Abstract
Failure to achieve optimal bone mineral accretion during childhood and adolescence results in subsequent suboptimal peak bone mass, contributing to osteoporosis risk later in life. To identify novel genetic factors that influence pediatric bone mass at discrete skeletal sites, we performed a sex-stratified genome-wide association study of areal bone mineral density (BMD) measured by dual energy X-ray absorptiometry at the 1/3 distal radius, spine, total hip and femoral neck in a cohort of 933 healthy European American children. We took forward signals with P<5x10−5 and minor allele frequency (MAF) >5% into an independent cohort of 486 European American children in search of replication. In doing so, we identified five loci that achieved genome wide significance in the combined cohorts (nearest genes: CPED1, IZUMO3, RBFOX1, SPBT, and TBPL2), of which the last four were novel and two were sex-specific (SPTB in females and IZUMO3 in males), with all of them yielding associations that were particularly strong at a specific skeletal site. Annotation of potential regulatory function, eQTL effects and pathway analyses identified several potential target genes at these associated loci. This study highlights the importance of sex-stratified analyses at discrete skeletal sites during the critical period of bone accrual, and identifies novel loci for further functional follow-up to pinpoint key genes and better understand the regulation of bone development in children.
Introduction
Osteoporosis is a common condition of aging affecting both men and women, which imposes a heavy burden on public health systems worldwide(1). Childhood and adolescence are considered critical periods for the determination of osteoporotic risk, since failure to achieve optimal bone mineral accretion during growth results in suboptimal peak bone mass, potentially contributing to lower bone mass later in life(2). Therefore, identifying the factors that influence bone mineral accretion during childhood and adolescence is pivotal for preventing this common, disabling disorder.
Family studies(3–5) and population ancestry differences(6) suggest that bone mineral density (BMD) and osteoporosis have a strong heritable component, but little is known about the genetic factors that regulate bone mineral accretion and bone mineral status during growth and development, and the timing of their effects. The genetic factors that affect pediatric bone acquisition may differ from those that impact bone loss later in life. Therefore, to fully understand risk factors for osteoporosis across the life cycle, it is useful to characterize the genetic factors operant during childhood.
Femoral neck and lumbar spine areal BMD measured by dual-energy X-ray absorptiometry (DXA) are the main diagnostic markers of osteoporosis(7,8), and genome-wide association studies (GWAS) have identified > 60 genetic loci associated with these traits in adults(9,10). However, despite recent progress(11), genetic influences specific to bone traits in childhood largely remain to be elucidated. To date, only four distinct pediatric bone density loci have been discovered (SP7, RIN3, EYA4 and 9p21.3)(12–14).
We have previously utilized a deeply phenotyped pediatric cohort (the Bone Mineral Density in Childhood Study, or BMDCS) with DXA measurements to identify genetic loci operating at specific bone sites relevant for osteoporosis and fracture risk in the pediatric context(14–18). In particular, we previously reported our findings from a trans-ethnic GWAS analysis at the distal radius only(14). The current study extends our analysis to three additional skeletal sites (i.e., total hip, femoral neck and spine) which are particularly relevant for osteoporotic risk later in life. We first performed a trans-ethnic analysis of the whole BMDCS cohort, which did not reveal any genome-wide significant loci for these three additional skeletal sites. We therefore restricted our analyses to the BMDCS children of European ancestry, since genetic loci in different populations may be tagged by different genetic markers leading to effect dilution in a trans-ethnic sample and the possibility of missing true positives signals. Additionally, because sexual dimorphism in bone strength, structure and accrual is well-recognized(19–22), we investigated the sex-specific effects of genetic loci operating during childhood and adolescence by performing sex-stratified analyses. Indeed, we have previously reported sex-specific effects in children for several known bone loci established in adults(14–18).
Materials and Methods
Discovery cohort sample
The Bone Mineral Density in Childhood Study (BMDCS) is a multi-center, multi-ethnic longitudinal study established to determine standards for BMC and BMD for American children aged 5 to 20 years old which has been previously described(6). Baseline measurements at the first visit were used for this study.
Replication cohort sample
Children of European descent aged 5 to 18 years (N=486) were subsequently enrolled as a replication cohort for a one-time visit in two US centers (Creighton and Cincinnati). All study procedures were the same as for the primary BMDCS cohort.
Skeletal phenotypes by bone densitometry
Hologic, Inc. (Bedford, MA) bone densitometers (QDR4500A, QDR4500W, Delphi A and Apex models) were used to obtain DXA scans of the 1/3 distal radius, spine, total hip and femoral neck. Both hip sites were included because total hip has a larger amount of cortical bone than the femoral neck, whereas the femoral neck has a greater proportion of trabecular bone. Hologic software (version Discovery 12.3 for baseline and Apex 2.1 for follow-up using the “compare” feature) was used for scan analysis at the DXA Core Laboratory (University of California, San Francisco, CA). Scans were adjusted for differences among devices and longitudinal drift in performance. Both BMD and bone mineral content (BMC) are routinely used to assess bone mineralization in children(19). BMD/BMC Z-scores were calculated using the BMDCS reference values(6) to account for nonlinear increases, increasing variability, and sex differences in BMD/BMC during growth. BMD/BMC Z-scores were adjusted for height-for-age Z-scores to minimize potential confounding by skeletal size, as previously described(6).
Genotyping
High-throughput genome-wide SNP genotyping was performed using the Illumina Infinium™ II OMNI Express plus Exome BeadChip technology (Illumina, San Diego, CA) at the Children’s Hospital of Philadelphia Center for Applied Genomics, as previously described(23).
Imputation
The PLINK software (v1.07) package(24) was employed to carry out quality control measures. Prior to imputation, we excluded individuals with incorrect sex assignments or whose sex could not be determined by genotype, and individuals with missing rate per person >5%. SNPs with a call rate <95%, and MAF <0.5% were removed. After quality control, 1,885 individuals who were genotyped at 739,284 SNPs were available for imputation. Genotypes were imputed to the 1000 Genome Phase I Integrated Release Version 3 reference panel(25). A two-step imputation process was performed using SHAPEIT(26) for haplotype phasing and IMPUTE2(27) for imputation, yielding a total of 39,345,920 imputed SNPs. For the X chromosome we used the specific options in SHAPEIT and IMPUTE2. Imputed genotypes were only used where directly assayed genotypes were unavailable.
Estimation of genomic ancestral components
Autosomal genotyped SNPs of all samples were pruned with PLINK(24), so that no pair of SNPs within a window of 200 markers was in linkage disequilibrium (LD r2=0.05). Based on these ~35,000 SNPs, we performed a maximum likelihood estimation of individual ancestries using ADMIXTURE software(28). This program models the probability of observed genotypes using ancestry proportions and ancestral population allele frequencies. The clustering method was set to group individuals in 3 ancestral populations (K = 3), corresponding to the expected main African, European and Asian ancestry components. Children were assigned to one of the three ancestry groups, based on their highest fraction (i.e., >0.50) of estimated ancestry proportions, using the HapMap Phase 2 population labels as a reference. Subjects were excluded from further analyses if none of the three ancestral populations reached this proportion of >0.5.
Statistical analysis
After estimation of genomic ancestral components, 933 children of European ancestry from the discovery cohort were included in the European-only analysis. For the trans-ethnic analysis, 1,399 children from the discovery cohort were included. We excluded SNPs with MAF <5%, imputation quality <0.4 or Hardy-Weinberg Equilibrium P-value<10−5; this yielded ~6.6 million SNPs for the analysis, including all autosomes and the X chromosome. The association between each SNP and the quantitative trait (BMD at multiple skeletal sites) was assessed using a univariate linear mixed model (as implemented in the software GEMMA(29)) to account for population stratification and relatedness, using the Wald test and the restricted maximum likelihood estimate (REML) of β. Our significance thresholds were P<5×10−5 in the discovery cohort, P<0.05 in the replication cohort and P<5×10−8 in the combined discovery and replication cohorts. We performed a sex-stratified analysis and included a covariate for study site in the model. We also included a covariate for discovery or replication cohort when performing analyses of the combined cohorts. The P-value for the test of sex heterogeneity was calculated as the right-tailed probability of the chi-squared distribution (βF − βM)2/(SEF2 + SEM2). The significance threshold was set at P<4×10−3 [alpha=0.05/(7 loci + 2 traits + 4 skeletal sites)]. For the conditional analysis, we included the imputed dosages of the sentinel SNP as a covariate in the linear mixed model. The genome-wide significance threshold was set at Pcond<5×10−8. We considered a secondary signal significant if Pcond <0.05 after Bonferroni correction for the number of SNPs tested in a 1 Mb region centered around the sentinel SNP.
Functional annotation, expression quantitative trait loci (eQTL) and Topologically Associated Domain (TAD) analyses
We annotated in-silico potential regulatory functions for all the genome-wide significant SNPs we observed, as well as all SNPs in high LD (r2>0.8), using HaploReg v 4.1(30). This is an online tool developed to explore non-coding variants on haplotype blocks at GWAS trait-associated loci. HaploReg performs variant annotation using information from publicly available databases (chromatin mark ChIP-seq tracks, DNase tracks, chromatin state segmentations from the Roadmap Epigenomics Project, conserved regions by GERP and SiPhy, and the SPP narrow peaks called by the ENCODE project on ChIP-seq experiments) and scores the SNPs’ effect on regulatory motifs(31).
To assess whether variants in the identified loci were involved in the regulation of messenger RNA levels via eQTLs, we queried publicly available genome-wide expression datasets using HaploReg, which also integrates multiple resources from the Genotype-Tissue Expression (GTEx) pilot study (Release 6) and published eQTL studies (http://www.broadinstitute.org/mammals/haploreg/data/eqtl_metadata_v4.1.tsv).
For the TAD analysis, we used the TAD boundaries reported in Dixon et al.(32). For each sentinel GWAS SNP, we retrieved the names of genes whose transcriptional starting site (TSS) was contained within the SNP’s TAD boundaries using custom MySQL scripts to query the RefSeq Genes track of the UCSC Genome Browser. We performed a literature search both by manually querying Google and PubMed using the “gene name AND bone” keywords and by using the Bioprofiler function of the Ingenuity Pathway Analysis software (QIAGEN).
Results
To identify novel genetic factors that influence pediatric bone phenotypes, we performed a sex-stratified GWAS of BMD Z-scores at four discrete skeletal sites (1/3 distal radius, spine, total hip and femoral neck) in a cohort of 933 European American children (488 girls and 445 boys). The study population is described in Table S1.
We tested signals with P < 5×10−5 for replication in an independent, similarly-aged cohort of 486 European American children (245 girls and 241 boys). The replication study population is described in Table S2. Summary statistics for the discovery and replication phases, plus the combined analysis, are reported in Table S3 (together with a similar analysis for BMC Z-scores). Phenotypic correlations among the analyzed traits are reported in Table S4.
The results showed appropriate control for population structure, with genomic inflation factors (λ) approaching unity and Q-Q plots revealing no early inflation of the test statistic (Figures S1 and S2).
In females, we identified two significant loci for radius BMD: a known bone locus nearest ‘cadherin-like and PC-esterase domain containing 1’ (CPED1) (top signal rs67991850, Pcombined= 8.91×10−12) and one novel locus, rs1957429 nearest ‘spectrin, beta, erythrocytic’ (SPTB) (Pcombined =5.97×10−9). In males, we identified a novel significant locus: rs184374109 nearest IZUMO family member 3 (IZUMO3) for spine BMD (Pcombined=1.18×10−8).
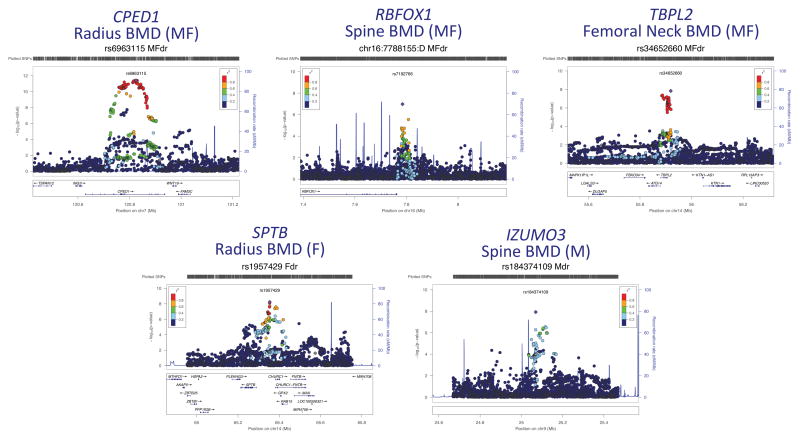
In analyses including both sexes, two loci were genome-wide significant: the same locus identified in females near CPED1 for radius BMD (top signal rs6963115, r2 with rs67991850=0.90, Pcombined =3.89×10−12) and a novel locus nearest ‘TATA-box binding protein like 2’ (TBPL2) for femoral neck BMD (top signal rs34652660, Pcombined=1.45×10−8). In addition, despite not reaching the threshold for the discovery cohort alone, chr16:7788155 nearest ‘RNA binding protein fox-1 homolog 1’ (RBFOX1) did achieve nominal significance in the replication cohort along with genome-wide significance in the analysis of both cohorts combined for spine BMD (Pcombined=4.06×10−8). The results are summarized in Table 1 and regional Manhattan plots are shown in Figure 1. Summary statistics for the significant loci at all four skeletal sites are reported in Table S5 (together with a similar analysis for BMC Z-scores).
Table 1. Five loci are associated with bone mineral density in children, with two loci (IZUMO3 and SPBT) showing sex-specific associations.
Summary statistics for top signals achieving genome-wide significance in the combined analysis (discovery plus replication) for BMD at four skeletal sites (1/3 distal radius, total hip, femoral neck and spine). Effect sizes (betas) refer to the minor allele. Genomic positions are reported in the GRCh37/hg19 build. The significance threshold for the test of sex heterogeneity is set at 4×10−3.
| Associated Loci | Females | Males | All | Het Test | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP (Locus, Pos) | Nearest Gene | Major Allele | Minor Allele | Trait | Stage | MAF | Beta | SE | P-value | N | MAF | Beta | SE | P-value | N | MAF | Beta | SE | P-value | N | P-value |
| rs6963115 | CPED1 | C | A | Radius | Discovery | 0.398 | −0.402 | 0.060 | 7.49×10−11 | 474 | 0.393 | −0.133 | 0.069 | 5.43×10−2 | 434 | 0.395 | −0.274 | 0.047 | 5.95×10−9 | 908 | 3.42×10−3 |
| 7q31.31 | Replication | 0.378 | −0.171 | 0.081 | 3.62×10−2 | 242 | 0.427 | −0.294 | 0.091 | 1.45×10−3 | 239 | 0.402 | −0.230 | 0.061 | 1.90×10−4 | 481 | 3.11×10−1 | ||||
| chr7:120825318 | Combined | 0.391 | −0.331 | 0.049 | 2.40×10−11 | 716 | 0.405 | −0.189 | 0.055 | 6.75×10−4 | 673 | 0.398 | −0.260 | 0.037 | 3.89×10−12 | 1389 | 5.48×10−2 | ||||
|
| |||||||||||||||||||||
| rs184374109 | IZUMO3 | C | A | Spine | Discovery | 0.042 | 0.114 | 0.160 | 4.76×10−1 | 485 | 0.057 | 0.727 | 0.147 | 1.02×10−6 | 442 | 0.050 | 0.447 | 0.110 | 5.53×10−5 | 927 | 4.81×10−3 |
| 9p21.3 | Replication | 0.028 | −0.279 | 0.325 | 3.91×10−1 | 240 | 0.037 | 0.760 | 0.254 | 3.11×10−3 | 239 | 0.033 | 0.322 | 0.203 | 1.13×10−1 | 479 | 1.17×10−2 | ||||
| chr9:25069552 | Combined | 0.038 | 0.020 | 0.145 | 8.92×10−1 | 725 | 0.050 | 0.737 | 0.128 | 1.18×10−8 | 681 | 0.044 | 0.411 | 0.097 | 2.57×10−5 | 1406 | 2.13×10−4 | ||||
|
| |||||||||||||||||||||
| chr16:7788155:D | RBFOX1 | CG | C | Spine | Discovery | 0.260 | −0.153 | 0.067 | 2.23×10−2 | 485 | 0.238 | −0.253 | 0.101 | 1.32×10−2 | 442 | 0.254 | −0.191 | 0.049 | 1.19×10−4 | 927 | 4.11×10−1 |
| 16p13.3 | Replication | 0.249 | −0.315 | 0.100 | 1.85×10−3 | 240 | 0.248 | −0.247 | 0.071 | 5.45×10−4 | 239 | 0.243 | −0.283 | 0.071 | 8.15×10−5 | 479 | 5.81×10−1 | ||||
| chr16:7788155 | Combined | 0.256 | −0.208 | 0.056 | 2.19×10−4 | 725 | 0.244 | −0.245 | 0.058 | 2.84×10−5 | 681 | 0.250 | −0.224 | 0.041 | 4.06×10−8 | 1406 | 6.41×10−1 | ||||
|
| |||||||||||||||||||||
| rs1957429 | SPTB | C | T | Radius | Discovery | 0.131 | 0.402 | 0.095 | 2.94×10−5 | 474 | 0.097 | −0.004 | 0.163 | 9.80×10−1 | 434 | 0.118 | 0.208 | 0.075 | 5.33×10−3 | 908 | 3.10×10−2 |
| 14q23.3 | Replication | 0.117 | 0.510 | 0.122 | 4.30×10−5 | 242 | 0.105 | −0.004 | 0.114 | 9.71×10−1 | 239 | 0.107 | 0.296 | 0.100 | 3.31×10−3 | 481 | 2.11×10−3 | ||||
| chr14:65354033 | Combined | 0.126 | 0.444 | 0.075 | 5.97×10−9 | 716 | 0.102 | −0.006 | 0.094 | 9.51×10−1 | 673 | 0.114 | 0.241 | 0.060 | 5.90×10−5 | 1389 | 1.77×10−4 | ||||
|
| |||||||||||||||||||||
| rs34652660 | TBPL2 | A | C | Femoral Neck | Discovery | 0.486 | −0.243 | 0.056 | 1.92×10−5 | 485 | 0.477 | −0.085 | 0.079 | 2.79×10−1 | 443 | 0.479 | −0.217 | 0.042 | 3.05×10−7 | 928 | 1.04×10−1 |
| 14q22.3 | Replication | 0.463 | −0.237 | 0.091 | 1.01×10−2 | 242 | 0.471 | −0.178 | 0.060 | 3.19×10−3 | 240 | 0.470 | −0.158 | 0.060 | 9.15×10−3 | 482 | 5.87×10−1 | ||||
| chr14:55919480 | Combined | 0.478 | −0.240 | 0.048 | 7.22×10−7 | 727 | 0.473 | −0.147 | 0.048 | 2.20×10−3 | 683 | 0.476 | −0.198 | 0.035 | 1.45×10−8 | 1410 | 1.70×10−1 | ||||
Figure 1.
Regional association plots of 5 genome-wide significant signals identified in this study (combined discovery plus replication analysis). Circles show P-values and positions of SNPs within each locus. The top SNPs are denoted by a purple diamond. Colors indicate varying degrees of pairwise linkage disequilibrium (HapMap CEU release 22) between the top SNP and all other SNPs.
To identify independent secondary signals, we performed conditional analysis accounting for the effect of the sentinel SNP for each trait. The results are shown in Table S6 and Figure S3. None of the conditional signals were genome-wide significant. The only locus showing a significant secondary peak in a 1 Mb region around the sentinel SNP was the CPED1 locus (rs2536180; Pcond=1.72×10−7). The secondary peak is not in LD with the primary signal (r2=0.01) and maps to the genomic region containing WNT16 and FAM3C.
We then assessed sex-specificity of the identified loci. A test of heterogeneity of the betas indicated sex-specificity for two loci implicated in the sex-stratified analyses only (Table 1; SPTB: rs1957429, P=1.9×10−4, radius BMD and IZUMO3: rs184374109, P=2.3×10−4, spine BMD).
To investigate the role of these pediatric-implicated loci in the adult setting, we performed a look-up of the key sentinel SNPs in the latest release of the The GEnetic Factors for OSteoporosis (GEFOS) Consortium data (http://www.gefos.org/?q=content/data-release-2015), which includes summary statistics for three DXA-derived sites (femoral neck, lumbar spine and forearm BMD)(33). We confirmed a role for the CPED1 locus at the forearm (rs6963115, β=−0.07, P=1.97×10−5) and to a lesser extent at the lumbar spine (β= −0.03, P=5.01×10−3) and the femoral neck (β=−0.02, P=3.49×10−2). The other loci did not show evidence of association [IZUMO3, rs184374109: Pforearm=0.78, Pls=0.45, Pfn=0.92; TBPL2, rs34652660: Pforearm=0.15, Pls=0.26, Pfn=0.69; SPTB, rs1957429: Pforearm=0.66, Pls=0.50, Pfn=0.75; RBFOX1, rs11640907 (proxy for chr16:7788155:D, R2=0.72): Pforearm=0.26, Pls=0.15, Pfn=0.58].
Expression Quantitative Trait Loci (eQTLs)
Subsequently, we investigated whether the pediatric-implicated variants we identified were correlated with eQTLs using HaploReg v4.1(30). We found evidence for two of the sentinel SNPs at the CPED1 locus (rs67991850 and rs6963115) influencing the expression of CPED1 in transformed fibroblasts(34), the sentinel SNP at the TBPL2 locus (rs34652660) influencing expression of a long intergenic noncoding RNA (lincRNA) (RP11-665C16.6) in testis(34) and the sentinel SNP at the SPTB locus (rs1957429) influencing SPTB expression in pancreas(34) and CHURC1 and RAB15 expression in lymphoblastoid cell lines(35). These tissues and cell lines are not primary sites with functional relevance to BMD and bone biology, and thus, further analyses in more relevant cell types (such as human osteoblasts) will be required to identify the target transcripts regulated by these GWAS signals.
Annotation of potential regulatory functions
All genome-wide significant SNPs, as well as all SNPs in high LD (r2>0.8), were non-coding (either intronic or intergenic). Therefore, we annotated in-silico potential regulatory functions for these SNPs using HaploReg v 4.1(30).
We did not observe any significant enrichment of these SNPs in regulatory elements in specific tissues. However, three out of five of our GWAS loci had SNPs in high LD (r2>0.8; SPTB, 2 SNPs; TBPL2, 12 SNPs; CPED1, 4 SNPs) that were located in either predicted enhancer or promoter regions in one of three bone-relevant cell types: osteoblast primary cells, mesenchymal stem cell derived chondrocyte cultured cells, and bone marrow derived mesenchymal cultured cells. Most of these regulatory regions are active in a variety of cell types and are not bone specific, with the exception of three SNPs in the CPED1 locus (rs1917118 and rs62469350, located in an enhancer that is active in mesenchymal stem cell-derived chondrocyte cultured cells and skeletal muscle tissues only, and rs6947494, in an enhancer active in mesenchymal stem cell-derived chondrocyte cultured cells, skeletal muscle and heart tissues).
Topologically Associated Domain (TAD) analysis
In order to identify potential candidate genes that might be regulated by our GWAS signals, we identified all the genes contained in each SNPs’ TAD (regions where SNP-promoter interactions are more likely to occur(32)) and performed a literature search to link these genes to pathways relevant to bone biology. The results are shown in Table S7. Besides WNT16, already known to play an important role in cortical bone homeostasis(36), we identified several genes with links to bone biology (ATG14, KTN1, GPX2 and LGALS3) which may represent attractive targets for further functional studies.
Discussion
Our GWAS performed in two cohorts of children of European ancestry identified five loci that associated with BMD at four discrete skeletal sites (one already reported: CPED1, and four novel: IZUMO3, RBFOX1, SPTB and TBPL2). Importantly, the associations involving variants near SPTB and IZUMO3 were sex-specific (Table 1). We also observed skeletal site specificity, with CPED1 and SPTB being more strongly associated at the radius, TBPL2 at the total hip and femoral neck, IZUMO3 and RBFOX1 at the spine, (Table S5).
The intronic GWAS signals harbored within CPED1 are in high LD (r2>0.8) with each other and are associated with radius BMD, with the association being stronger in females. This locus includes three genes in close proximity (CPED1, WNT16 and FAM3C) and has been established as a key genomic locus for pediatric bone density phenotypes(13,14,37). This locus is also associated with BMD in adults at various skeletal sites(38), particularly at the forearm(39), but with smaller effect sizes.
CPED1 (‘cadherin-like and PC-esterase domain containing 1’, also known as C7orf58) is a poorly characterized gene encoding a protein expressed in several tissues (most abundantly in the gastrointestinal mucosa) and containing an ATP-Grasp domain fused to a PC-esterase domain. FAM3C (‘family with sequence similarity 3 member C’) is an uncharacterized gene encoding a secreted protein with a GG domain that is expressed in most tissues. The neighboring gene, WNT16, is a member of the WNT gene family, implicated in many cellular developmental pathways such as cell fate determination, proliferation, differentiation, and apoptosis. The canonical or β-catenin WNT signaling pathway is involved in the regulation of bone formation and maintenance, and when activated leads to increased bone mass in mice and humans, inducing the differentiation of bone-forming osteoblasts and suppressing bone-resorbing osteoclasts(40). In mice, Wnt16 is derived from osteoblast-lineage cells and inhibits osteoclastogenesis, and Wnt16 deficiency causes spontaneous fractures in cortical bone(41). While the role of WNT16 in bone biology is well established, it remains to be shown whether the genetic variants identified by these GWAS actually influence the expression of WNT16 or another nearby gene (such as CPED1, FAM3C or others) in bone tissue.
None of the genes nearest to the novel signals identified by this study have been previously linked to bone biology, and available eQTL data do not implicate any obvious candidate genes which may be therapeutic targets for osteoporosis and bone strength. When we expanded our search for bone-related functions to genes residing in the TADs surrounding the signal SNPs, where SNP-promoter interactions are more likely to occur, we found four potentially interesting target genes: GPX2 (‘glutathione peroxidase 2’), a member of the glutathione peroxidase family which was found upregulated in adults affected with Kashin-Beck disease (KBD), an endemic, chronic osteochondropathy with unknown etiology diffused in China(42), near the SPTB locus (associated with radius BMD in females); ATG14 (‘autophagy related 14’), whose decreased expression promoted cisplatin-induced apoptosis in a drug-resistant osteosarcoma cell line in vitro(43); KTN1 (‘kinectin 1’), responsible for various functions related to tumorigenesis and found upregulated in giant cells tumors of bone (GCTB), benign bone tumors that cause osteolytic destruction(44); and LGALS3 (‘lectin, galactoside binding soluble 3’), a downstream regulator of matrix metalloproteinase-9 functioning during endochondral bone formation(45); these three genes all reside near the TBPL2 locus (associated with femoral neck BMD).
Among the newly identified loci, the strongest effect size (β=0.74) was found for a low-frequency SNP (rs184374109, MAF=5%), located in a gene desert on chromosome 9 flanked by IZUMO3 and TUSC1 (‘tumor suppressor candidate 1’). The effect size for this low-frequency variant is higher than those for recently described rare variants (MAF~1%) in EN1 and WNT16 in adults(33) and comparable to the effects size of the same rare EN1 variant we investigated in the BMDCS cohort(16). Similar to the EN1 variant (which was more strongly associated in females in the BMDCS), we found this association to be highly sex-specific, with the locus being associated with spine BMD in males only. Interestingly, the signal resides in the same region (9p21.3) in which we found signals associated with radius BMC – also in males only – in our previously published trans-ethnic analysis of the BMDCS cohorts (14), but these signals are not in LD (r2=0.0003). The radius BMC locus previously identified in our trans-ethnic analysis achieved only a suggestive P-value (rs7035284; P= 5.47×10−7) in the current analysis (using the European portion of the BMDCS cohort), due to a smaller sample size, in agreement with the observation of a very consistent magnitude of effect in both European American (β=0.32, SE=0.06, N=673) and African American (β=0.25, SE=0.10, N=164) males.
Conversely, the novel loci identified in this analysis (restricted to children of European ancestry only) achieved only suggestive P-values (P<5×10−5), but not genome-wide significance, in the trans-ethnic analysis combining children of all ethnicities (Table S8), suggesting that these loci are driven mainly by the European ancestry portion of the cohort. CPED1 was the only locus achieving genome-wide significance in the trans-ethnic analysis, although this signal, too, was principally from the European portion of the cohort (Table S8 and Chesi et al.(14)). The association of this locus (previously described in the literature(13,14,37)) with radius BMD was evident from both the trans-ethnic and the European-specific Q-Q plots (Figures S1 and S2).
These findings suggest that combining individuals of different ethnicities in the same analysis can either increase or decrease discovery power, depending on the locus and its LD structure in the different populations, and also validates our approach of performing both ethnicity-stratified and combined analyses. Our study also demonstrates the advantage of phenotyping discrete skeletal sites for bone genetic discovery, in agreement with previous studies(13); in fact, there appears to be a strong site specificity, with certain genetic loci acting preferentially at one or a few specific skeletal sites, such as the CPED1 locus acting mainly at the radius (Table S5).
Our GWAS in children (unlike adult studies) outlines two unexpected results – the sex-specificity of two loci and the large effect sizes. Sex-dependent genetic influences can be expected for BMD, a highly sexually-dimorphic trait. Bone size, strength and accrual differ greatly between sexes from birth to late adolescence, and it is conceivable that this dimorphism could be driven by distinct genetic influences. Although a well-powered meta-analysis for BMD in adults found no genome-wide significant evidence for gene-by-sex interaction(46), it is possible that sex-specific effects are stronger during childhood and adolescence, when bone accrual occurs. The signals identified in this study do not reside in the X chromosome, adding to the mounting evidence for autosomal sex-specific effects that recent GWAS have begun to uncover for different traits (cardiac-diseases related and anthropometric traits(47,48), type II diabetes(49) and asthma(50)). We cannot exclude that small sample size and potential sampling bias might affect the sex-specific findings in our study, although the two-step design we employed, with a discovery and a replication cohort, should mitigate the chance of spurious observations. Also, confounding arising from a potential collider bias between covariates and outcomes that differ between sexes should not be a concern, since the only covariates used in our model are site of visit plus sex and/or cohort in the combined analyses, which are not outcomes of either the exposures (SNPs) or the analyzed phenotype (BMD Z-scores).
The effect sizes we report for the GWAS associations in our study are higher (sometimes by an order of magnitude) than those reported in larger adult meta-analyses. While this could be an effect of the “winner’s curse” or Beavis effect due to the relatively small sample size of our study, it is plausible that genetic variation exerts a stronger influence in children (in whom cumulative exposure to environmental factors is lower) than in adults(51). This age-specificity may explain why we could not find strong evidence of replication for many of our pediatric signals in the publicly available datasets for BMD in adults (which also are not stratified by sex).
In summary, we report four novel loci for pediatric BMD at four discrete skeletal sites, plus the previously published 7q31.31 (CPED1/WNT16/FAM3C) locus. Since the two US sites where the replication cohort for this study was recruited also coincided with two of the recruitment sites for the discovery cohort, further studies will need to be conducted in larger pediatric cohorts in fully independent settings (once they become available) in order to validate our findings further.
In addition, functional studies in relevant tissues will be required to shed light on the molecular mechanisms linking these genetic signals to their key biological pathways.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health [grant numbers HD58886, HD076321]; the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) contracts (N01-HD-1-3228, -3329, -3330, -3331, -3332, -3333); and the CTSA program Grant 8 UL1 TR000077. Jonathan Mitchell was supported by K01 HL123612. Shana McCormack was supported by K23 DK102659. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. We appreciate the dedication of the study participants and their families, and the support of Dr. Karen Winer, Scientific Director of the Bone Mineral Density in Childhood Study.
Footnotes
Author Contributions
Study conception and design: AC, SG and BZ. Acquisition of data: SG, BZ, HK, JL, VG, SO and JS. Data analysis: AC, JM, JB and DC. Interpretation of data: AC, JM, HK, JB, JL, DC, SR, SM, VG, SO, HH, JS, AK, BZ and SG. Drafting manuscript: AC. Revising manuscript content: AC, JM, JB and DC. Interpretation of data: AC, JM, HK, JB, JL, DC, SR, SM, VG, SO, HH, JS, AK, BZ and SG. Approving final version of manuscript: AC, JM, JB and DC. Interpretation of data: AC, JM, HK, JB, JL, DC, SR, SM, VG, SO, HH, JS, AK, BZ and SG. AC, SG and BZ take full responsibility for the integrity of the data analysis.
References
- 1.Burge R, Dawson-Hughes B, Solomon DH, Wong JB, King A, Tosteson A. Incidence and economic burden of osteoporosis-related fractures in the United States, 2005–2025. J Bone Miner Res. 2007 Mar;22(3):465–75. doi: 10.1359/jbmr.061113. [DOI] [PubMed] [Google Scholar]
- 2.Heaney RP, Abrams S, Dawson-Hughes B, Looker A, Marcus R, Matkovic V, et al. Peak bone mass. Osteoporos Int. 2000;11(12):985–1009. doi: 10.1007/s001980070020. [DOI] [PubMed] [Google Scholar]
- 3.Gueguen R, Jouanny P, Guillemin F, Kuntz C, Pourel J, Siest G. Segregation analysis and variance components analysis of bone mineral density in healthy families. J Bone Miner Res. 1995 Dec;10(12):2017–22. doi: 10.1002/jbmr.5650101223. [DOI] [PubMed] [Google Scholar]
- 4.Seeman E, Hopper JL, Bach LA, Cooper ME, Parkinson E, McKay J, et al. Reduced bone mass in daughters of women with osteoporosis. N Engl J Med. 1989 Mar 2;320(9):554–8. doi: 10.1056/NEJM198903023200903. [DOI] [PubMed] [Google Scholar]
- 5.Soroko SB, Barrett-Connor E, Edelstein SL, Kritz-Silverstein D. Family history of osteoporosis and bone mineral density at the axial skeleton: the Rancho Bernardo Study. J Bone Miner Res. 1994 Jun;9(6):761–9. doi: 10.1002/jbmr.5650090602. [DOI] [PubMed] [Google Scholar]
- 6.Zemel BS, Kalkwarf HJ, Gilsanz V, Lappe JM, Oberfield S, Shepherd JA, et al. Revised reference curves for bone mineral content and areal bone mineral density according to age and sex for black and non-black children: results of the bone mineral density in childhood study. J Clin Endocrinol Metab. 2011 Oct;96(10):3160–9. doi: 10.1210/jc.2011-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummings SR, Black DM, Nevitt MC, Browner W, Cauley J, Ensrud K, et al. Bone density at various sites for prediction of hip fractures. The Study of Osteoporotic Fractures Research Group. Lancet. 1993 Jan 9;341(8837):72–5. doi: 10.1016/0140-6736(93)92555-8. [DOI] [PubMed] [Google Scholar]
- 8.Kroger H, Lunt M, Reeve J, Dequeker J, Adams JE, Birkenhager JC, et al. Bone density reduction in various measurement sites in men and women with osteoporotic fractures of spine and hip: the European quantitation of osteoporosis study. Calcif Tissue Int. 1999 Mar;64(3):191–9. doi: 10.1007/s002239900601. [DOI] [PubMed] [Google Scholar]
- 9.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014 Jan;42(Database issue):D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estrada K, Styrkarsdottir U, Evangelou E, Hsu Y, Duncan E, Ntzani E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat Rev Genet. 2012 Aug;13(8):576–88. doi: 10.1038/nrg3228. [DOI] [PubMed] [Google Scholar]
- 12.Timpson NJ, Tobias JH, Richards JB, Soranzo N, Duncan EL, Sims AM, et al. Common variants in the region around Osterix are associated with bone mineral density and growth in childhood. Hum Mol Genet. 2009 Apr 15;18(8):1510–7. doi: 10.1093/hmg/ddp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemp JP, Medina-Gomez C, Estrada K, St Pourcain B, Heppe DH, Warrington NM, et al. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet. 2014 Jun;10(6):e1004423. doi: 10.1371/journal.pgen.1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chesi A, Mitchell JA, Kalkwarf HJ, Bradfield JP, Lappe JM, McCormack SE, et al. A trans-ethnic genome-wide association study identifies gender-specific loci influencing pediatric aBMD and BMC at the distal radius. Hum Mol Genet. 2015 Sep 1;24(17):5053–9. doi: 10.1093/hmg/ddv210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell JA, Chesi A, Elci O, McCormack SE, Roy SM, Kalkwarf HJ, et al. Genetic Risk Scores Implicated in Adult Bone Fragility Associate With Pediatric Bone Density. J Bone Miner Res. 2016 Apr;31(4):789–95. doi: 10.1002/jbmr.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell JA, Chesi A, McCormack SE, Roy SM, Cousminer DL, Kalkwarf HJ, et al. Rare EN1 Variants and Pediatric Bone Mass. J Bone Miner Res. 2016 Aug;31(8):1513–7. doi: 10.1002/jbmr.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell JA, Chesi A, Elci O, McCormack SE, Kalkwarf HJ, Lappe JM, et al. Genetics of Bone Mass in Childhood and Adolescence: Effects of Sex and Maturation Interactions. J Bone Miner Res. 2015 Sep;30(9):1676–83. doi: 10.1002/jbmr.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell JA, Chesi A, Elci O, McCormack SE, Roy SM, Kalkwarf HJ, et al. Physical Activity Benefits the Skeleton of Children Genetically Predisposed to Lower Bone Density in Adulthood. J Bone Miner Res. 2016 May 12; doi: 10.1002/jbmr.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crabtree NJ, Arabi A, Bachrach LK, Fewtrell M, El-Hajj Fuleihan G, Kecskemethy HH, et al. Dual-energy X-ray absorptiometry interpretation and reporting in children and adolescents: the revised 2013 ISCD Pediatric Official Positions. J Clin Densitom. 2014 Apr-Jun;17(2):225–42. doi: 10.1016/j.jocd.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Ponrartana S, Aggabao PC, Dharmavaram NL, Fisher CL, Friedlich P, Devaskar SU, et al. Sexual Dimorphism in Newborn Vertebrae and Its Potential Implications. J Pediatr. 2015 Aug;167(2):416–21. doi: 10.1016/j.jpeds.2015.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey DA, Martin AD, McKay HA, Whiting S, Mirwald R. Calcium accretion in girls and boys during puberty: a longitudinal analysis. J Bone Miner Res. 2000 Nov;15(11):2245–50. doi: 10.1359/jbmr.2000.15.11.2245. [DOI] [PubMed] [Google Scholar]
- 22.Berger C, Goltzman D, Langsetmo L, Joseph L, Jackson S, Kreiger N, et al. Peak bone mass from longitudinal data: implications for the prevalence, pathophysiology, and diagnosis of osteoporosis. J Bone Miner Res. 2010 Sep;25(9):1948–57. doi: 10.1002/jbmr.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakonarson H, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–4. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pennisi E. Genomics. 1000 Genomes Project gives new map of genetic diversity. Science. 2010 Oct 29;330(6004):574–5. doi: 10.1126/science.330.6004.574. [DOI] [PubMed] [Google Scholar]
- 26.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012 Feb;9(2):179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 27.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009 Jun;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009 Sep;19(9):1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012 Jul;44(7):821–4. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012 Jan;40(Database issue):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011 May 5;473(7345):43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012 May 17;485(7398):376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng HF, Forgetta V, Hsu YH, Estrada K, Rosello-Diez A, Leo PJ, et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015 Oct 1;526(7571):112–7. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015 May 8;348(6235):648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013 Sep 26;501(7468):506–11. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gori F, Lerner U, Ohlsson C, Baron R. A new WNT on the bone: WNT16, cortical bone thickness, porosity and fractures. Bonekey Rep. 2015;4:669. doi: 10.1038/bonekey.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medina-Gomez C, Kemp JP, Estrada K, Eriksson J, Liu J, Reppe S, et al. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. 2012 Jul;8(7):e1002718. doi: 10.1371/journal.pgen.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009 Nov;41(11):1199–206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng HF, Tobias JH, Duncan E, Evans DM, Eriksson J, Paternoster L, et al. WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLoS Genet. 2012 Jul;8(7):e1002745. doi: 10.1371/journal.pgen.1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Canalis E. Wnt signalling in osteoporosis: mechanisms and novel therapeutic approaches. Nat Rev Endocrinol. 2013 Oct;9(10):575–83. doi: 10.1038/nrendo.2013.154. [DOI] [PubMed] [Google Scholar]
- 41.Moverare-Skrtic S, Henning P, Liu X, Nagano K, Saito H, Borjesson AE, et al. Osteoblast-derived WNT16 represses osteoclastogenesis and prevents cortical bone fragility fractures. Nat Med. 2014 Nov;20(11):1279–88. doi: 10.1038/nm.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen Y, Zhang F, Li C, He S, Tan W, Lei Y, et al. Gene expression analysis suggests bone development-related genes GDF5 and DIO2 are involved in the development of Kashin-Beck disease in children rather than adults. PLoS One. 2014;9(7):e103618. doi: 10.1371/journal.pone.0103618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Z, Tao L, Shen C, Liu B, Yang Z, Tao H. Silencing of Barkor/ATG14 sensitizes osteosarcoma cells to cisplatin induced apoptosis. Int J Mol Med. 2014 Feb;33(2):271–6. doi: 10.3892/ijmm.2013.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Babeto E, Conceicao AL, Valsechi MC, Peitl P, Junior, de Campos Zuccari DA, de Lima LG, et al. Differentially expressed genes in giant cell tumor of bone. Virchows Arch. 2011 Apr;458(4):467–76. doi: 10.1007/s00428-011-1047-4. [DOI] [PubMed] [Google Scholar]
- 45.Ortega N, Behonick DJ, Colnot C, Cooper DN, Werb Z. Galectin-3 is a downstream regulator of matrix metalloproteinase-9 function during endochondral bone formation. Mol Biol Cell. 2005 Jun;16(6):3028–39. doi: 10.1091/mbc.E04-12-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu CT, Estrada K, Yerges-Armstrong LM, Amin N, Evangelou E, Li G, et al. Assessment of gene-by-sex interaction effect on bone mineral density. J Bone Miner Res. 2012 Oct;27(10):2051–64. doi: 10.1002/jbmr.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilks WP, Abbott JK, Morrow EH. Sex differences in disease genetics: evidence, evolution, and detection. Trends Genet. 2014 Oct;30(10):453–63. doi: 10.1016/j.tig.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 48.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015 Feb 12;518(7538):187–96. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012 Sep;44(9):981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naumova AK, Al Tuwaijri A, Morin A, Vaillancourt VT, Madore AM, Berlivet S, et al. Sex- and age-dependent DNA methylation at the 17q12-q21 locus associated with childhood asthma. Hum Genet. 2013 Jul;132(7):811–22. doi: 10.1007/s00439-013-1298-z. [DOI] [PubMed] [Google Scholar]
- 51.Kemp JP, Medina-Gomez C, Tobias JH, Rivadeneira F, Evans DM. The case for genome-wide association studies of bone acquisition in paediatric and adolescent populations. Bonekey Rep. 2016;5:796. doi: 10.1038/bonekey.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.