Abstract
Alterations in the gut microbiome have been associated with changes in bone mass and microstructure, but the effects of the microbiome on bone biomechanical properties are not known. Here we examined bone strength under two conditions of altered microbiota: 1) an inbred mouse strain known to develop an altered gut microbiome due to deficits in the immune system (the toll-like receptor 5 deficient mouse, TLR5KO); and 2) disruption of the gut microbiota (ΔMicrobiota) through chronic treatment with selected antibiotics (ampicillin and neomycin). The bone phenotypes of TLR5KO and WT (C57Bl/6) mice were examined following disruption of the microbiota from 4 weeks to 16 weeks of age as well as without treatment (n = 7–16/group, 39 animals total). Femur bending strength was less in ΔMicrobiota mice than in untreated animals and the reduction in strength was not fully explained by differences in bone cross-sectional geometry, implicating impaired bone tissue material properties. Small differences in whole bone bending strength were observed between WT and TLR5KO mice after accounting for differences in bone morphology. No differences in trabecular bone volume fraction were associated with genotype or disruption of gut microbiota. Treatment altered the gut microbiota by depleting organisms from the phyla Bacteroidetes and enriching for Proteobacteria, as determined from sequencing of fecal 16S rRNA genes. Differences in splenic immune cell populations were also observed; B and T cell populations were depleted in TLR5KO mice and in ΔMicrobiota mice (p <0.001), suggesting an association between alterations in bone tissue material properties and immune cell populations. We conclude that alterations in the gut microbiota for extended periods during growth may lead to impaired whole bone mechanical properties in ways that are not explained by bone geometry.
Keywords: Biomechanics, Osteoporosis, Bone Matrix, Osteoimmunology
1.0 INTRODUCTION
The microbes that inhabit the gastrointestinal tract are known collectively as the gut microbiota. Alterations in the gut microbiota are associated with a number of conditions that cause bone loss or increase fracture risk including malnutrition (1,2), inflammatory bowel disease (3–5), obesity (6,7), and metabolic disease (8–10). The gut microbiota, therefore, have the potential to influence bone and contribute to differences in fracture risk among patient populations.
The gut microbiome is initially obtained at birth (11) and subsequently shaped by factors such as environment (12) and diet (13,14). Exposure to the gut microbiome is necessary for the proper education and development of the innate and adaptive immune systems (15). Dendritic cells, macrophages, granulocytes, T and B cells, and intestinal epithelial cells directly interact with the gut microbiome (15). Toll-like receptors are one set of receptors on immune cells that recognize the components of the gut microbiome and facilitate communication between the gut microbiome and the immune system (16). Alterations in the gut microbiota or improper communication between the immune system and gut microbiota can lead to chronic immune responses and disease (17).
The effects of the microbiome on bone structure and density have been studied in mice using two standard tools for manipulating the microbiome: germ-free animals and oral antibiotic treatments (18,19). The changes in bone following these manipulations of the gut flora differ considerably among studies. Germ-free mice (raised in the absence of live microbes) have been reported to display reduced bone mass(20), as well as increased bone mass (21) as compared to mice raised in conventional environments. Alterations in the gut microbiota through treatment with oral antibiotics have been reported to affect bone density in mice, but the findings have been mixed, possibly due to differences in animal age, sex, antibiotic used, dosing schedule and mouse genotype (22–25).
Genetic models are another tool for studying the effects of the microbiome on animal physiology. The Toll-like receptor 5 deficient mouse (TLR5KO) is a congenic mouse strain that has been used to study the effects of the gut microbiome on animal physiology and disease. Toll-like receptor 5 (TLR5) is the innate immune receptor for flagellin and does not have an endogenous ligand (26). Hence, phenotypic traits of the TLR5KO mouse are primarily due to alterations in host-microbe interactions (27). Failure of the TLR5KO mouse to respond to flagellin is associated with changes in the gut microbiome that lead to increases in intestinal and systemic inflammation and a metabolic syndrome-like phenotype characterized by mild obesity, insulin resistance, increased blood pressure and increased blood glucose (27,28). The metabolic syndrome-like phenotype of the TLR5KO mouse does not develop in mice raised in a germ-free environment and can be transferred to wild-type mice through transplantation of the gut microbiota, demonstrating that the phenotype depends on the gut flora (28).
While prior work has shown that the disruption or absence of the microbiome can influence bone, interpreting conflicting findings among studies is challenging because many prior studies use young animals of different ages (less than 12 weeks of age) or low resolution imaging techniques (mouse DXA). Comparing bone phenotypes in such young animals is not recommended because bone is changing rapidly during growth (29). Additionally, none of the previous studies have examined the effect of alterations in the gut microbiota on bone mechanical performance. In the present study, we tested the hypothesis that alterations in the gut microbiota can have an effect on whole bone biomechanical performance. Specifically, we determined changes in bone structure and strength associated with alterations in the gut microbiota caused by 1) genotypic alterations (the TLR5KO mouse) and 2) chronic treatment with antibiotics that target the gut microbiota.
2.0 MATERIAL AND METHODS
2.1 Study design
Animal procedures were approved by Cornell University’s Institutional Animal Care and Use Committee. Mice from the C57BL/6J inbred strain and the B6.129S1-Tlr5tm1Flv/J (TLR5KO) congenic strain were acquired from the Jackson Laboratory (Bar Harbor, ME) and each bred separately in conventional housing in our animal facility. C57BL/6J is the recommended control strain for TLR5KO (28,30). Animals were housed in plastic cages filled with 1/4-inch corn cob bedding (The Andersons’ Lab Bedding, Ohio), fed with standard laboratory chow (Teklad LM-485 Mouse/Rat Sterilizable Diet) and water ad libitum, and provided a cardboard refuge environmental enrichment hut (Ketchum Manufacturing; Brockville, Ontario). Male mice were divided into four groups: two groups treated to disrupt the gut microbiota (C57BL/6J: n=7, TLR5KO: n=8) and two untreated groups (C57BL/6J: n=12, TLR5KO: n=16). Mice with disrupted microbiota are referred to as “ΔMicrobiota.” Mice were housed in cages with other animals from the same genetic background/treatment group. Treated groups received broad-spectrum antibiotics (1.0 g/L ampicillin, 0.5 g/L neomycin) in their drinking water from weaning at 4 weeks of age until skeletal maturity (16 weeks of age) (28). Chronic antibiotics used in this manner causes consistent disruptions to the gut microbiota over a prolonged time period (31). Ampicillin and neomycin have poor bioavailability, thereby limiting extra-intestinal effects of treatment (28,32). Additionally, neomycin and ampicillin have never been associated with impaired bone growth. Animals were euthanized at 16 weeks of age. Femora, tibiae, epididymal fat pads, and spleen were collected immediately after euthanasia. Fecal pellets were collected one day prior to euthanasia to allow analysis of the microbiota.
2.2 Cortical bone mechanical testing
The right femora were harvested, wrapped in PBS-soaked gauze, and stored at −20°C prior to analysis. Femur length was measured from the greater trochanter to the lateral condyle using digital calipers. Images of the femoral diaphyseal cross-section were obtained by micro-CT with a voxel size of 25 μm (GE eXplore CT 120; 80 kVp, 32 μA, 100 ms integration time). Images were processed using a Gaussian filter to remove noise and a global threshold for each group was used to segment mineralized tissue from surrounding non-mineralized tissue. Femoral cross-sectional geometry was determined using a volume of interest extending 2.5% of total bone length and centered midway between the greater trochanter and lateral condyle (BoneJ, bonej.org, version 1.3.3) (33). Measurements included total area, cortical cross-sectional area, cortical thickness, marrow area, and moment of inertia about the medial-lateral axis.
Femora were thawed to room temperature and maintained hydrated during mechanical testing. Right femora were loaded to failure in three-point bending in the anterior-posterior direction at a rate of 0.1 mm/s using a span length of 6 mm between outer loading pins (858 Mini Bionix; MTS, Eden Prairie, MN, USA). Force and displacement were measured using a 10 lb. load cell (Transducer Techniques, SSM-100, Temecula, CA) and a linear variable differential transducer at a 100 Hz sampling rate. Bending stiffness was calculated as the slope of the linear portion of the force-displacement curve (29). Peak bending moment was calculated as half the peak load multiplied by half the span length (29). The peak bending moment is related to bone tissue material properties and bone midshaft geometry by the following equation(34):
where M is peak bending moment, σb is bone tissue material strength, I is the moment of inertia, and c is the distance from the neutral axis to bone surface. The term incorporates all geometrical properties that can influence peak bending moment. Differences in peak bending moment that are not explained by are caused by alterations in tissue material properties. Due to irregularities in force versus displacement data associated with motion some specimens were excluded from the biomechanical analysis (4 WT, 1 WT ΔMicrobiota, 3 TLR5KO).
2.3 Trabecular bone morphology
Images of the tibiae were collected using micro-computed tomography with 6 μm voxels (μCT35; Scanco Medical AG, Switzerland; 55 kVp, 145 μA, 600 ms integration time). The trabecular bone microarchitecture of the proximal tibial metaphysis was examined in a region extending from the growth plate to 10% of total bone length. Measurements included bone volume fraction (BV/TV), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), and cortical tissue mineral density (ct. TMD). A global threshold for each group was used to segment mineralized tissue from surrounding non-mineralized tissue. A randomized subset (n=8) was selected for analysis of trabecular bone morphology for the TLR5KO mice.
2.4 Gut Microbiome Analysis
2.4.1 DNA extraction
Gut microbiota analysis was performed on six samples per group. Isolation of DNA from feces was performed by using PowerSoil DNA Isolation Kit (MO BIO Laboratory Inc., Carlsbad, CA) according to manufacturers’ instructions. DNA concentration and purity were then evaluated using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE) at wavelengths of 230, 260, and 280 nm.
2.4.2 Quantitative PCR
The total bacterial load of fecal samples was determined using quantitative PCR (qPCR) as previously described (35). The total bacterial load was defined as the total number of 16S rRNA gene copies. Briefly, quantification of the 16S rRNA target DNA was achieved by using the forward: 5′-TGG AGC ATG TGG TTT AAT TCG A-3′, and reverse: 5′-TGC GGG ACT TAA CCC AAC A-3′) (36,37) Unibac primers, and 10-fold serial dilutions ranging from 100 to 107 plasmid copies of a plasmid DNA standard which was cloned in-house(35). Plasmid standards and feces samples were run in duplicates. The average of the cycle threshold value was used for calculation of the total bacterial load.
2.4.3 Next generation sequencing, and bioinformatics
Amplification of the 16S rRNA gene, library construction and bioinformatics were executed according to previously described methods (35). Briefly, for amplification of the V4 hypervariable region of the bacterial/archaeal 16S rRNA gene, primers 515F and 806R were used (38). The 5′-barcoded amplicons were generated in triplicate using 12–300 ng of template DNA, 2 X EconoTaq® Plus Green Master Mix (Lucigen®, Middleton, WI) and 10 μM of each primer. Replicate amplicons were pooled and purified using the Gel PCR DNA Fragment Extraction kit (IBI Scientific, Peosta, IA) and visualized by electrophoresis through 1.2% (weight/volume) agarose gel stained with 0.5 mg/ml ethidium bromide. Blank controls in which no DNA was added to the reaction were performed. Purified amplicon DNA was quantified using fluorometry (Quant-iT™ PicoGreen® from Life Technologies Corporation, Carlsbad, CA, USA).
Standardization of feces amplicon sample aliquots was performed to the same concentration and then pooled into one run according to individual barcode primers for the 16S rRNA gene. Final equimolar libraries were sequenced using the MiSeq reagent kit v2 (300 cycles) on the MiSeq platform (Illumina, Inc., San Diego, CA, USA).
Raw 16S rRNA gene sequences generated were demultiplexed using the open source software pipeline Quantitative Insights Into Microbial Ecology (QIIME, version 1.7.0-dev) (39). Sequences were filtered for quality using established guidelines (40). Taxonomy was assigned using UCLUST (www.drive5.com) consensus taxonomy assigner, against the Greengenes reference database(41). Low-abundance clusters were filtered, and chimeric sequences were removed using USEARCH (42). Additionally, we generated a species-level OTU table using the MiSeq Reporter Metagenomics Workflow. The MiSeq Reporter classification is based on the Greengenes database (http://greengenes.lbl.gov/), and the output of this workflow is a classification of reads at multiple taxonomic levels: kingdom, phylum, class, order, family, genus and species.
Shannon diversity index was performed (QIIME, version 1.7.0-dev). Before estimating the Shannon diversity index, all sample libraries were rarefied to an equal depth of 10,000 sequences (QIIME, version 1.7.0-dev).
2.5 Colon Histology
To evaluate gut inflammation, colons were collected at euthanasia and fixed in 10% neutral buffered formalin for 48 hours. Colons were embedded in paraffin, sectioned, and scored by the Cornell Animal Health Diagnostic Center. Each sample was scored based on 4 assays: lymphoid aggregate size, lymphoid aggregate density, apoptotic cells per high powered field, and presence of inflammation.
2.6 Flow Cytometry
Splenocytes were harvested from the spleen of three mice from each group immediately after euthanasia as described previously (43,44). The splenocytes were subsequently stained by incubation in 50 μL of FACS containing antibodies (1:500 dilution) for an hour. For analyzing B cells, Anti-CD20 antibody conjugated to Phycoerythrin (PE) (BD Pharmagen) was used and for T cells Anti-CD3 antibody conjugated to PE (BD Pharmagen) was used. The stained cells were rinsed twice with FACS buffer and re-suspended in 50 μL FACS buffer to be analyzed by BD Accuri C6 flow cytometer. The flow cytometer results were analyzed using FlowJo software (FlowJo LLC, Ashland, Oregon). Gut microbiota interact with and can be regulated by B and T cell populations (15,45). Therefore, we examined the relative percentages of B and T cells in spleens of these mice.
2.7 Statistical Analyses
Measures of bone were adjusted for body mass (unadjusted values are provided in Supplemental Table 1) (29). Homogenous variance was tested using Levene’s test and normality tested using the Shapiro-Wilk Test. If parametric assumptions were met, a one-way ANOVA followed by post-hoc Holm correction for multiple comparisons was performed to test for differences between groups. If parametric assumptions were violated, either data was submitted to a log transform to achieve homogenous variance and normality or a non-parametric ranked Dunn’s test followed by post-hoc Bonferroni adjustment for multiple comparisons was used.
To determine if genotype or treatment influenced whole bone strength in ways that were not explained by cross-sectional geometry, we performed an ANCOVA, implemented with a GLM model using as the covariate with genotype and treatment as fixed effects. Statistical tests were conducted using JMP Pro (v.9, 2013, SAS Institute Inc., Cary, NC, USA).
3.0 RESULTS
3.1 Body mass and TLR5KO phenotype
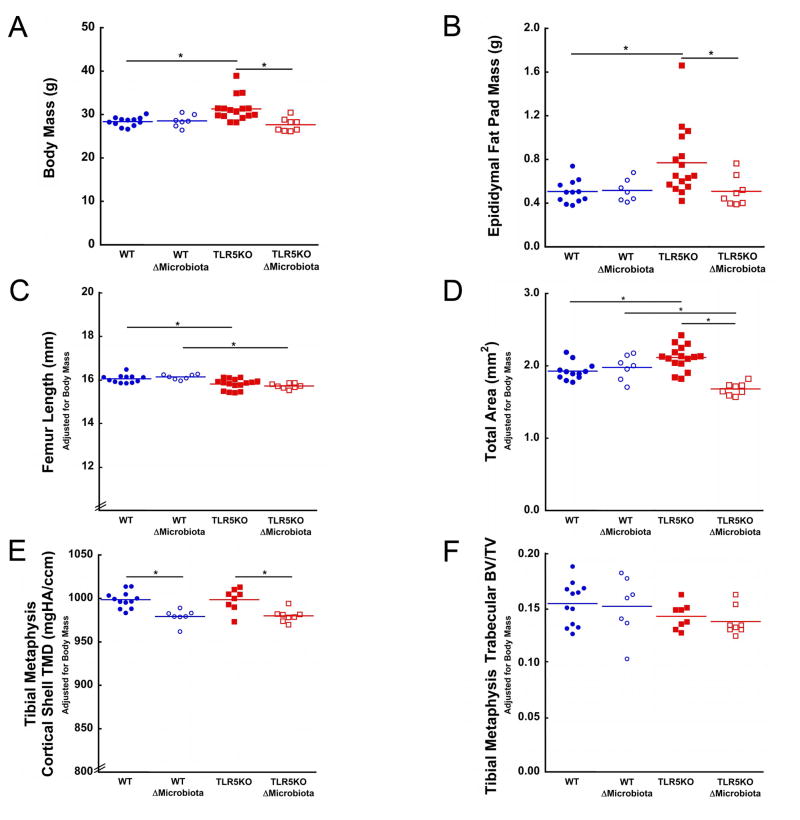
The TLR5KO mice showed a mild obesity phenotype with an average body mass 10.4% greater than WT (p <0.05; Fig 1A) and an average epididymal fat pad mass 52.0% greater than WT (p <0.05; Fig 1B). Body mass and fat pad mass in TLR5KO mice with disrupted microbiota and WT mice with disrupted microbiota were similar, as demonstrated in prior work (28). No differences in colon histological scoring were observed among groups. One TLR5KO mouse had elevated colon histological scores suggesting mild colitis, but did not display gross differences in bone morphology or body mass and was not excluded from the study (30).
Figure 1.
TLR5KO mice had greater body and fat pad mass. Disruption of the gut microbiota in TLR5KO mice prevented the development of increased body and fat pad mass. Disruption of the gut microbiota in WT mice had no effect on (A) body mass or (B) epididymal fat pad mass. (C) TLR5KO mice femur length was less than WT in both untreated and treated groups (D) Total area was increased in untreated TLR5KO mice compared to untreated WT mice. Disruption of gut microbiota led to a reduced total area in TLR5KO ΔMicrobiota mice. (E) Disruption of the gut microbiota in both genotypes was associated with a reduced tibial metaphysis cortical TMD. (F) No differences in tibial metaphysis BV/TV were observed between any groups. Solid colored lines on dot plots represent mean. Measures in Fig 1C, 1D, 1E, 1F are adjusted for body mass. * p < 0.05.
3.2 Femoral whole bone bending strength and geometry in TLR5KO mice
Bone morphology in TLR5KO mice differed from WT mice. Total cross-sectional area was larger in TLR5KO mice compared to WT mice (p < 0.05, Fig 1D). Marrow area, cortical area, and cortical thickness (Table 1) in TLR5KO mice were similar to that in WT mice. TLR5KO mice had a larger moment of inertia compared to WT mice (p < 0.05, Fig 2A). Femoral bone length was 1.5% smaller in TLR5KO mice compared to WT mice (p < 0.05, Fig 1C, Table 1).
Table 1.
Micro-computed tomography measures of cancellous and cortical bone and whole bone mechanical testing measures after adjustments for body mass are shown.
| Wild Type | TLR5KO | ||||
|---|---|---|---|---|---|
| Body Mass Adjusted Measure | Untreated | ΔMicrobiota | Untreated | ΔMicrobiota | |
| Proximal Tibia | Bone Volume Fraction | 0.15 ± 0.02 | 0.15 ± 0.03 | 0.14 ± 0.01 | 0.14 ± 0.01 |
| Trabecular Thickness (μm) | 0.045 ± 0.002 | 0.043 ± 0.005 | 0.041 ± 0.003 a | 0.038 ± 0.002$ | |
| Trabecular Separation (μm) | 0.197 ± 0.014 | 0.192 ± 0.008 | 0.187 ± 0.007 | 0.178 ± 0.007$ | |
| Cortical TMD (mg HA/cm3) | 999 ± 10 | 979 ± 8* | 999 ± 13 | 980 ± 7# | |
| Growth Plate Thickness (μm) | 558 ± 46 | 562 ± 41 | 550 ± 54 | 528 ± 21 | |
| Femoral Diaphysis | Cortical Area (mm2) | 0.87 ± 0.07 | 0.81 ± 0.09* | 0.94 ± 0.07 | 0.69 ± 0.04# |
| Marrow Area (mm2) | 1.05 ± 0.06 | 1.17 ± 0.10* | 1.17 ± 0.15 | 1.00 ± 0.08# | |
| Total Area (mm2) | 1.93 ± 0.12 | 1.98 ± 0.17 | 2.12 ± 0.17 a | 1.68 ± 0.08#,$ | |
| Cortical Thickness (μm) | 210 ± 10 | 189 ± 15* | 216 ± 13 | 172 ± 12#,$ | |
| Moment of Inertia (mm4) | 0.15 ± 0.02 | 0.14 ± 0.02 | 0.17 ± 0.03 a | 0.10 ± 0.01#,$ | |
| Moment of Inertia/c (mm3) | 0.22 ± 0.03 | 0.22 ± 0.03 | 0.25 ± 0.03 | 0.17 ± 0.01#,$ | |
| Whole Femur | Length (mm) | 16.04 ± 0.18 | 16.13 ± 0.12 | 15.80 ± 0.24a | 15.71 ± 0.12$ |
| Peak Bending Moment (N* mm) | 40.79 ± 2.71 | 37.18 ± 2.35* | 38.75 ± 2.37 | 30.22 ± 1.76#,$ | |
| Bending Stiffness (N/mm) | 193 ± 26 | 177 ± 21 | 175 ± 33 | 152 ± 7 | |
| Post Yield Displacement (mm) | 0.20 ± 0.17 | 0.35 ± 0.09 | 0.29 ± 0.11 | 0.42 ± 0.07 | |
Values are mean ± SD. TMD=Tissue mineral density
WT-ΔMicrobiota v. WT-untreated, p<0.05
TLR5KO-ΔMicrobiota v. TLR5KO-untreated, p<0.05
TL5KO-untreated v. WT-untreated, p<0.05
TLR5KO-ΔMicrobiota v. WT-ΔMicrobiota, p<0.05
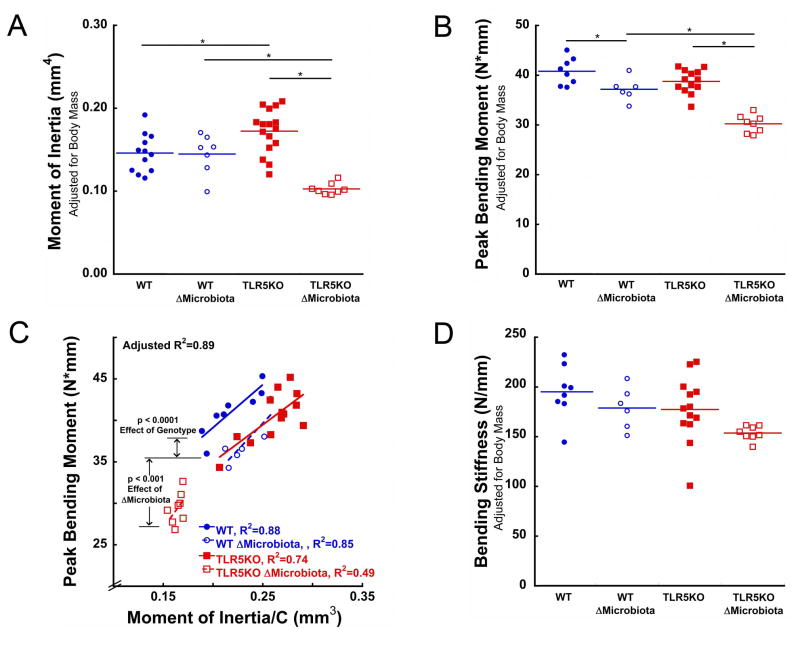
Figure 2.
Whole bone bending strength in mice with altered microbiota was less than would be expected from differences in cross-sectional geometry. (A) The moment of inertia was larger in TLR5KO mice. (B) Whole bone bending strength (peak bending moment) was less in ΔMicrobiota mice than in untreated animals. The peak bending moment in TLR5KO mice did not differ from that of WT mice. (C) Whole bone bending strength in TLR5KO mice was less than in WT mice after accounting for I/c (difference between solid red and blue lines). Bending strength in ΔMicrobiota mice was less than that in untreated animals (difference between dotted and solid lines indicates results of ANCOVA). (D). Disruption of the gut microbiota in both WT and TLR5KO mice showed a trend suggesting reduced whole bone femoral bending stiffness. Solid colored lines on dot plots represent mean. Measures in Fig 2A, 2B, and 2D are adjusted for body mass. * p < 0.05
The peak bending moment in untreated TLR5KO mice was similar to that in WT mice (Fig 2C), but the moment of inertia in TLR5KO mice was larger than in WT mice. Whole bone strength in TLR5KO mice was less than that in WT mice after accounting for differences in cross-sectional femoral geometry (ANCOVA, effect of genotype, p<0.0001, Fig 2C). No differences in post yield displacement (Table 1) or bending stiffness (Fig 2D) were observed between WT and TLR5KO mice.
3.3 Femoral whole bone bending strength and geometry in mice with a disrupted microbiota
Disruption of the gut microbiota resulted in differences in geometry in TLR5KO mice and in WT mice. Disruption of the gut microbiota in WT mice resulted in increased marrow area, decreased cortical area, and decreased cortical thickness compared to untreated WT mice (p < 0.05, Table 1). Disruption of the gut microbiota did not result in changes in total area, moment of inertia, or femoral length in WT mice (Fig 1C, Fig 1D, 2A, Table 1). Disruption of the gut microbiota in TLR5KO mice resulted in decreased total area, marrow area, cortical area, cortical thickness, and moment of inertia as compared to untreated TLR5KO mice (p < 0.05, Fig 1D, Fig 2A, Table 1). Disruption of the gut microbiota did not influence femoral length in TLR5KO mice (Fig 1C, Table 1). Femoral length was 2.6% smaller in TLR5KO ΔMicrobiota mice compared to WT ΔMicrobiota mice (p < 0.05, Fig 1C, Table 1).
Disruption of the gut microbiota was associated with reduced peak bending moment. Disruption of the gut microbiota in WT mice resulted in an average peak bending moment 9% less than in untreated WT mice (p < 0.05, Fig 2B). Disruption of the gut microbiota in TLR5KO mice led to a peak bending moment 22% less than in untreated TLR5KO mice (p < 0.05, Fig 2B). After accounting for differences in cross-sectional geometry, peak bending moment in mice with a disrupted microbiota was less than that in untreated mice (ANCOVA, effect of ΔMicrobiota, p < 0.0001, Fig 2C). The effect of disruption of the gut microbiota on bone tissue material properties appeared to differ between WT and TLR5KO mice (ANCOVA, ΔMicrobiota x genotype, p=0.09, Fig 2C). Disruption of the gut microbiota in both WT and TLR5KO mice showed a trend suggesting reduced whole bone femoral bending stiffness (p < 0.15, Fig 2D, Table 1). Disruption of the gut microbiota was not associated with differences in post yield displacement (Table 1).
3.4 Tibial trabecular microarchitecture and tissue mineral density
Cancellous bone volume fraction in the proximal tibia did not differ among groups (Fig 1F). No differences in tibial cortical bone tissue mineral density were observed between untreated WT and TLR5KO mice. Disruption of the gut microbiota was associated with reductions in cortical bone tissue mineral density in both strains of mice (p < 0.05, Fig 1F). The thickness of the growth plate in the proximal tibia did not differ among groups (Table 1).
3.5 Microbiome analysis
Sequences from feces microbiome assays were filtered for size, quality, and for the presence of chimeras and the total post-quality control number of sequences used in this study were 2,465,448. The average coverage was 102,727 ± 32,103 (mean ± SD) reads per sample. No differences in the mean number of reads for each group were observed (WT:112,309 ± 11,935; WT ΔMicrobiota: 88,325 ± 18,501; TLR5KO: 101,706 ± 39,625, and TLR5KO ΔMicrobiota: 108,568 ± 47,800) (p = 0.612).
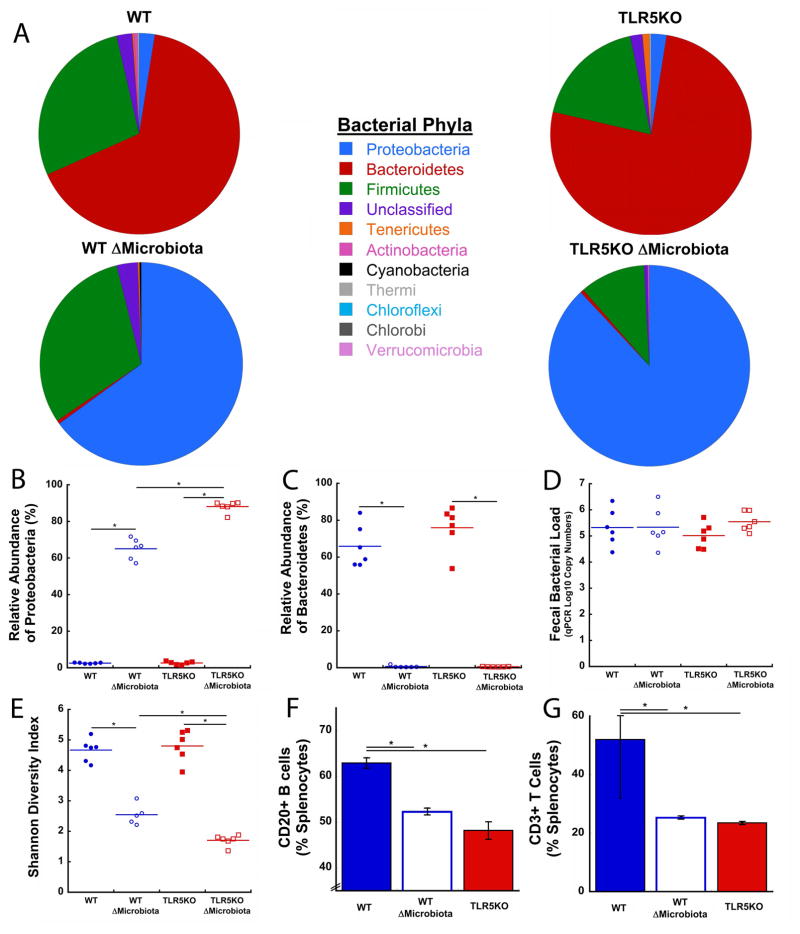
Although the total bacterial load did not differ among the four groups (Fig 3D), profound changes in the gut microbiota were observed. The gut microbiota composition at the phyla level differed among groups (Fig 3A). The gut microbiota in WT and TLR5KO mice was dominated by the Bacteroidetes phylum (Fig 3A, 3C). The gut microbiota in ΔMicrobiota mice was dominated by the Proteobacteria phylum (Fig 3A, 3B). Proteobacteria abundance was greater in TLR5KO ΔMicrobiota mice compared to WT ΔMicrobiota mice (p < 0.05, Fig 3B). The diversity of the gut microbiota, as measured by the Shannon Diversity Index, was reduced in groups with a disrupted gut microbiota (TLR5KO: 4.8 ± 0.5; TLR5KO ΔMicrobiota: 1.7 ± 0.2; WT: 4.7 ± 0.4; WT ΔMicrobiota: 2.5 ± 0.3) (p < 0.05, Fig 3E). Compared to untreated animals from the same genetic background, reductions in gut microbiota diversity in TLR5KO ΔMicrobiota mice were greater than those in WT ΔMicrobiota mice (p < 0.05, Fig 3E). One sample from the WT ΔMicrobiota mice was determined to be an outlier and removed (Fig 3E).
Figure 3.
Disruption of the gut microbiota with antibiotics did not alter total bacterial load, but had dramatic effects on gut microbiota composition and bacterial diversity, and immune cell count. (A) The relative composition of bacterial phyla shifted from a Bacteroidetes dominated phyla in untreated mice to one dominated by Proteobacteria in ΔMicrobiota mice (n=6/group). (B) Proteobacteria is enriched in ΔMicrobiota mice, especially in TLR5KO ΔMicrobiota mice (Bonferroni correction). (C) Bacteroidetes dominates gut microbiota composition in untreated WT and TLR5KO. (D) Total bacterial load was unaffected by antibiotic treatment. (E) Bacterial diversity was dramatically reduced in ΔMicrobiota mice. (F) The percentage of splenic CD20+ B cells was reduced in ΔMicrobiota mice and untreated TLR5KO mice (n=3/group). (G) The percentage of splenic CD3+ T cells in the spleen was reduced in ΔMicrobiota mice and untreated TLR5KO mice (n=3/group). Solid colored lines on dot plots represent mean. * p < 0.05 after adjusting for multiple comparisons
3.7 Splenoycte populations
The total percentage of CD20+ B cell splenocytes was reduced in TLR5KO mice and WT ΔMicrobiota mice compared to untreated WT mice (p < 0.05, Fig 3F). The percentage of CD3+ T cells in the spleen was reduced in TLR5KO and WT ΔMicrobiota mice compared to untreated WT mice (p < 0.05, Fig 3G). Splenocytes from TLR5KO ΔMicrobiota mice were not obtained due to user error.
4.0 DISCUSSION
Here we report the effects of an altered gut microbiota on bone mechanical properties in WT and TLR5KO mice. Disruption of the gut microbiota through long-term exposure to antibiotics led to reductions in whole bone bending strength that exceeded what could be explained by the associated changes in cross-sectional geometry, suggesting impairment of bone tissue material properties. Small differences in whole bone bending strength were observed between WT and TLR5KO mice after accounting for differences in bone morphology.
Together the differences in whole bone strength, cross-sectional geometry and tissue mineral density suggest that alterations in the gut microbiota changed the mechanical properties of the bone tissue itself. Whole bone strength in bending is determined by both cross-sectional geometry and tissue material properties. In bending, the ratio is the geometric measure that describes the entire effect of cross-sectional geometry on bending strength and is directly proportional to the maximum load an object can sustain in bending. Consistent with this relationship, the ratio was the single best predictor of whole bone strength, accounting for 71% of the variation in peak bending moment across groups. However, differences in the regression lines (Fig 2C) indicated that the ratio did not completely explain differences in strength among the four groups, a situation that implies alteration in bone tissue mechanical properties. Tissue mineral density (TMD) is a material property that can influence bone strength (46). TMD in the tibial metaphysis of mice with a disrupted microbiota was less than that of untreated mice. Although we did not measure TMD at the femoral midshaft directly, our findings in the tibia suggest that TMD may partially explain the reductions in femoral bone strength. Other factors such as collagen quality and non-collagenous proteins may also explain the reductions in femoral bone strength.
TLR5KO mice had larger total area than WT mice, but similar marrow area and cortical area. Increased total area without differences in marrow or cortical area at skeletal maturity has been associated with more rapid periosteal expansion during growth (29). The increased periosteal expansion in TLR5KO mice may be a mechanism employed by the skeleton to maintain whole bone strength despite impaired bone tissue material properties (47).
Disruption of the gut microbiota resulted in decreased cortical bone at the femoral diaphysis in both WT mice and TLR5KO mice. Disruption of the gut microbiota in WT mice was not associated with alterations in total area, but was associated with decreased cortical area and cortical thickness. Disruption of the gut microbiota in TLR5KO mice prevented the more rapid periosteal expansion that occurred in untreated TLR5KO mice, and resulted in smaller cortical area, marrow area, and cortical thickness. Though marrow area was smaller in TLR5KO ΔMicrobiota mice, marrow area was larger than would be expected from the associated changes in total area. Decreased cortical area and cortical thickness is often attributed to decreased accumulation of bone mass during growth (29).
Treatment with antibiotics had a larger effect on bone morphology and whole bone strength in TLR5KO mice than in WT mice. This observation has many potential explanations: First, disruption of the gut microbiota prevented the development of the mild obesity phenotype in TLR5KO mice. Obesity is associated with differences in bone morphology and mechanical performance(48). The bones in treated TLR5KO mice, therefore, not only have the effect of an impaired microbiota, but also reduced adiposity. Second, disruption of the gut microbiota in TLR5KO mice had a larger effect on the relative abundance of Proteobacteria and microbial diversity (the Shannon diversity index) than in WT mice, which could help explain the larger effect on the bone phenotype. Third, the immune system and immune responses are impaired in TLR5KO mice, leading to altered gene expression and activity by the gut microbiota (27).
The composition of gut microbiota in untreated and treated mice was consistent with prior work. The total bacterial load in fecal samples did not differ between antibiotic treated and untreated groups, consistent with previous reports that oral antibiotic treatment can cause a large initial reduction in a bacterial population that recovers over time to a newly stabilized population (23,49,50). The dominant phylum in untreated mice was Bacteroidetes, consistent with reports that Bacteroidetes are the predominant phylum throughout a healthy mouse’s lifespan (51). Disruption of the gut microbiota by chronic antibiotic treatment led to a gut microbiota population enriched by the phylum Proteobacteria (a minor component of the untreated mouse gut microbiota). The high relative abundance of Proteobacteria observed in mice with a disrupted microbiota at 16 weeks of age was similar to the immature and unstable gut microbiota typical of newborn mice (45). As a mouse matures, its immune system begins to regulate gut microbiota composition via B cell production of IgA antibodies that target Proteobacteria (45). The antibiotic treatment in the current study may have prevented the shift from Proteobacteria to Bacteroidetes that normally occurs in mice after weaning. Furthermore, the reduced splenic B cell count in mice with a disrupted microbiota is also consistent with the increased presence of Proteobacteria. The prevalence of members of the Proteobacteria phylum has been associated with increased incidence of microbial dysbiosis, metabolic disease, and inflammation, all factors known to influence host physiology and the immune system (16,52).
To understand the mechanisms linking changes in the microbiota to impaired bone tissue material properties it is useful to consider the three primary mechanisms through which the microbiome can influence organs distant from the gut: regulation of the immune system, regulation of nutrient absorption, and translocation of bacterial products across the epithelial barrier (53).
We consider the effects of the microbiota on the immune system to be a likely explanation for the differences in bone tissue material properties in the current study. Disruption of the gut microbiota with antibiotics reduced CD20+ B and CD3+ T cell populations and was correlated with reduced whole bone strength. Similarly, untreated TLR5KO mice also had reduced CD20+ B and CD3+ T cell populations. B and T cell populations have the potential to cause profound changes in bone remodeling and bone turnover (54–58). However, it is not yet clear how alterations in B and T cell populations would lead to changes in bone tissue material properties.
While we cannot ignore the possibility that alterations in nutritional absorption influenced our findings, we consider this explanation unlikely for several reasons: First, body mass and fat pad mass in the mice were all similar or greater than that in untreated wild type animals, suggesting an acceptable caloric intake. Second, trabecular bone volume fraction was not different among the groups, and femoral length only had small differences. Trabecular bone volume fraction and whole bone length are typically severely reduced in situations of nutritional deficiency (20,59). Trabecular bone is extremely responsive to impaired nutrition; animals submitted to short-term severe calcium and vitamin D deficiencies showed reductions in trabecular bone volume fraction of 24–58% (60,61), yet we did not observe reductions in trabecular bone volume fraction. Third, the reduction in peak bending moment seen in mice with a disrupted microbiota is not fully explained by changes in bone geometry or bone mass, whereas in animal models of reduced dietary calcium and vitamin D, reductions in whole bone strength are usually well described by changes in bone geometry, mass, and tissue mineral density (60,61). Lastly, examination of colon histology did not indicate intestinal inflammation in any of our groups, suggesting that treatment with antibiotics to disrupt the gut microbiota did not lead to increased gut inflammation that can impair nutritional absorption (62,63). Animal models with extensive intestinal inflammation commonly develop reduced body mass and dramatic trabecular bone loss, which, again, was not present in any of our treatment groups (64,65).
Translocation of bacterial products (or even live bacteria) across the gut endothelial barrier is another potential mechanism for gut microbiota to influence bone. Microbial products such as lipopolysaccharide and flagellin are capable of traveling through the bloodstream to distant organs and causing localized inflammation (66). Translocation of bacteria across the endothelial barrier is one of the mechanisms that explains the TLR5KO metabolic syndrome phenotype, so translocation may be involved in the observed differences in bone (27). While bone cells can respond to lipopolysaccharide and flagellin (53), how such a response would lead to changes in bone tissue mechanical properties is not clear.
A number of strengths of the current study are worth noting. First, the study is unique in examining the effect of alterations in the gut microbiome on whole bone mechanical performance. Previous studies in which the microbiota was modified focused solely on bone structure or bone mass and did not examine mechanical performance. Second, the current study examined the effects of prolonged disruption of the gut microbiota during growth on the bone phenotype achieved at skeletal maturity. Most of the prior studies of bone in mice under conditions of altered gut microbiota examined bone from young, rapidly growing animals (7–9 weeks of age)(53), and did not evaluate the bone phenotype at skeletally maturity. Differences in bone phenotype in growing animals sometimes indicate differences in growth rate and do not always imply changes in bone phenotype at skeletal maturity (29,67,68). As we only looked at skeletally mature mice, we could not assess differences in bone growth and acquisition, although differences in cross-sectional geometry such as total area suggest differences in rates of periosteal expansion (see above). Third, the current study provided both a detailed analysis of bone along with a full analysis of the constituents of the gut microbiome as determined using 16S rRNA sequencing and therefore provides differences in phyla, bacterial diversity, and total bacterial load along with a detailed bone morphological and biomechanical analysis. We are aware of only one prior study that provides both a detailed analysis of bone morphology and a detailed analysis of the microbiome (69).
Despite the novelty of the current study, some limitations must be considered when interpreting the results. The contents of the gut microbiota are dynamic and robust to external stimuli; short-term treatments (~1–2 weeks) with antibiotics generate a transient change in the gut microbiota that mostly returns to baseline when treatment was suspended (49). To examine a condition of sustained alterations in the gut microbiota during growth we treated mice with chronic antibiotics from the age of weaning until skeletal maturity. Although chronic antibiotic treatment is rarely applied to humans throughout growth and development, less drastic changes in the human gut microbiota do occur for prolonged periods of time as a result of diet or metabolic status (49). The study is further limited by not directly performing a compositional assessment of bone tissue. Direct measures of bone tissue material properties can help explain the mechanical phenotypes but more direct assays of mouse bone tissue mechanical properties than those performed here have additional limitations, especially in determination of tissue strength (see Supplementary Material from Jespen et al 2015 (29)). The current study does not include assessment of bone turnover. Recent findings, however, suggest that the relationship between the microbiota and bone remodeling is complex and dynamic. For example, mice treated with an antibiotic cocktail of ampicillin, vancomycin, metronidazole, and neomycin show changes to serum turnover markers after one week of treatment, but no detectable differences from untreated animals after one month of treatment (22). Understanding the effects of manipulation of the microbiome on bone remodeling would therefore require examination at many points during growth/treatment. Lastly, the current study uses the C57BL/6J as a control strain for the TLR5KO strain, despite the TLR5KO mice containing minor remnants of B6.129S1 genetics. However, the TLR5KO congenic strain is backcrossed for 11 generations to the C57BL/6J background to ensure the two strains are over 99.9% genetically identical, thus limiting potential effects of B6.129S1 genetics.
Despite the limitations of our study, our observations regarding changes in bone tissue mechanical properties suggest a new explanation to a long-standing clinical question. Fracture risk in some patient populations is much greater than expected from bone mineral density, a situation commonly attributed to impaired “bone quality”(70). Although the term bone quality encompasses many different characteristics of bone (71), impaired bone tissue mechanical properties are a well-recognized component. Changes in bone tissue mechanical properties are often cited as a contributor to fracture risk that exceeds what is explained by BMD in patients with obesity, diabetes, and inflammatory bowel disease – three chronic clinical conditions that are also associated with drastic changes in the gut microbiome. Our findings in mice suggest an intriguing possibility that alterations in gut microbiota may contribute to alterations in clinical fracture risk by regulating bone tissue mechanical properties, although further studies are required to confirm this hypothesis.
5.0 Conclusion
We conclude that alterations in the gut microbiota throughout growth can lead to changes in whole bone strength that are greater than expected from whole bone size or shape. These findings suggest that alterations in the gut microbiota can influence bone tissue mechanical properties.
Supplementary Material
Acknowledgments
This publication was supported in part by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (U.S) under Award Number AR068061 and by the Office of the Assistant Secretary of Defense for Health Affairs through the office of the Congressionally Directed Medical Research Programs (CDMRP) under Award No. W81XWH-15-1-0239. The content of the work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Defense. We thank Adrian Alepuz for his help in analyzing mechanical testing data.
Footnotes
Authors’ roles: Conceived and designed the experiments: JDG, REL, MCHM, SRG, CJH. Performed the experiments: JDG, MWH, FFF, TNS, ML, FA, SFL. Analyzed data: JDG, CJH, FA, AS, RCB, SFL. Wrote and Revised Manuscript: JDG, CJH, SFL. Critical revision and final approval of the manuscript: All authors.
Supplemental data have been included with the submission.
Disclosures
All authors state that they have no conflicts of interest.
References Cited
- 1.Lucas AR, Melton LJ, III, Crowson CS, O’Fallon WM. Long-term Fracture Risk Among Women With Anorexia Nervosa: A Population-Based Cohort Study. Mayo Clin Proc. 1999;74(10):972–7. doi: 10.4065/74.10.972. [DOI] [PubMed] [Google Scholar]
- 2.Kane AV, Dinh DM, Ward HD. Childhood Malnutrition and the Intestinal Microbiome Malnutrition and the microbiome. Pediatr Res. 2015;77(0):256–62. doi: 10.1038/pr.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostic AD, Xavier RJ, Gevers D. The Microbiome in Inflammatory Bowel Disease: Current Status and the Future Ahead. Gastroenterology. 2014;146(6):1489–99. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van staa T-P, Cooper C, Samuels Brusse L, Leufkens H, Javaid MK, Arden NK. Inflammatory bowel disease and the risk of fracture. Gastroenterology. 2003;125(6):1591–7. doi: 10.1053/j.gastro.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 5.Vestergaard P, Krogh K, Rejnmark L, Laurberg S, Mosekilde L. Fracture risk is increased in Crohn’s disease, but not in ulcerative colitis. Gut. 2000;46(2):176–81. doi: 10.1136/gut.46.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonnelli S, Caffarelli C, Nuti R. Obesity and fracture risk. Clin Cases Miner Bone Metab. 2014 Jan-Apr;11(1):9–14. doi: 10.11138/ccmbm/2014.11.1.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao JJ. Effects of obesity on bone metabolism. J Orthop Surg Res. 2011;6:30. doi: 10.1186/1749-799X-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivers RQ, Cumming RG, Mitchell P, Peduto AJ. Diabetes and Risk of Fracture: the Blue Mountains Eye Study. Diabetes Care. 2001;24(7):1198–203. doi: 10.2337/diacare.24.7.1198. [DOI] [PubMed] [Google Scholar]
- 9.Oei L, Rivadeneira F, Zillikens MC, Oei EHG. Diabetes, Diabetic Complications, and Fracture Risk. Curr Osteoporos Rep. 2015;13(2):106–15. doi: 10.1007/s11914-015-0260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oei L, Zillikens MC, Dehghan A, Buitendijk GHS, Castaño-Betancourt MC, Estrada K, et al. High Bone Mineral Density and Fracture Risk in Type 2 Diabetes as Skeletal Complications of Inadequate Glucose Control: the Rotterdam Study. Diabetes Care. 2013;36(6):1619–28. doi: 10.2337/dc12-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012 Jun 14;486(7402):222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips ML. Gut Reaction: Environmental Effects on the Human Microbiota. Environ Health Perspect. 2009;117(5):A198–A205. doi: 10.1289/ehp.117-a198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15(7):R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 15.Rescigno M. Intestinal microbiota and its effects on the immune system. Cell Microbiol. 2014 Jul;16(7):1004–13. doi: 10.1111/cmi.12301. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12(2):139–52. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009 May;9(5):313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ericsson AC, Franklin CL. Manipulating the Gut Microbiota: Methods and Challenges. ILAR Journal. 2015 Aug 31;56(2):205–17. doi: 10.1093/ilar/ilv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundberg R, Toft MF, August B, Hansen AK, Hansen CHF. Antibiotic-treated versus germ-free rodents for microbiota transplantation studies. Gut Microbes. 2016;7(1):68–74. doi: 10.1080/19490976.2015.1127463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 2016 Feb 19;351(6275):854–7. doi: 10.1126/science.aad8588. [DOI] [PubMed] [Google Scholar]
- 21.Sjögren K, Engdahl C, Henning P, Lerner UH, Tremaroli V, Lagerquist MK, et al. The gut microbiota regulates bone mass in mice. J Bone Miner Res. 2012;27(6):1357–67. doi: 10.1002/jbmr.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan J, Herzog JW, Tsang K, Brennan CA, Bower MA, Garrett WS, et al. Gut microbiota induce IGF-1 and promote bone formation and growth. PNAS. 2016 Nov 22;113(47):E7554–E63. doi: 10.1073/pnas.1607235113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012 Aug 30;488(7413):621–6. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158(4):705–21. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun Article. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410(6832):1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 27.Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14(5):571–81. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328(5975):228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jepsen KJ, Silva MJ, Vashishth D, Guo XE, van der Meulen MC. Establishing biomechanical mechanisms in mouse models: practical guidelines for systematically evaluating phenotypic changes in the diaphyses of long bones. J Bone Miner Res. 2015 Jun;30(6):951–66. doi: 10.1002/jbmr.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, Sitaraman SV, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007 Dec;117(12):3909–21. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laukens D, Brinkman BM, Raes J, De Vos M, Vandenabeele P. Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol Rev. 2016;40(1):117–32. doi: 10.1093/femsre/fuv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacGregor RR, Graziani AL. Oral Administration of Antibiotics: A Rational Alternative to the Parenteral Route. Clin Infect Dis. 1997 Mar 1;24(3):457–67. doi: 10.1093/clinids/24.3.457. [DOI] [PubMed] [Google Scholar]
- 33.Doube M, Klosowski MM, Arganda-Carreras I, Cordelieres FP, Dougherty RP, Jackson JS, et al. BoneJ: Free and extensible bone image analysis in ImageJ. Bone. 2010 Dec;47(6):1076–9. doi: 10.1016/j.bone.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner CH, Burr DB. Basic biomechanical measurements of bone: A tutorial. Bone. 1993 Jul 01;14(4):595–608. doi: 10.1016/8756-3282(93)90081-k. [DOI] [PubMed] [Google Scholar]
- 35.Lima SF, Teixeira AGV, Higgins CH, Lima FS, Bicalho RC. The upper respiratory tract microbiome and its potential role in bovine respiratory disease and otitis media. Sci Rep Article. 2016;6:29050. doi: 10.1038/srep29050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nonnenmacher C, Dalpke A, Mutters R, Heeg K. Quantitative detection of periodontopathogens by real-time PCR. J Microbiol Methods. 2004 Oct;59(1):117–25. doi: 10.1016/j.mimet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Boutin S, Graeber SY, Weitnauer M, Panitz J, Stahl M, Clausznitzer D, et al. Comparison of microbiomes from different niches of upper and lower airways in children and adolescents with cystic fibrosis. PLoS One. 2015;10(1):e0116029. doi: 10.1371/journal.pone.0116029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010 May;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013 Jan;10(1):57–9. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012 Mar;6(3):610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010 Oct;26(19):2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 43.Purwada A, Jaiswal MK, Ahn H, Nojima T, Kitamura D, Gaharwar AK, et al. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials. 2015;63:24–34. doi: 10.1016/j.biomaterials.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purwada A, Tian YF, Huang W, Rohrbach KM, Deol S, August A, et al. Self-Assembly Protein Nanogels for Safer Cancer Immunotherapy. Adv Healthc Mater. 2016;5(12):1413–9. doi: 10.1002/adhm.201501062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2014 Jan-Feb;5(1):28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernandez C, Beaupre G, Keller T, et al. The influence of bone volume fraction and ash fraction on bone strength and modulus. Bone. 2001 Jul;29(1):74–8. doi: 10.1016/s8756-3282(01)00467-7. [DOI] [PubMed] [Google Scholar]
- 47.Jepsen KJ, Andarawis-Puri N. The amount of periosteal apposition required to maintain bone strength during aging depends on adult bone morphology and tissue-modulus degradation rate. J Bone Miner Res. 2012;27(9):1916–26. doi: 10.1002/jbmr.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ionova-Martin SS, Wade JM, Tang S, Shahnazari M, Ager JW, 3rd, Lane NE, et al. Changes in cortical bone response to high-fat diet from adolescence to adulthood in mice. Osteoporos Int. 2011 Aug;22(8):2283–93. doi: 10.1007/s00198-010-1432-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012 Sep 13;489(7415):220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulfer A, Blaser MJ. Risks of Antibiotic Exposures Early in Life on the Developing Microbiome. PLoS Pathog. 2015 Jul;11(7):e1004903. doi: 10.1371/journal.ppat.1004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung H, Pamp Sünje J, Hill Jonathan A, Surana Neeraj K, Edelman Sanna M, Troy Erin B, et al. Gut Immune Maturation Depends on Colonization with a Host-Specific Microbiota. Cell. 149(7):1578–93. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015 Sep;33(9):496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 53.Hernandez CJ, Guss JD, Luna M, Goldring SR. Links Between the Microbiome and Bone. J Bone Miner Res. 2016 Sep;31(9):1638–46. doi: 10.1002/jbmr.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D’Amico L, Roato I. Cross-talk between T cells and osteoclasts in bone resorption. Bonekey Rep. 2012;1:82. doi: 10.1038/bonekey.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawai T, Matsuyama T, Hosokawa Y, Makihira S, Seki M, Karimbux NY, et al. B and T lymphocytes are the primary sources of RANKL in the bone resorptive lesion of periodontal disease. Am J Pathol. 2006 Sep;169(3):987–98. doi: 10.2353/ajpath.2006.060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manilay JO, Zouali M. Tight relationships between B lymphocytes and the skeletal system. Trends Mol Med. 2014;20(7):405–12. doi: 10.1016/j.molmed.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 57.Weitzmann MN, Pacifici R. The role of T lymphocytes in bone metabolism. Immunol Rev. 2005 Dec;208:154–68. doi: 10.1111/j.0105-2896.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 58.Zhao B, Ivashkiv LB. Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis Res Ther. 2011;13(4):234. doi: 10.1186/ar3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Onis M, Garza C, Victora CG. The WHO Multicentre Growth Reference Study: strategy for developing a new international growth reference. Forum Nutr. 2003;56:238–40. [PubMed] [Google Scholar]
- 60.Viguet-Carrin S, Hoppler M, Membrez Scalfo F, Vuichoud J, Vigo M, Offord EA, et al. Peak bone strength is influenced by calcium intake in growing rats. Bone. 2014 Nov;68:85–91. doi: 10.1016/j.bone.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 61.Donnelly E, Chen DX, Boskey AL, Baker SP, van der Meulen MC. Contribution of mineral to bone structural behavior and tissue mechanical properties. Calcif Tissue Int. 2010 Nov;87(5):450–60. doi: 10.1007/s00223-010-9404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Sullivan M, O’Morain C. Nutrition in inflammatory bowel disease. Best Pract Res Clin Gastroenterol. 2006;20(3):561–73. doi: 10.1016/j.bpg.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Bianchi ML. Inflammatory bowel diseases, celiac disease, and bone. Arch Biochem Biophys. 2010 Nov 1;503(1):54–65. doi: 10.1016/j.abb.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 64.Hamdani G, Gabet Y, Rachmilewitz D, Karmeli F, Bab I, Dresner-Pollak R. Dextran sodium sulfate-induced colitis causes rapid bone loss in mice. Bone. 2008 Nov;43(5):945–50. doi: 10.1016/j.bone.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 65.Irwin R, Lee T, Young VB, Parameswaran N, McCabe LR. Colitis-induced bone loss is gender dependent and associated with increased inflammation. Inflamm Bowel Dis. 2013 Jul;19(8):1586–97. doi: 10.1097/MIB.0b013e318289e17b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Potgieter M, Bester J, Kell DB, Pretorius E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol Rev. 2015 Jul;39(4):567–91. doi: 10.1093/femsre/fuv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanger TJ, Norgard EA, Pletscher LS, Bevilacqua M, Brooks VR, Sandell LJ, et al. Developmental and genetic origins of murine long bone length variation. J Exp Zool B Mol Dev Evol. 2011 Mar 15;316B(2):146–61. doi: 10.1002/jez.b.21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Price C, Herman BC, Lufkin T, Goldman HM, Jepsen KJ. Genetic variation in bone growth patterns defines adult mouse bone fragility. J Bone Miner Res. 2005 Nov;20(11):1983–91. doi: 10.1359/JBMR.050707. [DOI] [PubMed] [Google Scholar]
- 69.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351(6275) doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wallach S, Feinblatt JD, Carstens JH, Jr, Avioli LV. The bone “quality” problem. Calcif Tissue Int. 1992 Sep;51(3):169–72. doi: 10.1007/BF00334542. [DOI] [PubMed] [Google Scholar]
- 71.Hernandez CJ, Keaveny TM. A biomechanical perspective on bone quality. Bone. 2006;39(6):1173–81. doi: 10.1016/j.bone.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.