Abstract
Purpose
To assess the diagnostic weight of sequence-specific magnetic resonance features in characterizing clinically significant prostate cancers (csPCa).
Materials and methods
We used a prospective database of 262 patients who underwent T2-weighted, diffusion-weighted, and dynamic contrast-enhanced (DCE) imaging before prostatectomy. For each lesion, two independent readers (R1, R2) prospectively defined nine features: shape, volume (V_Max), signal abnormality on each pulse sequence, number of pulse sequences with a marked (S_Max) and non-visible (S_Min) abnormality, likelihood of extracapsular extension (ECE) and PSA density (dPSA). Overall likelihood of malignancy was assessed using a 5-level Likert score. Features were evaluated using the area under the receiver operating characteristic curve (AUC). csPCa was defined as Gleason ≥7 cancer (csPCa-A), Gleason ≥7(4+3) cancer (csPCa-B) or Gleason ≥7 cancer with histological extraprostatic extension (csPCa-C),
Results
For csPCa-A, the Signal1 model (S_Max+S_Min) provided the best combination of signal-related variables, for both readers. The performance was improved by adding V_Max, ECE and/or dPSA, but not shape. All models performed better with DCE findings than without.
When moving from csPCa-A to csPCa-B and csPCa-C definitions, the added value of V_Max, dPSA and ECE increased as compared to signal-related variables, and the added value of DCE decreased.
For R1, the best models were Signal1+ECE+dPSA (AUC = 0,805 [95%CI:0,757–0,866]), Signal1+V_Max+dPSA (AUC = 0.823 [95%CI:0.760–0.893]) and Signal1+ECE+dPSA [AUC = 0.840 (95%CI:0.774–0.907)] for csPCa-A, csPCA-B and csPCA-C respectively. The AUCs of the corresponding Likert scores were 0.844 [95%CI:0.806–0.877, p = 0.11], 0.841 [95%CI:0.799–0.876, p = 0.52]) and 0.849 [95%CI:0.811–0.884, p = 0.49], respectively.
For R2, the best models were Signal1+V_Max+dPSA (AUC = 0,790 [95%CI:0,731–0,857]), Signal1+V_Max (AUC = 0.813 [95%CI:0.746–0.882]) and Signal1+ECE+V_Max (AUC = 0.843 [95%CI: 0.781–0.907]) for csPCa-A, csPCA-B and csPCA-C respectively. The AUCs of the corresponding Likert scores were 0. 829 [95%CI:0.791–0.868, p = 0.13], 0.790 [95%CI:0.742–0.841, p = 0.12]) and 0.808 [95%CI:0.764–0.845, p = 0.006]), respectively.
Conclusion
Combination of simple variables can match the Likert score’s results. The optimal combination depends on the definition of csPCa.
Introduction
Multiparametric Magnetic Resonance (MR) imaging can detect clinically significant prostate cancer (csPCa) with good accuracy [1–7]. Unfortunately, its interpretation needs expertise. Indeed, prostate focal lesions can have very different appearances from one MR pulse sequence to another, and it may be difficult to distinguish, within the large number of combinations of shape and signal abnormalities, those that are benign from those that are malignant.
Because it is impossible to definitely characterize as benign or malignant all prostate focal lesions, the use of a 5-point subjective score has been widely encouraged to describe the level of suspicion of a given lesion [8, 9]. This so-called Likert score is a highly significant predictor of the malignant nature of prostate focal lesions [3, 10–13]. However, because there are no descriptions of specific criteria to be used in the scoring process, the Likert score relies heavily on the reader’s experience. Therefore, some research groups tried to set up more objective scoring systems to improve inter-reader agreement [14–17].
In 2012, the European Society of Urogenital Radiology (ESUR) endorsed the Prostate Imaging Reporting and Data System (PIRADS) score [18]. It was shown to be a significant predictor of malignancy and aggressive behavior [19, 20]. However, it did not outperform the Likert score, at least for experienced readers, and did not improve inter-reader agreement either [11, 21, 22]. Two main limitations were identified. First, it gave the same diagnostic weight to all pulse sequences, and second the interpretation of dynamic contrast-enhanced (DCE) imaging relied mostly on the shape of the enhancement curve that was shown to be a poor predictor of malignancy [11].
In 2015, the ESUR and the American College of Radiology endorsed the so-called PIRADS v2 score [23, 24] that introduced the concept of a dominant pulse sequence bearing most of the diagnostic weight (diffusion-weighted [DW] imaging for the peripheral zone [PZ]; T2-weighted [T2W] imaging for the transition zone [TZ]) and that limited the role of DCE imaging. This PIRADS v2 score gave good results in characterizing prostate focal lesions [25, 26], but, again, some limitations were pointed out [27]. It does not seem to improve inter-reader agreement as compared to the PIRADS v1 score, even after training [28, 29], results in a high false positive rate [30] and was outperformed by alternative in-house scores [17, 28]. These results have led some authors to suggest that there might be structural limits to the ability of any score based on MR imaging to allow detection of prostate cancer with high specificity [28].
The PIRADS scores were not based on an analysis of a large dataset, but rather were a result of expert opinion. As others [17], we hypothesize that processing large prospective databases detailing sequence-specific findings may help further refining prostate MR scoring systems. We therefore undertook this study to assess the relative diagnostic weight of sequence-specific MR features in characterizing aggressive cancers in PZ, using a prospectively acquired radiologic-pathologic database of patients treated by prostatectomy.
Material and methods
Radiologic-pathologic database
As of September 2008, all patients who underwent prostate multiparametric MR imaging before radical prostatectomy at our institution (Hospices Civils de Lyon) were proposed to have their radiologic and pathologic data entered in a prospective database approved by our Institutional Review Board (Comité de Protection des Personnes Sud-Est IV). All patients gave written informed consent. The database was used for other studies evaluating prostate cancer detection rates [3] and accuracy of tumor volume estimation [31] at multiparametric MR imaging, existing MR scoring systems [11] and diagnostic accuracy of quantitative MR parameters [32–34]. These studies do not overlap with the present one that aims at understanding the relative weight of non-quantitative sequence-specific findings in the characterization of aggressive prostate cancers, in order to improve current MR scoring systems.
MR image analysis
MR examinations comprised at least T2W, DW and DCE imaging at 1.5T and 3T, but protocol parameters varied as per the standard of care at the time of the examination (S1 Table). Upon inclusion, preoperative MR examinations were prospectively analyzed by two senior readers blinded to clinical and histological data. Reader 1 (R1, OR) and 2 (R2, FB) had respectively 11 years and 1 year of experience at the start of the database in 2008. Readers independently described all prostate suspicious focal lesions. They took into account all PZ lesions showing low-signal intensity on T2W images and/or on apparent diffusion coefficient (ADC) maps, and/or showing early enhancement at visual inspection of DCE images.
First, readers delineated each lesion on all three MR pulse sequence images using the Osirix software (Osirix imaging software, Geneva, Switzerland). This allowed the calculation of the lesion volume on each MR pulse sequence.
Second, readers specified the lesion shape on the three MR pulse sequences images, using the following list: not visible; ill-defined area with indistinct margins; linear lesion perpendicular to the capsule; linear lesion parallel to the capsule; triangular lesion; nodular lesion without mass effect on the adjacent TZ or capsule; nodular lesion with mass effect on the adjacent TZ or capsule.
Third, they separately noted the degree of signal abnormality on T2W, DW and DCE images using the following qualitative 4-level scale: 0, not visible; 1, mild abnormality; 2, moderate abnormality; 3, marked abnormality.
Fourth, they assessed the likelihood of extracapsular extension (ECE) using a subjective 5-level score: 1, definitely absent; 2, likely absent; 3, indeterminate; 4, likely present; 5, definitively present.
Finally, they assessed the likelihood of malignancy of each lesion, using the following subjective 5-level Likert score: 1, definitely benign; 2, likely benign; 3, indeterminate; 4, likely malignant; 5, definitely malignant. Because a Likert score of 1/5 was used only in areas with normal appearance on all pulse sequences, focal lesions had, by definition, a Likert score ≥2/5.
Comparison of MR and histopathologic findings
Whole-mount sections of prostatectomy specimens were obtained every 3 mm according to guidelines [35]. A single uropathologist (FML) with 10 years of experience at the start of the database and blinded to the readers’ assessements, assigned individual Gleason scores to all cancer foci and delineated them on the glass cover of whole-mount sections. Then, the readers and the uropathologist compared MR and histopathologic findings. The pathologist decided which MR lesions matched the positions of histologic cancers. These matching lesions were considered true positives only if their largest diameter was 50–150% of the diameter of the corresponding cancer to minimize chance detection [36, 37].
Assessment of variables and combination of variables in characterizing csPCa-A in PZ
We first assessed the performance of the 9 variables decribed in Table 1 in characterizing in PZ csPCa defined as Gleason ≥7 cancers, i.e. as cancers with an International Society of Urological Pathology (ISUP) grade group ≥2 (csPCa-A) [38]. The variables comprised the PSA density (dPSA) and eight variables describing MR lesions (S_T2, S_DW, S_DCE, S_Max, S_Min, Shape, ECE and Vmax).
Table 1. Assessed individual variables.
| Description | Possible values | |
|---|---|---|
| S_T2 | Degree of signal abnormality on T2W | not visible, mild, moderate, marked |
| S_DW | Degree of signal abnormality on DW | not visible, mild, moderate, marked |
| S_DCE | Degree of signal abnormality on DCE | not visible, mild, moderate, marked |
| S_Max | Number of MR pulse sequences showing a marked signal abnormality | 0, 1, 2, 3 |
| S_Min | Number of MR pulse sequences on which the lesion was not visible | 0, ≥1 |
| Shape | Shape of the lesion on T2W (1) | not visible, ill-defined area, linear perpendicular to the capsule, linear parallel to the capsule, triangular, nodular without mass effect, nodular with mass effect (2) |
| ECE | Extracapsular extension score | 1/5, 2/5, 3/5, 4/5 or 5/5 |
| V_Max | Largest of the lesion’s volumes measured on the three MR pulse sequences (mL) | Continuous variable |
| dPSA | PSA density (ng/mL/mL) | Continuous variable |
T2W: T2-weighted imaging; DW: diffusion-weighted imaging; DCE: dynamic contrast-enhanced imaging
(1) If the lesion was not visible on T2W images, the shape on DW images was taken into consideration. If the lesion was not visible also on DW images, DCE images were used to describe the shape of the lesion.
(2) Ill-defined areas and linear lesions perpendicular to the capsule were grouped in a single category for statistical analysis.
Then we assessed the performance of three combinations of variables related to the lesions’ signal. Signal1 model comprised S_Max and S_ Min, Signal2 model comprised S_Max, S_ Min and S_DW, and Signal3 model comprised S_T2, S_DW, S_DCE. Signal1a, Signal2a and Signal3a models used the three MR pulse sequences. Signal1b, Signal2b and Signal3b models did not use DCE imaging. In Signal2a model, S_Max and S_Min used only the results of T2W and DCE imaging since DW findings were coded by S_DW. In Signal2b model, S_Max and S_Min used only the results of T2W imaging. These models were compared to S_Max as a stand-alone, with (S_Maxa) and without (S_Maxb) use of DCE imaging.
Signal1 model was selected for the rest of the analysis. The variables Shape, ECE, Vmax and dPSA were sequentially added to it. Each resulting multivariable model was assessed first using the findings of the three MR pulse sequences and then without DCE imaging.
Alternative definitions of csPCa in PZ
The 9 individual variables, the Signal1, Signal2 and Signal3 models, and the combinations of the Signal1 model and the variables Shape, ECE, Vmax and dPSA were also assessed using two alternative definitions for csPCa: (i) csPCa-B: Gleason ≥7(4+3) (ISUP grade group ≥3) and (ii) csPCa-C: Gleason ≥7 (ISUP Grade group ≥2) and histologically-proven extraprostatic extension (pathological stage ≥pT3a).
In the database, extraprostatic extension was assessed at the lesion level (i.e., it was specified whether each lesion showed features of extraprostatic extension at pathological examination). This allowed combining the Gleason score and extraprostatic extension features at the lesion level.
Statistical analysis
The analysis units were the lesions identified by each reader. The probability that a lesion corresponded to a csPCa was modelled using a logistic regression for each of the 9 studied variables, and for the different combinations of variables described above. Receiver operating characteristic (ROC) curves were built using the probabilities of csPCa predicted by the different models. The diagnostic performance of each variable and combination of variables was quantified using the area under the ROC curve (AUC). Because the same data were used for developing the models and assessing their performance, the AUC can be overestimated. This is often referred to as optimism. We therefore used a bootstrap procedure to estimate a corrected AUC as proposed by Harrel [39] (S1 Appendix). To take account of the clustered structure of the data, the bootstrap procedures used a clustered resampling at the patient level. A similar bootstrap procedure was used for model-to-model comparison and for constructing confidence intervals. All interval estimations given in this paper are 95% confidence intervals (95%CI). Analyses were performed using R software version 3.2.4 (http://cran.r-project.org).
Results
Study population
At the time of analysis, the database contained 262 patients imaged between September 2008 and February 2013. MR imaging was performed at 1.5T on scanner A (n = 72) or at 3T on scanner B (n = 113) or C (n = 77). The patients median age, PSA level and PSA density at the time of imaging were 62 years (interquartile range (IQR), 58–66), 6.5 ng/mL (IQR, 5.0–9.9) and 0.16 ng/mL/mL (IQR, 0.12–0.25) respectively.
R1 described 474 PZ lesions in 250 patients. Two lesions were excluded because of artefacts on DW imaging, leaving 472 lesions (204 Gleason ≥7 cancers, 79 Gleason ≤6 cancers and 189 benign findings). R2 described 392 PZ lesions in 242 patients. Three lesions were excluded because of artefacts on DW imaging leaving 389 lesions (191 Gleason ≥7 cancers, 64 Gleason ≤6 cancers and 134 benign findings; Fig 1). In total, 21.4% (101/472) and 26% (101/389) of the lesions described by R1 and R2 respectively showed histologically-proven extracapsular extension.
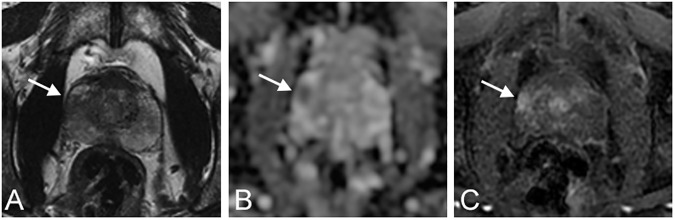
Fig 1. Axial multiparametric MR images acquired on scanner B at 3T, in a 58 year-old patient with a PSA density of 0.28 ng/mL/mL.
A) T2-weighted image. B) Apparent diffusion coefficient map. C) Dynamic contrast-enhanced image. One suspicious lesion was described by both readers in the right peripheral zone (A-C, arrow). The lesion was noted as nodular without mass effect by both readers. S_T2, S_DW and S_DCE were respectively marked, marked and moderate for both readers. V_Max was 2.0 cc and 2.1 cc for readers 1 and 2 respectively. The ECE and Likert scores were respectively 2/5 and 5/5 for both readers. Analysis of the prostatectomy specimen showed a matching Gleason 9 (4+5) cancer with a histological volume of 1.6 cc.
The percentage of csPCa-A was 43.2% (204/472) for R1 and 48.7% (191/389) for R2. The percentage of csPCa-B was 21.8% (103/471) for R1 and 25.8% (101/392) for R2. The percentage of csPCa-C was 20.8% (98/471) for R1 and 24.5% (96/392) for R2.
Assessment of individual variables in characterizing csPCa-A in PZ
S2 Table shows the distribution of individual variables for tissue classes and readers. S_Max obtained the highest AUC for both readers (Table 2). For R1, it was significantly different from that of S_T2 (p = 0.002), S_DW (p = 0.01) and S_DCE (p = 0.005). For R2, it was significantly different from that of S_T2 (p = 0.001) and S_DCE (p = 0.011) but not from that of S_DW (p = 0.094)
Table 2. Performance of individual variables in characterizing csPCa-A in PZ.
| AUC (95%CI) | ||
|---|---|---|
| Reader 1 | Reader 2 | |
| S_T2 | 0.709 (0.647–0.769) | 0.631 (0.567–0.700) |
| S_DW | 0.731 (0.672–0.793) | 0.701 (0.639–0.769) |
| S_DCE | 0.686 (0.630–0.751) | 0.675 (0.600–0.745) |
| S_Max | 0.785 (0.727–0.839) | 0.759 (0.688–0.829) |
| S_Min | 0.604 (0.555–0.652) | 0.566 (0.53–0.603) |
| Shape | 0.654 (0.594–0.724) | 0.667 (0.589–0.745) |
| ECE | 0.735 (0.685–0.788) | 0.702 (0.634–0.77) |
| V_Max | 0.649 (0.579–0.718) | 0.654 (0.577–0.732) |
| dPSA | 0.581 (0.473–0.649) | 0.603 (0.498–0.68) |
AUC: area under the receiver operating characteristic curve with optimism correction; 95%CI: 95% confidence intervals
Assessment of signal-related models in characterizing csPCa-A in PZ
Table 3 shows the results of S_Maxa-b, Signal1a-b, Signal2a-b and Signal3a-b models. For both readers, all models performed better with use of DCE imaging, and the difference was statistically significant for S_Max and Signal1 models for R1. As compared to S_Max, the Signal1-3 models either performed worse or yielded only marginal improvement. The highest AUC was obtained by Signal1a model for both readers. However the AUC differences (ΔAUC) with S_Maxa were only +0.009 for R1 and +0.010 for R2, and not statistically significant (p = 0.49 for R1 and p = 0.14 for R2).
Table 3. Performance of models based on combinations of signal abnormalities in characterizing csPCa-A in PZ.
| Description | Reader 1 | Reader 2 | |||||
|---|---|---|---|---|---|---|---|
| AUC (95%CI) | P (1) | AUC (95%CI) | P(1) | ||||
| With DCE | Without DCE | With DCE | Without DCE | ||||
| S_Max | 0.785 (0.727–0.839) | 0.742 (0.681–0.801) | 0.015 | 0.759 (0.688–0.829) | 0.725 (0.654–0.789) | 0.107 | |
| Signal1 | S_Max + S_Min | 0.794 (0.734–0.851) | 0.748 (0.683–0.808) | 0.015 | 0.769 (0.698–0.835) | 0.725 (0.658–0.791) | 0.060 |
| Signal2 | S_Max + S_Min + S_DW | 0.750 (0.700–0.822) | 0.719 (0.657–0.790) | 0.057 | 0.760 (0.687–0.830) | 0.703 (0.637–0.771) | 0.061 |
| Signal3 | S_T2 + S_DW + S_DCE | 0.784 (0.736–0.848) | 0.757 (0.695–0.821) | 0.051 | 0.762 (0.702–0.840) | 0.720 (0.656–0.794) | 0.088 |
DCE: dynamic contrast-enhanced imaging; AUC: area under the receiver operating characteristic curve with optimism correction; 95%CI: 95% confidence intervals
(1) P value comparing the performances of the model with and without DCE imaging.
Assessment of other multivariable models in characterizing csPCa-A in PZ
Table 4 shows the results of the models associating Signal1 model and other variables. When DCE imaging was used, the addition of other variables either decreased the diagnostic performance or slightly improved it, with a maximal ΔAUC of +0.011 for R1 and +0.021 for R2. When DCE imaging was not used, almost all multivariable models performed better than Signal1b model with a maximum ΔAUC of +0.037 for R1 and +0.040 for R2.
Table 4. Performances of multivariable models in characterizing csPCa-A in PZ.
| AUC (95%CI)–Reader 1 | AUC (95%CI)–Reader 2 | |||
|---|---|---|---|---|
| With DCE | Without DCE | With DCE | Without DCE | |
| Signal1 | 0.794 (0.734–0.851) | 0.748 (0.683–0.808) | 0.769 (0.698–0.835) | 0.725 (0.658–0.791) |
| Signal1 + Shape | 0.784 (0,729–0,848) | 0.750 (0.687–0.818) | 0.762 (0.705–0.837) | 0.723 (0.666–0.806) |
| Signal1 + ECE | 0.802 (0,748–0,864) | 0.781 (0.724–0.843) | 0.776 (0.712–0.845) | 0.762 (0.700–0.834) |
| Signal1 + Vmax | 0.797 (0,741–0,854) | 0.763 (0.701–0.823) | 0.790 (0.729–0.856) | 0.761 (0.705–0.832) |
| Signal1 + dPSA | 0.800 (0,745–0,858) | 0.761 (0.702–0.826) | 0.780 (0.718–0.851) | 0.746 (0.681–0.820) |
| Signal1 + Shape + ECE | 0.785 (0,735–0,855) | 0.767 (0.708–0.836) | 0.758 (0.702–0.840) | 0.741 (0.689–0.824) |
| Signal1 + Shape + Vmax | 0.784 (0,734–0,849) | 0.754 (0.697–0.819) | 0.775 (0.722–0.851) | 0.747 (0.691–0.828) |
| Signal1 + Shape + dPSA | 0.789 (0,738–0,855) | 0.756 (0.698–0.829) | 0.766 (0.711–0.845) | 0.735 (0.678–0.812) |
| Signal1 + ECE + Vmax | 0.797 (0,746–0,864) | 0.773 (0.714–0.839) | 0.777 (0.720–0.850) | 0.765 (0.712–0.838) |
| Signal1 + ECE + dPSA | 0.805 (0,757–0,866) | 0.785 (0.735–0.847) | 0.777 (0.719–0.847) | 0.765 (0.710–0.834) |
| Signal1 + Vmax + dPSA | 0.795 (0,744–0,855) | 0.764 (0.708–0.827) | 0.790 (0.731–0.857) | 0.763 (0.705–0.832) |
| Signal1 + Shape + ECE + Vmax | 0.780 (0,733–0,851) | 0.762 (0.705–0.832) | 0.762 (0.710–0.845) | 0.748 (0.693–0.828) |
| Signal1 + Shape + ECE + dPSA | 0.789 (0,745–0,858) | 0.772 (0.714–0.841) | 0.759 (0.705–0.842) | 0.746 (0.696–0.825) |
| Signal1 + Shape + Vmax + dPSA | 0.785 (0,736–0,849) | 0.757 (0.698–0.826) | 0.776 (0.720–0.851) | 0.748 (0.695–0.824) |
| Signal1 + ECE + Vmax + dPSA | 0.800 (0,752–0,861) | 0.779 (0.725–0.842) | 0.779 (0.727–0.850) | 0.764 (0.715–0.834) |
| Signal1 + Shape + ECE + Vmax + dPSA | 0.785 (0,738–0,855) | 0.767 (0.712–0.838) | 0.763 (0.710–0.843) | 0.747 (0.697–0.828) |
DCE: dynamic contrast-enhanced imaging; AUC: area under the receiver operating characteristic curvewith optimism correction; 95%CI: 95% confidence intervals.
For each reader, the models achieving the highest AUC value were highlighted in green. The models achieving an AUC value that was inferior to the highest AUC value by 0.01 or less were highlighted in yellow.
For all models, AUC values were higher when DCE imaging was used, with a maximal ΔAUC of +0.039 for both readers.
For R1, the Signal1a+ECE+dPSA model obtained the highest AUC (0.805 [95%CI: 0.757–0.866]). It performed better with use of DCE imaging, but the difference was not statistically significant (p = 0.104). As compared to Signal 1a model, Signal1a+ECE+dPSA model achieved a ΔAUC of +0.011, but the difference was not significant (p = 0.167).
For R2, the Signal1a+Vmax+dPSA model obtained the highest AUC (0.790 [95%CI: 0.731–0.857]). It performed better with use of DCE imaging, but the difference was not statistically significant (p = 0.104). As compared to Signal 1a model, Signal1a+Vmax+dPSA model achieved a ΔAUC of +0.021, but the difference was not significant (p = 0.215).
Comparison of the best multivariable models to the Likert scores in characterizing csPCa-A in PZ
The AUC of the Likert score was 0.844 (95%CI: 0.806–0.877) for R1 and 0.829 (95%CI: 0.791–0.868) for R2. The ΔAUC with the best multivariable model (Signal1a+ECE+dPSA for R1 and Signal1a+Vmax+dPSA for R2) did not reach statistical significance (p = 0.121 for R1, and p = 0.121 for R2).
Assessment of single variables and multivariable models in characterizing csPCa-B in PZ
Table 5 shows the AUC values achieved by the 9 variables and the different multivariable models developed in the previous steps in characterizing csPCa-B.
Table 5. Performances of multivariable models in characterizing csPCa-B in PZ.
| AUC (95%CI)–Reader 1 | AUC (95%CI)–Reader 2 | |||
|---|---|---|---|---|
| With DCE | Without DCE | With DCE | Without DCE | |
| ST2 | 0.692 (0.604–0.767) | 0.616 (0.554–0.685) | ||
| SDWI | 0.734 (0.658–0.816) | 0.708 (0.647–0.769) | ||
| SDCE | 0.726 (0.655–0.793) | 0.675 (0.604–0.746) | ||
| S_max | 0.808 (0.741–0.871) | 0.767 (0.692–0.837) | 0.777 (0.711–0.845) | 0.726 (0.660–0.794) |
| S_min | 0.604 (0.556–0.652) | 0.551 (0.519–0.581) | ||
| Shape | 0.712 (0.633–0.784) | 0.696 (0.619–0.784) | ||
| ECE | 0.732 (0.659–0.808) | 0.720 (0.638–0.801) | ||
| Vmax | 0.718 (0.643–0.786) | 0.695 (0.613–0.774) | ||
| dPSA | 0.610 (0.447–0.694) | 0.623 (0.455–0.707) | ||
| Signal1 | 0.809 (0.741–0.876) | 0.764 (0.687–0.838) | 0.775 (0.711–0.847) | 0.721 (0.658–0.793) |
| Signal2 | 0.734 (0.665–0.835) | 0.739 (0.663–0.827) | 0.787 (0.720–0.855) | 0.747 (0.687–0.810) |
| Signal3 | 0.785 (0.720–0.874) | 0.754 (0.683–0.841) | 0.774 (0.703–0.851) | 0.730 (0.664–0.805) |
| Signal1 + Shape | 0.803 (0.738–0.878) | 0.775 (0.702–0.853) | 0.786 (0.718–0.870) | 0.760 (0.690–0.844) |
| Signal1 + ECE | 0.801 (0.737–0.877) | 0.776 (0.705–0.854) | 0.787 (0.715–0.866) | 0.780 (0.715–0.855) |
| Signal1 + Vmax | 0.821 (0.757–0.891) | 0.795 (0.721–0.872) | 0.813 (0.746–0.882) | 0.790 (0.723–0.865) |
| Signal1 + dPSA | 0.820 (0.761–0.885) | 0.785 (0.705–0.859) | 0.791 (0.723–0.869) | 0.759 (0.687–0.839) |
| Signal1 + Shape + ECE | 0.789 (0.728–0.872) | 0.772 (0.694–0.854) | 0.777 (0.707–0.866) | 0.764 (0.703–0.850) |
| Signal1 + Shape + Vmax | 0.813 (0.751–0.886) | 0.795 (0.724–0.874) | 0.798 (0.727–0.880) | 0.777 (0.709–0.866) |
| Signal1 + Shape + dPSA | 0.811 (0.753–0.883) | 0.786 (0.714–0.865) | 0.793 (0.723–0.875) | 0.773 (0.704–0.856) |
| Signal1 + ECE + Vmax | 0.808 (0.748–0.884) | 0.786 (0.714–0.865) | 0.799 (0.730–0.871) | 0.784 (0.721–0.864) |
| Signal1 + ECE + dPSA | 0.811 (0.755–0.881) | 0.785 (0.717–0.861) | 0.791 (0.725–0.868) | 0.783 (0.724–0.863) |
| Signal1 + Vmax + dPSA | 0.823 (0.760–0.893) | 0.797 (0.722–0.875) | 0.809 (0.744–0.884) | 0.788 (0.722–0.869) |
| Signal1 + Shape + ECE + Vmax | 0.799 (0.740–0.880) | 0.782 (0.715–0.868) | 0.780 (0.711–0.868) | 0.767 (0.703–0.853) |
| Signal1 + Shape + ECE + dPSA | 0.799 (0.744–0.879) | 0.777 (0.712–0.860) | 0.779 (0.714–0.867) | 0.773 (0.710–0.857) |
| Signal1 + Shape + Vmax + dPSA | 0.815 (0.755–0.888) | 0.796 (0.732–0.878) | 0.795 (0.731–0.879) | 0.777 (0.711–0.865) |
| Signal1 + ECE + Vmax + dPSA | 0.811 (0.754–0.887) | 0.787 (0.718–0.867) | 0.798 (0.734–0.874) | 0.783 (0.726–0.868) |
| Signal1 + Shape + ECE + Vmax + dPSA | 0.801 (0.747–0.882) | 0.782 (0.721–0.872) | 0.778 (0.714–0.869) | 0.768 (0.705–0.860) |
DCE: dynamic contrast-enhanced imaging; AUC: area under the receiver operating characteristic curve with optimism correction; 95%CI: 95% confidence intervals.
For each reader, the models achieving the highest AUC value were highlighted in green. The models achieving an AUC value that was inferior to the highest AUC value by 0.01 or less were highlighted in yellow.
When DCE imaging was used, the addition of other variables to the Signal1a model tended to increase the AUC, with a maximal ΔAUC of +0.014 for R1 and +0.038 for R2. When DCE was not used, all multivariable models performed better than Signal1b model with a maximum ΔAUC of +0.033 for R1 and +0.069 for R2.
For all models, AUC values were higher when DCE imaging was used, with a maximal ΔAUC of +0.035 for R1 and +0.032 for R2.
The highest AUC values were obtained by the Signal1a+Vmax+dPSA model for R1 (0.823 [95%CI: 0.760–0.893]) and by the Signal1a+Vmax for R2 (0.813 [95%CI: 0.746–0.882]).
The AUC of the Likert score was 0.841 (95%CI: 0.799–0.876) for R1 and 0.790 (95%CI: 0.742–0.841) for R2. The ΔAUC between the AUC of the best multivariable model and the Likert score was not significant, neither for R1 (p = 0.52) nor for R2 (p = 0.12).
Assessment of single variables and multivariable models in characterizing csPCa-C in PZ
When DCE imaging was used, the addition of other variables to the Signal 1a model tended to increase the AUC, with a maximal ΔAUC of +0.031 for R1 and +0.076 for R2 (Table 6). When DCE was not used, all multivariable models performed better than Signal1b model with a maximum ΔAUC of +0.065 for R1 and +0.133 for R2.
Table 6. Performances of multivariable models in characterizing csPCa-C in PZ.
| AUC (95%CI)–Reader 1 | AUC (95%CI)–Reader 1 | |||
|---|---|---|---|---|
| With DCE | Without DCE | With DCE | Without DCE | |
| ST2 | 0.734 (0.662–0.794) | 0.628 (0.560–0.693) | ||
| SDWI | 0.751 (0.670–0.819) | 0.678 (0.617–0.742) | ||
| SDCE | 0.714 (0.640–0.791) | 0.660 (0.584–0.735) | ||
| S_max | 0.793 (0.713–0.863) | 0.752 (0.679–0.823) | 0.749 (0.674–0.823) | 0.706 (0.628–0.781) |
| S_min | 0.624 (0.583–0.666) | 0.562 (0.537–0.587) | ||
| Shape | 0.685 (0.611–0.770) | 0.685 (0.596–0.779) | ||
| ECE | 0.809 (0.727–0.880) | 0.824 (0.757–0.892) | ||
| Vmax | 0.752 (0.680–0.830) | 0.750 (0.670–0.828) | ||
| dPSA | 0.594 (0.448–0.675) | 0.614 (0.477–0.698) | ||
| Signal1 | 0.809 (0.731–0.875) | 0.768 (0.699–0.837) | 0.763 (0.690–0.829) | 0.710 (0.636–0.783) |
| Signal2 | 0.769 (0.694–0.842) | 0.766 (0.680–0.840) | 0.762 (0.692–0.829) | 0.709 (0.638–0.785) |
| Signal3 | 0.802 (0.739–0.878) | 0.782 (0.716–0.851) | 0.751 (0.685–0.831) | 0.706 (0.635–0.783) |
| Signal1 + Shape | 0.798 (0.719–0.875) | 0.770 (0.690–0.851) | 0.770 (0.703–0.850) | 0.729 (0.659–0.821) |
| Signal1 + ECE | 0.836 (0.768–0.903) | 0.825 (0.766–0.898) | 0.835 (0.776–0.894) | 0.833 (0.774–0.898) |
| Signal1 + Vmax | 0.831 (0.763–0.896) | 0.806 (0.742–0.880) | 0.818 (0.757–0.886) | 0.804 (0.732–0.877) |
| Signal1 + dPSA | 0.816 (0.748–0.880) | 0.784 (0.713–0.855) | 0.780 (0.710–0.854) | 0.742 (0.664–0.824) |
| Signal + Shape + ECE | 0.819 (0.747–0.895) | 0.814 (0.749–0.887) | 0.816 (0.756–0.885) | 0.814 (0.754–0.886) |
| Signal1 + Shape + Vmax | 0.820 (0.752–0.890) | 0.800 (0.737–0.877) | 0.806 (0.748–0.881) | 0.788 (0.723–0.867) |
| Signal1 + Shape + dPSA | 0.801 (0.730–0.875) | 0.780 (0.708–0.858) | 0.775 (0.711–0.859) | 0.742 (0.673–0.834) |
| Signal1 + ECE + Vmax | 0.839 (0.774–0.906) | 0.829 (0.771–0.899) | 0.839 (0.784–0.902) | 0.843 (0.781–0.907) |
| Signal1 + ECE + dPSA | 0.840 (0.774–0.907) | 0.833 (0.776–0.902) | 0.836 (0.779–0.897) | 0.835 (0.777–0.901) |
| Signal1 + Vmax + dPSA | 0.829 (0.764–0.895) | 0.806 (0.743–0.881) | 0.816 (0.757–0.887) | 0.802 (0.733–0.874) |
| Signal1 + Shape + ECE + Vmax | 0.825 (0.757–0.902) | 0.818 (0.760–0.893) | 0.824 (0.770–0.893) | 0.823 (0.763–0.897) |
| Signal1 + Shape + ECE + dPSA | 0.820 (0.754–0.899) | 0.817 (0.757–0.894) | 0.817 (0.755–0.888) | 0.815 (0.758–0.887) |
| Signal1 + Shape + Vmax + dPSA | 0.817 (0.752–0.891) | 0.801 (0.736–0.881) | 0.804 (0.746–0.880) | 0.786 (0.723–0.868) |
| Signal1 + ECE + Vmax + dPSA | 0.839 (0.775–0.905) | 0.833 (0.778–0.902) | 0.838 (0.786–0.904) | 0.839 (0.779–0.905) |
| Signal1 + Shape + ECE + Vmax + dPSA | 0.823 (0.755–0.902) | 0.819 (0.763–0.895) | 0.823 (0.767–0.897) | 0.822 (0.763–0.896) |
DCE: dynamic contrast-enhanced imaging; AUC: area under the receiver operating characteristic curve with optimism correction; 95%CI: 95% confidence intervals.
For each reader, the models achieving the highest AUC value were highlighted in green. The models achieving an AUC value that was inferior to the highest AUC value by 0.01 or less were highlighted in yellow.
Bold characters indicate models that performed better without DCE imaging.
AUC values tended to be higher when DCE imaging was used, with a maximal ΔAUC of +0.032 for R1 and +0.041 for R2. However, two models performed better without DCE imaging for reader 2, including the model providing the best AUC value.
The highest AUC values were obtained by the Signal1a+ECE+dPSA model for R1 (0.840 [95%CI: 0.777–0.907]) and by the Signal1b+ECE+Vmax for R2 (0.843 [95%CI: 0.781–0.907]).
The AUC of the Likert score was 0.849 (95%CI: 0.811–0.884) for R1 and 0.808 (95%CI: 0.764–0.845) for R2. The ΔAUC between the AUC of the best multivariable model and the Likert score was not significant for R1 (p = 0.49) but was significant for R2 (p = 0.006).
Evolution of diagnostic performances of models and Likert score as a function of definition of csPCa
Table 7 summarizes the changes in the model performances with changes in csPCa definition. When moving from definition csPCa-A to definition csPCA-C, the ΔAUC between the Signal 1 model and the best multivariate models tended to increase, and the ΔAUC between models with and without DCE tended to decrease. These trends were more pronounced for R2 than for R1.
Table 7. Evolution of diagnostic performances of models and Likert scores in PZ as a function of csPCa definition.
| Reader | Signal 1a model | Best multivariate model | Likert score | Signal 1a vs multivariable models | Signal 1b vs multivariable models | Multivariable models with vs without DCE | ||
|---|---|---|---|---|---|---|---|---|
| AUC (95%CI) | Type | AUC (95%CI) | AUC (95%CI) | Median ΔAUC(1) [IQR] | Median ΔAUC(1) [IQR] | Median ΔAUC(2) [IQR] | ||
| R1 | csPCa-A | 0.794 | Signal1+ECE+dPSA | 0.805 | 0.844 | -0.005 | 0.016 | 0.024 |
| (0.734–0.851) | (0.757–0.866) | (0.806–0.877) | [-0.009; 0.005] | [0.011; 0.025] | [0.019 ; 0.032] | |||
| csPCa-B | 0.809 | Signal 1 + Vmax + dPSA | 0.823 | 0.841 | 0.002 | 0.021 | 0.024 | |
| (0.741–0.876) | (0.760–0.893) | (0.799–0.876) | [-0.008 ; 0.005] | [0.016 ; 0.027] | [0.019 ; 0.026] | |||
| csPCa-C | 0.809 | Signal 1 + ECE + dPSA | 0.840 | 0.849 | 0.014 | 0.046 | 0.011 | |
| (0.731–0.875) | (0.774–0.907) | (0.811–0.884) | [0.009 ; 0.025] | [0.033 ; 0.054] | [0.007 ; 0.022] | |||
| R2 | csPCa-A | 0.769 | Signal1+Vmax+dPSA | 0.790 | 0.829 | 0.007 | 0.023 | 0.017 |
| (0.698–0.835) | (0.731–0.857) | (0.791–0.868) | [-0.007; 0.009] | [0.021; 0.038] | [0.014; 0.029] | |||
| csPCa-B | 0.775 | Signal 1 + Vmax | 0.813 | 0.790 | 0.016 | 0.056 | 0.015 | |
| (0.711–0.847) | (0.746–0.882) | (0.742–0.841) | [0.008 ; 0.023] | [0.047 ; 0.062] | [0.012 ; 0.021] | |||
| csPCa-C | 0.763 | Signal 1 + ECE + Vmax | 0.843 | 0.808 | 0.054 | 0.104 | 0.002 | |
| (0.690–0.829) | (0.781–0.907) | (0.764–0.845) | [0.042 ; 0.067] | [0.077 ; 0.118] | [0.001 ; 0.018] | |||
AUC: area under the receiver operating characteristic curve with optimism correction; ΔAUC: difference between two AUC values; 95%CI: 95% confidence intervals; R1: reader 1; R2: reader 2.
(1) A positive difference indicates a better performance of the multivariable models.
(2) A positive difference indicates a better performance of the model using DCE imaging.
Discussion
The purpose of this study was not to build a new scoring system that could compete with existing ones, but rather to define the most informative MR features in characterizing csPCa in PZ, and to assess the relative diagnostic weight of these features, and how they could be optimally combined. Answering these questions is indeed mandatory if one wants to improve existing scores in the future.
To achieve this purpose, we used a radiologic-pathologic database containing a detailed description of sequence-specific features of all lesions visible at pre-operative prostate multiparametric MR imaging. This description was made prospectively by two independent readers and was then compared to prostatectomy specimens findings used as reference.
We used several definitions for csPCa since there is currently no consensus on this matter. As a primary objective, we defined csPCa as cancers with a Gleason score ≥7 because the metastatic and lethal potential of Gleason 6 cancers is low [40], and because this definition is commonly used [41]. Using this definition, we built the best multivariate models through a stepwise approach. Then, the models were assessed using more stringent definitions for csPCa.
Unsurprisingly, S_DW gave consistently better results than S_T2, and S_DCE for both readers, whatever the definition used for csPCa. This confirms that DW imaging is the most informative pulse sequence in PZ. Nonetheless, combining the results of the three pulse sequences into S_Max resulted in a substantial improvement in the diagnostic performance for both readers. This is in line with the good results obtained at the National Institute of Health (NIH) with an in-house score taking into account only the number of positive pulse sequences [16, 42], and points out that signal abnormalities remain the most informative features and should play a central role in any scoring system.
The Signal1-3 models consisted in three different combinations of the signal-based variables. They provided only marginal improvement as compared to S_Max. Particularly, the Signal2 model, that was an attempt to increase the diagnostic weight of DW imaging among the signal-based variables, failed to improve the characterization of csPCa. Thus, although DW imaging is the most informative pulse sequence in PZ, its optimal combination with the other pulse sequences remains to be defined.
Because the Signal1 model tended to give the best results, it was selected for the next step that assessed the added value of variables that were not related to signal abnormalities. Three variables (dPSA, ECE and V_Max) showed consistent added value and all best multivariable models included at least one of them. Their added value over signal-based variables (i.e. the ΔAUC between the best multivariable model and the Signal 1 model) tended to increase when DCE imaging was not used and when more stringent definitions of csPCa were used. Unsurprisingly, the diagnostic weight of ECE increased when the definition of csPCa included extraprostatic extension (csPCa-C), and the diagnostic weight of V_Max increased when the definition of csPCa was restricted to more aggressive tumors (csPCa-B instead of csPCa-A). Thus, an international consensus on the definition of csPCa is becoming crucial, since this will impact the scoring system to be used on multiparametric MR imaging. Interestingly, good results have recently been reported with a refinement of the NIH score combining ECE features and the number of positive pulse sequences [28]. Our results are in line and suggest that ECE features not only assess prostate cancer extracapsular extension, but also help characterizing the nature of the lesion. dPSA was the only non-MR variable included in this study. The fact that it consistently provided independent information to MR features is in line with a recent study that found that combining the PIRADS v2 score with dPSA improved prostate lesion characterization [43]. If the aim of scoring systems is to assess the likelihood of presence of csPCa, it might therefore be necessary to associate MR features and clinical or biochemical features in the future.
Taking into account the shape of the lesions decreased the performance of almost all models. This strongly suggests that shape is not a good predictor of csPCa, even if the PIRADS v2 and other scoring systems [15, 17] use it to characterize focal lesions in PZ. The poor diagnostic value of shape had already been found in another study [14] and may be due to the fact that it remains a very subjective feature.
There is currently a controversy about the added value of DCE imaging, as compared to T2W and DW imaging. DCE imaging may indeed help detect small cancers, but may also increase the number of false positive findings [23, 44–47]. Several groups failed to find clear added value for DCE imaging when MR images were interpreted visually or using scoring systems [25, 48–50]. Nonetheless, in most quantitative studies aimed at characterizing prostate lesions [34, 51–54], DCE-derived parameters were part of the best final models. Our study is in line with these quantitative studies. When we removed DCE findings, we observed a systematic decrease in the diagnostic performances of nearly all models, for both readers, suggesting that DCE imaging does provide information. The best way to incorporate this information into a scoring system remains to be defined. Interestingly, the added value of DCE imaging tended to decrease for both readers when more stringent definitions of csPCa were used.
In the last part of our study, we compared the best models to the Likert score prospectively assigned to each lesion. For the most experienced reader, the Likert score constantly outperformed the best model, even if the difference was never statistically significant and tended to decrease as more stringent definitions of csPCa were used. For the least experienced reader, however, the best model outperformed the Likert score for csPCa-B and csPCa-C, and the difference was statistically significant for csPCa-C. Most of the image features we used were subjective and this may be seen as a contradiction with our initial goal to obtain more objective scoring systems. Unless an entirely quantitative approach is used for all pulse sequences, subjective assessment of images remains unavoidable. Existing scoring systems share the same limitation, as shown by the PIRADS v2 score that distinguishes indistinct hypointense (score 2), mildly/moderately hypointense (score 3) or markedly hypointense (scores 4–5) lesions on ADC maps. However, our results suggest that breaking down the diagnostic process into separate features, and combining these features into predefined models may help less experienced readers better characterise MR lesions.
Our study has some limitations.The description of the sequence-specific features was done prospectively. Although this could be seen as a strength of the study, it also induced two limitations. First, we were not able to compare the best models with the PIRADS v2 score that was not launched at the start of the study. However, our main purpose was not to compare our models to existing scores, but to understand the features’ relative weight in characterizing csPCa. Second, the features noted in the database for TZ lesions were mostly based on signal abnormalities. Features as homogeneous pattern, presence of a capsule, apical or anterior location, that are now known as major predictors of malignancy in TZ [55, 56] were not prospectively recorded. As a result we chose not to use the database for assessing diagnostic models in TZ. Another limitation is due to the fact that both readers were from the same institution and may have characterized prostate lesions in a similar way. The best multivariable models may have been different with readers from other institutions. Finally, our study mixed patients imaged at different field strengths with varying protocols. Although this resulted in a heterogeneous population, it may better reflect daily routine population.
Conclusion
The number of pulse sequences showing marked signal abnormality for a given lesion (S_max) was one the most informative variable for both readers, whatever the definition used for csPCa. A moderate improvement could be obtained by taking into account, in addition to S_max, the number of negative pulse sequences, the presence of extracapsular extension features, the volume of the lesion and the PSA density. The added value of the three latter variables depended on the definition used for csPCa and tended to increase when more stringent definitions were used. Removing DCE findings decreased performance in nearly all models, but the difference decreased when more stringent definitions were used for csPCa. Finally the Likert score outperformed the best multivariable models for the most experienced reader whatever the definition used for csPCa. For the other reader, the multivariable models outperformed the Likert score for csPCa-B and csPCa-C definitions, and the difference was significant for csPCa-C. This suggests that scoring systems based on semi-objective variables may help less-experienced radiologists.
Supporting information
TR: Repetition Time; TE: Echo time; PPA: pelvic phased array; T2w: T2 weighted imaging; Dw: Diffusion weighted imaging; DCE: dynamic contrast enhanced imaging.
(DOCX)
Sd: standard deviation; IQR: interquartile range.
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Turkbey B, Pinto PA, Mani H, Bernardo M, Pang Y, McKinney YL, et al. Prostate cancer: value of multiparametric MR imaging at 3 T for detection—histopathologic correlation. Radiology. 2010;255(1):89–99. doi: 10.1148/radiol.09090475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vargas HA, Akin O, Shukla-Dave A, Zhang J, Zakian KL, Zheng J, et al. Performance characteristics of MR imaging in the evaluation of clinically low-risk prostate cancer: a prospective study. Radiology. 2012;265(2):478–87. Epub 2012/09/07. doi: 10.1148/radiol.12120041 ; PubMed Central PMCID: PMC3480819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratan F, Niaf E, Melodelima C, Chesnais AL, Souchon R, Mege-Lechevallier F, et al. Influence of imaging and histological factors on prostate cancer detection and localisation on multiparametric MRI: a prospective study. Eur Radiol. 2013;23(7):2019–29. Epub 2013/03/16. doi: 10.1007/s00330-013-2795-0 . [DOI] [PubMed] [Google Scholar]
- 4.Kim JY, Kim SH, Kim YH, Lee HJ, Kim MJ, Choi MS. Low-risk prostate cancer: the accuracy of multiparametric MR imaging for detection. Radiology. 2014;271(2):435–44. Epub 2014/02/04. doi: 10.1148/radiol.13130801 . [DOI] [PubMed] [Google Scholar]
- 5.Futterer JJ, Briganti A, De Visschere P, Emberton M, Giannarini G, Kirkham A, et al. Can Clinically Significant Prostate Cancer Be Detected with Multiparametric Magnetic Resonance Imaging? A Systematic Review of the Literature. Eur Urol. 2015;68(6):1045–53. Epub 2015/02/07. doi: 10.1016/j.eururo.2015.01.013 . [DOI] [PubMed] [Google Scholar]
- 6.Valerio M, Donaldson I, Emberton M, Ehdaie B, Hadaschik BA, Marks LS, et al. Detection of Clinically Significant Prostate Cancer Using Magnetic Resonance Imaging-Ultrasound Fusion Targeted Biopsy: A Systematic Review. Eur Urol. 2015;68(1):8–19. Epub 2014/12/03. doi: 10.1016/j.eururo.2014.10.026 . [DOI] [PubMed] [Google Scholar]
- 7.Schoots IG, Roobol MJ, Nieboer D, Bangma CH, Steyerberg EW, Hunink MG. Magnetic Resonance Imaging-targeted Biopsy May Enhance the Diagnostic Accuracy of Significant Prostate Cancer Detection Compared to Standard Transrectal Ultrasound-guided Biopsy: A Systematic Review and Meta-analysis. Eur Urol. 2015;68(3):438–50. Epub 2014/12/07. doi: 10.1016/j.eururo.2014.11.037 . [DOI] [PubMed] [Google Scholar]
- 8.Dickinson L, Ahmed HU, Allen C, Barentsz JO, Carey B, Futterer JJ, et al. Magnetic resonance imaging for the detection, localisation, and characterisation of prostate cancer: recommendations from a European consensus meeting. Eur Urol. 2011;59(4):477–94. doi: 10.1016/j.eururo.2010.12.009 [DOI] [PubMed] [Google Scholar]
- 9.Rosenkrantz AB, Pujara AC, Taneja SS. Use of a Quality Improvement Initiative to Achieve Consistent Reporting of Level of Suspicion for Tumor on Multiparametric Prostate MRI. AJR Am J Roentgenol. 2016;206(5):1040–4. Epub 2016/04/23. doi: 10.2214/AJR.15.15768 . [DOI] [PubMed] [Google Scholar]
- 10.Habchi H, Bratan F, Paye A, Pagnoux G, Sanzalone T, Mege-Lechevallier F, et al. Value of prostate multiparametric magnetic resonance imaging for predicting biopsy results in first or repeat biopsy. Clin Radiol. 2014;69(3):e120–8. Epub 2013/12/18. doi: 10.1016/j.crad.2013.10.018 . [DOI] [PubMed] [Google Scholar]
- 11.Vache T, Bratan F, Mege-Lechevallier F, Roche S, Rabilloud M, Rouviere O. Characterization of prostate lesions as benign or malignant at multiparametric MR imaging: comparison of three scoring systems in patients treated with radical prostatectomy. Radiology. 2014;272(2):446–55. Epub 2014/06/18. doi: 10.1148/radiol.14131584 . [DOI] [PubMed] [Google Scholar]
- 12.Mozer P, Roupret M, Le Cossec C, Granger B, Comperat E, de Gorski A, et al. First round of targeted biopsies using magnetic resonance imaging/ultrasonography fusion compared with conventional transrectal ultrasonography-guided biopsies for the diagnosis of localised prostate cancer. BJU Int. 2015;115(1):50–7. Epub 2014/02/21. doi: 10.1111/bju.12690 . [DOI] [PubMed] [Google Scholar]
- 13.Costa DN, Lotan Y, Rofsky NM, Roehrborn C, Liu A, Hornberger B, et al. Assessment of Prospectively Assigned Likert Scores for Targeted Magnetic Resonance Imaging-Transrectal Ultrasound Fusion Biopsies in Patients with Suspected Prostate Cancer. J Urol. 2016;195(1):80–7. Epub 2015/07/21. doi: 10.1016/j.juro.2015.07.080 . [DOI] [PubMed] [Google Scholar]
- 14.Rouviere O, Papillard M, Girouin N, Boutier R, Rabilloud M, Riche B, et al. Is it possible to model the risk of malignancy of focal abnormalities found at prostate multiparametric MRI? Eur Radiol. 2012;22(5):1149–57. Epub 2012/01/10. doi: 10.1007/s00330-011-2343-8 . [DOI] [PubMed] [Google Scholar]
- 15.Puech P, Rouviere O, Renard-Penna R, Villers A, Devos P, Colombel M, et al. Prostate Cancer Diagnosis: Multiparametric MR-targeted Biopsy with Cognitive and Transrectal US-MR Fusion Guidance versus Systematic Biopsy—Prospective Multicenter Study. Radiology. 2013;268(2):461–9. Epub 2013/04/13. doi: 10.1148/radiol.13121501 . [DOI] [PubMed] [Google Scholar]
- 16.Rais-Bahrami S, Siddiqui MM, Turkbey B, Stamatakis L, Logan J, Hoang AN, et al. Utility of multiparametric magnetic resonance imaging suspicion levels for detecting prostate cancer. J Urol. 2013;190(5):1721–7. Epub 2013/06/04. doi: 10.1016/j.juro.2013.05.052 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rastinehad AR, Waingankar N, Turkbey B, Yaskiv O, Sonstegard AM, Fakhoury M, et al. Comparison of Multiparametric MRI Scoring Systems and the Impact on Cancer Detection in Patients Undergoing MR US Fusion Guided Prostate Biopsies. PloS one. 2015;10(11):e0143404 Epub 2015/11/26. doi: 10.1371/journal.pone.0143404 ; PubMed Central PMCID: PMC4659614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barentsz JO, Richenberg J, Clements R, Choyke P, Verma S, Villeirs G, et al. ESUR prostate MR guidelines 2012. Eur Radiol. 2012;22(4):746–57. Epub 2012/02/11. doi: 10.1007/s00330-011-2377-y ; PubMed Central PMCID: PMC3297750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamoen EH, de Rooij M, Witjes JA, Barentsz JO, Rovers MM. Use of the Prostate Imaging Reporting and Data System (PI-RADS) for Prostate Cancer Detection with Multiparametric Magnetic Resonance Imaging: A Diagnostic Meta-analysis. Eur Urol. 2015;67(6):1112–21. Epub 2014/12/04. doi: 10.1016/j.eururo.2014.10.033 . [DOI] [PubMed] [Google Scholar]
- 20.de Cobelli O, Terracciano D, Tagliabue E, Raimondi S, Bottero D, Cioffi A, et al. Predicting Pathological Features at Radical Prostatectomy in Patients with Prostate Cancer Eligible for Active Surveillance by Multiparametric Magnetic Resonance Imaging. PloS one. 2015;10(10):e0139696 Epub 2015/10/09. doi: 10.1371/journal.pone.0139696 ; PubMed Central PMCID: PMC4596627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenkrantz AB, Kim S, Lim RP, Hindman N, Deng FM, Babb JS, et al. Prostate Cancer Localization Using Multiparametric MR Imaging: Comparison of Prostate Imaging Reporting and Data System (PI-RADS) and Likert Scales. Radiology. 2013;269(2):482–92. Epub 2013/06/22. doi: 10.1148/radiol.13122233 . [DOI] [PubMed] [Google Scholar]
- 22.Renard-Penna R, Mozer P, Cornud F, Barry-Delongchamps N, Bruguiere E, Portalez D, et al. Prostate Imaging Reporting and Data System and Likert Scoring System: Multiparametric MR Imaging Validation Study to Screen Patients for Initial Biopsy. Radiology. 2015;275(2):458–68. Epub 2015/01/20. doi: 10.1148/radiol.14140184 . [DOI] [PubMed] [Google Scholar]
- 23.Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, et al. PI-RADS Prostate Imaging—Reporting and Data System: 2015, Version 2. Eur Urol. 2016;69(1):16–40. Epub 2015/10/03. doi: 10.1016/j.eururo.2015.08.052 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barentsz JO, Weinreb JC, Verma S, Thoeny HC, Tempany CM, Shtern F, et al. Synopsis of the PI-RADS v2 Guidelines for Multiparametric Prostate Magnetic Resonance Imaging and Recommendations for Use. Eur Urol. 2016;69(1):41–9. Epub 2015/09/12. doi: 10.1016/j.eururo.2015.08.038 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vargas HA, Hotker AM, Goldman DA, Moskowitz CS, Gondo T, Matsumoto K, et al. Updated prostate imaging reporting and data system (PIRADS v2) recommendations for the detection of clinically significant prostate cancer using multiparametric MRI: critical evaluation using whole-mount pathology as standard of reference. Eur Radiol. 2015. Epub 2015/09/24. doi: 10.1007/s00330-015-4015-6 ; PubMed Central PMCID: PMC4803633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY, Jung DC, Oh YT, Cho NH, Choi YD, Rha KH, et al. Prostate Cancer: PI-RADS Version 2 Helps Preoperatively Predict Clinically Significant Cancers. Radiology. 2016:151133 Epub 2016/02/03. doi: 10.1148/radiol.16151133 . [DOI] [PubMed] [Google Scholar]
- 27.Rosenkrantz AB, Oto A, Turkbey B, Westphalen AC. Prostate Imaging Reporting and Data System (PI-RADS), Version 2: A Critical Look. AJR Am J Roentgenol. 2016:1–5. Epub 2016/02/26. doi: 10.2214/AJR.15.15765 . [DOI] [PubMed] [Google Scholar]
- 28.Muller BG, Shih JH, Sankineni S, Marko J, Rais-Bahrami S, George AK, et al. Prostate Cancer: Interobserver Agreement and Accuracy with the Revised Prostate Imaging Reporting and Data System at Multiparametric MR Imaging. Radiology. 2015;277(3):741–50. Epub 2015/06/23. doi: 10.1148/radiol.2015142818 ; PubMed Central PMCID: PMC4666087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenkrantz AB, Ginocchio LA, Cornfeld D, Froemming AT, Gupta RT, Turkbey B, et al. Interobserver Reproducibility of the PI-RADS Version 2 Lexicon: A Multicenter Study of Six Experienced Prostate Radiologists. Radiology. 2016:152542 Epub 2016/04/02. doi: 10.1148/radiol.2016152542 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mertan FV, Greer MD, Shih JH, George AK, Kongnyuy M, Muthigi A, et al. Prospective Evaluation of the Prostate Imaging Reporting and Data System Version 2 for Prostate Cancer Detection. J Urol. 2016;196(3):690–6. Epub 2016/04/23. doi: 10.1016/j.juro.2016.04.057 . [DOI] [PubMed] [Google Scholar]
- 31.Bratan F, Melodelima C, Souchon R, Hoang Dinh A, Mege-Lechevallier F, Crouzet S, et al. How accurate is multiparametric MR imaging in evaluation of prostate cancer volume? Radiology. 2015;275(1):144–54. Epub 2014/11/26. doi: 10.1148/radiol.14140524 . [DOI] [PubMed] [Google Scholar]
- 32.Niaf E, Lartizien C, Bratan F, Roche L, Rabilloud M, Mege-Lechevallier F, et al. Prostate focal peripheral zone lesions: characterization at multiparametric MR imaging—influence of a computer-aided diagnosis system. Radiology. 2014;271(3):761–9. Epub 2014/03/07. doi: 10.1148/radiol.14130448 . [DOI] [PubMed] [Google Scholar]
- 33.Hoang Dinh A, Souchon R, Melodelima C, Bratan F, Mege-Lechevallier F, Colombel M, et al. Characterization of prostate cancer using T2 mapping at 3T: a multi-scanner study. Diagnostic and interventional imaging. 2015;96(4):365–72. Epub 2014/12/31. doi: 10.1016/j.diii.2014.11.016 . [DOI] [PubMed] [Google Scholar]
- 34.Hoang Dinh A, Melodelima C, Souchon R, Lehaire J, Bratan F, Mege-Lechevallier F, et al. Quantitative Analysis of Prostate Multiparametric MR Images for Detection of Aggressive Prostate Cancer in the Peripheral Zone: A Multiple Imager Study. Radiology. 2016;280(1):117–27. Epub 2016/02/10. doi: 10.1148/radiol.2016151406 . [DOI] [PubMed] [Google Scholar]
- 35.Samaratunga H, Montironi R, True L, Epstein JI, Griffiths DF, Humphrey PA, et al. International Society of Urological Pathology (ISUP) Consensus Conference on Handling and Staging of Radical Prostatectomy Specimens. Working group 1: specimen handling. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24(1):6–15. Epub 2010/09/14. doi: 10.1038/modpathol.2010.178 . [DOI] [PubMed] [Google Scholar]
- 36.Dhingsa R, Qayyum A, Coakley FV, Lu Y, Jones KD, Swanson MG, et al. Prostate cancer localization with endorectal MR imaging and MR spectroscopic imaging: effect of clinical data on reader accuracy. Radiology. 2004;230(1):215–20. doi: 10.1148/radiol.2301021562 [DOI] [PubMed] [Google Scholar]
- 37.Hom JJ, Coakley FV, Simko JP, Qayyum A, Lu Y, Schmitt L, et al. Prostate cancer: endorectal MR imaging and MR spectroscopic imaging—distinction of true-positive results from chance-detected lesions. Radiology. 2006;238(1):192–9. Epub 2005/12/24. doi: 10.1148/radiol.2381041675 . [DOI] [PubMed] [Google Scholar]
- 38.Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. The American journal of surgical pathology. 2016;40(2):244–52. Epub 2015/10/23. doi: 10.1097/PAS.0000000000000530 . [DOI] [PubMed] [Google Scholar]
- 39.Harrell FE Jr., Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics in medicine. 1996;15(4):361–87. Epub 1996/02/28. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4 . [DOI] [PubMed] [Google Scholar]
- 40.Eggener SE, Badani K, Barocas DA, Barrisford GW, Cheng JS, Chin AI, et al. Gleason 6 Prostate Cancer: Translating Biology into Population Health. J Urol. 2015;194(3):626–34. Epub 2015/04/08. doi: 10.1016/j.juro.2015.01.126 ; PubMed Central PMCID: PMC4551510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moldovan PC, Van den Broeck T, Sylvester R, Marconi L, Bellmunt J, van den Bergh RC, et al. What Is the Negative Predictive Value of Multiparametric Magnetic Resonance Imaging in Excluding Prostate Cancer at Biopsy? A Systematic Review and Meta-analysis from the European Association of Urology Prostate Cancer Guidelines Panel. Eur Urol. 2017. Epub 2017/03/25. doi: 10.1016/j.eururo.2017.02.026 . [DOI] [PubMed] [Google Scholar]
- 42.Turkbey B, Mani H, Aras O, Ho J, Hoang A, Rastinehad AR, et al. Prostate cancer: can multiparametric MR imaging help identify patients who are candidates for active surveillance? Radiology. 2013;268(1):144–52. Epub 2013/03/08. doi: 10.1148/radiol.13121325 ; PubMed Central PMCID: PMC3689450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Washino S, Okochi T, Saito K, Konishi T, Hirai M, Kobayashi Y, et al. Combination of prostate imaging reporting and data system (PI-RADS) score and prostate-specific antigen (PSA) density predicts biopsy outcome in prostate biopsy naive patients. BJU Int. 2017;119(2):225–33. Epub 2016/03/05. doi: 10.1111/bju.13465 . [DOI] [PubMed] [Google Scholar]
- 44.Girouin N, Mege-Lechevallier F, Tonina Senes A, Bissery A, Rabilloud M, Marechal JM, et al. Prostate dynamic contrast-enhanced MRI with simple visual diagnostic criteria: is it reasonable? Eur Radiol. 2007;17(6):1498–509. doi: 10.1007/s00330-006-0478-9 [DOI] [PubMed] [Google Scholar]
- 45.Perdona S, Di Lorenzo G, Autorino R, Buonerba C, De Sio M, Setola SV, et al. Combined magnetic resonance spectroscopy and dynamic contrast-enhanced imaging for prostate cancer detection. Urol Oncol. 2013;31(6):761–5. Epub 2011/09/13. doi: 10.1016/j.urolonc.2011.07.010 . [DOI] [PubMed] [Google Scholar]
- 46.Puech P, Sufana-Iancu A, Renard B, Lemaitre L. Prostate MRI: Can we do without DCE sequences in 2013? Diagnostic and interventional imaging. 2013;94(12):1299–311. Epub 2013/11/12. doi: 10.1016/j.diii.2013.09.010 . [DOI] [PubMed] [Google Scholar]
- 47.Tan CH, Hobbs BP, Wei W, Kundra V. Dynamic contrast-enhanced MRI for the detection of prostate cancer: meta-analysis. AJR Am J Roentgenol. 2015;204(4):W439–48. Epub 2015/03/21. doi: 10.2214/AJR.14.13373 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delongchamps NB, Rouanne M, Flam T, Beuvon F, Liberatore M, Zerbib M, et al. Multiparametric magnetic resonance imaging for the detection and localization of prostate cancer: combination of T2-weighted, dynamic contrast-enhanced and diffusion-weighted imaging. BJU Int. 2011;107(9):1411–8. Epub 2010/11/04. doi: 10.1111/j.1464-410X.2010.09808.x . [DOI] [PubMed] [Google Scholar]
- 49.Vilanova JC, Barcelo-Vidal C, Comet J, Boada M, Barcelo J, Ferrer J, et al. Usefulness of prebiopsy multifunctional and morphologic MRI combined with free-to-total prostate-specific antigen ratio in the detection of prostate cancer. AJR Am J Roentgenol. 2011;196(6):W715–22. Epub 2011/05/25. doi: 10.2214/AJR.10.5700 . [DOI] [PubMed] [Google Scholar]
- 50.Isebaert S, Van den Bergh L, Haustermans K, Joniau S, Lerut E, De Wever L, et al. Multiparametric MRI for prostate cancer localization in correlation to whole-mount histopathology. J Magn Reson Imaging. 2013;37(6):1392–401. Epub 2012/11/23. doi: 10.1002/jmri.23938 . [DOI] [PubMed] [Google Scholar]
- 51.Langer DL, van der Kwast TH, Evans AJ, Trachtenberg J, Wilson BC, Haider MA. Prostate cancer detection with multi-parametric MRI: logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI. Journal of magnetic resonance imaging. 2009;30(2):327–34. doi: 10.1002/jmri.21824 [DOI] [PubMed] [Google Scholar]
- 52.Hambrock T, Vos PC, Hulsbergen-van de Kaa CA, Barentsz JO, Huisman HJ. Prostate cancer: computer-aided diagnosis with multiparametric 3-T MR imaging—effect on observer performance. Radiology. 2013;266(2):521–30. Epub 2012/12/04. doi: 10.1148/radiol.12111634 . [DOI] [PubMed] [Google Scholar]
- 53.Riches SF, Payne GS, Morgan VA, Dearnaley D, Morgan S, Partridge M, et al. Multivariate modelling of prostate cancer combining magnetic resonance derived T2, diffusion, dynamic contrast-enhanced and spectroscopic parameters. Eur Radiol. 2015;25(5):1247–56. Epub 2015/03/10. doi: 10.1007/s00330-014-3479-0 . [DOI] [PubMed] [Google Scholar]
- 54.Vos EK, Kobus T, Litjens GJ, Hambrock T, Hulsbergen-van de Kaa CA, Barentsz JO, et al. Multiparametric Magnetic Resonance Imaging for Discriminating Low-Grade From High-Grade Prostate Cancer. Invest Radiol. 2015;50(8):490–7. Epub 2015/04/14. doi: 10.1097/RLI.0000000000000157 . [DOI] [PubMed] [Google Scholar]
- 55.Chesnais AL, Niaf E, Bratan F, Mege-Lechevallier F, Roche S, Rabilloud M, et al. Differentiation of transitional zone prostate cancer from benign hyperplasia nodules: evaluation of discriminant criteria at multiparametric MRI. Clin Radiol. 2013;68(6):e323–30. Epub 2013/03/27. doi: 10.1016/j.crad.2013.01.018 . [DOI] [PubMed] [Google Scholar]
- 56.Hoeks CM, Hambrock T, Yakar D, Hulsbergen-van de Kaa CA, Feuth T, Witjes JA, et al. Transition Zone Prostate Cancer: Detection and Localization with 3-T Multiparametric MR Imaging. Radiology. 2013;266(1):207–17. Epub 2012/11/13. doi: 10.1148/radiol.12120281 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TR: Repetition Time; TE: Echo time; PPA: pelvic phased array; T2w: T2 weighted imaging; Dw: Diffusion weighted imaging; DCE: dynamic contrast enhanced imaging.
(DOCX)
Sd: standard deviation; IQR: interquartile range.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.