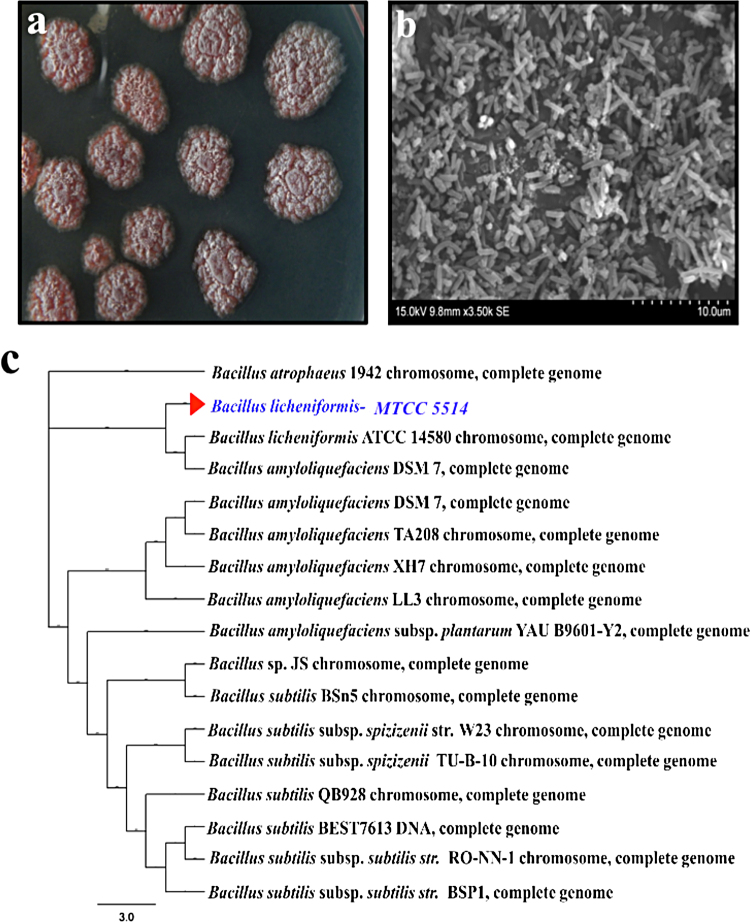
Fig. 1(a–c) illustrates the morphology, SEM image and phylogenic profile of the isolate MTCC 5514 employed in the present study. The bacterial colony has irregular margin, rough surface with pink pigmentation. The staining studies revealed the Gram +ve nature of the isolate and the SEM analysis suggested the short rod nature of the isolate. The phylogenic profile infers the isolate MTCC 5514 belongs to Bacillus licheniformis. The distance matrix showed the genetic distance value between MTCC 5514 with B. licheniformis ATCC 14580 was 0.004.
Fig. 1.
(a) Plate morphology of marine isolate MTCC 5514 grown in Zobell marine agar. (b) Scanning electron micrograph analysis of marine isolate MTCC 5514. (c) Phylogenic analysis of marine isolate MTCC 5514 constructed by neighbor-joining method, bar 3.0 substitutions per nucleotide position.
3.2. Degradation studies
Anthracene biodegradation study carried out at 37 °C under shaking conditions using MTCC 5514 displayed an interesting observation. The physical observations made during the growth suggested that from day 1 to till day 7 most of the anthracene molecules (irrespective of the concentrations studied) were settled at the bottom of the flask, despite, much turbidity in the external medium due to the growth of the organisms. However, after day 15, deposition of only fewer anthracene molecules at lower concentration than higher concentrations was observed. Further, after 22 days, no deposits were found at lower concentration, however, a fewer deposits were at higher concentration.
Samples withdrawn at scheduled time intervals (10, 16 and 22 days) were subjected to various analyses after extracting with ethyl acetate. However, before extraction, analysis such as pH, biomass and surface activity were made for all the concentrations. The percentage of degradation of anthracene was calculated based on the absorption displayed in UV–visible spectral analysis at 254 nm and using standard graph.
3.3. Growth profile
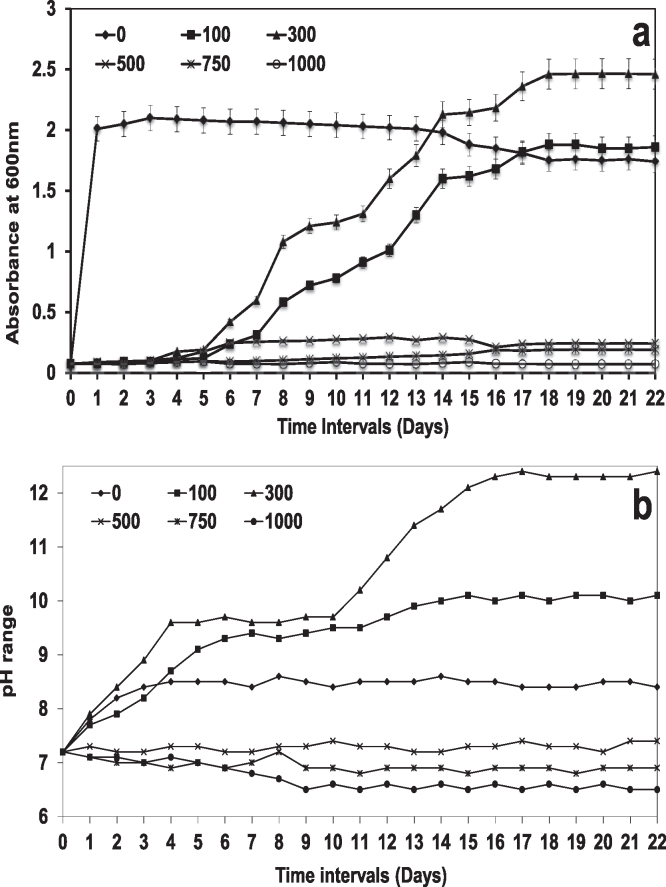
Fig. 2a displays the growth profile of the isolate MTCC 5514 in the presence of anthracene at 100–1000 ppm concentration. The chosen isolate MTCC 5514 showed a bi-phasic growth profile in the presence of anthracene at 100 and 300 ppm concentration. At 500 ppm concentration, the organism showed tolerance till 15 days with no bi-phasic growth. This kind of tolerance could be reasoned to the presence of inbuilt stress proteins of Gram +ve bacteria. However, with 750 and 1000 ppm concentration, no growth was observed. On comparing the growth of MTCC 5514 in the presence of 100 and 300 ppm concentration, growth was more pronounced with 300 ppm than with 100 ppm, suggested the effective metabolism of anthracene molecule. Till 7 days, the growth OD was less than 0.5 (at 600 nm), whereas, after 15 days, the growth OD increases to more than 1.0 and maintained till 18 days, and after that the growth OD slowly increases to 2.2 and again maintained till 22 days. And between day 18 and day 22 a stationary phase has been reached.
Fig. 2.
(a) Growth profile of marine isolate MTCC 5514 grown in the presence of increasing concentration of anthracene (0–1000 ppm). (b) pH profile of the cell free broth measured at different time intervals during the incubation of MTCC 5514 in the presence of different concentration of antheracene (0–1000 ppm).
The pH of the external medium, an important variable in the degradation studies was determined and Fig. 2b displays the pH profile with reference to incubation days. The pH of the external medium showed a slow increase from the initial pH of 7.2 ± 0.2 to 8.2 ± 0.4 for the control sample, and rose to >9.0 ± 0.2 after 15 days of incubation for both 100 and 300 ppm concentration. On further increasing the incubation period, pH of the medium also increased in the experimental samples compared to control and the final pH of >12.0 ± 0.4 was observed after 22 days of incubation at 300 ppm concentration, whereas, it was only less than 10 ± 0.2 at 100 ppm concentration. For other concentrations, the pH was around 7.0 ± 0.2 and it even decreased to 6.5 ± 0.2.
Surface activity measurements of the external medium displayed the maximum activity of 28 ± 4 mN/m throughout the experimental period of 22 days for the control samples as well as the samples of 100 and 300 ppm concentration of anthracene indented. Though characterization of surface active agents (results not shown) reveal more than 75% similarity with the commercially available surfactin, however, the non-hemolytic and non-ionic behavior of surfactant of MTCC 5514 demonstrated the difference. Thus, the identified biosurfactant was named as ‘Microsurf’.
3.4. Fingerprinting of degraded products
The preliminary TLC analysis of the ethyl acetate extraction (after 15 days of incubation) of the extracellular medium displayed more than 7 spots with different Rf values. And from HPLC analysis five fractions were received and GC–MS analysis of the fractions reveals the nature of the degraded products.
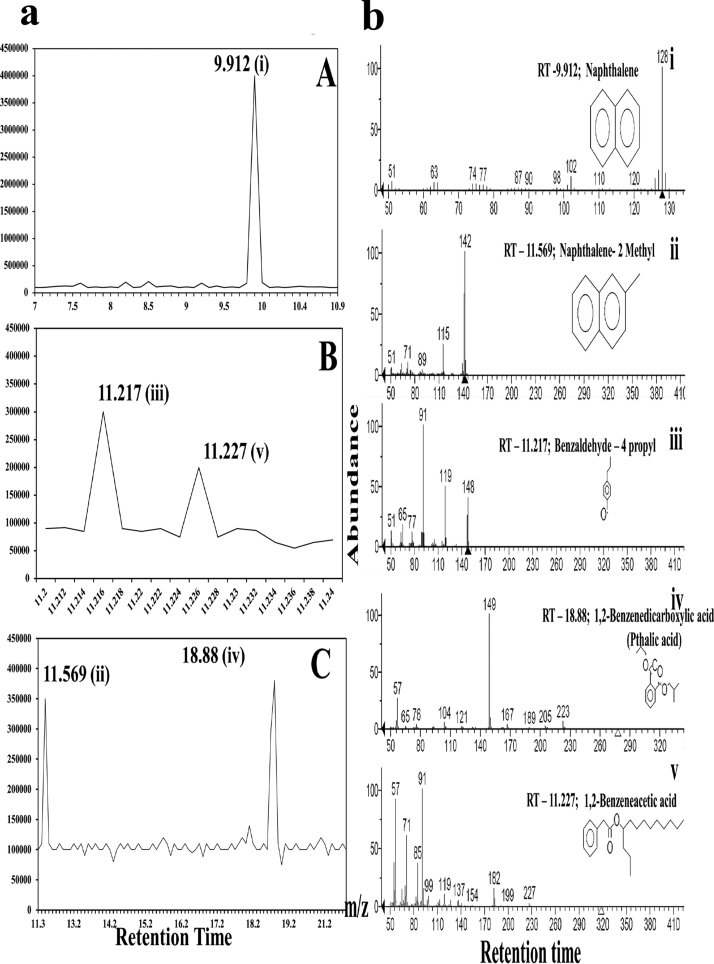
Fig. 3a (A–C) illustrates the GC – chromatogram followed by Fig. 3b (i–v) on MS analyses. Mass spectral analyses and the library details suggested that (i) naphthalene (m/z-128), (ii) naphthalene-2-methyl (m/z-142), (iii) benzaldehyde-4-propyl (m/z-148), (iv) 1,2, benzene di-carboxylic acid (m/z-167) and (v) benzene acetic acid (m/z-137) were the major degraded products detected. All the spectral analyses displayed more than 95% similarity with the mass databases. The percentage degradation analysis suggested that more than 95% of anthracene molecule degraded when the concentration of anthracene was at 300 ppm.
Fig. 3.
(a) GC analysis of metabolites obtained during the incubation of marine isolate in the presence of anthracene [(i) naphthalene; (ii) naphthalene-2-methyl; (iii) benzaldehyde-4-propyl; (iv) 1,2, benzene dicarboxylic acid; (v) benzeneacetic acid]. (b) MS analysis of metabolites obtained during the incubation of marine isolate in the presence of anthracene. [(i) naphthalene; (ii) naphthalene-2-methyl; (iii) benzaldehyde-4-propyl; (iv) 1,2, benzene dicarboxylic acid; (v) benzeneacetic acid].
3.5. Encoding of genes
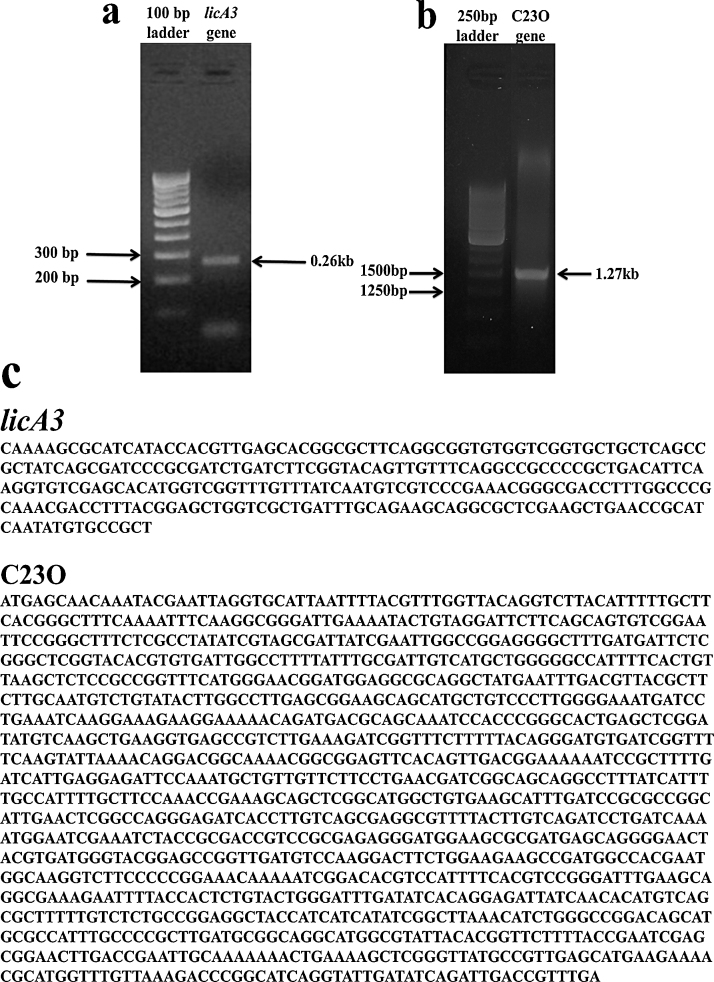
Followed by the identification of the metabolites, the study has been reversed back to examine the isolate for the specific genes responsible for the anthracene catabolism. As described in Section 1, the presence of dissolution agents is the primary requirement of the microorganisms to attack or encounter the lipophilic molecule. Though, the isolate displayed surface-active agents during the growth, the gene responsible for the production of surface-active agent was examined using molecular techniques. Fig. 4a illustrates the PCR amplified product of licA3 gene determined with 0.26 kb and Fig. 4b depicts the PCR amplified product of catechol 2,3 dioxygenase (C23O) gene obtained using primers designed specific for hydrocarbon degradation yielded an amplified product of the expected size of 1.27 kb respectively. Conserved regions of MTCC 5514 were selected to design oligonucleotide primers for detection of the genes. Thus, it has been confirmed that the chosen isolates catabolize anthracene through dioxygenase pathway.
Fig. 4.
(a) PCR – amplification of licA3 genes of MTCC5514. (b) PCR – amplification of C23O genes of MTCC5514. (c) Aligned sequences details of the PCR amplified product respective to licA3 and C23O.
The sequences of the PCR products obtained were verified in the NCBI databases for the gene/species confirmation and thus validating the presence of the genes in the selected strains of Bacillus. Fig. 4c depicts the aligned sequence of PCR products respective to licA3 and C23O genes encoded for surface active agent and degradative enzyme of MTCC 5514.
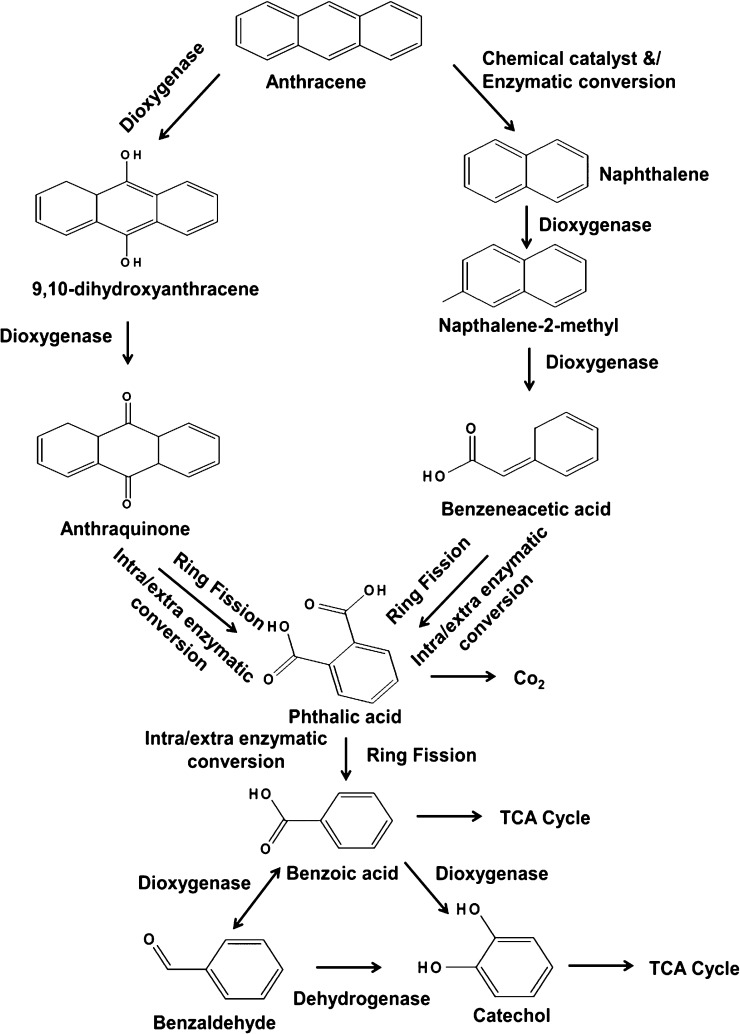
3.6. Elucidation of anthracene degradation pathway
Fig. 5 depicts the proposed degradation pathway elucidated based on the metabolites identified. The indented anthracene molecule may be degraded in two different ways. The left hand side pathway suggested that the primary attack of anthracene after day 15 (because synthesize of catabolizing enzymes triggers only after nutrient depletion) was through a dioxygenase enzyme system, which leads to the formation of di-hydroxy anthracene, which, further and immediate attack by the same enzyme system transformed to anthraquinone.
Fig. 5.
Proposed anthracene degradation pathway exhibited by MTCC 5514 elucidated based on the finger printing of the degraded products identified.
However, the right side reactions demonstrated that, the generation of phthalic acid via naphthalene (as evidenced from GC–MS analysis) and may further degraded as shown and enter in to TCA cycle.
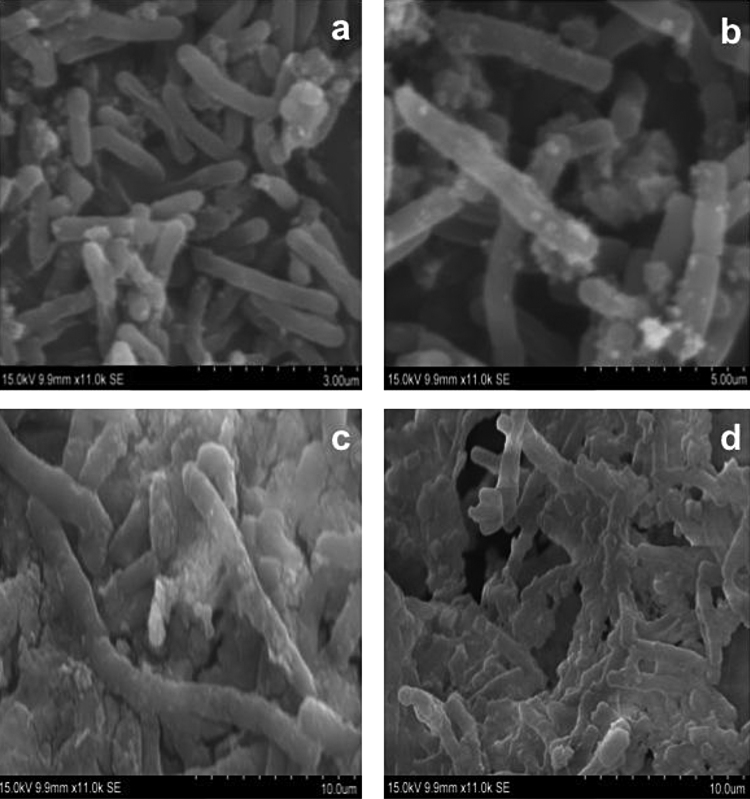
3.7. Cell surface morphology analysis after degradation of anthracene
Fig. 6 depicts the SEM micrograph of biomass obtained at scheduled time intervals of 10, 16 and 22 days showed interesting observations. The filamentous growth was extensive with increased cell volume with reference to the incubation period and in the presence of the test compound anthracene. The maximum increase in cell volume was observed on day 16 samples, and further on day 22, high filamentous growth leads to aggregation of cells in the form of biofilm and showed a clumsy mass.
Fig. 6.
Scanning electron microscopic analysis of potent marine isolate MTCC 5514 in the presence of anthracene at different incubation periods.
4. Discussion
In the present study, a potential marine isolate MTCC 5514 was tested for its anthracene degradation efficacy and the results of the study further confirmed the degradation of anthracene. The isolate MTCC 5514 displayed the production of surface-active agents and it showed tolerance up to pH 12.0 during the degradation process. In general, Gram +ve bacteria, especially the Bacillus species are tolerant to toxic chemicals and solvents, were the added advantages to these genera [34].
Results on bi-phasic growth pattern suggests, the chosen isolate metabolize anthracene at very slow and steady state and the stationary phase like observation made after day 7 to day 18 and after 18 to 22 days, could be due to the time taken for the solubilization of the degraded products for further availability to the organisms.
Further, an increase in pH of the external medium for the control sample reasoned to the alkaliphilic nature of the isolate MTCC 5514. However, meager reports were on the increase in pH of the medium in the presence of PAHs like anthracene, whereas, Zaidi et al. [35] observed an increase in pH in the presence of PAHs like naphthalene, pyrene, phenanthrene and further interpreted that even a small shift in pH play a dramatic change in the degradation of PAHs in oligotrophic environment. With regard to the surface activity measurements, high surface activity and the alkaline pH increase the solubility of the intended anthracene molecules and also enhance the selective permeability of the molecules. Mahanty et al. [17] reported that the emulsification activity of surface-active agents was high at alkaline pH. Since, the adherence of a bacterial cell to hydrocarbon–water interface was more important, in the present study, it was affected through the surface-active agents. In the present study, the surface-active agent ‘Microsurf’, displayed an extensive applications including the removal of chromium VI [11]. Moreover, because of the transport of various molecules, the change in membrane fluidity accelerates the biosynthesis of phospholipids and could be the reason for the sustainability in the concentration and activity of surface-active agent of MTCC 5514 throughout the experimental period.
The presence of both, licA3 and C23O gene in MTCC 5514 correlates well with the literatures reported. Though biosurfactant helps to solubilize or mediate the interaction between the organism and the compound, the catabolic reactions observed in the present study has been executed by the dioxygenase genes as observed from the amplified product of 1.27 kb. This gene was identified as an important gene responsible for catabolizing low molecular weight as well as high molecular weight PAHs [15].
According to Nievas et al. [21], both, dioxygenase and monooxygenase enzymes were considered as major degrading enzymes in the degradation of PAHs. Ahmed et al. [1] observed the formation of anthrone by alkaliphilic bacteria at C9 and C10 position and further leads to the formation of quinone product of PAHs. According to Cerniglia [5] and Ye et al. [33], anthraquinone is the common oxidation products of PAH degradation. Ring opening of anthraquinone molecule by the presence of extracellular and intracellular enzyme systems generates phthalic acid, which, further undergo ring fission reaction by the enzymatic system release benzoic acid and then transformed to benzaldehyde, which, then converted to catechol by the same enzyme system. According to Evans et al. [10] and Cerniglia and Yang [4], similar to naphthalene degradation pathway, catechol also degraded to simple aliphatic compounds. Though naphthalene has been identified as one of the degraded products in the present study, the presence of di-hydroxy anthracene and anthraquinone reveals that the catabolism has been realized through dioxygenase system of the isolate. The initial enzymatic attack at C-1 and C-2 position observed in the present study showed similarity with the naphthalene dioxygenase system. Though complete degradation of anthracene by Pseudomonas, Sphingomonas, Nocardia, Beijerinckia, Rhodococcus and Mycobacterium [9], [10], [19] in the presence of external surface-active agent, nevertheless, in the present study, in situ production of surface-active agent mediates the degradation as observed.
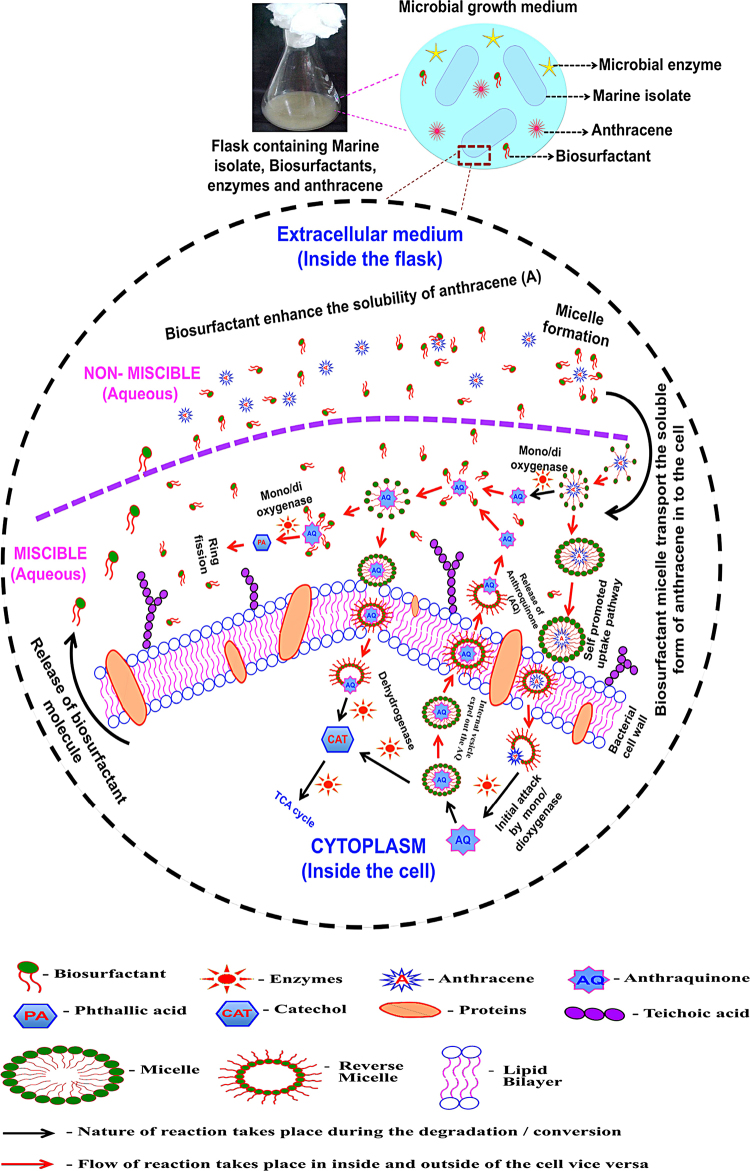
Further, the presence of anthracene and the process of degradation tremendously altered the cell volume. The modification of cell surface morphology with reference to external stress was observed in both Gram −ve bacteria and Gram +ve bacteria. An extensive filamentous growth of B. licheniformis was observed when grown in the presence of organic solvents and a toxic compound [28] and suggested that this kind of filamentation of a bacterial cell reduces the environmental stress and also helps in communicating and exchange the information. However, the observations made in the present study suggested that the continuous flow of the molecules by selective permeability of cell membrane of MTCC 5514 and the micelle and reverse micellar aggregations occurs in the lipid bilayer as shown schematically (Scheme 1), reflected as increase in cell volume, however, the said hypothesis, further needs explorations. In addition, the increase in cell volume may also be reasoned to the chemotaxis behavior of the isolate MTCC 5514.
Scheme 1.
Hypothetical representation of various possibilities of degradation of anthracene in the presence of in situ production of biosurfactants and enzymes, which provides tolerance and nutrients to the marine isolate MTCC 5514.
4.1. Hypothesis derived from the present study
Though, the degradation was ascertained based on the release of degradation of products, the actual degradation mechanism can be explained schematically. Since, it has been observed that, biosurfactant, pH, intra/extra cellular and degradative enzymes, temperature, shaking condition and concentration of the test compound played the significant role in the degradation observed, Scheme 1 convey the actual steps followed during the degradation studies. In brief, once the target molecule intended to the external medium, the presence of surface-active agents result with the formation of micelles and by selective permeability, micelles containing the anthracene molecule make an entry into the lipid bi-layer. Formation of reverse micelle in the lipid bi-layer enhances the flow of molecule and finally enters to the cytosol or in the membrane itself, where, degradation by the actual dioxygenase enzyme system starts with the release of degraded products namely, naphthalene/hydroxylated anthracene, which, further oxidized to anthraquinone. The degraded products of first step may then expel out from the membrane/cytosol through the internal surface-active agents. Once, these products came out, the alkali pH, the available enzyme system and the surface-active agents facilitate the flow of the molecule inside the membrane. This kind of transport of molecules from inside to outside and vice versa occurs till the realization of complete degradation. The time taken for the entry and exit of each molecule result with the biphasic growth profile as observed in the present study. Further, an increase in the average volume of the cell may also be reasoned to the continuous opening and closing of the bi-layer as shown schematically.
5. Conclusion
In the present study, marine alkaliphile MTCC 5514, degrade the anthracene molecule up to 300 ppm concentration in an aqueous media through its in-built genes responsible for the surface active agent (licA3) production and catabolic degradative enzyme (C23O) system. Further, this organism displayed tolerance up to 500 ppm of anthracene concentration. The adoption period of less than 7 days suggested that the isolate might have pre-exposure to the target molecule and the triggering of de nova synthesis of the enzyme leads to the degradation of anthracene.
Acknowledgement
The authors acknowledge Council of Scientific and Industrial Research, New Delhi, for the financial assistance provided in the form of network project (CSC 0127) under 12th Five Year Plan.
Footnotes
Available online 17 October 2014
References
- 1.Ahmed A., Choi C.H., Choi M.C., Kim S. Mechanisms behind the generation of protonated ions for polyaromatic hydrocarbons by atmospheric pressure photoionization. Anal. Chem. 2012;84:1146–1151. doi: 10.1021/ac202858k. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodour A.A., Miller-Maier R.M. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing microorganisms. J. Microbiol. Met. 1998;32:273–280. [Google Scholar]
- 4.Cerniglia C.E., Yang S.K. Stereoselective metabolism of anthracene and phenanthrene by the fungus Cunninghamella elegans. Appl. Environ. Microbiol. 1984;47:119–124. doi: 10.1128/aem.47.1.119-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerniglia C.E. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 1992;3:351–368. [Google Scholar]
- 6.Chi B.K., Kobayashi K., Albrecht D., Hecker M., Antelmann H. The paralogous MarR/DUF24-family repressors YodB and CatR control expression of the catechol dioxygenase CatE in Bacillus subtilis. J. Bacteriol. 2010;192:4571–4581. doi: 10.1128/JB.00409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole J.R., Wang Q., Cardenas E., Fish J., Chai B., Farris R.J., Kulam Syed Mohideen A.S., McGarrell D.M., Marsh T., Garrity Tiedje J.M. The ribosomal database project improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das P., Mukherjee S., Sen R. Improved bioavailability and biodegradation of a model polyaromatic hydrocarbon by a biosurfactant producing bacterium of marine origin. Chemosphere. 2008;72:1229–1234. doi: 10.1016/j.chemosphere.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Dean-Ross D., Cerniglia C.E. Degradation of pyrene by Mycobacterium flavescens. Appl. Microbiol. Biotechnol. 1996;46:307–312. doi: 10.1007/s002530050822. [DOI] [PubMed] [Google Scholar]
- 10.Evans W.C., Fernley H.N., Griffiths E. Oxidative metabolism of phenanthrene and anthracene by soil Pseudomonads. Biochem. J. 1965;95:819–831. doi: 10.1042/bj0950819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnanamani A., Kavitha V., Radhakrishnan N., Rajakumar G.S., Sekaran G., Mandal A.B. Microbial products (biosurfactant and extracellular chromate reductase) of marine microorganism are the potential agents reduce the oxidative stress induced by toxic heavy metals. Colloids Surf. B: Biointer. 2010;79:334–339. doi: 10.1016/j.colsurfb.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Guieysse B., Cirne M.D., Mattiasson B. Microbial degradation of phenanthrene and pyrene in a two-liquid phase partitioning bioreactor. Appl. Microbiol. Biotechnol. 2001;56:796–802. doi: 10.1007/s002530100706. [DOI] [PubMed] [Google Scholar]
- 13.Hafez E.E., Rashad M., Abd-Elsalamand H.E., EL-Hanafy A.A. The polyaromatic hydrocarbons as a serious environmental pollutants and the role of bioremediation to overcome this problem. In: Basu S.K., Datta Banik S., editors. Environment, Health and Nutrition – Global Issues. APH Publishing Corporation; New Delhi, India: 2008. [Google Scholar]
- 14.Juhasz A.L., Stanley G.A., Britz M.L. Microbial degradation and detoxification of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia strain VUN 10003. Lett. Appl. Microbiol. 2000;30:396–401. doi: 10.1046/j.1472-765x.2000.00733.x. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y.H., Freeman J.P., Moody J.D., Engesser K.H., Cerniglia C.E. Effects of pH on the degradation of phenanthrene and pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Microbiol. Biotechnol. 2005;67:275–285. doi: 10.1007/s00253-004-1796-y. [DOI] [PubMed] [Google Scholar]
- 16.Lai C.C., Huang C.C., Wei Y.H., Chang J.S. Biosurfactant enhanced removal of total petroleum hydrocarbons from contaminated soil. J. Hazard. Mater. 2009;167:609–614. doi: 10.1016/j.jhazmat.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Mahanty B., Pakshirajan K., Dasu V.V. Production and properties of a biosurfactant applied to polycyclic aromatic hydrocarbon solubilisation. Appl. Biochem. Biotechnol. 2006;134:129–141. doi: 10.1385/abab:134:2:129. [DOI] [PubMed] [Google Scholar]
- 18.Maneerat S. Biosurfactants from marine microorganisms. J. Sci. Technol. 2000;27:1263–1272. [Google Scholar]
- 19.Moody J.D., Freeman J.P., Doerge D.R., Cerniglia C.E. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 2001;67:1476–1483. doi: 10.1128/AEM.67.4.1476-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni Chadhain S.M., Norman R.S., Pesce K.V., Kukor J.J., Zylstra G.J. Microbial dioxygenase gene population shifts during polycyclic aromatic hydrocarbon biodegradation. Appl. Environ. Microbiol. 2006;72:4078–4087. doi: 10.1128/AEM.02969-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nievas M.L., Commendatore M.G., Olivera N.L., Esteves J.L., Bucala V. Biodegradation of bilge waste from Patagonia with an indigenous microbial community. Bioresour. Technol. 2006;97:2280–2290. doi: 10.1016/j.biortech.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 22.Plante C.J., Coe K.M., Plante R.G. Isolation of surfactant-resistant bacteria from natural, surfactant-rich marine habitats. Appl. Environ. Microbiol. 2008;74:5093–5099. doi: 10.1128/AEM.02734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruma R., Ray R., Chowdhury R., Bhattacharya P. Degradation of polyaromatic hydrocarbons by mixed culture isolated from oil contaminated soil. A bioprocess engineering study. Ind. J. Biotechnol. 2007;6:107–113. [Google Scholar]
- 24.Santos E.C., Jacques R.J.S., Bento F.M., Peralba M.C.R., Selbach P.A., Sa E.L.S., Cameroon F.A.O. Anthracene biodegradation and surface activity by an iron stimulated Pseudomonas sp. Bioresour. Technol. 2008;99:2644–2649. doi: 10.1016/j.biortech.2007.04.050. [DOI] [PubMed] [Google Scholar]
- 25.Shin K., Kim K., Seagren E. Combined effects of pH and biosurfactant addition on solubilization and biodegradation of phenanthrene. Appl. Microbiol. Biotechnol. 2004;6:336–343. doi: 10.1007/s00253-004-1561-2. [DOI] [PubMed] [Google Scholar]
- 26.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., William H.M., Remmert M., Soding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson D.R., Natraj N.R., McInerney M.J., Duncan K.E. Biosurfactant-producing Bacillus are present in produced brines from Oklahoma oil reservoirs with a wide range salinities. Appl. Microbiol. Biotechnol. 2011;91:1083–1093. doi: 10.1007/s00253-011-3326-z. [DOI] [PubMed] [Google Scholar]
- 28.Torres C.E., Lenon G., Craperi D., Wilting W., Blanco A. Enzymatic treatment for preventing biofilm formation in the paper industry. Appl. Microbiol. Biotechnol. 2011;929:95–103. doi: 10.1007/s00253-011-3305-4. [DOI] [PubMed] [Google Scholar]
- 29.Turner S., Pryer K.M., Miao V.P.W., Palmer J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Euk. Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 30.Vater J., Kablitz B., Wilde C., Franke P., Mehta N., Cameotra S.S. Matrix assisted laser desorption ionization time of right mass spectrometry of lipopeptide biosurfactants in whole cells and culture filtrates of Bacillus subtilis C-1 isolated from petroleum sludge. Appl. Environ. Microbiol. 2002;68:6210–6219. doi: 10.1128/AEM.68.12.6210-6219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vinas M., Sabate J., Espuny M.J., Solanas A.M. Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl. Environ. Microbiol. 2005;71:7008–7018. doi: 10.1128/AEM.71.11.7008-7018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson S.C., Jones K.C. Bioremediation of soil contaminated with polynuclear aromatic hydrocarbons (PAHs): a review. Environ. Pollut. 1993;81:229–249. doi: 10.1016/0269-7491(93)90206-4. [DOI] [PubMed] [Google Scholar]
- 33.Ye J.S., Yin H., Qianga J., Penga H., Qin M.H., Zhang N., He B.Y. Biodegradation of anthracene by Aspergillus fumigates. J. Hazard. Mater. 2011;185:174–181. doi: 10.1016/j.jhazmat.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Zahir Z., Seed K.D., Dennis J.J. Isolation and characterization of novel organic solvent-tolerant bacteria. Extremophiles. 2006;10:129–138. doi: 10.1007/s00792-005-0483-y. [DOI] [PubMed] [Google Scholar]
- 35.Zaidi B.R., Stucki G., Alexander M. Low chemical concentration and pH as factors limiting the success of inoculation to enhance biodegradation. Environ. Toxicol. Chem. 1988;7:143–151. doi: 10.1021/es00177a005. [DOI] [PubMed] [Google Scholar]