Abstract
Pancreatic ductal adenocarcinoma (PDA) is the third leading cause of cancer related deaths in the U.S., while colorectal cancer (CRC) is the third most common cancer. The RNA binding protein HuR (ELAVL1), supports a pro-oncogenic network in gastrointestinal (GI) cancer cells through enhanced HuR expression. Using a publically available database, HuR expression levels were determined to be increased in primary PDA and CRC tumor cohorts as compared to normal pancreas and colon tissues, respectively. CRISPR/Cas9 technology was successfully used to delete the HuR gene in both PDA (MIA PaCa-2 and Hs 766T) and CRC (HCT116) cell lines. HuR deficiency has a mild phenotype, in vitro, as HuR-deficient MIA PaCa-2 (MIA.HuR-KO(−/−)) cells had increased apoptosis when compared to isogenic wild-type (MIA.HuR-WT(+/+)) cells. Using this isogenic system, mRNAs were identified that specifically bound to HuR and were required for transforming a 2D culture into 3D (i.e., organoids). Importantly, HuR-deficient MIA PaCa-2 and Hs 766T cells were unable to engraft tumors in vivo compared to control HuR-proficient cells, demonstrating a unique xenograft lethal phenotype. While not as a dramatic phenotype, CRISPR knockout HuR HCT116 colon cancer cells (HCT.HuR-KO(−/−)) showed significantly reduced in vivo tumor growth compared to controls (HCT.HuR-WT(+/+)). Finally, HuR deletion affects KRAS activity and controls a subset of pro-oncogenic genes.
Implications
The work reported here supports the notion that targeting HuR is a promising therapeutic strategy to treat GI malignancies.
Keywords: HuR, CRISPR, Cas9, Pancreatic ductal adenocarcinoma, Colorectal cancer
Introduction
Pancreatic ductal adenocarcinoma (PDA), with an overall 5-year survival rate of roughly 7 percent, is one of the deadliest solid tumors (1,2). Colorectal cancer (CRC) is the third most common cancer in the United States and the fourth most common cause of death worldwide (3). Both CRC and PDA tumorigenesis are associated with numerous sequential genetic changes (4). However, methods to target common mutations (KRAS, p16/DSKN2A, TP53) in PDA and CRC have been particularly challenging and shown limited therapeutic value (5,6). Genetic mutations and copy number changes are critical regulatory mechanisms promoting the initiation and progression of tumorigenesis. Beyond driving mutations, it is becoming clearer that post-transcriptional gene regulatory mechanisms play an important role in controlling the available transcriptome (7). Post-transcriptional gene regulation characteristically involves trans-acting microRNAs (miRNAs) and RNA-binding proteins (RBPs) that are potent regulators controlling the abundance of specific pro-oncogenic proteins (8,9).
Hu proteins are a family of trans-acting RBPs that recognize and bind to cis-acting AU-rich RNA elements (AREs) within the mRNA 3′ untranslated regions (3′ UTRs) (10). The RNA-binding protein HuR (ELAVL1) is expressed ubiquitously and is extensively involved in mRNA stability and translation. HuR targets, which include proto-oncogenes, cytokines, growth and invasion factors, implicate an important role of HuR in cancer development (11). HuR is abundant in both PDA and CRC specimens compared to normal adjacent tissue with cytoplasmic subcellular localization associated with increased tumor stage (12,13). As tumor cells are under stress in the tumor microenvironment (14) (e.g., nutrient deprivation and hypoxia) (15,16), HuR translocates from the nucleus, binds to pro-oncogenic mRNA transcripts (e.g. WEE1, PIM1, COX-2, VEGF), promoting cancer cell survival and tumorigenesis (14,17,18).
To further depict the role of HuR in GI cancer biology, we genetically deleted HuR in PDA and CRC cells by using clustered, regularly interspaced, short palindromic repeat (CRISPR)/Cas9 technology. The system efficiently targets a specific genomic sequence and then creates disruptive double strand breaks (DSB). The DSB is repaired via non-homologous end joining and thus insertions/deletions (INDELs) are randomly placed at the sites of repair (19). These INDELs can lead to deleterious frameshift mutations and ultimately premature stop codons to effectively disrupt the targeted gene on one (+/−) or both (−/−) alleles, and thus allowing an efficient way to generate an isogenic cell culture model in which to study the importance of HuR in GI cancer cells.
Materials and Methods
Evaluation of HuR mRNA in Pancreas Clinical Specimens
Previously reported microarray expression (20) data were downloaded from GEO (accession number: GSE71729). This dataset contains 46 normal pancreatic tissue samples, 145 primary pancreatic adenocarcinomas, 42 normal colon and 286 primary colon adenocarcinomas. The relative expression levels for HuR was extracted from the downloaded matrix and used for comparisons between the three groups of samples.
Cell Culture, Transfections and Treatments
Cell culture conditions
MIA PaCa-2, Hs 766T and HCT116 cells were purchased from ATCC. Cells were cultured in DMEM supplemented with 10% FBS (Gibco/Invitrogen), 1% L-glutamine (Gibco/Invitrogen), and 1% penicillin-streptomycin (Invitrogen) at 37°C in 5% humidified CO2 incubators.
Transfections
MIA PaCa-2 and Hs 766T cells were seeded in 6-well plate at the density of 500,000 cells per well for 24 h. Cells transfected with approximately 5 μg of CRISPR control and HuR knockout plasmids using Lipofectamine 2000 (Gibco/Invitrogen) as previously described (14). Cells were harvested after 48 h of transfection and GFP positive cells were single sorted into 96-well plates using FACS Calibur flow cytometer (BD Biosciences, East Rutherford, NJ).
MIA.HuR-KO(−/−) cells were seeded in 6-well plate at the density of 500,000 cells per well and cells were transfected with HuR overexpression plasmid as previously described (21).
Treatments
Cells were treated with the IC50 values of the DNA damaging agent MMC (mitomycin C; Sigma, St. Louis, MO), gemcitabine (Eli Lilly) and oxaliplatin (Sigma) for 24 h as previously described (14) by adding directly into the culture medium.
Note: All experiments using PDA CRISPR transfected cells were performed on cells passage between 10 – 25.
Plasmid Design
CRISPR knockout in MIA PaCa-2 and Hs 766T PDA cell lines
Three different guide RNAs of HuR fused with CRISPR-cas9 and GFP protein were designed and purchased from Sigma (St. Louis, MO; Fig. S1A and Supplementary Table S1). Guide RNA 1 was located on exon 2 (sense), guide RNA 2 was also located on exon 2 (antisense) and guide RNA 3 was located on exon 3 (sense) (Supplementary Table S1). The CRISPR Universal Negative Control plasmid was purchased from Sigma (Cat. No. CRISPR06-1EA). Plasmids were purified using Qiagen’s plasmid mini-purification kit following manufacture’s protocol (Qiagen, Hilden, Germany).
Note: In the figures, the MIA.HuR-WT(+/+) clone is ‘MIA.HuR’, MIA.HuR-KO(+/+) clone is ‘Clone.4’, MIA.HuR-KO(−/−).1 clone is ‘Clone.11’, MIA.HuR-KO(−/−).2 clone is ‘Clone.14’ and MIA.HuR-KO(−/−) + HOE is ‘Clone.11 stably transfected with an HuR overexpression plasmid’.
CRISPR knockout in HCT116 colon cancer cell line
CRISPR/Cas9-mediated knockout of ELAVL1 in the human colon cancer cell line HCT116 (ATCC; Manassas, VA) was accomplished using HuR CRISPR/Cas9 KO plasmid (sc-400141; Santa Cruz Biotechnology), followed by homology-directed repair (HDR) insertion of a puromycin resistance gene/red fluorescent protein (RFP) cassette (sc-400141-HDR; Santa Cruz Biotechnology) into the double strand break according to the vendor’s protocol. Transfected cells were selected in normal growth medium containing 0.5 μg/mL puromycin (ThermoFisher Scientific) for 2–3 weeks. Individual clones were isolated using cloning cylinders, and HuR knockout clones screened by western blotting for HuR and genotyped by PCR to verify exon insertion of the Puro/RFP cassette. For stable cell maintenance, the puromycin concentration was reduced to 0.2 μg/mL.
Sequencing Analysis
Sanger sequencing was performed using sequencing primers listed in Table S2 on genomic DNA of the clones. The Sequencher software (Gene Codes Corporation, Ann Arbor, MI) was used to analyze the chromatogram and identify mutation sites. Sequence changes were classified according to the HGVS nomenclature recommendation and based on the NCBI reference sequence: NM_001419.2. Clone sequencing and expression data are provided in Supplementary table S4 and Fig. S1B and S1C. For simplicity, the clones employed in our experiments will be referred to as MIA.HuR-KO(−/−).1 (Clone.11, created with Guide RNA 3), MIA.HuR-KO(−/−).2 (Clone.14, created with Guide RNA 3), MIA.HuR-KO(+/+) (Clone.4, created with Guide RNA 1) and HsT.HuR-KO(−/−) (created with Guide RNA 3). These clones correlate with NM_001419[c.211delCG];[c.211delCG], NM_001419[c.204delGTGACCGCGA];[c.206delGACCGCGAAGGATGCAG], NM_001419 Wild Type and NM_001419[c.211_212insATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTAC] ;[c.211_212insATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTAC] mutation sites, respectively.
Whole Cell Extracts and SDS-PAGE/Western Blotting
Whole cell lysates were isolated using RIPA lysis buffer (Invitrogen) by incubating the cell pellets on ice for 10 minutes followed by centrifugation at 13,000 *g for 15 minutes at 4°C as previously described (14). Samples were then mixed 4:1 with 5X Laemmli buffer and boiled for 5 minutes. Proteins were measured with Pierce BCA kit (ThermoFisher) and approximately 50 μg was separated using a 12% Bis-Tris polyacrylamide gel and transferred to PVDF membrane (Invitrogen), blocked in 1:1 Licor Odyssey blocking buffer and incubated overnight at 4°C with α-Tubulin (Santa Cruz; #sc-5286) and HuR (Santa Cruz; #sc-5261) antibodies. Protein complexes were then visualized with Licor Odyssey.
Immunofluorescence
Approximately 50,000 cells were plated on coverslips per well in 24-well plate. After 24 h, cells were treated with MMC, gemcitabine and oxaliplatin for 24 h. Cells were then washed 2 times with 1X PBS, fixed with 4% paraformaldehyde for 10 minutes at room temperature, permeabilized with 0.25% Triton-X for 30 minutes, blocked with 5% goat serum for 1 h, and incubated overnight with HuR antibody, as previously described (14). DAPI was used to stain cell nuclei and coverslips were mounted with DAPI ProLong Gold Antifade (Invitrogen). Slides were visualized with a Zeiss LSM-510 Confocal Laser Microscope. All images were taken at 40X magnification with oil. Images were cropped using AIM browser.
Flow Cytometry Analysis
Cleaved caspase 3
Cells were seeded at 300,000 cells per well in 6-well plates for 24 h. Apoptosis was measured by incubating the cells for 30 min with CellEvent™ Caspase-3/7 Green Flow Cytometry Assay Kit (ThermoFisher Scientific), according to the manufacturer’s instructions, and analyzed using a FACS Calibur flow cytometer (BD Biosciences, East Rutherford, NJ). Calculated relative mean fluorescence intensity (MFI) was produced using FlowJo software (Tree Star, Inc., Ashland, OR).
Cell cycle
Cells were seeded at 500,000 cells per well in 6-well plates for 24 hours. Cells were pulse chased with BrdU (Amersham, Piscataway, NJ, USA) by adding directly to actively growing cells for 1 h as previously described (14) and analyzed on FACS Calibur flow cytometer. Graphs were generated on Excel (Microsoft).
Soft Agar Assay
A bottom layer of 0.75% agar was prepared in complete medium in 60 mm tissue culture dishes. A top layer of 0.36% agar supplemented with 10,000 cells/mL was added as previously described _ENREF_10(14). Pictures were taken using Floid Cell Imaging Station (Life Technologies) and colonies were counted using ImageJ (http://imagej.nih.gov/ij/).
Organoid Cultures
Cell lines were obtained from 2D culture (standard cell culture) and washed with cold organoid basic media. Cells are then spun at 1,200 rpm for 5 minutes and washed again. The pellet was suspended in matrigel and 50 μl of cell-matrigel mixture is added to each well of a pre-warmed 24 well plate. The plate was then placed in a standard tissue culture incubator until the matrigel solidified. 500 μl of pre-warmed complete organoid feeding media is then added to each well. The plates are placed in a tissue culture incubator and inspected on a daily basis as described in Boj et al and Baker et al (22,23). For all organoid cultures and studies, we validated HuR knockout for the CRISPR lines via qRT-PCR analysis (data not shown).
Image Analysis performed using Paint Shop Pro (Version 7.04, Corel Corporation, Ottawa ON, Canada) and R (Version 3.3.2, the R Foundation for statistical computing). Microscopic images of identical magnification were digitally acquired at three time points (1280×960 Pixels). Images were centered and cropped to 800×800 pixels. Organoid objects maps of the images were created using gray scale value thresholding and also visually assessed and corrected for merging and segmentation errors. The produced maps were assessed for size and number of objects per map using the EBImage: Image processing and analysis toolbox for R (http://rdrr.io/bioc/EBImage/). Objects with surface area smaller than 10 pixels were excluded from the analysis. Cell line organoid sizes were assessed for normality of distribution with the Kolmogorov-Smirnov test (24). Area size was compared using Mann-Whitney test for non-parametric distributions and t-test for normal distributions(25). P values < 0.05 were considered as statistically significant.
Serum Starvation Assay
MIA.HuR-WT(+/+) and MIA.HuR-KO(−/−) cells growing in log phase in normal growth medium (RPMI 1640 + 10% FBS) were serum starved (0.1% FBS) for 16 h or 72 h. Cells were homogenized in lysis buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 10% glycerol, 10 mM MgCl2, 1× phosphatase and protease inhibitors (HALT, Thermo) and protein concentration was quantified by BCA assay (Pierce). A Ras-GTP pulldown was performed according to previously established protocols (26,27). Western blot analysis was performed on the pulldown and straight protein lysate samples blotting for K-Ras and K-Ras-GTP (Calbiochem; #OP24), AKT (Cell Signaling; #4691), pAKT (Cell Signaling; #4060), MEK (Cell Signaling; #8727), pMEK (Cell Signaling; #9154) and Vinculin (Sigma; #V9131). Protein bands were visualized by Luminata ECL reagents (Millipore) on x-ray film or a FluorChem M (Protein Simple) imager. Protein bands were analyzed by densitometry, normalized for protein load against vinculin and relative to MIA.HuR-WT(+/+) signals.
MTT Assay
Cells were seeded at 5,000 cells/well in 96-well tissue culture plates. Cell growth was assayed using the MTT-based cell growth determination kit (Sigma-Aldrich) as previously described (28). Cell growth was calculated as relative absorbance (A570) normalized to cells after 1 day of growth. Data are represented as average of 3 independent experiments ± SEM.
Short-term Drug Sensitivity Assay
Cells were seeded at 1,000 cells/well in 96-well tissue culture plates in triplicates. Cells were treated 24 h later and exposed to only one dose. After 7-day of treatment, cells were washed, lysed, stained with PicoGreen (Invitrogen) and fluorescence intensity was read as previously described (29). Graph was generated using GraphPad (San Diego, CA). Data are represented as average of 3 independent experiments ± SEM.
RNP-IP and qPCR
Cells were plated at 50% confluency in 100 mm dishes. Cells were either left untreated or treated with 1 μM oxaliplatin for 24 h. Immunoprecipitation was performed using either anti-HuR or IgG control antibodies as previously described (14). Total RNA was isolated and WEE1, IDH1 and PARP1 mRNA binding was validated by RT-qPCR.
In Vivo Experiment
MIA PaCa-2 and Hs 766T xenografts
The experiments involving mice received the approval of the Thomas Jefferson University Institutional Animal Care regulations and Use Committee (IACUC). Six week-old, female, athymic nude mice were purchased from Harlan Laboratories and 5 × 106 cells were injected subcutaneously in the left and right flanks, respectively (5 mice per group). Cells were prepared in 100 μL solution comprised of 70% DPBS and 30% Matrigel. Tumor volumes and body weight were measured three times per week using a caliper, and tumor volumes were calculated using the formula Volume = Length × Width2/2. Upon termination of the experiment, mice were euthanized using carbon dioxide inhalation followed by cervical dislocation, and tumors were harvested.
HCT116 Xenografts
Athymic nude (Nu/Nu) mice were purchased from Jackson Laboratories and maintained under sterile conditions in cage micro-isolators according to approved IACUC guidelines. Parental HCT116 and HuR knockout clones 1 and 2 (2 × 106 cells) used between passages 14 – 23 were resuspended in PBS containing 50% Matrigel (Corning) and injected into the dorsal subcutaneous tissue. Tumor growth was assayed as described (28,30).
Results
HuR mRNA is more abundant in primary GI tumors compared to normal tissue
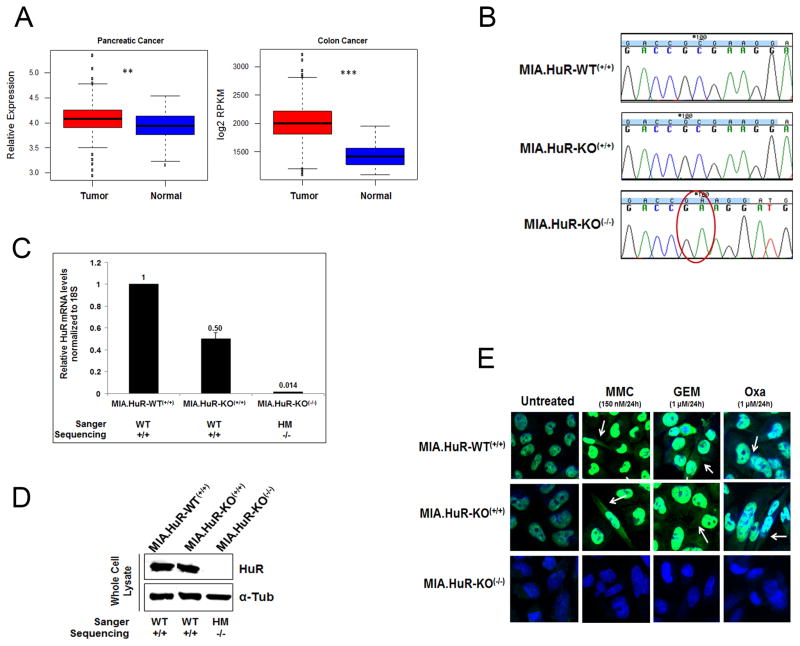
We have previously demonstrated that HuR is upregulated in CRC cells (13). To determine HuR expression levels in pancreatic samples, we analyzed previously reported microarray data (20) of HuR mRNA levels in normal pancreatic tissues (n=46) and primary tumors (n=145) as well as in colon normal tissues (n=42) and primary tumors (n=286). We detected that HuR mRNA levels is enhanced slightly in both pancreatic (p<0.005) and colon (p<0.0005) primary tumors compared to normal tissues (Fig. 1A).
Figure 1. CRISPR-cas9 system inhibits HuR expression and localization in MIA PaCa-2 PDA cells.
A, Shown are boxplots of HuR expression levels across 519 samples comprised of normal pancreatic tissue (n=46), primary pancreatic adenocarcinomas (n=145), normal colon tissue (n=42) and colorectal cancer (n=286). For each boxplot, the black line represents the median expression value and the boxes are the 1st and 3rd quartiles. ** p<0.005; *** p<−0.005. B, Chromatograms from Sanger sequencing of HuR CRISPR-transfected purified PCR products showing the homozygous mutant [MIA.HuR-KO(−/−).1] and homozygous wild type [MIA.HuR-KO(+/+)] clones. C, Relative mRNA HuR expression was assessed in different clones by qPCR. (Note: only three clones are shown here. Complete list of clones are shown in Fig. S1B). D, Relative protein expression of HuR was assessed in different clones by immunoblot. (Note: this is a photoshop cropped image of three clones shown in Fig. S1C). E, HuR nuclear and cytoplasmic translocation was assessed by immunofluorescence upon mitomycin C (MMC), gemcitabine (GEM) and oxaliplatin (Oxa) stressors. MIA.HuR-WT(+/+) in Figure B–E is a product from MIA PaCa-2 cells with CRISPR control transfection. WT: Wild type; HM: Homozygous mutant. Note: in Fig. B–E, the MIA.HuR-WT(+/+) clone is ‘MIA.HuR’,
Successful generation of HuR knockout PDA cells using the CRISPR-Cas9 system
We transfected the PDA cell line MIA PaCa-2 cells, known to harbor common genetic lesions found in most PDAs (K-RAS, p53 and p16) (31), with three different guide RNAs designed to target exon 2 and 3 of human HuR (ELAVL1; Supplementary Table S1). A plasmid containing Cas9 protein fused with GFP and guide RNA expressed from a single vector was transfected to MIA PaCa-2 cells (Fig. S1A). As a control, a non-specific guide RNA in this vector was also transfected. Forty-eight hours post-transfection, cells were single sorted with FACS into 96-well plates. The mutation was confirmed by Sanger sequencing in expanded single cell colonies (Supplementary Table S2, S3, S4 and Fig. 1B). Only guide RNA 3 targeting the exon 3 of HuR resulted in producing 5 stable clones (Fig. 1C, 1D; Supplementary Figs. S1B, S1C; Supplementary Tables S3 and S4). HuR mRNA levels were analyzed by qPCR (Fig. 1C and Supplementary Fig. S1B) and protein expression was detected by immunoblotting (Fig. 1D and Supplementary Fig. S1C), to demonstrate HuR gene expression correlated to the genotype. Both MIA.HuR-WT(+/+) (wild-type parental CRISPR control transfected cells showing homozygous wild type genotype) and MIA.HuR-KO(+/+) (HuR-CRISPR transfected cells showing homozygous wild type genotype; Supplementary Tables S3 and S4) clones displayed similar levels of HuR nucleo-cytoplasmic expression upon mitomycin C, gemcitabine and oxaliplatin stressors by immunofluorescence (Fig. 1E), indicating functional HuR translocation to the cytoplasm from the nucleus upon specific stress (14). The MIA.HuR-KO(−/−) (HuR-CRISPR transfected cells showing homozygous knockout/mutant genotype; Supplementary Tables S3 and S4) cells showed no sign of HuR expression (Fig. 1E). We also created a CRISPR HuR knockout in another PDA cell line Hs 766T using guide RNA 3. The mutation in HuR was again confirmed by Sanger sequencing (Supplementary Fig. S1D and Supplementary Table S4) and functional inactivation of HuR was confirmed by immunoblot analysis (Supplementary Fig. S1E). These data demonstrate that the HuR gene was successfully knocked out using CRISPR-Cas9 system in multiple PDA cell lines.
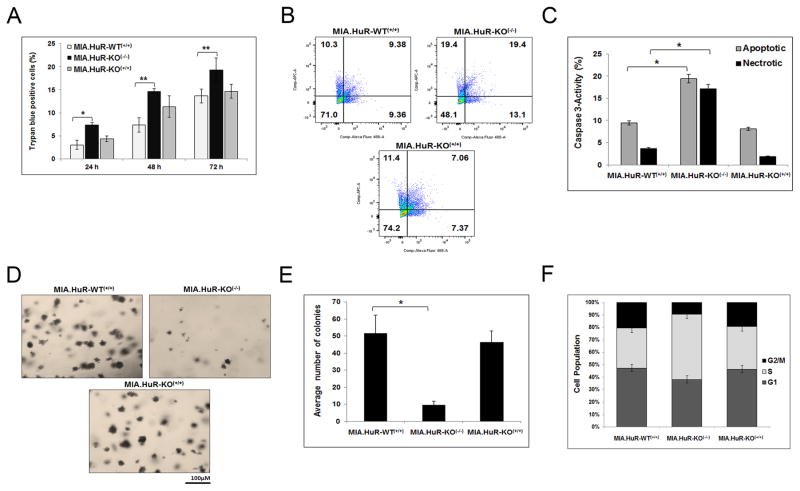
HuR-deficient PDA cells have a mild phenotype in vitro
To determine the effect of HuR deletion has on the cancer phenotype, HuR knockout clone MIA.HuR-KO(−/−).1 along with controls, MIA.HuR-WT(+/+) and MIA.HuR-KO(+/+) were analyzed for in vitro growth viability with a trypan blue exclusion assay. Results demonstrated that MIA.HuR-KO(−/−).1 induced higher amounts of cell death compared to controls (Fig. 2A). In addition, cleaved caspase 3 activity, a marker of cell death, was measured with FACS and similar results were obtained (Fig. 2B–C), indicating HuR’s role in inhibiting apoptotic cell death. Taken together, these data demonstrate that the complete loss of HuR induces more cell death consistent with our previous work (32).
Figure 2. HuR-deficient cells induce more cell death.
A, Cells were stained with Trypan blue and number of dead cells was counted after 24, 48 and 72 hours for one clone depicting each mutation. Graphs are average of three individual experiments. * p<0.003; ** p<0.004. B and C, Cell death was also assessed by cleaved caspase 3-activity after 24 hours of cell seeding via FACS. Graphs are average of three individual experiments. * p<0.0004. D and E, Long-term cell survival assay was assessed by colony formation soft agar assay. Graphs are average of two individual experiments. * p<0.003. F, Cell kinetics was measured by pulse chasing the cells by BrdU followed by FACS. Graphs are average of two individual experiments.
We have performed drug sensitivity assay (short-term cell survival) using a chemotherapeutic drug mitomycin C (MMC) and under glucose deprivation and observed that MIA.HuR-KO(−/−) cells are sensitive compared to controls (Supplementary Fig. S2A and B). To determine the long-term cell survival and anchorage independence growth, soft agar assays were performed. Results showed that loss of HuR (in the MIA.HuR-KO(−/−).1 model) resulted in approximately 5-fold decrease in colony formation compared to controls (Fig. 2D and E). Lastly, the effects of HuR on cell cycle were analyzed by pulse-labeling the cells with bromodeoxyuridine (BrdU). Shown in Fig. 2F, MIA.HuR-KO(−/−).1 cells are primarily arrested in S phase with reduced number of cells in the G2/M phase, indicating that cells were cycling through mitosis and most likely prematurely dying.
In an effort to study a malignant characteristic of the HuR-deficient cells and how they would establish under the stressful transition from moving into a 3D culture, we evaluated the ability of each cell line’s capacity to transform into organoids (22). After 7–10 days of plating, MIA.HuR-KO(+/+) cells formed spheroids easily in 3D cultures within in a passage, while MIA.HuR-KO(−/−).2 cells only formed single cell sheets and were unable to form substantial spheroids even after 4 weeks of plating (Supplementary Figs. S2C and S2D). Image analysis of the spheroid structures in each group revealed the MIA.HuR-KO(+/+) spheroids to be significantly larger than the MIA.HuR-KO(−/−).2 spheroids (5–10 fold increase in cross-section area, P<0.01, Supplementary Fig. S2D). Furthermore, during the course of two weeks the MIA.HuR-KO(+/+) continued to grow and doubled in size (P<0.01) compared to the MIA.HuR-KO(−/−).2 spheroids. Similarly, when HsT.HuR-WT(+/+) and HsT.HuR-KO(−/−) cells were plated in 3D cultures the HsT.HuR-WT(+/+) demonstrated more robust spheroid formation and growth than the HsT.HuR-KO(−/−) cells (data not shown). These data suggest that HuR is critical for initiation of growth in a stressful transition to 3D and also viability in a more relevant in vitro, 3D culture (22).
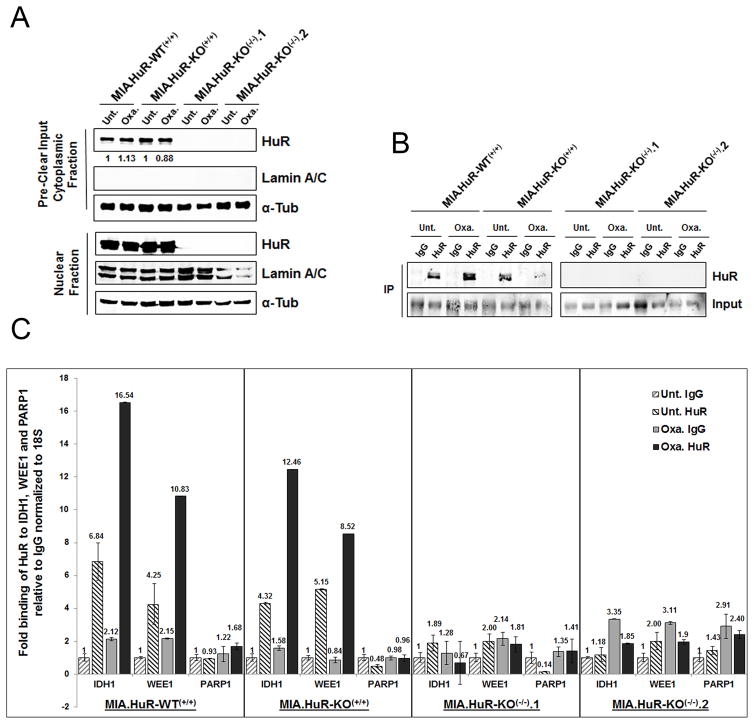
Using isogenic MIA.HuR-WT(+/+) and MIA.HuR-KO(−/−) PDA cells to identify HuR targets
It was previously published that HuR translocates to the cytoplasm upon specific stress where it binds to its mRNA targets and stabilizes their transcript and/or increases the protein translation (33). We and others have demonstrated that Ribonucleotide immunoprecipitation (RNP-IPs) assays (34) are a reliable and efficient way to capture HuR-bound mRNA targets (14,17,32,33,35). To evaluate binding of HuR to target mRNAs, two MIA.HuR-KO(−/−) (MIA.HuR-KO(−/−).1 and MIA.HuR-KO(−/−).2) cell lines along with controls MIA.HuR-WT(+/+) and MIA.HuR-KO(+/+) cells were treated with oxaliplatin (1 μM) for 24 hours to induce cytoplasmic translocation (Fig. 3A). Cytoplasmic lysates were subjected to HuR immunoprecipitation (Fig. 3B), followed by analysis of HuR targets IDH1 and WEE1 by qPCR. Shown in Fig. 3C, PDA cells deficient in HuR showed background levels of IDH1 and WEE1 mRNA, whereas control cells showed increased binding of HuR to these targets upon oxaliplatin stress. PARP1, a non-target of HuR, showed no significant binding in knockout as well as in controls samples (Fig. 3C).
Figure 3. HuR-deficient cells failed to bind to its targets.
A and B, Ribonucleoprotein immunoprecipitation (RNP-IP) assay was performed to determine the binding of HuR to its mRNA targets. Cells were treated with oxaliplatin (1 μM) for 24 hour and immunoprecipitated either using control IgG or HuR antibodies. Western blot showing HuR expression in cytoplasmic and nuclear lysates and immunoprecipitated lysates. C, HuR binding to WEE1 and IDH1 mRNA was analyzed by qPCR. PARP1 is a non-target of HuR.
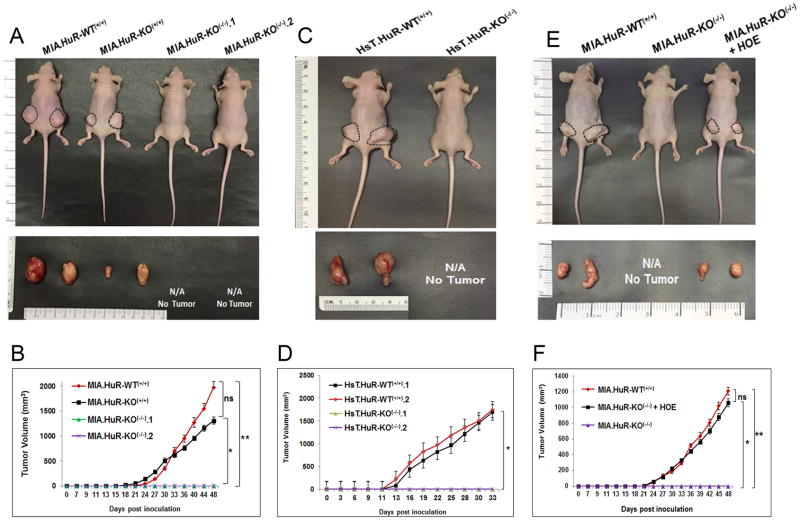
HuR is required for PDA tumor growth
Since we were able to obtain viable HuR-deficient cancer cell lines (see Fig. 1B–E) and the in vitro phenotype was not dramatic for the MIA PaCa-2 line (Fig. 2), we evaluated if loss of HuR would disrupt cancer cell viability and growth in vivo. To test the effect of HuR knockout on in vivo PDA tumor growth, we first performed two individual experiments utilizing nude mice subcutaneously injected in their hind flanks with equal numbers of MIA.HuR-WT(+/+), MIA.HuR-KO(+/+) and two of different MIA.HuR-KO(−/−) cells (MIA.HuR-KO(−/−).1 and MIA.HuR-KO(−/−).2). Tumor volumes and body weight were measured three times per week and harvested after 48 days of tumor growth, Shown in Fig. 4A, tumors derived from the MIA.HuR-WT(+/+) and MIA.HuR-KO(+/+) groups were 1976.7 mm3 and 1305.8 mm3, respectively. The mean tumor volumes generated from MIA.HuR-KO(+/+) cells were not significantly smaller than those from MIA.HuR-WT(+/+) cells (P<0.716; Fig. 4B and P<0.589; Supplementary Fig. S3A). However, this is in sharp contrast to both MIA.HuR-KO(−/−).1 and MIA.HuR-KO(−/−).2 groups, where tumor growth was not observed (Figs. 4A and B) and reproducible in an independent experiment (Supplementary Figs. S3A and B). To then validate these results, we performed the same experiment in Hs 766T HuR knockout cell line. Nude mice were subcutaneously injected with HsT.HuR-WT(+/+) and HsT.HuR-KO(−/−) cells showed similar growth results as in MIA PaCa-2 cell line (Figs. 4C and D).
Figure 4. HuR knockout by CRISPR inhibits the PDA xenograft growth.
A, Representative images of subcutaneous tumors on the flanks of nude female mice and representative pictures of corresponding excised tumors (day 48). B, Tumor growth curves of MIA.HuR-WT(+/+), MIA.HuR-KO(+/+), MIA.HuR-KO(−/−).1 and MIA.HuR-KO(−/−).2 xenografts in nude mice shown in figure A. Data represent means ± SEM of n = 5. ns, non-significant,* p<0.01; ** p<0.003. C, Representative images of subcutaneous tumors on the flanks of nude female mice and their corresponding excised tumors injected with CRISPR HuR (HsT.HuR-KO(−/−)) and CRISPR Control (HsT.HuR-WT(+/+)) cells in Hs 766T cell line (Day 33). D, Tumor growth curves in HsT.HuR-WT(+/+) and HsT.HuR-KO(−/−) xenografts in nude mice shown in figure C. Data represent means ± SEM of n = 4. ns, non-significant, * p<0.0003. E, Representative images of subcutaneous tumors on the flanks of nude female mice and representative pictures of corresponding excised tumors (day 48). F, Tumor growth curves of MIA.HuR-WT(+/+), MIA.HuR-KO(−/−).1, and MIA.HuR-KO(−/−).1 + HOE xenografts in nude mice shown in figure E. Data represent means ± SEM of n = 5. ns, non-significant,* p<0.005; ** p<0.006.
Adding HuR in a HuR-null background rescues the tumor growth phenotype
To validate that loss of HuR was responsible for the observed lack of tumor growth, we created a stable cell line by overexpressing HuR in MIA.HuR-KO(−/−).1 cells and assaying xenograft tumor growth. Results demonstrated that xenografts with MIA.HuR-KO(−/−).1 cells overexpressing HuR [labeled as (MIA.HuR-KO(−/−)) + HOE] restored the tumor formation (1061.4 mm3 vs. 0 mm3; P <0.0002) demonstrating that HuR plays a vital role in PDA growth (Fig. 4E and F). These findings using a PDA xenograft model demonstrate that targeted genetic disruption of HuR may prove to be a novel therapeutic strategy to inhibit tumor growth as a treatment for PDA.
HuR knockout suppresses xenograft growth in a colon cancer model
In order to determine the effect of HuR knockout on cancer models other than PDA, we created HuR knockouts in the colorectal carcinoma HCT116 cell line. Previously, we have demonstrated that HuR is upregulated in HCT116 and other CRC cell lines (13,18). A similar CRISPR/Cas9-mediated approach was taken, except that a puromycin resistance gene/red fluorescent protein (RFP) cassette was inserted into cleavage sites within ELAVL1 exons 3, 4, or 5 by homology-directed repair (HDR). Clones were screened for puromycin resistance and RFP expression and then selected loss of HuR expression (Fig. 5A). Confirmation of cassette insertion by PCR-based genotyping identified 2 HuR knockout clones (HCT.HuR-KO(−/−).1 and HCT.HuR-KO(−/−).2). Similar to our PDA model, short-term in vitro growth assays of HCT.HuR-KO(−/−) cells displayed attenuated growth compared to the control (Fig. 5B). To determine the effects of HuR loss in colon cancer xenografts, we subcutaneously injected nude mice in their hind flanks with equal number of HCT.HuR-WT(+/+) and HCT.HuR-KO(−/−) cells. Tumor volumes were measured on alternate days and harvested after 35 days. Results showed that tumors in the HCT.HuR-KO(−/−) group were dramatically smaller than control HCT.HuR-WT(+/+) (Figs. 5C, D and E), consistent to what was observed with PDA cells. Similar results were also observed in HCA7 colon cancer cells with CRISPR/Cas9-mediated deletion of HuR (data not shown). These results demonstrate that targeted genetic disruption of HuR inhibit cancer growth in both PDA and CRC models.
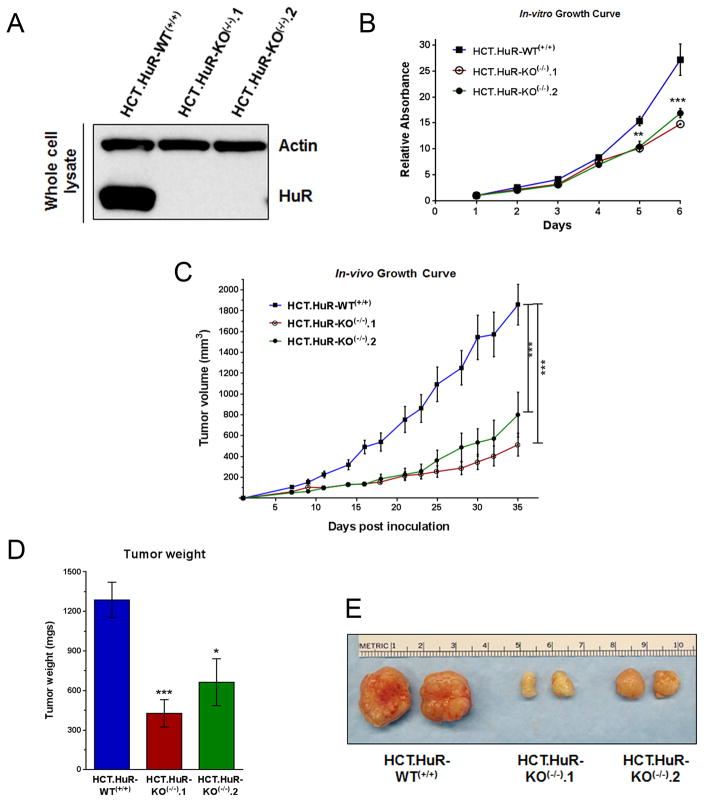
Figure 5. CRISPR/Cas9 knockout of HuR in colon cancer cells retards cell and tumor growth.
A, Western blot analysis of HuR expression in parental HCT116 cells (labeled HCT.HuR-WT(+/+)) and HuR knockout clones (HCT.HuR-KO(−/−).1 and HCT.HuR-KO(−/−).2). Total protein extracts were analyzed by SDS-PAGE and probed for HuR; actin served as a loading control. B, In vitro cell growth was measured by MTT assay. Relative absorbance (A570) was normalized to cells after 1 day of growth. Cells were used between passages 5 to 12. Data is represented as average of 3 independent experiments ± SEM. C, In vivo tumor growth of HCT116 cells and HuR knockout clones 1 and 2 (passages 23, 14, and 16, respectively) xenografts in nude mice. Tumor volume (mm3) is represented as average of 7 tumors ± SEM. D and E, Average tumor weight ± SEM and representative tumors excised at day 35 are shown. * P <0.05; ** P <0.01; *** P <0.001.
HuR-deficient PDA cells have decreased K-Ras activation
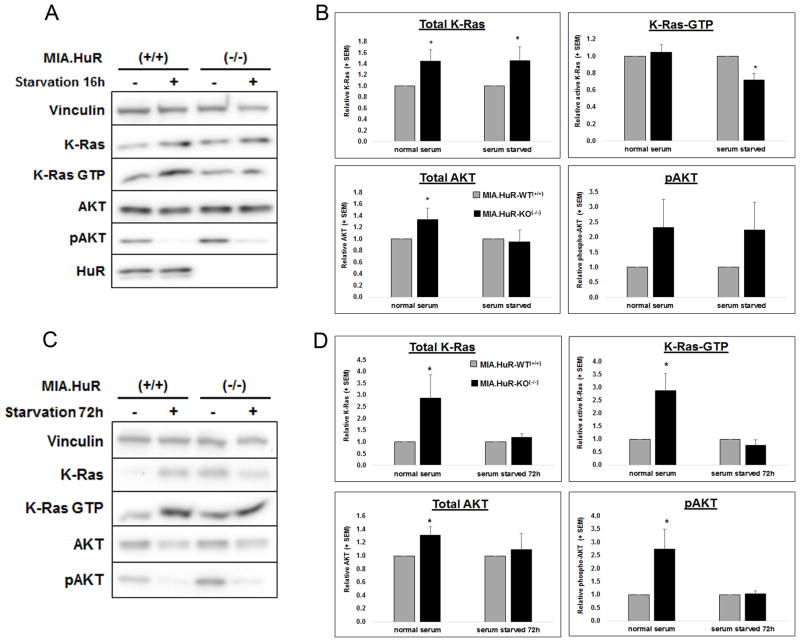
Since disrupted K-Ras signaling is a hallmark of GI cancers and previously deletion of K-ras caused a similar dramatic in vivo phenotype (36), we sought to define the role HuR has on K-Ras activation. Under normal growth conditions, a loss of HuR results in an increase in K-Ras and AKT proteins, but interestingly, not a concomitant significance increase in their respective activations (Figs. 6A and B). Under acute (16 hr) serum deprivation, MIA.HuR-WT(+/+) cells respond by increasing K-Ras activation, while HuR knockout cells can no longer mount this stress response, and show a significant decline in GTP loading of K-Ras.
Figure 6. Loss of HuR diminishes stress-induced K-Ras activation.
Immunoblot depiction of total and activated (GTP-bound) K-Ras and AKT protein in MIA PaCa-2 cells lacking HuR under 16 (A) or 72 h (C) serum deprivation, respectively. Densitometric analysis of immunoblots for acute or (B) or 72 h (D), * p<0.05 HuR(−/−) compared to MIA.HuR.WT(+/+), n=3–6 independent experiments. Protein bands are normalized to vinculin and are and relative to MIA.HuR.WT(+/+).1 signals.
Upon extended (72 hr; Figs. 6C and D) serum deprivation, MIA.HuR-KO(−/−).1 cells no longer maintained the high levels of compensatory K-Ras expression seen in the acute serum starvation, and again K-Ras activation was blunted (unlike HuR competent cells, where K-Ras-GTP was increased approximately 6 ±2-fold in 72 hr starved versus normal growth conditions). While MIA.HuR-KO(−/−).1 cells were capable of augmenting AKT expression and activation in normal growth conditions, this survival pathway activation was diminished under chronic serum starvation. These data suggest that in PDA, survival mechanisms that are in part mitigated by oncogenic, activated KRas can be regulated by HuR function under stressed conditions. Thus, proteins responsible for proper KRas activation (i.e. post-translational modification, trafficking, GEF/GAP activity ratio, etc.) may be targets of HuR binding and mRNA stabilization.
Discussion
CRISPR/Cas9-mediated HuR knockout is a novel way to investigate HuR biology since previous studies relied on RNAi-based approaches using shRNA inducible knockdowns or siRNA transient transfections (33), where leakiness in the constructs and incomplete knockdown can lead to inconsistent results. If amenable, a genomic deletion is an effective and unbiased way to evaluate gene function at a more fundamental level. Furthermore, this approach allows for identifying off-target effects of molecularly targeted therapies where the target is not present in the cell-based model. Using this approach, we deleted HuR in MIA PaCa-2, Hs 766T and HCT116 cells, and demonstrated an ablation of HuR protein expression compared to wild-type and CRISPR control cells. These commonly used cell models of GI cancer will advance the field for studying HuR biology in PDA and CRC, along with facilitating current drug discovery where HuR is a target in pre-clinical models (14,17,18,21,32,33,35,37–39). However, it should be noted that at the time of acceptance, we found that the later passaged MIA.HuR-KO(−/−).1 line seemed to also contain a population having an HuR (Wild type) allele (as determined by Sanger sequencing). We have re-validated the HuR depletion at early passages where experiments herein where performed (data not shown), but we cannot rule out the possibility of a small contamination by other single cell clones (e.g., a heterogeneous population emerged of +/+, +/− cells).
It has previously been shown that complete genetic deletion of HuR is embryonically lethal in mouse models and this deletion leads to lethality within 10 days (40). Interestingly, mice with conditional HuR deletion in a cell/tissue-specific manner are viable as observed in the GI tract (41,42), lung epithelium (43,44), brain (45), T-cells (46), and macrophages (47), and suggested that deletion of HuR in GI cancer cells would be feasible. Our findings demonstrate that PDA and CRC HuR-null cells are able to establish and grow in vitro, however, when cells become stressed with chemotoxic agents (Supplementary Fig. S2A), growth in soft agar (Fig. 2D and E) and in nude mice they unable to survive (Fig. 4A and B, Supplementary Fig. S3A and B, Fig. 4C and D). In principle, the in vivo microenvironment simulates a more stressful, hypoxic and glucose deprived environment as compared to an in vitro environment containing high nutrient and normoxic conditions.
This work strongly supports the ongoing notion that HuR is a viable target in PDA and CRC. Currently, the best-characterized small molecule inhibitor of HuR is the chrysanthone-like compound MS-444 that inhibits HuR homodimerization and its cytoplasmic translocation (30,48). There have been promising pre-clinical studies that show MS-444 has potent affects in CRC mouse models (30) and other studies have demonstrated that MS-444 sensitizes PDA cells to oxaliplatin and 5-fluorouracil in relevant tumor microenvironment conditions, but issues with stability and bioavailability of MS-444 may hinder first human applications (17). Recently, there have been success in identifying other small molecules that disrupt the HuR-mRNA interactions, leading to impaired HuR function and premature decay of mRNAs that encode proteins related to tumor processes (37). There has also been innovative work conjugating siHUR to a 3D nanocarrier in order to target HuR in ovarian tumor–bearing mice, resulting in the suppression of tumor growth and ascites development, and ultimately prolonging lifespan (37,39). A limitation of the models analyzed herein is the ability to assess the specificity of targeting HuR in GI cancer cells. Although previous work has shown that HuR is abundant in GI cancer cells compared to normal adjacent cells, future strategies will focus on targeting HuR expression and function specifically in cancer cells. Still, these findings indicate that post-transcriptional regulation is a promising target in cancer cells and future studies utilizing these cell culture models of PDA and CRC should be a valuable resource for advancing the field. It is our hope that HuR inhibitors can eventually become a viable therapeutic option for patients either as a monotherapy or in combination of other current strategies.
Supplementary Material
Acknowledgments
Grant Support This work was supported by the National Institutes of Health [R21 CA182692 (JRB), R01 CA134609 (DAD)], NIH/NCI Cancer Center Support Grant P30 CA168524 (DAD), PhRMA Foundation Translational Medicine Grant (PMC), and fellowship awards from Prevent Cancer Foundation and K-INBRE P20 GM103418 (RP). We also wish to acknowledge the Mary Halinski Pancreatic Cancer Research Fund; Fund A Cure; Gail Coleman Fund, and Research supported by the 2015 Pancreatic Cancer Action Network-AACR Research Acceleration Network Grant, Grant Number 15-90-25-BROD. Also we are supported by the Cancer Center Support Grant 5P30CA056036-17 and used the shared resource (the Core Genomics Laboratory).
Footnotes
Disclosure of Potential Conflicts of Interest: The authors have no conflicts of interest to disclose.
Authors’ Contributions:
Conception and design: S. Lal, E. C. Cheung, J. R. Brody
Development of methodology: S. Lal, E. C. Cheung, R. Preet, M. Zarei
Acquisition of data: S. Lal, E. Cheung, M. Zarei, R. Preet, N.C. Mambelli-Lisboa., S. N. Chand, M. C. Stout, C. Romeo, A. Goetz, C. Y. Lowder, E. Londin
Analysis and interpretation of data: S. Lal, E. C. Cheung, P. M. Campbell, A. Nevler, J. M. Winter, D. A. Dixon, J. R. Brody
Writing, review and/or revision of the manuscript: S. Lal, E. C. Cheung, M. Zarei, P. M. Campbell, J. M. Winter, C. J. Yeo, D. A. Dixon, J. R. Brody
Administrative, technical, or material support: E. Londin, J. M. Winter, C. J. Yeo, D. A. Dixon, J. R. Brody
Study supervision: J. M. Winter, C. J. Yeo, J. R. Brody
References
- 1.Oberstein PE, Olive KP. Pancreatic cancer: why is it so hard to treat? Therap Adv Gastroenterol. 2013;6:321–37. doi: 10.1177/1756283X13478680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A Cancer Journal for Clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Haggar FA, Boushey RP. Colorectal Cancer Epidemiology: Incidence, Mortality, Survival, and Risk Factors. Clinics in Colon and Rectal Surgery. 2009;22:191–7. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12:319–34. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- 6.Linnekamp JF, Wang X, Medema JP, Vermeulen L. Colorectal Cancer Heterogeneity and Targeted Therapy: A Case for Molecular Disease Subtypes. Cancer Research. 2015;75:245–9. doi: 10.1158/0008-5472.CAN-14-2240. [DOI] [PubMed] [Google Scholar]
- 7.Krueger KE, Srivastava S. Posttranslational protein modifications: current implications for cancer detection, prevention, and therapeutics. Mol Cell Proteomics. 2006;5:1799–810. doi: 10.1074/mcp.R600009-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Wurth L, Gebauer F. RNA-binding proteins, multifaceted translational regulators in cancer. Biochim Biophys Acta. 2015;1849:881–6. doi: 10.1016/j.bbagrm.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Dixon DA, Kaplan CD, McIntyre TM, Zimmerman GA, Prescott SM. Post-transcriptional Control of Cyclooxygenase-2 Gene Expression: THE ROLE OF THE 3′-UNTRANSLATED REGION. Journal of Biological Chemistry. 2000;275:11750–7. doi: 10.1074/jbc.275.16.11750. [DOI] [PubMed] [Google Scholar]
- 10.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–81. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci. 2013;14:10015–41. doi: 10.3390/ijms140510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards NG, Rittenhouse DW, Freydin B, Cozzitorto JA, Grenda D, Rui H, et al. HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients. Ann Surg. 2010;252:499–505. doi: 10.1097/SLA.0b013e3181f1fd44. discussion -6. [DOI] [PubMed] [Google Scholar]
- 13.Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–79. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lal S, Burkhart RA, Beeharry N, Bhattacharjee V, Londin ER, Cozzitorto JA, et al. HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells. Cancer Res. 2014;74:1128–40. doi: 10.1158/0008-5472.CAN-13-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chand S, O’Hayer K, Blanco FF, Winter JM, Brody JR. The Landscape of Pancreatic Cancer Therapeutic Resistance Mechanisms. Int J Biol Sci. 2016;12:273–82. doi: 10.7150/ijbs.14951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen R, Neuzillet C, Tijeras-Raballand A, Faivre S, de Gramont A, Raymond E. Targeting cancer cell metabolism in pancreatic adenocarcinoma. Oncotarget. 2015;6:16832–47. doi: 10.18632/oncotarget.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, et al. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene. 2015;35(19):2529–41. doi: 10.1038/onc.2015.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. Journal of Clinical Investigation. 2001;108:1657–65. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 20.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–78. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jimbo M, Blanco FF, Huang YH, Telonis AG, Screnci BA, Cosma GL, et al. Targeting the mRNA-binding protein HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells. Oncotarget. 2015;6:27312–31. doi: 10.18632/oncotarget.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boj SF, Hwang C-I, Baker LA, Chio IIC, Engle DD, Corbo V, et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell. 2015;160:324–38. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker LA, Tiriac H, Clevers H, Tuveson DA. Modeling Pancreatic Cancer with Organoids. Trends in Cancer. 2016;2(4):176–190. doi: 10.1016/j.trecan.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolmogorov AN. Sulla determinazione empirica di una legge di distribuzione. Giornale dell’Istituto Italiano degli Attuari. 1993;4:83–91. [Google Scholar]
- 25.Fay MP, Proschan MA. Wilcoxon-Mann-Whitney or t-test? On assumptions for hypothesis tests and multiple interpretations of decision rules. Stat Surv. 2010;4:1–39. doi: 10.1214/09-SS051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras Promotes Growth Transformation and Invasion of Immortalized Human Pancreatic Cells by Raf and Phosphatidylinositol 3-Kinase Signaling. Cancer Research. 2007;67:2098–106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- 27.Stout MC, Asiimwe E, Birkenstamm JR, Kim SY, Campbell PM. Analyzing Ras-Associated Cell Proliferation Signaling. In: Noguchi E, Gadaleta CM, editors. Cell Cycle Control: Mechanisms and Protocols. New York, NY: Springer New York; 2014. pp. 393–409. [DOI] [PubMed] [Google Scholar]
- 28.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–65. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lal S, Zarei M, Chand SN, Dylgjeri E, Mambelli-Lisboa NC, Pishvaian MJ, et al. WEE1 inhibition in pancreatic cancer cells is dependent on DNA repair status in a context dependent manner. Scientific Reports. 2016;6:33323. doi: 10.1038/srep33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco FF, Preet R, Aguado A, Vishwakarma V, Stevens LE, Vyas A, et al. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget. 2016;7(45):74043–74058. doi: 10.18632/oncotarget.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deer EL, Gonzalez-Hernandez J, Coursen JD, Shea JE, Ngatia J, Scaife CL, et al. Phenotype and Genotype of Pancreatic Cancer Cell Lines. Pancreas. 2010;39:425–35. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romeo C, Weber MC, Zarei M, DeCicco D, Chand SN, Lobo AD, et al. HuR Contributes to TRAIL Resistance by Restricting Death Receptor 4 Expression in Pancreatic Cancer Cells. Molecular Cancer Research. 2016;7:599–611. doi: 10.1158/1541-7786.MCR-15-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pineda DM, Rittenhouse DW, Valley CC, Cozzitorto JA, Burkhart RA, Leiby B, et al. HuR’s post-transcriptional regulation of death receptor 5 in pancreatic cancer cells. Cancer Biology & Therapy. 2012;13:946–55. doi: 10.4161/cbt.20952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cozzitorto JA, Jimbo M, Chand S, Blanco F, Lal S, Gilbert M, et al. Studying RNA-binding protein interactions with target mRNAs in eukaryotic cells: native ribonucleoprotein immunoprecipitation (RIP) assays. Methods Mol Biol. 2015;1262:239–46. doi: 10.1007/978-1-4939-2253-6_14. [DOI] [PubMed] [Google Scholar]
- 35.Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, et al. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res. 2009;69:4567–72. doi: 10.1158/0008-5472.CAN-09-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–7. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 37.Wu X, Lan L, Wilson DM, Marquez RT, Tsao W-c, Gao P, et al. Identification and Validation of Novel Small Molecule Disruptors of HuR-mRNA Interaction. ACS chemical biology. 2015;10:1476–84. doi: 10.1021/cb500851u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burkhart RA, Pineda DM, Chand SN, Romeo C, Londin ER, Karoly ED, et al. HuR is a post-transcriptional regulator of core metabolic enzymes in pancreatic cancer. RNA Biol. 2013;10:1312–23. doi: 10.4161/rna.25274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, et al. Delivery of Therapeutics Targeting the mRNA-Binding Protein HuR Using 3DNA Nanocarriers Suppresses Ovarian Tumor Growth. Cancer Res. 2016;76:1549–59. doi: 10.1158/0008-5472.CAN-15-2073. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh M, Aguila HL, Michaud J, Ai Y, Wu M-T, Hemmes A, et al. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. The Journal of Clinical Investigation. 2009;119:3530–43. doi: 10.1172/JCI38263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, Gritz D, Parent CA. PKCβII acts downstream of chemoattractant receptors and mTORC2 to regulate cAMP production and myosin II activity in neutrophils. Molecular Biology of the Cell. 2014;25:1446–57. doi: 10.1091/mbc.E14-01-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giammanco A, Blanc V, Montenegro G, Klos C, Xie Y, Kennedy S, et al. Intestinal Epithelial HuR Modulates Distinct Pathways of Proliferation and Apoptosis and Attenuates Small Intestinal and Colonic Tumor Development. Cancer Research. 2014;74(18):5322–35. doi: 10.1158/0008-5472.CAN-14-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herjan T, Yao P, Qian W, Li X, Liu C, Bulek K, et al. HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. Journal of immunology (Baltimore, Md : 1950) 2013;191:640–9. doi: 10.4049/jimmunol.1203315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sgantzis N, Yiakouvaki A, Remboutsika E, Kontoyiannis DL. HuR controls lung branching morphogenesis and mesenchymal FGF networks. Developmental Biology. 2011;354:267–79. doi: 10.1016/j.ydbio.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 45.Kraushar ML, Thompson K, Wijeratne HRS, Viljetic B, Sakers K, Marson JW, et al. Temporally defined neocortical translation and polysome assembly are determined by the RNA-binding protein Hu antigen R. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E3815–E24. doi: 10.1073/pnas.1408305111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gubin MM, Techasintana P, Magee JD, Dahm GM, Calaluce R, Martindale JL, et al. Conditional Knockout of the RNA-Binding Protein HuR in CD4(+) T Cells Reveals a Gene Dosage Effect on Cytokine Production. Molecular Medicine. 2014;20:93–108. doi: 10.2119/molmed.2013.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yiakouvaki A, Dimitriou M, Karakasiliotis I, Eftychi C, Theocharis S, Kontoyiannis DL. Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis. The Journal of Clinical Investigation. 2012;122:48–61. doi: 10.1172/JCI45021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat Chem Biol. 2007;3:508–15. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.