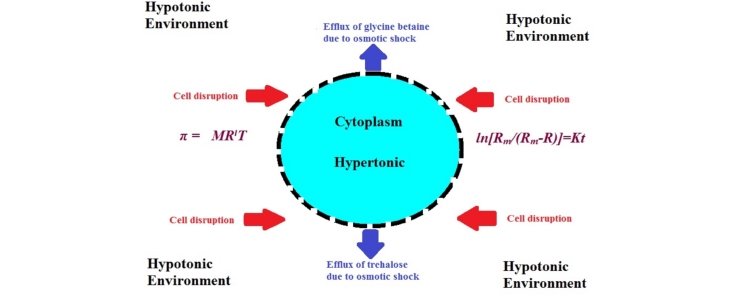
Graphical abstract
Keywords: Osmotic shock, Morse equation, Actinopolyspora halophila
Highlights
-
•
Various cell disruption methods evaluated on an extreme halophile, Actinopolyspora halophila.
-
•
Osmotic shock was found to be the best suited and can be industrially scaled up.
-
•
Cell bursting pressure was calculated using Morse equation.
-
•
Osmotic shock in A. halophila was found to follow first order release rate kinetics.
Abstract
Actinopolyspora halophila produces glycine betaine and trehalose intracellularly in considerable quantities. These biomolecules are commercially important as they have applications in food, pharmaceuticals, and agricultural sector. Development of an efficient cell disruption technique is an important step for the release of these biomolecules. In this study, various cell disruption methods such as chemical, enzymatic, physico-mechanical and physical methods were evaluated. Cell disruption by osmotic shock was found to be the best suited method for A. halophila which also has a potential to be industrially scaled up. Cell bursting pressure that is generated during osmotic shock in A. halophila was computed using Morse equation and was found to be π = 238.37 ± 29.54 atm or 2.35 ± 0.29 kPa. In addition, it was found that osmotic shock followed a first order release rate kinetics in A. halophila. The findings can be used for commercially important biomolecules from other halophilic and/or halotolerant microbes.
1. Introduction
Glycine betaine and trehalose are found in many organisms, mainly as osmoprotectants, which protect them in an environment of high salt concentration [1]. Glycine betaine is a quaternary ammonium compound which is amphoteric in nature and is metabolically synthesized from choline or in some cases, by glycine [2]. Trehalose is a disaccharide that is metabolically synthesized from UDP glucose and glucose-6-phosphate [3]. Glycine betaine and trehalose are economically significant molecules as they have a multitude of applications in a variety of fields [4], [5], [6], [7], [8], [9]. Natural glycine betaine is industrially isolated from sugar beet molasses and Ascophyllum nodosum through various bioseparation techniques [10], [11]. Although glycine betaine is chemically produced, the use of natural glycine betaine like Betafin® is advocated over synthetic glycine betaine due to its natural origin and absence of chemical contaminants [12]. Commercial production of trehalose is carried out through fermentation [13], but the potential for newer organisms need to be continuously explored. Hence, their separation and isolation are of commercial importance.
Actinopolyspora halophila produces glycine betaine and trehalose intracellularly [1]. Previous work from our laboratories has shown significantly high production of glycine betaine (9.07 ± 0.25 g/L) and trehalose (2.49 ± 0.14 g/L) by A. halophila in acid whey as a growth medium [14]. The cell wall of A. halophila is very unique and different from the cell walls of other actinomycetes [15]. A. halophila requires at least 12% (w/v) NaCl in liquid medium for its survival [16]. Yamaguchi has classified actinomycetes into five different types depending upon their cell wall composition [17]. However, actinomycetes are generally not classified on the basis of their resistance to salinity [18], [19]. As compared to other actinomycetes which have higher concentration of amino acids other than that required for peptidoglycan synthesis in their cell wall composition [20], A. halophila cell wall constitutes 70% of peptidoglycan of the total cell wall weight [15]. A. halophila cell wall comprises of glutamic acid, alanine, and diaminopimelic acid in a 1:2:1 molar ratio, lipids, d-galactose and d-arabinose [15], [16]. Due to its unique cell wall composition, an effective method of cell disruption for substantial release of glycine betaine and trehalose is of utmost importance.
Cell disruption methods have been extensively studied in yeasts, Escherichia coli and Bacillus subtilis that have industrial importance due to the large volumes of intracellular substances recovered from them, both from the native as well as in genetically modified forms [21]. Very few cell disruption studies have been conducted on microorganisms other than the above mentioned microbes [21]. There are many methods of cell disruption on a lab scale [22], but industrially only few mechanical methods of cell disruption like bead mill, high pressure homogenizer and Hughes press are used extensively due to ease in scale up of the operations and cost effectiveness [23]. Another reason of preferring mechanical methods like bead mill over chemical and enzymatic methods for cell disruption at industrial scale is to avoid the increase in unit operation steps during downstream processing. The enzymes or chemicals used to achieve cell disruption have to be removed from the cell lysate during the purification of the product of interest, thus increasing the cost of production. Physical methods of cell disruption like osmotic shock, freeze–thaw, liquid nitrogen freezing with grinding and nitrogen bomb have the potential to be used on an industrial scale but have limited applicability [21], [22], [24], [25]. In many studies, the extent of cell disruption and the release of intracellular materials have been measured by using indirect methods like protein estimation, carbohydrate estimation, conductance and colorimetric measurements [23], [26], [27], [28] instead of measuring the actual product of interest. The best way to estimate the extent of cell disruption is by directly estimating the product of interest for which the cell disruption is being performed [28].
An efficient method of 70% ethanol lysis [29], which has been successfully utilized in E. coli for release of intracellular glycine betaine was also found to be a good method in A. halophila. Cell lysis using 70% ethanol was therefore, used as a benchmark to compare other cell disruption methods. The aim was to identify a cell disruption method for A. halophila that can be industrially scaled up and should be energy efficient (in turn cost-effective). These criteria were found in an osmotic shock process. Besides, the kinetics of cell disruption by osmotic shock was studied which has not been previously documented. The approximate intracellular cell bursting pressure that is generated during the osmotic shock was also calculated by measuring the amount of NaCl in the cell lysate (which is the major salt in case of A. halophila).
2. Material and methods
2.1. Materials
Materials used in the formulation of media were bought from HiMedia Ltd. (NaCl, NH4Cl, K2HPO4, CaCl2·2H2O, MnSO4·H2O and glycine) Mumbai, India; Merck (MgSO4·7H2O and FeSO4·7H2O), India; Sigma–Aldrich (corn steep liquor and SeCl4), India. Solvents and other chemicals were bought from S.D. Fine Chemicals Limited, Mumbai, India. Enzymes used were papain (RM058-HiMedia, India, having an activity of 31,734 TU/g of protein), trypsin (204013-Sisco Research Laboratories, India, having an activity of 2500 × 103 NFU/g of protein), protease (Protex 6L-Genecor, Denmark, having an activity of 580,000 DU/g protein), pancreatin (P-1750, Sigma–Aldrich, India, having an activity of 106,261 U/g protein), and lipase (L3126, Sigma–Aldrich, India, having an activity of 340,745 U/g of protein).
2.2. Biomass production of A. halophila and measurement of dry cell weight (DCW)
Acid whey was provided by small scale paneer (cottage cheese) industry in Mumbai, India. Media composition: 21.9 (% w/v) NaCl, 3.38 (% w/v) NH4Cl, 0.1 M MgSO4·7H2O, 0.1 M K2HPO4, 10 mM FeSO4·7H2O, 5 mM CaCl2·2H2O, 5 mM MnSO4·H2O, 6.21 mM SeCl4, 1.5 (% w/v) glycine, 3 (% w/v) corn steep liquor, 94.07 (% v/v) whey and pH of media was adjusted to 8.0. Media with whey was processed [30] with few modifications. Whey media was kept at 100 °C for 20 min. It was centrifuged at 3857 × g for 30 min at 4 °C and then passed through sterile filter of pore size 0.2 μm to filter sterilize the media under sterile conditions. 50 mL of sterile media were transferred into sterile 250 mL Erlenmeyer flasks under sterile conditions. These flasks were incubated at 37 °C, 180 rpm for 96 h. Inoculum was prepared in the same media as mentioned above, and 2% of 72 h old inoculum with 105 cells/mL was inoculated into these flasks. The biomass was separated after 96 h from the broth by centrifuging at 3857 × g for 20 min at 4 °C. Cells were then washed twice with a 21.9 (% w/v) salt solution (same concentration as used in media) to get rid of cellular debris. These cells of A. halophila were then used for disruption studies. 2 mL of broth was taken in 2 mL Eppendroff tubes (weighed) and centrifuged at 9615 × g. The cell pellet was vortexed and washed with 21.9 (%w/v) sodium chloride solutions. This was repeated twice, and the difference in weight was measured for dry cell weight (DCW) after drying the tubes in 60 °C hot air oven for 48 h, where a constant and persistent weight was obtained.
2.3. Analysis of glycine betaine, trehalose and sodium
After disrupting the cells by various methods, the cell lysates were centrifuged and dried. Known amounts of this dried cell lysate were dissolved in distilled water (d/w) or methanol (depending upon the analysis that has to be performed). This solution was then quantified for glycine betaine by a modified UV spectrometric method [31], where samples were subjected to periodide reaction by using 0.5 mL of concentrated sulfuric acid instead of 10 drops and then centrifuged in a swinging bucket centrifuge at 2450 × g for 20 min at 0 °C. Graduated and tipped centrifuge tubes of Borosil glass (15 mL) were used to carry out the periodide reaction. A standard curve developed over a concentration range of 0–50 μg glycine betaine gave a regression equation y = 0.0119× (R2 = 0.9957), where y was the optical density at 365 nm and x was the concentration of GB in microgram. Trehalose was quantified by a well-established HPTLC method [32]. Sodium analysis was performed in order to calculate NaCl concentration and in turn the total cell bursting pressure by Morse equation for the optimized osmotic shock method. Sodium content of the cell lysate solution was analyzed by Inductively Coupled Plasma-Atomic Emission Spectrometer (ICP-AES) (ARCOS from M/s. Spectro, Germany) using standard curve of sodium (y = 52058×, R2 = 0.9996, where y was the intensity (cps) at 589.59 nm, and x was the concentration of Na in ppm). Cell lysate solutions were diluted appropriately for ICP-AES analysis and prepared in deionized d/w.
| (1) |
where π is cell rupturing pressure generated due to osmotic shock (units in atm or kPa), M is molar concentration of NaCl, RI is 0.0821 L atm/K mol, and T is absolute temperature in °K at which the osmotic shock was performed. Osmotic pressure online calculator hosted by Georgia State University was used for calculation of osmotic shock (http://hyperphysics.phy-astr.gsu.edu/hbase/kinetic/ospcal.html)
2.4. Chemical cell disruption
Tween 80, a detergent and ethyl acetate were mixed with 10% (w/v) of A. halophila cells separately at varying concentrations and were kept for 2 h at room temperature (RT 27 ± 2 °C).
2.5. Enzymatic cell disruption
All the enzyme mixes of trypsin (pH 8.0, 37 °C), protease (pH 9.5, 60 °C), papain (pH 7.0, 60 °C), pancreatin (pH 8.1, 37 °C) and lipase (pH 7.4, 37 °C) were prepared in 100 mM phosphate buffer saline, and the reactions were carried out in their respective optimal pH and temperature. In all these samples, A. halophila cell density was adjusted to 10% (w/v).
2.6. Physico-mechanical cell disruption
Mechanical methods of cell disruption were carried out in combination with osmotic shock. In all the physico-mechanical methods, the cell density of A. halophila was adjusted to 10% (w/v). Deionized d/w was used for osmotic shock. 10 mL of the cell suspension was made with varying ratio of glass beads in 15 mL Falcon tubes. This was vortexed for 3 min at RT (27 ± 2 °C). Probe sonication was carried out at various time intervals with acoustic power (50 W) and duty cycle (70%) in a Branson ultrasonic probe sonicator. The temperature could not be maintained as sonication itself leads to rise in temperature. Range of temperature observed was around 27–55 °C (with varying time interval). Bath sonication was performed at 60 °C at various time intervals in a bath sonicator of local make.
2.7. Physical cell disruption
Freeze–thaw cycles were tried along with osmotic shock. One cycle comprised of freezing 10 mL of 10% (w/v) A. halophila cell suspension in 15 mL Falcon tubes with deionized d/w at −20 °C for 2 h followed by thawing at RT (27 ± 2 °C). Osmotic shock was performed by taking 10% (w/v) A. halophila cell concentration in deionized d/w for 30 min at various temperatures. Osmotic shock method was optimized for time interval and cell density parameters at 85 °C by following one factor at a time optimization process.
2.8. Cell disruption kinetics by osmotic shock method
Osmotic shock was applied to A. halophila cells in the optimized parameters of 85 °C and 20% cell density at varying time intervals of 0, 2, 6, 10 and 14 min, using deionized d/w. In order to check if the release of intracellular products viz. glycine betaine and trehalose by osmotic shock followed first order kinetics, a graph of ln [Rm/(Rm − R)] vs. time, t, was plotted by using the first order release rate kinetic equation:
| (2) |
where ln is natural logarithm, Rm is the maximum amount of glycine betaine or trehalose available for release, R is the amount of glycine betaine or trehalose released, k is the release rate constant (min−1), and t is the time interval.
2.9. Protein and DNA analysis of the cell lysate
Cell lysate was passed through a 0.2 μm nylon filter to obtain a clear solution. This solution was used for analysis of protein and DNA. Protein quantification was carried out by Bradford’s assay [33]. Qualitative confirmation of DNA in cell lysate obtained after following optimized osmotic shock protocol, was carried out by performing agarose gel electrophoresis in presence of ethidium bromide [34]. Rough estimate of DNA and protein was carried out by measuring the absorbance ratio (A260/280) of DNA and protein at 260 nm and 280 nm [34], respectively, using HITACHI U-2001 spectrometer.
2.10. Statistical analysis
Experiments were performed in triplicates to get standard deviation. Mean, standard deviation and slopes were calculated using Microsoft Excel 2010.
3. Results and discussion
3.1. Chemical cell disruption
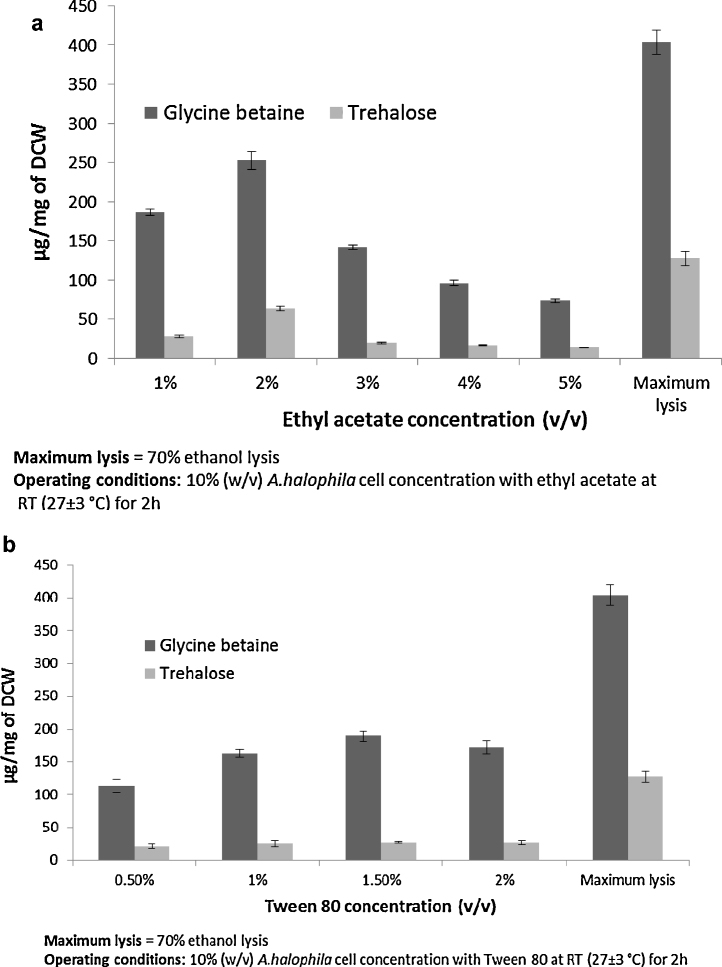
Cell lysis using 70% ethanol was found to be better than ethyl acetate and Tween 80 for cell disruption (Fig. 1). Although 2% (v/v) ethyl acetate showed a marginally higher release of glycine betaine and trehalose, no such phenomenon was seen with Tween 80 (Fig. 1a and b). The ineffectiveness of Tween 80 on the release of intracellular metabolites may be due to the ability of A. halophila to produce extracellular lipases, which can hydrolyze the Tween 80 [16]. A. halophila contains large amounts of peptidoglycan in its cell wall [15] and hence, may have a resistance or tolerance mechanism for ethyl acetate as described by Torres et al. for other microbes [35]. This tolerance or resistance mechanism is indicated by the comparatively low release of the glycine betaine and trehalose.
Fig. 1.
Cell disruption by chemical methods (a) ethyl acetate and (b) by Tween 80.
3.2. Enzymatic cell disruption
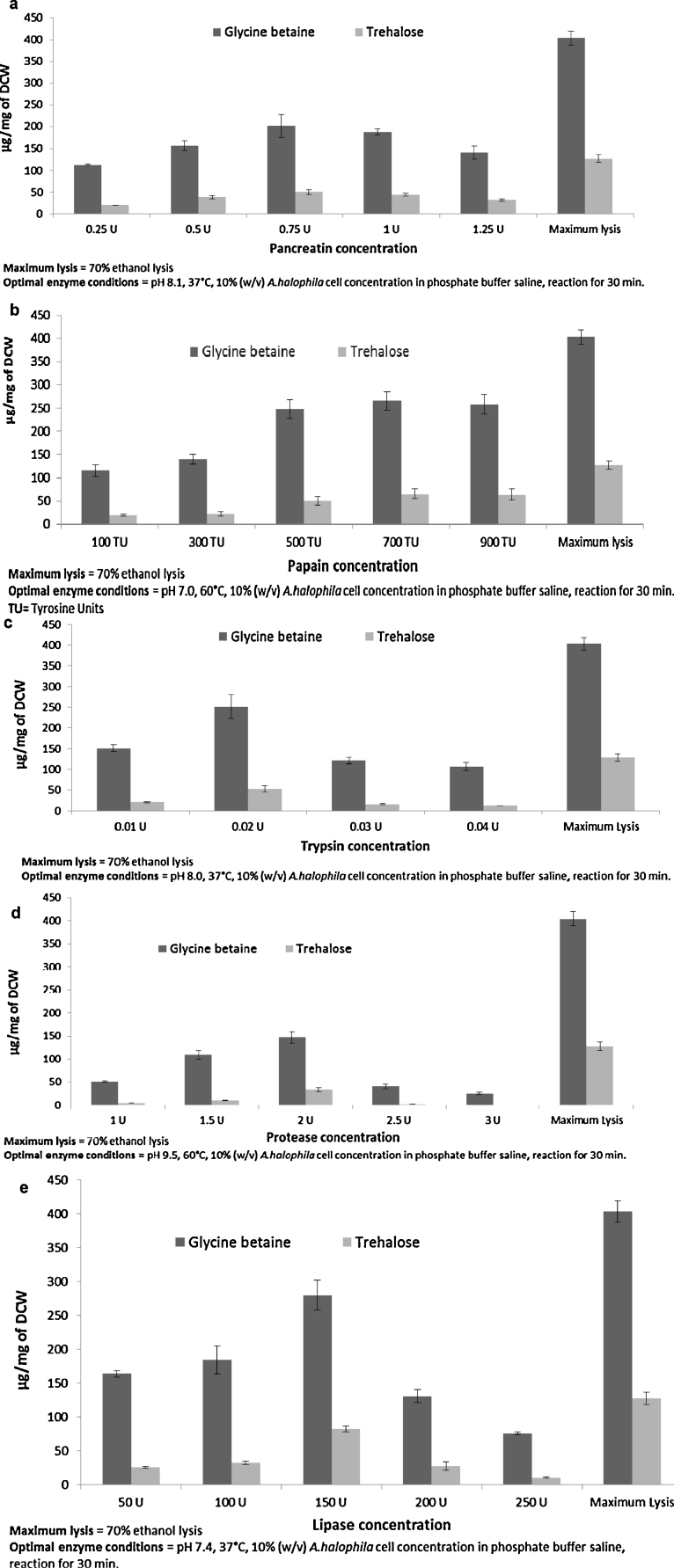
A good cell disruption was observed with 700 TU of papain, 0.02 U of trypsin and 150 U of lipase as compared to protease and pancreatin which was evident from the release of glycine betaine and trehalose (Fig. 2). However, none of the enzymes could lyse cells as effectively as 70% ethanol (Fig. 2). A. halophila has a mucopeptide cell wall [16] which makes it sensitive to degradation by lysozyme and Myxobacter AL-I protease [15], [16]. Lysozyme is a costly enzyme as it is widely used in food and pharmaceutical industry [36]. Due to the cost factor associated with the use of the lysozyme itself and also the increase in the downstream processing cost associated with the usage of enzymes (these enzymes have to be separated from product of interest during purification) [23], we refrained from using lysozyme. Use of lysozyme in A. halophila will not be feasible industrially due to cost factors associated with it. Hence, inexpensive enzyme sources and enzymes which have never been used on A. halophila like papain (plant source), protease (bacterial source), lipase, trypsin and pancreatin (porcine source) were used in our studies. Lipase and protease are produced extracellulary in high quantities by A. halophila [16], [37]. A. halophila may be source specific for cell disruption action of protease [15] as comparatively lower release of glycine betaine and trehalose was seen when compared to papain, trypsin and lipase (Fig. 2b–e). Compared to other enzymes, lipase showed relatively higher release of glycine betaine and trehalose (Fig. 2e) suggesting the presence of high lipoprotein content in A. halophila cell wall. Further research is necessary on cell wall composition to confirm this finding.
Fig. 2.
Enzymatic cell disruption by using (a) pancreatin (b) papain (c) trypsin (d) protease and (e) lipase.
3.3. Physico-mechanical cell disruption
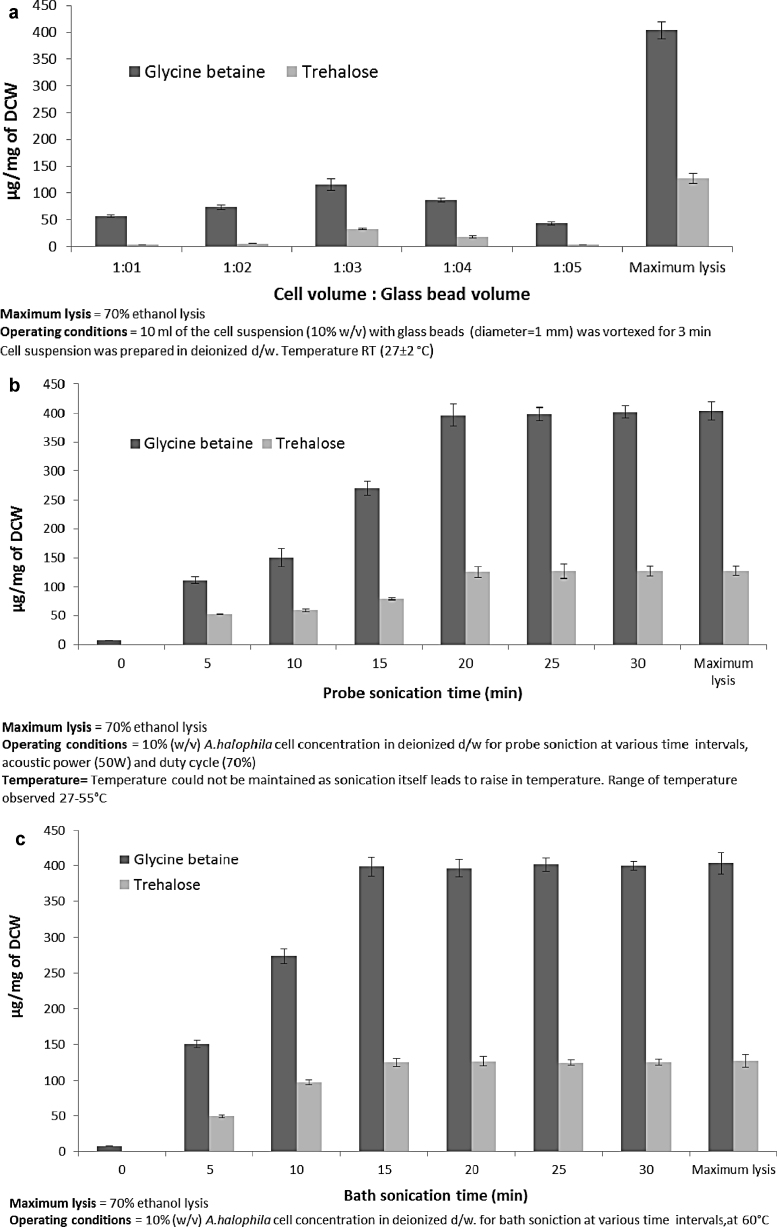
Cell disruption by glass beads mimics the industrial bead mill process on a lab scale. Disruption by glass beads takes place by shear force which is non-specific and generalized in action [25]. Bead volume to cell volume ratio is a very important factor during glass bead cell disruption [38]. Hence, experiments were carried out by varying the volume of glass beads while keeping the cell volume constant. In addition to this shear force, osmotic shock was also applied by carrying out the process in deionized d/w. Disruption by glass beads along with osmotic shock did not work out well (Fig. 3a). This may be due to the entire process being non-specific [25] and/or the time of action for the osmotic shock being too low at 3 min.
Fig. 3.
Physico-mechanical cell disruption methods, with apllication of osmotic shock along with various mechanical cell disruption (a) glass bead disruption (b) probe sonication and (c) bath sonication.
Disruption by probe sonication with osmotic shock at 20 min and bath sonication with osmotic shock at 15 min was at par with 70% ethanol lysis (Fig. 3b and c). Rough net energy consumption was computed and found to be 0.634 kW h and 0.871 kW h for probe sonication at 20 min and bath sonication at 15 min, respectively (Supplementary file 1). In addition to this high kilowatt hour net energy consumption, ultrasonication methods are not suitable for large scale industrial use [23]. Ultrasonication creates small cell debris which creates difficulty in downstream processing [39], and large cell volumes cannot be disrupted by ultrasonication [40].
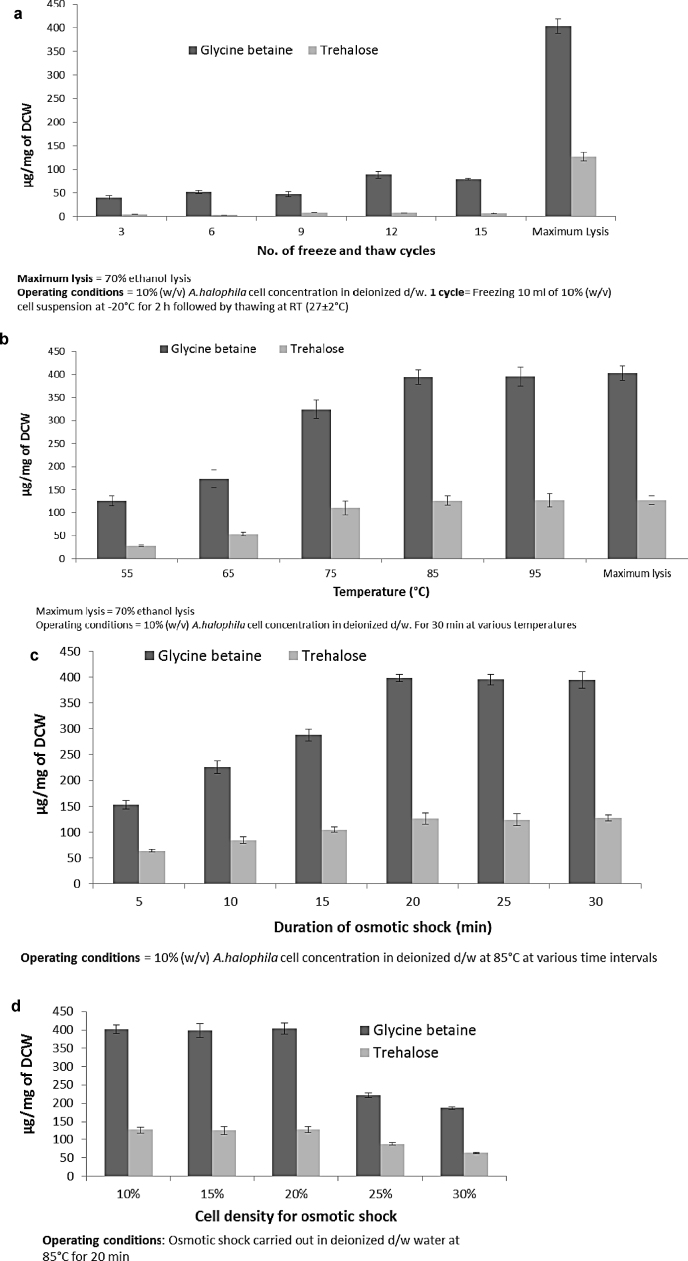
3.4. Physical cell disruption
Freezing results in ice crystals that break the cell wall of microorganisms. Freeze and thaw method of cell disruption is an ideal method for heat labile products and is a gentle method for extraction of intracellular enzymes [41]. From Fig. 4a, it is clearly seen that even after 15 cycles of freezing and thawing along with osmotic shock, the release of glycine betaine and trehalose did not take place efficiently. This failure of freeze and thaw along with osmotic shock may be due to two reasons. Firstly, osmotic shock is a function of temperature, which is clearly seen from Morse Eq. (1). At lower temperature, i.e., during freezing, osmosis slows down; and during thawing, the energy of rise in temperature is utilized in melting of ice crystals rather than for osmosis. Secondly, glycine betaine and trehalose are both reported cryoprotectants [42], and they may be protecting the A. halophila cells from freeze–thaw shock.
Fig. 4.
Physical cell disruption methods (a) freeze and thaw along with osmotic shock, (b) osmotic shock at differen temperatures. Optimization of osmotic shock for (c) time of application and (d) A. halophila cell density.
Osmotic shock has been successfully applied in Halomonas elongata in a pilot scale continuous process called ‘bacterial milking’ for efficient release of ectoine [43]. However, no such data is available for A. halophila which is an extremely halophilic microorganism that can grow in 30% w/v NaCl [16]. When a varied range of temperature was applied to enhance osmotic shock for a constant time of 30 min, an efficient release of glycine betaine and trehalose was achieved at 85 °C which was at par with 70% ethanol lysis (Fig. 4b). When rough net energy consumption was computed of osmotic shock for 30 min at 85 °C, it was found to be 0.591 kW h, which is much lower as compared to 0.634 kW h and 0.871 kW h for probe sonication at 20 min and bath sonication at 15 min, respectively (Supplementary file 1, Pg. 2–4). Hence, osmotic shock was found to be an ideal cell disruption method and was optimized further by optimizing time and cell density by keeping temperature constant at 85 °C. The final optimized parameters that were obtained were 85 °C for 20 min to 20% w/v cell density of A. halophila. When the rough net energy consumption of optimized osmotic shock was computed, it was found to be 0.340 kW h, which is very low, and the whole process can be easily scaled up industrially for a profitable use.
3.5. Cell bursting pressure and first order release kinetics
Bradford’s method [33] was used for protein estimation after cell disruption by osmotic shock. However, absurd results were obtained due to the quaternary ammonium nature of glycine betaine which may have interfered with Bradford’s assay. Hence, agarose gel electrophoresis was performed to check for the release of DNA as a qualitative confirmation of cell disruption (Supplementary file 1, Pg. 5). In addition to this A260/280 ratio of 1.69 ± 0.03 of the fresh cell lysate obtained after following optimized osmotic shock protocol indicated approximately 75% proteins and 25% DNA presence in the cell lysate [34]. The presence of DNA in the cell lysate clearly indicated rupture of A. halophila cells to release glycine betaine and trehalose and not due to mechanosensitive release, which is indeed the case with many other microorganisms [44]. However, further detailed research on presence of mechanisms and special protein channels of mechanosensitive release [44] in A. halophila under ambient conditions is necessary. Therefore, cell disruption was quantified in terms of release of the products of interest i.e., glycine betaine and trehalose [28]. In organisms where the cell wall has high mechanical strength and low content of intracellular sodium chloride, osmotic shock does not disrupt the cell wall due to inadequate generation of osmotic pressure, as is the case with Sarcina lutea [44]. This necessitated knowing the intracellular bursting pressure that was generated during the optimized osmotic shock. Hence, ICP-AES analysis was performed to find out sodium concentration and in turn calculate NaCl concentration after the cells were lysed by the optimized osmotic shock. NaCl concentration was found to be 4.68 ± 0.58 g/L (0.08 ± 0.01 M) in the lysate solution. Although there are other salts present intracellularly, NaCl is present in large quantities as 21.9% w/v of NaCl is used for growth of A. halophila. Hence the efflux of NaCl will generate the maximum pressure as compared to other salts. The pressure resulting due to efflux of NaCl was calculated to be π = 238.37 ± 29.54 atm or 2.35 ± 0.29 kPa using Eq. (1) and Georgia State University online osmotic pressure calculator. This osmotic pressure value is very high and can easily cause cell disruption in A. halophila cells. This pressure is very large as compared to 20 atm pressure required for Sarcina lutea, which is halotolerant in nature [45]. Harsh cell disruption methods like sonication and homogenization are required to rupture S. lutea cell wall which has been compared to reinforced concrete [23], [45]. This fact gave us an idea of the strength of A. halophila cell wall. Further research on cell wall thickness of A. halophila by performing freeze fracture technique may reveal its true nature of cell wall.
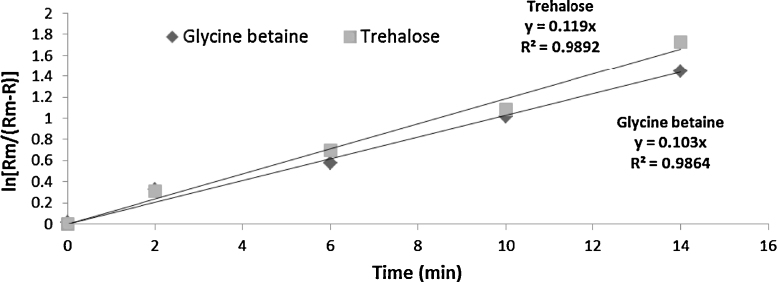
No literature on the kinetics of osmotic shock was found in the scientific literature, although extensive work has been performed with respect to first order release rate kinetics on ultrasonication and bead mill [23]. In Eq. (2) and Fig. 5, t (time) represents x-axis and ln [Rm/(Rm − R)] represents y-axis. If osmotic shock followed first order release rate kinetics, then a straight line should be anticipated which was found to be the case with R2 = 0.9864 and 0.9892 for release of glycine betaine and trehalose, respectively (Fig. 5). The release rate constant, k, can be easily deduced by finding the slope of the lines and was found to be 0.103 for glycine betaine and 0.119 for trehalose. Therefore, the release rate constant for glycine betaine was 103 × 10−3 min−1 and that of trehalose was 119 × 10−3 min−1. The k values indicated that trehalose was released at much faster rate than glycine betaine during osmotic shock in A. halophila.
Fig. 5.
Release kinetics of glycine betaine and trehalose from A. halophila by osmotic shock.
4. Conclusions
Investigations of various methods of cell disruption on A. halophila cells for efficient release of glycine betaine and trehalose were compared to lysis with 70% ethanol. Cell disruption by osmotic shock was found to be the most suitable and industrially scalable and hence, was optimized. Intracellular bursting pressure in A. halophila was computed by using Morse equation. For the first time, we have shown that osmotic shock followed a first order release rate kinetics in A. halophila. Hence, efficient cell lysis of A. halophila by osmotic shock could release two important biomolecules, glycine betaine and trehalose. This approach can also be used for other organisms for release of different biomolecules.
Acknowledgment
Authors are grateful to University Grants Commission, India for the financial support provided for this research.
Footnotes
Available online 18 December 2014
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.btre.2014.12.005.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Severin J., Wohlfarth A., Galinski E.A. The predominant role of recently discovered tetrahydropyrimidines for the osmoadaptation of halophilic eubacteria. J. Gen. Microbiol. 1992;138:1629–1638. [Google Scholar]
- 2.Nyyssola A., Leisola M. Actinopolyspora halophila has two separate pathways for betaine synthesis. Arch. Microbiol. 2001;176:294–300. doi: 10.1007/s002030100325. [DOI] [PubMed] [Google Scholar]
- 3.Styrvold O.B., Strøm A.R. Synthesis, accumulation, and excretion of trehalose in osmotically stressed Escherichia coli K-12 strains: influence of amber suppressors and function of the periplasmic trehalase. J. Bacteriol. 1991;173:1187–1192. doi: 10.1128/jb.173.3.1187-1192.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lever M., Slow S. The clinical significance of betaine, an osmolyte with a key role in methyl group metabolism. Clin. Biochem. 2010;43:732–744. doi: 10.1016/j.clinbiochem.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Mäkelä P., Jokinen K., Kontturi M., Peltonen-Sainio P., Pehu E., Somersalo S. Foliar application of glycinebetaine—a novel product from sugar beet—as an approach to increase tomato yield. Ind. Crops Prod. 1998;7:139–148. [Google Scholar]
- 6.Park E.-J. Exogenous application of glycinebetaine increases chilling tolerance in tomato plants. Plant Cell Physiol. 2006;47:706–714. doi: 10.1093/pcp/pcj041. [DOI] [PubMed] [Google Scholar]
- 7.Eklund M., Bauer E., Wamatu J., Mosenthin R. Potential nutritional and physiological functions of betaine in livestock. Nutr. Res. Rev. 2005;18:31. doi: 10.1079/NRR200493. [DOI] [PubMed] [Google Scholar]
- 8.Roser B. Trehalose drying: a novel replacement for freeze-drying. BioPharm. 1991;4:47–53. [Google Scholar]
- 9.Kidd G., Devorak J. Trehalose is a sweet target for agrobiotech. J. Biotechnol. 1994;12:1328–1329. [Google Scholar]
- 10.H.O. Heikkila, J.A. Melaja, D.E.D. Millner, J.J. Virtanen, Betaine recovery process, Google Patents (1982). http://www.google.co.in/patents/US4359430.
- 11.Whapham C.A., Blunden G., Jenkins T., Hankins S.D. Significance of betaines in the increased chlorophyll content of plants treated with seaweed extract. J. Appl. Phycol. 1993;5:231–234. [Google Scholar]
- 12.http://www.in-cosmetics.com/__novadocuments/7930, (accessed 07.08.14).
- 13.Schiraldi C., Di Lernia I., De Rosa M. Trehalose production: exploiting novel approaches. Trends Biotechnol. 2002;20:420–425. doi: 10.1016/s0167-7799(02)02041-3. [DOI] [PubMed] [Google Scholar]
- 14.J.R. Kar, J.E. Hallsworth, R.S. Singhal, Fermentative production of glycine betaine and trehalose from acid whey using Actinopolyspora halophila (MTCC 263), Environ. Technol. Innov. in press.
- 15.Johnson K.G., Lanthier P.H., Gochnauer M.B. Cell walls from Actinopolyspora halophila, an extremely halophilic actinomycete. Arch. Microbiol. 1986;143:365–369. [Google Scholar]
- 16.Gochnauer M.B., Leppard G.G., Komaratat P., Kates M., Novitsky T., Kushner D.J. Isolation and characterization of Actinopolyspora halophila, gen. et sp. nov., an extremely halophilic actinomycete. Can. J. Microbiol. 1975;21:1500–1511. doi: 10.1139/m75-222. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi T. Comparison of the cell–wall composition of morphologically distinct actinomycetes. J. Bacteriol. 1965;89:444–453. doi: 10.1128/jb.89.2.444-453.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tresner H.D., Hayes J.A., Backus E.J. Differential tolerance of streptomycetes to sodium chloride as a taxonomic aid. Appl. Microbiol. 1968;16:1134–1136. doi: 10.1128/am.16.8.1134-1136.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlieb D. General considerations and implications of the actinomycetales. In: Sykes G., Skinner F.A., editors. Actinomycetales. Academic Press; New York: 1973. pp. 1–10. [Google Scholar]
- 20.Deweese M.S., Gerencser M.A., Slack J.M. Quantitative analysis of actinomyces cell walls. Appl. Microbiol. 1968;16:1713–1718. doi: 10.1128/am.16.11.1713-1718.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geciova J., Bury D., Jelen P. Methods for disruption of microbial cells for potential use in the dairy industry—a review. Int. Dairy J. 2002;12:541–553. [Google Scholar]
- 22.Brown R.B., Audet J. Current techniques for single-cell lysis. J. R. Soc. Interface. 2008;5:S131–S138. doi: 10.1098/rsif.2008.0009.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chisti Y., Moo-Young M. Disruption of microbial cells for intracellular products. Enzyme Microb. Technol. 1986;8:194–204. [Google Scholar]
- 24.Autuori F., Brunk U., Peterson E., Dallner G. Fractionation of isolated liver cells after disruption with a nitrogen bomb and sonication. J. Cell Sci. 1982;57:1–13. doi: 10.1242/jcs.57.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Middelberg A.P.J. Process-scale disruption of microorganisms. Biotechnol. Adv. 1995;13:491–551. doi: 10.1016/0734-9750(95)02007-p. [DOI] [PubMed] [Google Scholar]
- 26.Liu D., Zeng X.-A., Sun D.-W., Han Z. Disruption and protein release by ultrasonication of yeast cells. Innov. Food Sci. Emerg. Technol. 2013;18:132–137. [Google Scholar]
- 27.Silva Galdino T., Menna-Barreto R.F.S., Britto C., Samudio F., Brandão A., Kalume D.E. Cell disruption using a different methodology for proteomics analysis of Trypanosoma cruzi strains. Anal. Biochem. 2014;448:1–8. doi: 10.1016/j.ab.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Middelberg A.P.J. 2 microbial cell disruption by high-pressure homogenization. In: Desai M.A., editor. Downstream Processing of Proteins. Humana Press; Totowa, NJ: 2000. pp. 11–21. http://link.springer.com/10.1007/978-1-59259-027-8_2 (accessed 21.09.14) [Google Scholar]
- 29.Ghoul M., Bernard T., Cormier M. Evidence that Escherichia coli accumulates glycine betaine from marine sediments. Appl. Environ. Microbiol. 1990;56:551–554. doi: 10.1128/aem.56.2.551-554.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dlamini A.M., Peiris P.S. Production of exopolysaccharide by Pseudomonas sp. ATCC 31461 (Pseudomonas elodea) using whey as fermentation substrate. Appl. Microbiol. Biotechnol. 1997;47:52–57. [Google Scholar]
- 31.Barak A.J., Tuma D.J. A simplified procedure for the determination of betaine in liver. Lipids. 1979;14:860–863. doi: 10.1007/BF02534129. [DOI] [PubMed] [Google Scholar]
- 32.Ranganathan T., Kulkarni P.R. A simple method for the analysis of trehalose using HPTLC. Food Chem. 2002;77:263–265. [Google Scholar]
- 33.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 35.Torres S., Pandey A., Castro G.R. Organic solvent adaptation of Gram positive bacteria: applications and biotechnological potentials. Biotechnol. Adv. 2011;29:442–452. doi: 10.1016/j.biotechadv.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Proctor V.A., Cunningham F.E., Fung D.Y.C. The chemistry of lysozyme and its use as a food preservative and a pharmaceutical. CRC Crit. Rev. Food Sci. Nutr. 1988;26:359–395. doi: 10.1080/10408398809527473. [DOI] [PubMed] [Google Scholar]
- 37.Johnson K.G., Lanthier P.H., Gochnauer M.B. Studies of two strains of Actinopolyspora halophila, an extremely halophilic actinomycete. Arch. Microbiol. 1986;143:370–378. [Google Scholar]
- 38.Asenjo J.A., editor. Marcel Dekker; New York: 1990. Separation Processes in Biotechnology. [Google Scholar]
- 39.Wang D.I.-c., editor. Wiley; New York: 1979. Fermentation and Enzyme Technology. [Google Scholar]
- 40.Lambert P.W. Industrial enzyme production and recovery from filamentous fungi. In: Smith J.E., Berry D.R., Kristiansen B., editors. The Filamentous Fungi. Arnold Publishers; London: 1983. pp. 210–237. [Google Scholar]
- 41.Mayerhoff Z.D.V.L., Franco T.T., Roberto I.C. A study of cell disruption of Candida mogii by glass bead mill for the recovery of xylose reductase. Sep. Purif. Technol. 2008;63:706–709. [Google Scholar]
- 42.Cleland D., Krader P., McCree C., Tang J., Emerson D. Glycine betaine as a cryoprotectant for prokaryotes. J. Microbiol. Methods. 2004;58:31–38. doi: 10.1016/j.mimet.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Sauer T., Galinski E.A. Bacterial milking: a novel bioprocess for production of compatible solutes. Biotechnol. Bioeng. 1998;57:306–313. [PubMed] [Google Scholar]
- 44.Booth I.R., Blount P. The MscS and MscL families of mechanosensitive channels act as microbial emergency release valves. J. Bacteriol. 2012;194:4802–4809. doi: 10.1128/JB.00576-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wimpenny J.W.T. Breakage of microorganisms. Proc. Biochem. 1967;2:41–44. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.