Summary
Fat storage‐inducing transmembrane protein 2 (FIT2) is an endoplasmic reticulum (ER)‐localized protein that plays an important role in lipid droplet (LD) formation in animal cells. However, no obvious homologue of FIT2 is found in plants. Here, we tested the function of FIT2 in plant cells by ectopically expressing mouse (Mus musculus) FIT2 in Nicotiana tabacum suspension‐cultured cells, Nicotiana benthamiana leaves and Arabidopsis thaliana plants. Confocal microscopy indicated that the expression of FIT2 dramatically increased the number and size of LDs in leaves of N. benthamiana and Arabidopsis, and lipidomics analysis and mass spectrometry imaging confirmed the accumulation of neutral lipids in leaves. FIT2 also increased seed oil content by ~13% in some stable, overexpressing lines of Arabidopsis. When expressed transiently in leaves of N. benthamiana or suspension cells of N. tabacum, FIT2 localized specifically to the ER and was often concentrated at certain regions of the ER that resembled ER‐LD junction sites. FIT2 also colocalized at the ER with other proteins known to be involved in triacylglycerol biosynthesis or LD formation in plants, but not with ER resident proteins involved in electron transfer or ER‐vesicle exit sites. Collectively, these results demonstrate that mouse FIT2 promotes LD accumulation in plants, a surprising functional conservation in the context of a plant cell given the apparent lack of FIT2 homologues in higher plants. These results suggest also that FIT2 expression represents an effective synthetic biology strategy for elaborating neutral lipid compartments in plant tissues for potential biofuel or bioproduct purposes.
Keywords: endoplasmic reticulum, fat storage‐inducing transmembrane protein 2, lipid droplets, lipid storage, triacylglycerol, lipid partitioning
Introduction
Lipid droplets (LDs) are organelles found in all types of plant cells where they function to compartmentalize neutral lipids such as triacylglycerols (TAGs), steryl esters (SEs) and even isoprenoids (e.g. carotenoids, rubber) (Chapman and Ohlrogge, 2012; Murphy, 2001). Once considered as simply a repository for lipid storage molecules, these organelles are becoming more appreciated for their roles in other dynamic cellular processes, including signalling, trafficking and membrane remodelling (Brasaemle and Wolins, 2012; Chapman et al., 2012). LDs have a ‘half‐unit’ membrane of phospholipids covering a hydrophobic core of nonpolar lipids, and in different cell types, different proteins localize to the LD surface (Murphy, 2012). Perhaps best studied in plants are the oleosins associated with LDs in seed tissues (Chapman et al., 2012; Frandsen et al., 2001; Huang, 1996), but a number of other proteins are located on the surface of LDs as well, including the recently identified lipid droplet‐associated proteins (LDAPs) that appear to stabilize LDs in many nonseed (i.e. vegetative) cell types (Chapman et al., 2012; Gidda et al., 2016; Horn et al., 2013; Kim et al., 2016).
Cytosolic LDs, in both seeds and nonseed tissues, have also been referred to in the literature as lipid bodies, oil bodies, oleosomes and spherosomes, but more recently, the term lipid droplet has been used to emphasize a more cohesive and evolutionary relationship with neutral‐lipid‐containing organelles from various cell types and organisms (Murphy, 2012). Cytosolic LDs in plant cells are distinctly different in location and composition from plastoglobuli, which are LDs that originate from the thylakoid membrane and accumulate in the plastid stroma (Austin et al., 2006; Nacir and Bréhélin, 2013). By contrast, cytosolic LDs are believed to originate from the endoplasmic reticulum (ER), where the neutral lipids are synthesized and partitioned somehow between the membrane bilayer leaflets before being packaged for release from the ER surface (Wanner et al., 1981). In plants, storage lipid accumulation into LDs has mostly been studied in seed tissues (Chapman et al., 2012), and transcriptional regulation of lipid synthesis and compartmentation is modulated through several key transcription factors, including WRINKLED1 (WRI1) and LEAFY COTYLEDON2 (LEC2) (Sreenivasulu and Wobus, 2013). Although a number of specific proteins in yeast, invertebrates and mammals have been identified that participate in LD ontogeny (Beller et al., 2010; Gao and Goodman, 2015; Pol et al., 2014), relatively little is known mechanistically about how plant cells form LDs at the ER and facilitate their release into the cytosol in seeds or nonseed tissues.
There are several models for cytosolic LD biogenesis in eukaryotic cells, and these all involve the ER in some manner. A widely accepted ‘lens’ model indicates that TAGs collect within the ER membrane bilayer and then bud from the ER surface into the cytosol to form an individual LD (Choudhary et al., 2015). In another model, pre‐existing vesicles are filled with neutral lipids from the ER to form LDs (Walther and Farese, 2009). Yet another model suggests that LDs that are formed from ER‐derived TAGs remain attached to the ER, perhaps even being interconnected with the ER lumen, and enlarging or reducing in size and prevalence based on cellular needs (Farese and Walther, 2009; Jacquier et al., 2011; Mishra et al., 2016). Others have suggested the possibility that LDs grow by small LDs fusing together to form large LDs (Wilfling et al., 2014). And finally, an ‘egg‐in‐cup’ model has been proposed where LDs originate at a particular location on the cytosolic surface of the ER membrane, whereby the ER partially surrounds the developing LD in such a way that it resembles an LD‐’egg’ inside an ER‐membrane ‘cup’ (Robenek et al., 2006). Although these various models represent different mechanistic views of how the LD organelle is produced, one shared feature is that LDs are formed in association with the ER. Indeed, the ways in which LDs form in cells likely include several mechanisms that are cell type‐ or stage‐specific and may not be mutually exclusive (Chapman et al., 2012; Pol et al., 2014; Wilfling et al., 2014).
The ER is the subcellular location for the majority of glycerolipid assembly within cells and includes enzymes for the synthesis of the neutral lipid LD core, as well as the phospholipid monolayer of the LD surface (Chapman and Ohlrogge, 2012; Chapman et al., 2012). In addition, a number of membrane‐bound proteins, mostly identified in animals and yeast, have been shown to be involved in the biogenesis of LDs and localized at the ER, often at ER‐LD junctions, or even on nascent LDs (Brasaemle and Wolins, 2012; Pol et al., 2014; Walther and Farese, 2009; Wang et al., 2016; Wilfling et al., 2014). One of the proteins that has been implicated in the sequestration of TAG for LD formation is the fat storage‐inducing transmembrane protein (FIT) (Kadereit et al., 2008). There are two isoforms of the FIT protein in mammals, namely FIT1 and FIT2. FIT1 is found in skeletal muscle and is less evolutionarily conserved than FIT2. FIT2 homologues, on the other hand, are found in organisms ranging from Saccharomyces cerevisiae to humans and in mammals are expressed predominantly in adipose tissue (Gross et al., 2010; Miranda et al., 2014). Ectopic overexpression of FIT2 in mammals induced the accumulation of large‐sized LDs at higher levels than are normally found and in tissues that are not normally LD‐rich (Gross et al., 2010; Kadereit et al., 2008). FIT2 overexpression did not, however, increase the synthesis of TAGs directly, but rather promoted the more efficient partitioning of neutral lipids from the ER membrane into nascent LDs (Gross et al., 2011; Kadereit et al., 2008). Notably, modifications of the FIT2 polypeptide sequence that reduced or enhanced TAG‐binding affinity resulted in the production of less or more LDs, respectively (Gross et al., 2011). Further, knock‐down of FIT2 expression in adipose cells, zebrafish and mice reduced the accumulation of LDs (Goh et al., 2015; Gross et al., 2011; Kadereit et al., 2008; Miranda et al., 2014).
FIT2 was shown recently to be essential for the budding of nascent LDs from the ER into the cytosol in yeast and mammalian cells (Choudhary et al., 2015), and disruption of FIT2 in Caenorhabditis elegans was lethal (Choudhary et al., 2015). Interestingly, no homologues of FIT2 have been detected in the genomes of plants (Chapman et al., 2012). The importance of FIT2 in promoting LD formation in other eukaryotes prompted us to ask whether mammalian FIT2 would/could function to promote LD biogenesis in a plant cell context. Towards that end, we expressed the FIT2 protein from mouse (Mus musculus) in two transient plant cell systems, Nicotiana tabacum suspension‐cultured cells and Nicotiana benthamiana leaves, as well as via stable expression in transgenic Arabidopsis thaliana plants. Overall, our results indicate that FIT2, as it does in other eukaryotes, localizes to the ER in plant cells, including regions of the ER that appear to be involved in TAG biosynthesis and LD formation. FIT2 also promotes the accumulation of LDs in plant cells that normally do not accumulate substantial amounts of LDs, like those in leaves. We conclude that FIT2 likely interacts with the native LD biosynthetic machinery in plant cells to promote enhanced neutral lipid compartmentalization and the budding of LDs from ER, and its ectopic expression represents a novel strategy to elaborate the neutral lipid compartment in plants for various biotechnology‐oriented applications.
Results
Mouse FIT2 induces the accumulation of cytosolic LDs in transiently transformed tobacco leaves
The FIT2 coding sequence from mouse was expressed in Agrobacterium‐infiltrated tobacco leaves to evaluate the effect of FIT2 expression on LD proliferation in a plant cell context (Figure 1). As shown in Figure 1a, confocal images of BODIPY 493/503‐stained LDs in mock (infiltration buffer only)‐transformed leaves or leaves that were expressing only the P19 viral protein (serving as an enhancer of transgene expression and included in all infiltrations [Petrie et al., 2010]) showed few LDs distributed throughout the cytosol (Figure 1a), which is typical of mesophyll cells in N. benthamiana leaves (Cai et al., 2015; Gidda et al., 2016). Similarly, chlorophyll autofluorescence in mock‐ and P19‐transformed cells revealed the expected distribution of chloroplasts, which mostly surrounded the cell's large central vacuole (Figure 1a). By contrast, expression of FIT2 in leaf cells resulted in a marked increase in both the numbers and sizes of LDs (Figure 1a, quantified in Figure 1b), all of which appeared to be localized within the cytosol and not in chloroplasts, indicating they were distinct from plastoglobuli. Further, expression of a FIT2 variant (FIT2‐N[80]A) known to have reduced TAG‐binding affinity compared to native FIT2 (Gross et al., 2011) resulted in an increase in the number of LDs in comparison with mock or P19 controls, but relatively fewer medium (3–6 μm2), large (6–10 μm2) or so‐called super‐sized (>10 μm2) LDs in comparison with cells expressing the native FIT2 protein. Expression of a FIT2 variant with enhanced TAG‐binding activity (i.e. FIT2‐FLL[157‐159]AAA; Gross et al., 2011), on the other hand, resulted in a further increase in super‐sized LDs in comparison with native FIT2 (Figure 1a and b).
Figure 1.
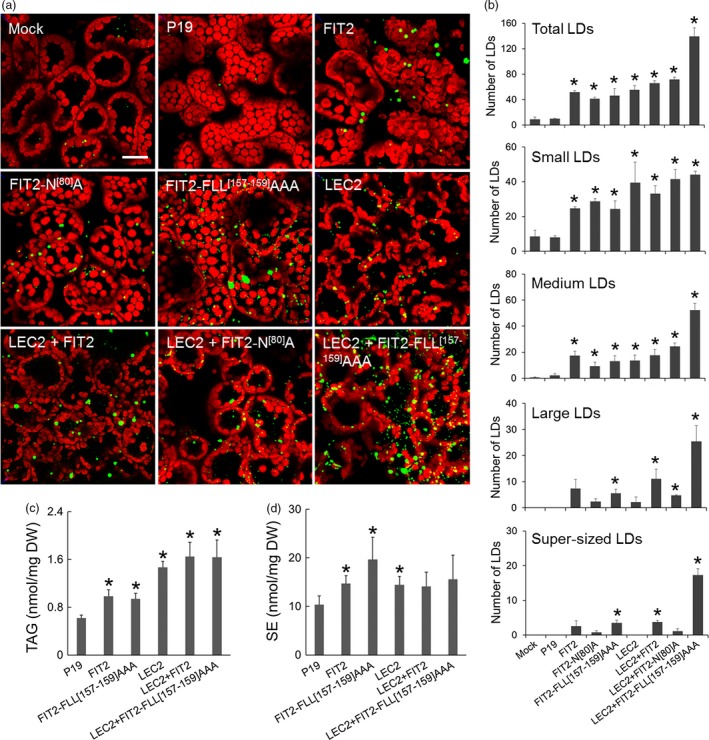
FIT2 expression in Nicotiana benthamiana leaves elaborates a neutral lipid compartment. (a) Representative confocal images of N. benthamiana leaves expressing FIT2 or mutated FIT2 proteins (i.e. FIT2‐N[80]A or FIT2‐FLL [157‐159] AAA) in the presence or absence of LEC2. Cells were (co)transformed as indicated by the panel label, and LDs (green) were visualized by staining with BODIPY 493/503. Red colour corresponds to chloroplast autofluorescence. Images are 2D projections of Z‐stacks of 50 optical sections. Bar = 20 μm. (b) Number of total LDs and LDs in different size categories per image area. The numbers and sizes of LDs were quantified by ImageJ as BODIPY‐stained lipid area in 2D projections of Z‐stacks (see Experimental procedures for details). Small LDs: BODIPY‐stained lipid area <3 μm2. Medium LDs: 3–6 μm2. Large LDs: 6–10 μm2. Super‐sized LDs: >10 μm2. Averages and SDs were calculated from three biological replicates (three Z‐stacks from each replicate). Asterisks indicate significant difference relative to the mock control at P ≤ 0.05 determined by Student's t‐test. (c) and (d) TAG and SE content in infiltrated N. benthamiana leaves. Averages and SDs were plotted from four biological replicates. Asterisks indicate significant difference in comparison with the P19 control at P ≤ 0.05 determined by Student's t‐test.
As shown also in Figure 1a, ectopic expression of the LEC2 transcription factor, which induces genes for seed oil like synthesis in leaves (Santos Mendoza et al., 2005), resulted in an increase in LD proliferation, as expected (Cai et al., 2015; Gidda et al., 2016). While the total numbers of LDs induced by LEC2 expression were similar to that of FIT2, the numbers of large and super‐sized LDs were conspicuously greater with FIT2 in comparison with LEC2 (Figure 1b). These data suggest that FIT2 promotes the formation of larger‐sized LDs in plant cells. Indeed, combined expression of LEC2 and FIT2, particularly with the ‘enhanced’ FIT2 variant (FLL[157‐159]AAA), exaggerated further the total numbers of LDs in leaves, with a greater proportion of large and super‐sized LDs, as well as medium‐sized LDs, than with either FIT2 alone or LEC2 alone (Figure 1a and b). Moreover, FIT2 cooperated with the LEC2‐induced machinery to package ~2.5 times more neutral lipids (i.e. TAGs and some SEs) in these leaves (Figure 1c and d). Detailed analysis of the TAG and SE molecular species by conventional, direct‐infusion ESI‐MS/MS (electrospray ionization tandem mass spectrometry) (Figure S1) indicated that, generally, the TAGs and SEs that are normally present in leaves were all increased upon FIT2 expression. These same molecular species of TAG and SE were exaggerated further by co‐expression of FIT2 and LEC2, making the overall increase in TAGs and SE even more obvious (Figure S1). Notably, there was no further increase in TAG and SE in leaves co‐expressing LEC2 and the enhanced FIT2 variant (FLL[157‐159]AAA) in comparison with amounts observed in leaves co‐expressing LEC2 and native FIT2 (Figure 1c), despite a nearly twofold difference in the total number of LDs (Figure 1b). These observations would be consistent, however, with a primary role for FIT2 in the binding and partitioning of TAG into LDs, rather than a role in stimulating TAG synthesis (Gross et al., 2011; Kadereit et al., 2008).
FIT2 localizes to the ER in plant cells and is enriched in ER subdomains involved in TAG assembly and LD biogenesis
The proliferation of LDs in plant cells following FIT2 expression suggested that it was functioning at the subcellular level in a manner similar to that in animal and yeast cells. Indeed, as shown in Figure 2, confocal imaging revealed that when GFP‐tagged FIT2 was expressed in N. benthamiana leaves, it localized exclusively to the ER, as evidenced by colocalization with a co‐expressed ER marker protein, CFP‐HDEL (Szymanski et al., 2007). Notably, GFP‐FIT2 and CFP‐HDEL co‐expressing cells also displayed a marked increase in the abundance of Nile red‐stained LDs when compared to cells expressing CFP‐HDEL alone (Figure 2), consistent with the induction of LDs in tobacco leaves transiently expressing FIT2 (Figure 1a and b). GFP‐tagged versions of FIT2‐N[80]A or FIT2‐FLL[157‐159]AAA also localized to the ER in tobacco leaf cells (Figure 2), and the elaboration of LDs in these cells was distinct from each other and that of wild‐type GFP‐FIT2, similar to results obtained using their non‐GFP‐tagged counterparts (Figure 1). That is, leaf cells expressing GFP‐FIT2‐N[80]A or GFP‐FIT2‐FLL[157‐159]AAA, relative to those expressing GFP‐FIT2, appeared to possess fewer or more larger‐sized LDs, respectively (Figure 2). Cells expressing GFP‐FIT2‐FLL[157‐159]AAA also displayed several conspicuous alterations in ER morphology, whereby the fusion protein (and CFP‐HDEL) localized not only throughout the normal, reticular‐like ER network, but also accumulated in specific regions of the ER that appeared globular in shape and were not observed in cells expressing CFP‐HDEL alone (Figure 2; refer to arrowheads in bottom row). These ER structures typically colocalized with or next to Nile red‐stained LDs, suggesting that these might be ER‐LD junction sites that formed as a result of excessive TAG binding and/or inefficient release of the nascent LDs from the ER, or simply might be a reflection of a normal process that was exaggerated by ectopic expression. It is possible also that the LDs with TAG may be trapped in the ER lumen at these locations (Mishra et al., 2016) or wrapped in ER membranes (Choudhary et al., 2015), but the resolution of light microscopy prevents more conclusive statements to be made about the nature of these structures.
Figure 2.
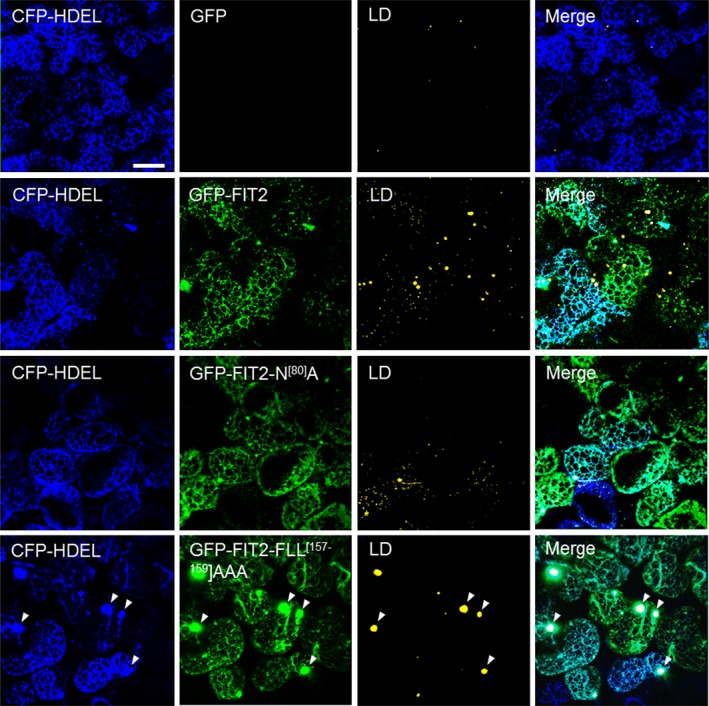
FIT2 and mutated FIT2 GFP‐tagged fusion proteins localize to the ER in Nicotiana benthamiana leaf mesophyll cells. As indicated by panel labels, ER, GFP‐FIT2/FIT2 variants and LDs were visualized (via confocal microscopy) in mesophyll cells based on fluorescence signals of CFP‐HDEL (blue), N‐terminal‐GFP‐tagged FIT2 or FIT2 variants (i.e. FIT2‐N[80]A or FIT2‐FLL [157‐159] AAA) (green) and Nile red (yellow), respectively). Shown also are the corresponding merged images. Note that no GFP fluorescence signal was detectable in the cell expressing CFP‐HDEL alone (top row), as expected. Arrowheads in the bottom row indicate examples of regions of the ER with altered morphology, where both GFP‐FIT2‐FLL [157‐159] AAA and Nile red‐stained LDs appear to accumulate. All images are 2D projections of Z‐stacks of ~30 slices. Bar = 20 μm.
We tested next whether mouse FIT2 could colocalize with other proteins known to be involved in TAG synthesis and/or LD formation in plant cells. For this purpose, we employed tobacco (N. tabacum) Bright Yellow 2 (BY‐2) suspension‐cultured cells, which serve as a well‐established model system for protein localization studies (Brandizzi et al., 2003; Miao and Jiang, 2007) and, given their relatively large size, are amenable to microscopic analysis of ER subdomains (Gidda et al., 2011; Hanton et al., 2007; Shockey et al., 2006). The proteins tested for colocalization included Vernicia fordii (tung tree) diacylglycerol acyltransferase 2 (DGAT2), which is known to be involved in TAG synthesis and localized to ER subdomains (Shockey et al., 2006), and Arabidopsis SEIPIN1, which localizes to ER‐LD sites where it is thought to promote the compartmentalization of neutral lipids into nascent, large‐sized LDs from the ER (Cai et al., 2015). As shown in Figures 3a and S2, and similar to the results presented above for GFP‐FIT2 in N. benthamiana leaf cells (Figure 2), transient expression of Cherry‐FIT2 or GFP‐FIT2 in BY‐2 cells resulted in a marked increase in monodansylpentane (MDH)‐stained LDs in comparison with mock‐transformed control cells, and the fusion protein localized throughout concanavalin A (ConA)‐stained ER. Unlike control cells, however, the ER of cells expressing FIT2 was generally more punctate and globular in nature (Figure 3a), which is often observed for transiently expressed proteins that target to ER subdomains. Co‐expression of Cherry‐tagged FIT2 with either myc‐epitope‐tagged DGAT2 or GFP‐tagged SEIPIN1 showed clear colocalizations in specific regions of ER (Figure 3b). These associations appeared to be subdomain specific, as Cherry‐FIT2 did not colocalize with co‐expressed GFP‐tagged Arabidopsis SEC24, a soluble protein that localizes to subdomains of ER that serve as transport‐vesicle exit sites (Gidda et al., 2011; Hanton et al., 2007), or with a GFP‐tagged isoform of tung tree cytochrome b 5 (Cb5), which is known to induce ER membrane ‘whorls’ or ‘karmallae’ when ectopically overexpressed (Hwang et al., 2004; Snapp et al., 2003) (Figure 3b). Control experiments are shown to help illustrate that each of the respective proteins is targeted to ConA‐stained ER subdomains, regardless of FIT2 co‐expression (Figure 3c). While we recognize that caution should be exercised in the interpretation of these results, the data suggest that mouse FIT2 colocalizes specifically with machinery in plant cells that is devoted to TAG biosynthesis and LD formation, and not likely indiscriminately localized to regions involved in other ER processes (e.g. vesicle export) or due to nonspecific membrane protein aggregation.
Figure 3.
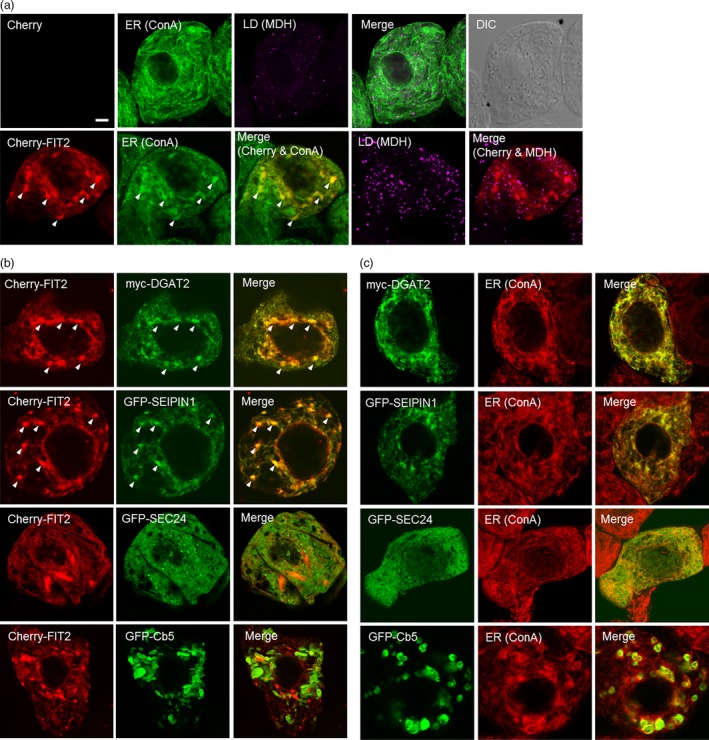
FIT2 expressed in Nicotiana tabacum suspension‐cultured BY‐2 cells is enriched in ER domains that also accumulate proteins involved in TAG biosynthesis and LD biogenesis. BY‐2 cells were transiently (co)transformed with the indicated proteins, and then, cells were visualized using confocal microscopy. (a) Representative confocal images showing the ER and LDs in mock‐transformed BY‐2 cells (top row), whereby the ER was stained using fluor‐conjugated ConA and LDs were stained using MDH (false‐coloured magenta). Shown also is the corresponding merged and differential interference contrast (DIC) images. The bottom row shows the fluorescence‐staining pattern of transiently expressed Cherry‐FIT2 in comparison with ConA‐stained ER and MDH‐stained LDs. Arrowheads in the bottom row indicate examples of Cherry‐FIT2 enriched in specific regions of ConA‐stained ER. Note that no Cherry fluorescence signal was detected in the mock‐transformed cell (top row), as expected. Bar = 5 μm. (b) Confocal images showing the localizations of Cherry‐FIT2 and various other co‐expressed ER‐localized proteins, including myc‐tagged DGAT2 or GFP‐tagged SEIPIN1, SEC24 or Cb5. Shown also are the corresponding merged images for each pair of co‐expressed proteins. Arrowheads illustrate examples of colocalization of FIT2 with DGAT2 and SEIPIN1. (c) Confocal images showing the localization of individually expressed Myc‐DGAT2 or GFP‐tagged SEIPIN1, SEC24 or Cb5 at the ConA‐stained ER. Shown also are the corresponding merged images for each protein and ConA‐stained ER.
Stable expression of FIT2 in Arabidopsis increases neutral lipid accumulation in both leaf and seed tissues
While mouse FIT2 appeared to localize to ER subdomains and promote LD formation in transient plant expression assays (Figures 1, 2, 3), we next determined how this protein would function in stably transformed plants. Towards this end, we generated several independent transgenic Arabidopsis lines ectopically expressing GFP‐tagged or nontagged mouse FIT2. Transgene expression was verified using reverse transcriptase (RT)‐PCR (Figure S3a). As shown in Figure 4, leaves of mature, 24‐day‐old transgenic plants showed significant increases in the numbers of LDs compared to nontransformed, wild‐type plants (Figure 4a and b), consistent with results obtained for transient expression in tobacco leaves and suspension cells (Figures 1, 2, 3). Further, in one line (i.e. FIT2‐OE‐C6), in addition to increased numbers of LDs, the LDs were significantly larger than those in wild‐type plants or other FIT2 lines (Figure 4c).
Figure 4.
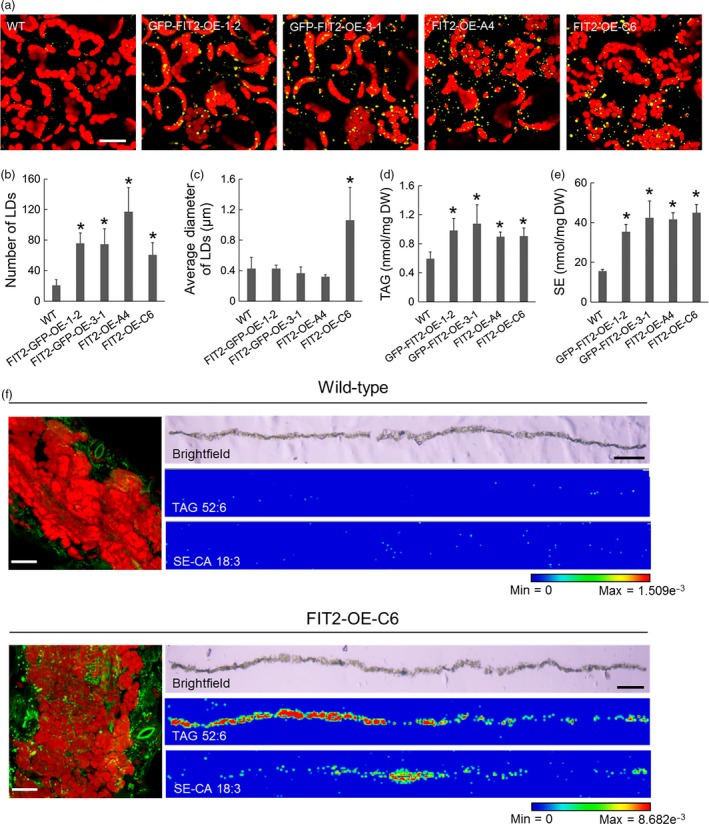
Stable expression of FIT2 in Arabidopsis increases LD number and neutral lipid content in leaf tissues. (a) Representative confocal images of wild‐type and FIT2 transgenic Arabidopsis leaves. Red colour indicates autofluorescence attributable to chloroplasts. LDs were stained with Nile red (false‐coloured yellow). Images are projections of Z‐stacks of 30 optical sections. (b) and (c) Numbers and sizes of LDs per view (image area) of Arabidopsis leaf mesophyll. LDs were quantified as Nile red‐stained lipid area in 2D images (as single optical slices). Averages and SDs were calculated based on nine images from at least three Arabidopsis plants. Asterisks indicate significant difference at P ≤ 0.05 determined by Student's t‐test. (d) and (e) TAG and SE content in wild‐type and FIT2 transgenic Arabidopsis leaves. Averages and SDs are calculated from four biological replicates. Asterisks indicate significant difference at P ≤ 0.05 determined by Student's t‐test. (f) Spatial distribution of BODIPY‐stained LDs (green), one major TAG species (TAG 52:6, m/z 889.668) and one major SE species (campesterol [CA] 18:3, m/z 699.547) in wild‐type and transgenic (i.e. FIT2‐OE‐C6) Arabidopsis leaf cross sections. Left panels are representative confocal images of BODIPY‐stained LDs (green) in wild‐type and transgenic Arabidopsis leaf tissue. Autofluorescence of chloroplasts in confocal images are shown in red colour. Confocal images are projections of Z‐stacks. Bar = 20 μm. Right panels are bright field microscopy images of leaf sections and corresponding spatial distribution of TAG 52:6 and CA 18:3 detected by MALDI. Bar = 0.5 mm. The blue‐to‐red scale bar is used to indicate minimum and maximum ion intensity of potassiated TAG 52:6 and potassiated CA 18:3 molecules.
Figure 4 shows also that the neutral lipid content was increased in FIT2 transgenic Arabidopsis plant leaves (with or without GFP), with significant increases in both TAG and SE quantified by ESI‐MS/MS, in comparison with wild‐type plants (Figure 4d and e; and Figure S4; to 0.1% TAG and 3% SE on a dry weight basis). Recently, it has become possible to visualize lipid species directly in tissues by MALDI‐MS (Horn and Chapman, 2014a; Sturtevant et al., 2016). Hence, we confirmed in situ by MALDI‐MS (matrix‐assisted laser desorption/ionization MS) imaging of cryosectioned leaves that two major neutral lipid molecular species, one TAG (TAG 52:6, m/z 889.668) and one SE (Campesterol [CA] 18:3, m/z 699.547), were in fact elevated and distributed in the mesophyll tissues of FIT2 transgenic leaves, that is FIT2‐OE‐C6 (Figure 4f). Curiously, there seemed to be a somewhat heterogeneous distribution of the TAG species relative to the SE species in the leaves of this line (Figure 4g), suggesting that there is a difference in the location in leaves for the synthesis and/or accumulation of these two neutral lipid classes, but this possibility, and how FIT2 might participate in this process, needs to be examined in more detail. Nevertheless, this general distribution of elevated amounts of neutral lipids throughout the mesophyll tissues of leaves, compared to wild‐type leaves, was consistent with an increase in BODIPY 493/503‐stained LDs (Figure 4f, left) or Nile red‐stained LDs (Figure 4a) observed in mesophyll cells expressing FIT2 in Arabidopsis transgenic lines. We observed also that LDs were more prevalent in young and mature leaves (i.e. the 7th true leaf and the 5th true leaf from the bottom of the plant, respectively) compared with older leaves (i.e. the 3rd true leaf from the bottom of the plant) (Figure S3b), perhaps reflecting an influence of FIT2 in more metabolically active tissues where neutral lipids are likely to be synthesized. Further, transgenic leaves exhibited quantifiable increases in both TAGs and SEs, and most major molecular species of these neutral lipids increased significantly (Figure S4), indicating that FIT2 promoted a general elevation in neutral lipids that were already present in leaves, and not a specific accumulation of new or selected species. The composition of PC molecular species was not altered in a substantial way by stable, ectopic expression of FIT2 (Figure S5).
While there was a significant increase in the numbers of LDs and contents of neutral lipids in vegetative tissues of Arabidopsis plants stably expressing mouse FIT2 (Figure 4), we also asked whether FIT2 might enhance the accumulation of TAGs in lipid‐rich seed tissues in these same lines. As shown in Figure 5a, confocal microscopy of parenchyma cells in embryo cotyledons suggested that the size and distribution of Nile red‐stained LDs was somewhat different in transgenic seeds compared to wild‐type, with larger‐sized LDs often observed within the interior of these cells (Figure 5a). Furthermore, while seed size and dry weight were not significantly altered by FIT2 expression (Figure 5b and c), there was a modest but significant increase in seed oil content in some, but not all, FIT2 transgenic lines (measured by time‐domain 1H‐NMR), and also in the amounts of total fatty acids per seed measured by gas chromatography‐flame ionization detection (GC‐FID) (Figure 5d and e). More specifically, when plants were grown under identical conditions, some FIT2‐expressing lines displayed an increase in seed oil content over wild‐type by as much as 13% (e.g. 34% oil versus 30% oil on a dry weight basis for FIT2‐OE‐C6 versus WT, respectively) (Figure 5d), suggesting that FIT2 expression can enhance overall oil content in seed tissues, as well as in nonseed (vegetative) tissues of plants. Furthermore, fatty acid composition of the seed oil was mostly similar between wild‐type and FIT2 lines, with only some subtle differences due to an increase in 18:1 and decrease in 20:1 acyl moieties (Figure 5f).
Figure 5.
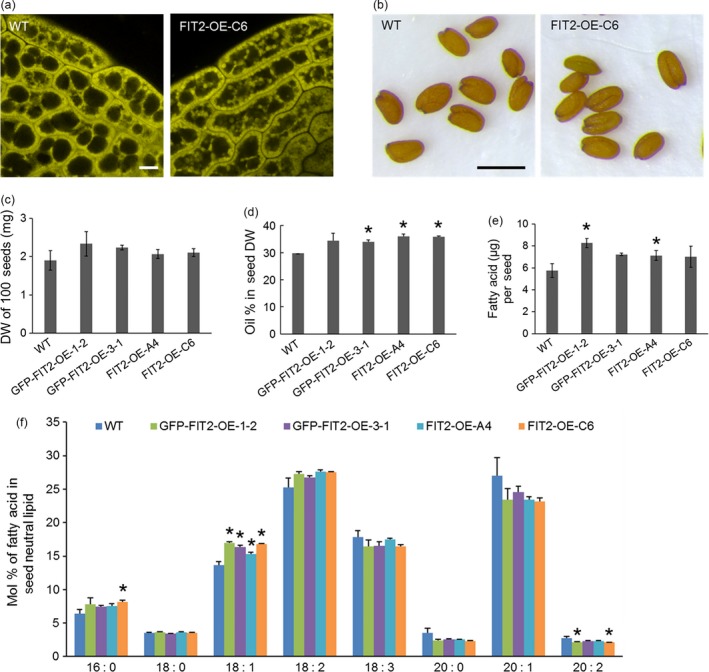
Stable FIT2 expression increases oil content in Arabidopsis seeds. (a) Representative confocal images of LDs in wild‐type and FIT2 transgenic (i.e. FIT2‐OE‐C6 line) Arabidopsis seeds. LDs were stained with Nile red (false‐coloured yellow). Bar = 5 μm. (b) Mature seeds from wild‐type and FIT2 transgenic Arabidopsis plants. Bar = 0.5 mm. (c) to (f) Seed size and seed oil content of wild‐type and FIT2 transgenic Arabidopsis. Averages and SDs were calculated from three biological replicates. Seeds were collected from Arabidopsis plants grown under the same conditions. Asterisks indicate significant difference at P ≤ 0.05 determined by Student's t‐test. (c) Weight of 100 dry seeds. (d) Oil content in dry seeds on a weight basis measured by NMR. (e) Oil content per seed on a fatty acid basis quantified by GC‐FID. (f) Fatty acid composition in mature seeds.
GFP‐tagged FIT2 was also visualized in Arabidopsis leaves and seeds to confirm protein expression and evaluate the localization of FIT2 relative to LDs in these tissues (Figure S6). Similar to results in tobacco leaves and suspension cells (Figures 2 and 3), in Arabidopsis leaves, GFP‐tagged FIT2 showed accumulations of concentrated fluorescence often in the proximity of Nile red‐stained LDs (Figure S6). In seeds, the GFP fluorescence signal was relatively weak but indicated that GFP‐FIT2 was associated with the LDs in places (Figure S6); however, the ER organization is rearranged in desiccated seed tissues (Hsieh and Huang, 2004; Wang et al., 2010), and perhaps the residual FIT2 persisting through seed desiccation was localized to LDs or regions of ER intimately surrounding the LDs.
Discussion
FIT2 has been described as part of an evolutionarily conserved family of proteins that are important for the subcellular compartmentalization of neutral lipids in mammals, yeast, insects and C. elegans (Choudhary et al., 2015; Goh et al., 2015; Gross et al., 2010; Kadereit et al., 2008). It is curious, therefore, that this essential protein would not be present in all eukaryotes that form LDs from the ER, as no obvious homologues have been identified in plant genomes. Cytosolic LD biogenesis from the ER membrane shares conserved features at the cellular level across kingdoms, and there are likely overlapping sets of proteins that cooperate in this process, perhaps encoded by distantly related genes. Indeed, SEIPIN genes were recently identified in plant genomes, and three isoforms in Arabidopsis were shown to participate in the biogenesis of LDs in plant cells, similar to their function in other eukaryotes (Cai et al., 2015). In some cases, a shared cellular process might also be carried out by distinctly different proteins, such as the oleosins and LDAPs in plants and the perilipins in mammals, all of which bind the LD surface and stabilize LDs in the cytosol, yet share no obvious sequence similarity (Chapman et al., 2012; Gidda et al., 2016).
Here, we have asked whether a protein without any known homologues in plants might still function in a plant cell context to influence LD biogenesis. FIT2 functions, at least in part, by binding to TAG (Gross et al., 2011), and thus, its role in LD biogenesis is likely dependent on biophysical properties of cellular membranes that might also be present in plant cells. Notably, similar approaches were used to explore the targeting and function of oleosins, caleosins and perilipins in LD formation in yeast cells, which otherwise lack these proteins endogenously (Beaudoin and Napier, 2002; Froissard et al., 2009; Jacquier et al., 2013; Mishra and Schneiter, 2015; Rowe et al., 2016). Clearly, ectopic expression of mouse FIT2 in plant cells, similar to results observed in yeast and animal cells (Choudhary et al., 2015; Gross et al., 2010; Kadereit et al., 2008), increases production of LDs and associated TAGs (Figures 1, 2, 4 and 5). From the results presented here, it also appears that FIT2, due to its inherent ability to bind TAGs (Gross et al., 2011), localizes to TAG‐forming domains of the ER in plant cells where it functions to promote LD formation. SEs were also elevated in leaves expressing FIT2 (Figures 1 and 4; Figures S1 and S6), however, suggesting that FIT2 engages in and enhances a general coordination of neutral lipid synthesis and LD formation. Indeed, when co‐expressed with DGAT2 or SEIPIN1, FIT2 colocalized with both proteins (Figure 3), consistent with the premise that FIT2 associates with domains involved in both TAG synthesis and nascent LD biogenesis (Choudhary et al., 2015). The activity of FIT2 in plant cells suggests it might be useful as a probe for further studying the molecular nature of LD‐forming domains in plants, possibly identifying other, as yet unidentified proteins involved in LD biogenesis.
The ectopic expression of a seed‐specific transcription factor such as LEC2 in leaves has been shown by us and others to promote the accumulation of TAGs in leaf tissues (Mu et al., 2008; Sanjaya et al., 2013; Slocombe et al., 2009), and this is accentuated when additional proteins that promote TAG assembly, like DGAT, or proteins that bind to cytosolic LDs, like oleosins, are coproduced (Andrianov et al., 2010; Vanhercke et al., 2013, 2014; Winichayakul et al., 2013). Together, these engineering approaches have been described in a ‘push, pull and protect’ strategy to overproduce TAG in vegetative tissues (Vanhercke et al., 2014; Xu and Shanklin, 2016). In this model, seed‐specific transcription factors up‐regulate the fatty acid biosynthetic machinery (Grimberg et al., 2015), effectively ‘pushing’ reduced photosynthetic carbon into lipid, up‐regulation of DGAT promotes the synthesis of TAGs at the ER, ‘pulling’ reduced carbon into TAG, and oleosins ‘protect’ the sequestered TAG from the active metabolic pool and prevent the hydrolysis of TAGs and turnover of LDs. In fact, remarkable amounts of TAG have been accumulated in leaves of transgenic tobacco plants—up to more than 15% dry weight of leaves—using a combination of these engineering processes (Vanhercke et al., 2014). The increase in the energy density of leaves (i.e. increased lipid content relative to carbohydrate) will likely find multiple applications in agriculture and bioenergy, such as higher calorie and nutritionally balanced feeds, or as a source of oleochemicals and biodiesel feedstock (Horn and Benning, 2016). Indeed, it has been suggested that an increase in vegetative biomass of lipid up to 10% dry weight would result in a 30% increase in usable liquid fuels, through a combination of both biodiesel and lignocellulosic‐derived fuels (Ohlrogge and Chapman, 2011). The production of large amounts of lipids in vegetative tissues makes possible the separation of calories in edible oilseeds needed for human nutrition from a potential bioenergy feedstock derived from vegetative biomass, preventing renewable sources of energy from competing with food production. What remains to be explored is how proteins involved in packaging TAG into LDs, such as SEIPIN, FIT2 (or its yet‐to‐be identified functionally equivalent plant orthologue), might complement ongoing approaches to enhance the energy content of plant vegetative tissues. Certainly, it seems that co‐expression of LEC2 with variants of FIT2 in plant leaves can result in marked increase of compartmentalized LDs and neutral lipid content above that seen with either LEC2 or FIT2 alone (Figure 1), and so perhaps some component of LD ‘release’ would add significantly to the ‘push–pull–protect’ theory of increasing lipid accumulation in plant vegetative tissues.
Experimental procedures
Plant material, growth conditions and transformation
A. thaliana (Columbia‐0) plants were grown in growth chambers on either one‐half‐strength MS‐media‐containing plates (Murashige and Skoog, 1962) or in soil at 21 °C under a 16/8‐h light/dark cycle with 50 μE/m2/s light intensity. Nicotiana benthamiana plants were grown in soil at 28 °C under a 14/10‐h light/dark cycle. Plants used in the same experiments were grown concurrently, in the same conditions. Plasmids were transformed into Agrobacterium tumefaciens (GV3101) by electroporation (Bio‐Rad), and stable transformation of Arabidopsis plants was carried out using the floral dip method (Clough and Bent, 1998). Gene expression was evaluated in transgenic Arabidopsis seedlings (progeny) using RT‐PCR, and independent lines were selected for further study. Infiltration of N. benthamiana leaves was carried out as described previously (Cai et al., 2015; Petrie et al., 2010). Tomato Bushy Stunt Virus (TBSV) P19 was included in all infiltrations to suppress transgene silencing and intensify transgene expression (Petrie et al., 2010); Arabidopsis LEC2 was included in some co‐infiltrations to promote seed‐like lipid synthesis in leaves (Cai et al., 2015; Gidda et al., 2016; Santos Mendoza et al., 2005). Nicotiana tabacum Bright Yellow (BY)‐2 suspension‐cultured cells were maintained and prepared for (co)transformation via biolistic particle bombardment using a Bio‐Rad PDS system 1000/HE, as described previously (Lingard et al., 2008).
Plasmid construction
The cDNA of M. musculus FIT2 (EMBL Accession No. BAE37420.1) was kindly donated by David Silver (Duke—National University of Singapore). The FIT2 ORF was subcloned into the plant binary expression plasmids pMDC32 and pMDC43 [Curtis and Grossniklaus, 2003; ] using restriction enzymes AscI and PacI, yielding pMDC32/FIT2 and pMDC43/GFP‐FIT2, respectively. Both pMDC32 and pMDC43, as well as all other plant expression plasmids used in this study, contain the CaMV 35S constitutive promoter (Benfey et al., 1989). Mutated versions of FIT2 (i.e. FIT2‐N[80]A and FIT2‐FLL[157‐159]AAA) were generated using PCR‐fusion cloning procedures (Atanassov et al., 2009) and then subcloned into pMDC32 and pMDC43. pRTL2/Cherry‐FIT2 was constructed by subcloning the FIT2 ORF into the transient expression plasmid pRTL2/Cherry‐MCS, which contains the Cherry fluorescent protein coding sequence (Gidda et al., 2011). Plant binary expression plasmids containing Arabidopsis LEC2 (pORE04‐LEC2) and Tomato Bushy Stunt Virus P19 (pORE04‐P19) were provided by Qing Liu (CSIRO) (Petrie et al., 2010). Other plant expression plasmids, including pMDC32/CFP‐HDEL, pMDC43/GFP‐SEIPIN1, pRTL2/GFP‐SEC24, pRTL2/Myc‐DGAT2 and pRTL2/GFP‐Cb5, have been described elsewhere (Cai et al., 2015; Gidda et al., 2011; Hwang et al., 2004; Shockey et al., 2006).
Rt‐pcr
RNA was extracted from 4‐week‐old Arabidopsis plants using the RNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA), and DNA contamination was eliminated using DNase (Promega, Madison, WI, USA). Approximately 100 ng of total RNA was used for RT‐PCRs, which were performed with TaKaRa One Step RT‐PCR Kit following the manufacturer's instructions. The PCR program was as follows: 42 °C for 15 min, 95 °C for 5 min, 35 amplification cycles (94 °C for 30 s, 55 °C for 30 s, 72 °C 30–60 s), and 72 °C for 7 min. Primers used to amplify Arabidopsis EF1α were those used elsewhere (Cai et al., 2015). Primers used to amplify mouse FIT2 were FIT2RTF 5′‐ATGGAGCACCTGGAGCGC‐3′ and FIT2RTR 5′‐ TCATTTCTTGTAAGTATCTCGCTTCAAAG‐3′.
Lipid analysis
For neutral lipid content and compositional analysis, ~45 mg dry weight of N. benthamiana leaves, 15 mg of Arabidopsis leaves (dry weight) or 10 mg of Arabidopsis seeds were used in each replicate. Sample tissues were homogenized with glass beads in a bead beater (BioSpec Mini‐Beadbeater‐16) with 70 °C isopropanol. TAG (tri‐17:0) standard (Avanti) was added to Arabidopsis seed samples, while TAG (tri‐15:0) standard and cholesterol ester (C13:0) standard (Nu‐Chek Prep) were added to leaf samples. Total lipid was extracted as described by Cai et al. (2015). The neutral lipid fraction and polar lipid fraction were separated from total leaf lipid extract by solid‐phase extraction using 6‐mL silica columns (Sigma‐Aldrich, St. Louis, MO, USA). Hexane and diethyl ether (4:1) were used to elute neutral lipids, and chloroform and methanol (1:2) were used to elute polar lipids. PC (di‐14:0) (Avanti) standard was added to the polar lipid fraction of Arabidopsis leaf samples. Lipid content in Arabidopsis (TAG, SE and PC) and N. benthamiana (TAG and SE) leaves was analysed by electrospray ionization mass spectrometry (ESI‐MS) on an API3000 triple quadrupole mass spectrometer (Applied Biosystems/Sciex, Framingham, MA), and data were processed using Metabolite Imager (v.1.0) to quantify the TAG and SE content (Horn and Chapman, 2014b; Li et al., 2014) and using LipidomeDB Data Calculation Environment to quantify the PC content (Welti et al., 2002; Zhou et al., 2011). Lipid extract from Arabidopsis seeds was transesterified with methanolic HCl, and the fatty acid methyl esters were analysed using gas chromatography–flame ionization detection (GC‐FID, Hewlett Packard, HP 5890 II plus GC). Conditions for both ESI‐MS and GC‐FID were as described in James et al. (2010). Total oil content of Arabidopsis seeds was quantified using time‐domain, pulse‐field 1H‐NMR with a Bruker Minispec mq20 (Bruker optics), as described by Chapman et al. (2008), except calibrated for small volumes of Arabidopsis seeds.
Spatial distributions of TAG and SE in Arabidopsis leaves were determined in situ by matrix‐assisted laser desorption/ionization mass spectrometry imaging (MALDI‐MSI). Arabidopsis leaves were embedded in porcine gelatin and then frozen to −80 °C, before equilibration to −20 °C. Tissues were cut into 40‐μm‐thick cross sections on a cryostat (Leica CM1950) at −18 °C and lyophilized overnight before application of matrix (2,5‐dihydroxybenzoic acid) by sublimation. A MALDI Orbitrap hybrid mass spectrometer (MALDI LTQ Orbitrap‐XL; Thermo Fisher Scientific, Waltham, MA, USA) was used to scan the tissue sections at 40 micron steps, and the MS data were used to reconstruct false‐colour MALDI‐MSI images based on ion counts of selected lipid species (Horn and Chapman, 2014b). Lipid species were identified by exact mass comparisons with the LIPID MAPS database (http://www.lipidmaps.org/). Detailed procedures for data acquisition and analyses were described previously (Horn et al., 2012).
Microscopy
Confocal images of Arabidopsis leaves and seeds and N. benthamiana leaves were obtained using a Zeiss LSM 710 confocal laser‐scanning microscope (CLSM). Images of N. tabacum (BY‐2) cells were obtained using either a Leica DM RBE microscope equipped with a Leica TCS SP2 scanning head or a Leica SP5 CLSM system equipped with a Radius 405‐nm laser. Arabidopsis leaves and seeds, as well as tobacco leaves, were processed for CLSM imaging, including staining of LDs with the neutral lipid stains BODIPY 493/503 (Invitrogen) or Nile red (Sigma‐Aldrich), as described previously (Cai et al., 2015; Gidda et al., 2016; Park et al., 2013). The fifth leaf from the bottom of 24‐day‐old Arabidopsis plants was collected for analysis. Arabidopsis leaves used in comparison with young, mature and older mature leaves were the seventh, fifth and third true leaf from the bottom of the plant, respectively. Three fully expanded leaves from the top of the N. benthamiana plants (4‐week‐old) were used in the transient expression experiments. BY‐2 cells were processed for CSLM as described previously (Gidda et al., 2011, 2016; Lingard et al., 2008). Briefly, cells ~4–6 h following biolistic bombardment were fixed in paraformaldehyde (Electron Microscopy Sciences), permeabilized and then incubated with the ER stain fluor‐conjugated concanavalin A (ConA) (Sigma‐Aldrich) and/or the neutral lipid stain MDH (Abgent). Cherry was excited with a 543‐nm laser, and emission signals were collected from 590 to 640 nm. BODIPY 493/503, GFP and Nile red, as well as chlorophyll, were excited with a 488‐nm laser, and emission signals were acquired from 500 to 540 nm (BODIPY and GFP), 560 to 620 nm (Nile red) and 640 to 720 nm (chlorophyll), respectively. CFP and MDH were excited with a 405‐nm laser, and the emission signal was collected from 450 to 490 nm and 420 to 480 nm, respectively. All fluorophore emissions were collected sequentially in double‐ or triple‐labelling experiments; single‐labelling experiments showed no detectable crossover at the settings used for image acquisition. Depending on the plant material, images were acquired as a Z‐series (leaves of Arabidopsis and N. benthamiana) or single optical sections (Arabidopsis seeds and BY‐2 cells). All fluorescence images of cells shown are representative of at least two separate experiments, including at least three independently infiltrated leaves from three N. benthamiana plants or 20 transiently transformed BY‐2 cells. The numbers and sizes of LDs in Arabidopsis and N. benthamiana leaves were quantified as BODIPY‐ or Nile red‐stained lipid area according to Cai et al. (2015) using the Analyze Particles function at ImageJ (version 1.43). All the significance assessments in this study were performed using Student's t‐test.
Supporting information
Figure S1 Composition of TAG (a) and SE (b) in Nicotiana benthamiana leaves expressing FIT2 or FIT2‐FLL[157‐159]AAA in the presence or absence of LEC2.
Figure S2 Localization of GFP‐FIT2 to the ER in N. tabacum suspension‐cultured BY‐2 cells.
Figure S3 Expression of FIT2 in transgenic Arabidopsis leaves and influence of ectopically‐expressed FIT2 on LDs in different‐aged leaves of Arabidopsis plants.
Figure S4 Composition of TAG (a) and SE (b) in wild‐type and FIT2 transgenic (i.e., FIT2‐OE‐C6) Arabidopsis leaves.
Figure S5 Composition of PC molecular species in wild‐type and FIT2 transgenic Arabidopsis leaves.
Figure S6 Visualization of GFP‐FIT2 and LDs in leaf and seed tissues of stable transgenic Arabidopsis.
Acknowledgements
Research with FIT2 and LD formation in plants was supported initially by a grant from the U.S. Department of Energy (DOE), BER Division, DE‐FG02‐09ER64812, and completed with funds from the U.S. DOE, Office of Science, BES‐Physical Biosciences program (DE‐SC0016536). Support was also provided from the Natural Sciences and Engineering Research Council of Canada (NSERC), including an NSERC Undergraduate Student Research Award to Thuy Nguyen. Some of the experimental results reported here were initiated as part of a research‐based undergraduate course, BIOL 3900, at UNT with the support of HHMI grant #52006955. Students deserving special recognition include Kevin Mutore, Charles McDaniel, Tucker Burke, Kathryn Haydon, Fancine Mascarenhas, Kyle Current and Reed Archer Killingsworth. Dr. Charlene Case assisted with manuscript preparation. Chris James assembled two of the binary vectors and generated some Arabidopsis transgenic lines for proof of concept. The authors declare no conflict of interest.
References
- Andrianov, V. , Borisjuk, N. , Pogrebnyak, N. , Brinker, A. , Dixon, J. , Spitsin, S. , Flynn, J. et al (2010) Tobacco as a production platform for biofuel: overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 8, 277–287. [DOI] [PubMed] [Google Scholar]
- Atanassov, I.I. , Etchells, J.P. and Turner, S.R. (2009) A simple, flexible and efficient PCR‐fusion/Gateway cloning procedure for gene fusion, site‐directed mutagenesis, short sequence insertion and domain deletions and swaps. Plant Meth. 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, J.R. , Frost, E. , Vidi, P.A. , Kessler, F. and Staehelin, L.A. (2006) Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell, 18, 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin, F. and Napier, J.A. (2002) Targeting and membrane‐insertion of a sunflower oleosin in vitro and in Saccharomyces cerevisiae: the central hydrophobic domain contains more than one signal sequence, and directs oleosin insertion into the endoplasmic reticulum membrane using a signal anchor sequence mechanism. Planta, 215, 293–303. [DOI] [PubMed] [Google Scholar]
- Beller, M. , Bulankina, A.V. , Hsiao, H.H. , Urlaub, H. , Jäckle, H. and Kühnlein, R.P. (2010) PERILIPIN‐dependent control of lipid droplet structure and fat storage in Drosophila . Cell Metab. 12, 521–532. [DOI] [PubMed] [Google Scholar]
- Benfey, P.N. , Ren, L. and Chua, N.H. (1989) The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue‐specific expression patterns. EMBO J. 8, 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi, F. , Irons, S. , Kearns, A. and Hawes, C. (2003) BY‐2 cells: culture and transformation for live cell imaging. Curr. Protoc. Cell Biol., Chapter 1, Unit. 1.7. [DOI] [PubMed] [Google Scholar]
- Brasaemle, D.L. and Wolins, N.E. (2012) Packaging of fat: an evolving model of lipid droplet assembly and expansion. J. Biol. Chem. 287, 2273–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y. , Goodman, J.M. , Pyc, M. , Mullen, R.T. , Dyer, J.M. and Chapman, K.D. (2015) Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell, 27, 2616–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, K.D. and Ohlrogge, J.B. (2012) Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287, 2288–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, K.D. , Neogi, P.B. , Hake, K.D. , Stawska, A.A. , Speed, T.R. , Cotter, M.Q. , Garrett, D.C. et al (2008) Reduced oil accumulation in cottonseeds transformed with a Brassica non‐functional allele of a Delta‐12 Fatty Acid Desaturase (FAD2). Crop Sci. 48, 1470–1481. [Google Scholar]
- Chapman, K.D. , Dyer, J.M. and Mullen, R.T. (2012) Biogenesis and functions of lipid droplets in plants. J. Lipid Res. 53, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, V. , Ojha, N. , Golden, A. and Prinz, W.A. (2015) A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 211, 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D. and Grossniklaus, U. (2003) A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese, R.V. and Walther, T.C. (2009) Lipid droplets finally get a little R‐E‐S‐P‐E‐C‐T. Cell 139, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen, G.I. , Mundy, J. and Tzen, J.T. (2001) Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant. 112, 301–307. [DOI] [PubMed] [Google Scholar]
- Froissard, M. , D'andréa, S. , Boulard, C. and Chardot, T. (2009) Heterologous expression of AtClo1, a plant oil body protein, induces lipid accumulation in yeast. FEMS Yeast Res. 9, 428–438. [DOI] [PubMed] [Google Scholar]
- Gao, Q. and Goodman, J.M. (2015) The lipid droplet‐a well‐connected organelle. Front Cell Dev. Biol. 3, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda, S.K. , Shockey, J.M. , Falcone, M. , Kim, P.K. , Rothstein, S.J. , Andrews, D.W. , Dyer, J.M. et al (2011) Hydrophobic‐domain‐dependent protein‐protein interactions mediate the localization of GPAT enzymes to ER subdomains. Traffic 12, 452–472. [DOI] [PubMed] [Google Scholar]
- Gidda, S.K. , Park, S. , Pyc, M. , Yurchenko, O. , Cai, Y. , Wu, P. , Andrews, D.W. et al (2016) Lipid droplet‐associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 170, 2052–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, V.J. , Tan, J.S. , Tan, B.C. , Seow, C. , Ong, W.Y. , Lim, Y.C. , Sun, L. et al (2015) Postnatal deletion of fat storage‐inducing transmembrane protein 2 (FIT2/FITM2) causes lethal enteropathy. J. Biol. Chem. 290, 25686–25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg, Å. , Carlsson, A.S. , Marttila, S. , Bhalerao, R. and Hofvander, P. (2015) Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 15, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, D.A. , Snapp, E.L. and Silver, D.L. (2010) Structural insights into triglyceride storage mediated by fat storage‐inducing transmembrane (FIT) protein 2. PLoS ONE, 5, e10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, D.A. , Zhan, C. and Silver, D.L. (2011) Direct binding of triglyceride to fat storage‐inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc. Natl Acad. Sci. USA, 108, 19581–19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton, S.L. , Matheson, L.A. , Chatre, L. and Brandizzi, F. (2007) Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. Plant J. 57, 963–974. [DOI] [PubMed] [Google Scholar]
- Horn, P.J. and Benning, C. (2016) The plant lipidome in human and environmental health. Science 353, 1228–1232. [DOI] [PubMed] [Google Scholar]
- Horn, P.J. and Chapman, K.D. (2014a) Lipidomics in situ: insights into plant lipid metabolism from high resolution spatial maps of metabolites. Prog. Lipid Res. 54, 32–52. [DOI] [PubMed] [Google Scholar]
- Horn, P.J. and Chapman, K.D. (2014b) Metabolite imager: customized spatial analysis of metabolite distributions in mass spectrometry imaging. Metabolomics, 10, 337–348. [Google Scholar]
- Horn, P.J. , Korte, A.R. , Neogi, P.B. , Love, E. , Fuchs, J. , Strupat, K. , Borisjuk, L. et al (2012) Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell, 24, 622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, P.J. , James, C.N. , Gidda, S.K. , Kilaru, A. , Dyer, J.M. , Mullen, R.T. , Ohlrogge, J.B. et al (2013) Identification of a new class of lipid droplet‐associated proteins in plants. Plant Physiol. 162, 1926–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, K. and Huang, A.H. (2004) Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol. 136, 3427–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A. (1996) Oleosins and oil bodies in seeds and other organs. Plant Physiol. 110, 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, Y.T. , Pelitire, S.M. , Henderson, M.P. , Andrews, D.W. , Dyer, J.M. and Mullen, R.T. (2004) Novel targeting signals mediate the sorting of different isoforms of the tail‐anchored membrane protein cytochrome b5 to either endoplasmic reticulum or mitochondria. Plant Cell, 16, 3002–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier, N. , Choudhary, V. , Mari, M. , Toulmay, A. , Reggiori, F. and Schneiter, R. (2011) Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae . J. Cell Sci. 124, 2424–2437. [DOI] [PubMed] [Google Scholar]
- Jacquier, N. , Mishra, S. , Choudhary, V. and Schneiter, R. (2013) Expression of oleosin and perilipins in yeast promotes formation of lipid droplets from the endoplasmic reticulum. J. Cell Sci. 126, 5198–5209. [DOI] [PubMed] [Google Scholar]
- James, C.N. , Horn, P.J. , Case, C.R. , Gidda, S.K. , Zhang, D. , Mullen, R.T. , Dyer, J.M. et al (2010) Disruption of the Arabidopsis CGI‐58 homologue produces Chanarin‐Dorfman‐like lipid droplet accumulation in plants. Proc. Natl Acad. Sci. USA, 107, 17833–17838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit, B. , Kumar, P. , Wang, W.J. , Miranda, D. , Snapp, E.L. , Severina, N. , Torregroza, I. et al (2008) Evolutionarily conserved gene family important for fat storage. Proc. Natl Acad. Sci. USA, 105, 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E.Y. , Park, K.Y. , Seo, Y.S. and Kim, W.T. (2016) Arabidopsis small rubber particle protein homolog SRPs play dual roles as positive factors for tissue growth and development and in drought stress responses. Plant Physiol. 170, 2494–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Baughman, E. , Roth, M.R. , Han, X. , Welti, R. and Wang, X. (2014) Quantitative profiling and pattern analysis of triacylglycerol species in Arabidopsis seeds by electrospray ionization mass spectrometry. Plant J. 77, 160–172. [DOI] [PubMed] [Google Scholar]
- Lingard, M.J. , Gidda, S.K. , Bingham, S. , Rothstein, S.J. , Mullen, R.T. and Trelease, R.N. (2008) Arabidopsis PEROXIN11c‐e, FISSION1b, and DYNAMIN‐RELATED PROTEIN3A cooperate in cell cycle‐associated replication of peroxisomes. Plant Cell 20, 1567–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, Y. and Jiang, L. (2007) Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat. Protoc. 2, 2348–2353. [DOI] [PubMed] [Google Scholar]
- Miranda, D.A. , Kim, J.H. , Nguyen, L.N. , Cheng, W. , Tan, B.C. , Goh, V.J. , Tan, J.S. et al (2014) Fat storage‐inducing transmembrane protein 2 is required for normal fat storage in adipose tissue. J. Biol. Chem. 289, 9560–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, S. and Schneiter, R. (2015) Expression of perilipin 5 promotes lipid droplet formation in yeast. Comm. Integr. Biol. 8, e1071728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, S. , Khaddaj, R. , Cottier, S. , Stradalova, V. , Jacob, C. and Schneiter, R. (2016) Mature lipid droplets are accessible to ER luminal proteins. J. Cell Sci. 129, 3803–3815. [DOI] [PubMed] [Google Scholar]
- Mu, J. , Tan, H. , Zheng, Q. , Fu, F. , Liang, Y. , Zhang, J. , Yang, X. et al (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T. and Skoog, F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 15, 473–497. [Google Scholar]
- Murphy, D.J. (2001) The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40, 325–438. [DOI] [PubMed] [Google Scholar]
- Murphy, D.J. (2012) The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249, 541–585. [DOI] [PubMed] [Google Scholar]
- Nacir, H. and Bréhélin, C. (2013) When proteomics reveals unsuspected roles: the plastoglobule example. Front Plant Sci. 4, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge, J. and Chapman, K. (2011) The seeds of green energy: expanding the contribution of plant oils as biofuels. Biochemist 33, 34–38. [Google Scholar]
- Park, S. , Gidda, S.K. , James, C.N. , Horn, P.J. , Khuu, N. , Seay, D.C. , Keereetaweep, J. et al (2013) The α/β Hydrolase CGI‐58 and Peroxisomal Transport Protein PXA1 Coregulate Lipid Homeostasis and Signaling in Arabidopsis. Plant Cell, 25, 1726–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie, J.R. , Shrestha, P. , Liu, Q. , Mansour, M.P. , Wood, C.C. , Zhou, X.R. , Nichols, P.D. et al (2010) Rapid expression of transgenes driven by seed‐specific constructs in leaf tissue: DHA production. Plant Meth. 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol, A. , Gross, S.P. and Parton, R.G. (2014) Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 204, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robenek, H. , Hofnagel, O. , Buers, I. , Robenek, M.J. , Troyer, D. and Severs, N.J. (2006) Adipophilin‐enriched domains in the ER membrane are sites of lipid droplet biogenesis. J. Cell Sci. 119, 4215–4224. [DOI] [PubMed] [Google Scholar]
- Rowe, E.R. , Mimmack, M.L. , Barbosa, A.D. , Haider, A. , Isaac, I. , Ouberai, M.M. , Thiam, A.R. et al (2016) Conserved amphipathic helices mediate lipid droplet targeting of perilipins 1‐3. J. Biol. Chem. 291, 6664–6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjaya, Miller, R. , Durrett, T.P. , Kosma, D.K. , Lydic, T.A. , Muthan, B. , Koo, A.J. et al (2013) Altered lipid composition and enhanced nutritional value of Arabidopsis leaves following introduction of an algal diacylglycerol acyltransferase 2. Plant Cell, 25, 677–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Mendoza, M. , Dubreucq, B. , Miquel, M. , Caboche, M. and Lepiniec, L. (2005) LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 579, 4666–4670. [DOI] [PubMed] [Google Scholar]
- Shockey, J.M. , Gidda, S.K. , Chapital, D.C. , Kuan, J.C. , Dhanoa, P.K. , Bland, J.M. , Rothstein, S.J. et al (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell, 18, 2294–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocombe, S.P. , Cornah, J. , Pinfield‐Wells, H. , Soady, K. , Zhang, Q. , Gilday, A. , Dyer, J.M. et al (2009) Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J. 7, 694–703. [DOI] [PubMed] [Google Scholar]
- Snapp, E.L. , Hegde, R.S. , Francolini, M. , Lombardo, F. , Colombo, S. , Pedrazzini, E. , Borgese, N. et al (2003) Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu, N. and Wobus, U. (2013) Seed‐development programs: a systems biology‐based comparison between dicots and monocots. Annu. Rev. Plant Biol. 64, 189–217. [DOI] [PubMed] [Google Scholar]
- Sturtevant, D. , Lee, Y.J. and Chapman, K.D. (2016) Matrix assisted laser desorption/ionization‐mass spectrometry imaging (MALDI‐MSI) for direct visualization of plant metabolites in situ . Curr. Opin. Biotechnol. 37, 53–60. [DOI] [PubMed] [Google Scholar]
- Szymanski, K.M. , Binns, D. , Bartz, R. , Grishin, N.V. , Li, W.P. , Agarwal, A.K. , Garg, A. et al (2007) The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl Acad. Sci. USA, 104, 20890–20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhercke, T. , El Tahchy, A. , Shrestha, P. , Zhou, X.R. , Singh, S.P. and Petrie, J.R. (2013) Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 587, 364–369. [DOI] [PubMed] [Google Scholar]
- Vanhercke, T. , El Tahchy, A. , Liu, Q. , Zhou, X.R. , Shrestha, P. , Divi, U.K. , Ral, J.P. et al (2014) Metabolic engineering of biomass for high energy density: oilseed‐like triacylglycerol yields from plant leaves. Plant Biotechnol. J. 12, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C. and Farese, R.V. (2009) The life of lipid droplets. Biochim. Biophys. Acta 1791, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Ren, Y. , Liu, X. , Jiang, L. , Chen, L. , Han, X. , Jin, M. et al (2010) OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. Plant J. 64, 812–824. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Becuwe, M. , Housden, B.E. , Chitraju, C. , Porras, A.J. , Graham, M.M. , Liu, X.N. et al (2016) Seipin is required for converting nascent to mature lipid droplets. Elife, 5, e16582. doi: 10.7554/eLife.16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner, G. , Formanek, H. and Theimer, R.R. (1981) The ontogeny of lipid bodies (spherosomes) in plant cells: Ultrastructural evidence. Planta, 151, 109–123. [DOI] [PubMed] [Google Scholar]
- Welti, R. , Li, W. , Li, M. , Sang, Y. , Biesiada, H. , Zhou, H.E. , Rajashekar, C.B. et al (2002) Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing‐induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002. [DOI] [PubMed] [Google Scholar]
- Wilfling, F. , Haas, J.T. , Walther, T.C. and Farese, R.V. (2014) Lipid droplet biogenesis. Curr. Opin. Cell Biol. 29, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winichayakul, S. , Scott, R.W. , Roldan, M. , Hatier, J.H. , Livingston, S. , Cookson, R. , Curran, A.C. et al (2013) In vivo packaging of triacylglycerols enhances Arabidopsis leaf biomass and energy density. Plant Physiol. 162, 626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. and Shanklin, J. (2016) Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu. Rev. Plant Biol. 67, 179–206. [DOI] [PubMed] [Google Scholar]
- Zhou, Z. , Marepally, S.R. , Nune, D.S. , Pallakollu, P. , Ragan, G. , Roth, M.R. , Wang, L. et al (2011) LipidomeDB data calculation environment: online processing of direct‐infusion mass spectral data for lipid profiles. Lipids 46, 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Composition of TAG (a) and SE (b) in Nicotiana benthamiana leaves expressing FIT2 or FIT2‐FLL[157‐159]AAA in the presence or absence of LEC2.
Figure S2 Localization of GFP‐FIT2 to the ER in N. tabacum suspension‐cultured BY‐2 cells.
Figure S3 Expression of FIT2 in transgenic Arabidopsis leaves and influence of ectopically‐expressed FIT2 on LDs in different‐aged leaves of Arabidopsis plants.
Figure S4 Composition of TAG (a) and SE (b) in wild‐type and FIT2 transgenic (i.e., FIT2‐OE‐C6) Arabidopsis leaves.
Figure S5 Composition of PC molecular species in wild‐type and FIT2 transgenic Arabidopsis leaves.
Figure S6 Visualization of GFP‐FIT2 and LDs in leaf and seed tissues of stable transgenic Arabidopsis.


