Abstract
UV radiation exposure from sunlight and artificial tanning beds is the major risk factor for the development of skin cancer and skin photoaging. UV-induced skin damage can trigger a cascade of DNA damage response signaling pathways, including cell cycle arrest, DNA repair, and, if damage is irreparable, apoptosis. Compensatory proliferation replaces the apoptotic cells to maintain skin barrier integrity. Disruption of these processes can be exploited to promote carcinogenesis by allowing the survival and proliferation of damaged cells. UV radiation also induces autophagy, a catabolic process that clears unwanted or damaged proteins, lipids, and organelles. The mechanisms by which autophagy is activated following UV exposure, and the functions of autophagy in UV response are only now being clarified. Here, we summarize the current understanding of the mechanisms governing autophagy regulation by UV, the roles of autophagy in regulating cellular response to UV-induced photodamage, and the implications of autophagy modulation in the treatment and prevention of photoaging and skin cancer.
Keywords: Autophagy, ultraviolet radiation, UV, skin, cancer, aging
Introduction
Exposure to ultraviolet (UV) radiation through sunlight and indoor tanning beds is the major risk factor for skin cancer development and skin photoaging. UV radiation is divided into three major types by wavelength: UVA (315-400 nm), UVB (280-315 nm), and UVC (100-280 nm). Of these, UVA is the most abundant, as it accounts for about 95% of solar UV radiation, and indoor tanning beds emit UVA at doses up 12-fold higher than the sun1. UVB accounts for the remaining 5% of solar UV radiation, while UVC is filtered out by the ozone2.
Absorption of UV radiation, and consequently the extent of skin damage, is dependent on a number of factors, including skin color/type, time of exposure, latitude, altitude, season, and wavelength3. UVA penetrates deep into the dermis4, while UVB reaches only the epidermis5.However, UVA is much less efficient in causing direct DNA damage than UVB, instead inducing oxidative DNA damage through reactive oxygen species (ROS)6. Both UVA and UVB have been shown to cause cell proliferation, growth arrest, and apoptosis, although these responses can be context-dependent. These effects of UVA and UVB on skin will be reviewed here. UVC, although it is the most mutagenic type of UV radiation7, will not be discussed here, as it is not a major relevant source of skin-damaging radiation.
Macroautophagy (hereafter autophagy) is an essential, homeostatic cellular process of “self-eating.” Through this process, cells clear unwanted or damaged proteins, lipids, and other cellular components, and in doing so regulate the availability of a number of cell signaling factors. Furthermore, autophagy-mediated recycling of cytoplasmic contents facilitates cell survival and adaptation during starvation, genotoxic stress, and oxidative stress in normal cells8. Autophagy can also provide nutrients to sustain high rates of proliferation in times of growth. Dysregulation of autophagy can therefore contribute to the development of a number of skin diseases, including skin cancer. UVA, UVB, and UVC have all been reported to induce autophagosome formation and upregulation of autophagy markers9–11. However, given the varying effects of UV radiation, it is likely autophagy plays a number of context-dependent roles in UV response. Here, we will examine our current understanding of the regulation and function of autophagy in photodamage response, as well as the implications for UV-induced skincancer and aging.
Mechanisms of Autophagy
Autophagy Induction
Genotoxic stress can be induced by a number of pharmacological and environmental factors, including UV radiation. Genotoxic stress induces autophagy to mitigate the effects of DNA damage12. Defects in autophagy are associated with increased DNA damage, gene amplification, and aneuploidy13. These effects are likely due to insufficient metabolic precursors in the absence of autophagy13. Genotoxic stress regulates autophagy at least in part through the stabilization of p53 (Figure 1, 2 and 3), which regulates the transcription of various autophagy and lysosomal genes14.
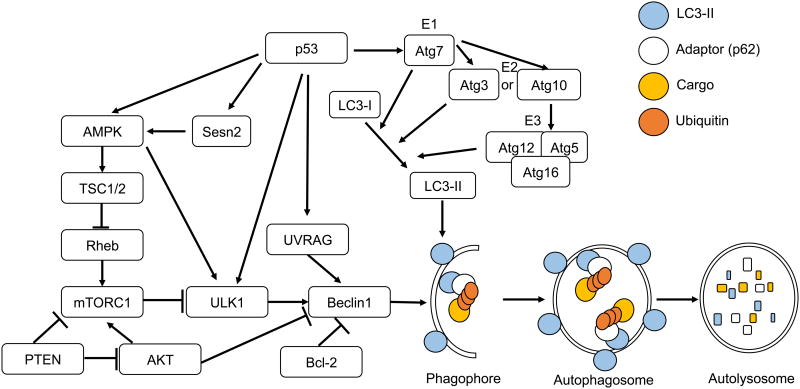
Figure 1. Mechanisms of Autophagy.
Autophagy is initiated upon activation of the AMPK signaling pathway. AMPK can be activated by p53 and Sesn2 in response to cellular stress. AMPK then activates TSC1/2 to inhibit mTORC1. mTORC1 is further negatively regulated by PTEN to activate autophagy. ULK1 is activated by inhibition of mTORC1, direct phosphorylation by AMPK, or p53-mediated transcriptional upregulation. ULK1 then forms a complex which activates Beclin1. Beclin1, which binds to Bcl-2 in normal conditions, dissociates from Bcl-2 and binds to UVRAG. UVRAG, a p53 target gene, promotes the formation of the Beclin1 complex to initiate phagophore nucleation. Two ubiquitin-like conjugation systems facilitate the lipidation of LC3-I to form LC3-II, and elongation of the burgeoning phagophore. E1-like activating enzyme Atg7 leads to the activation of E2-like enzyme Atg3 (System 1) or Atg10 (System 2), subsequently the activation of the E3-link enzyme complex Atg5-Atg12 conjugate with Atg16, and subsequently the generation of LC3-II. LC3-II binds to an adaptor protein or autophagy receptor, such as p62, which in turn binds to cargo with modifications such as polyubiquitination. The autophagosome encloses the cargo and subsequently fuses with a lysosome to form an autolysosome. Adaptor and cargo are degraded at the autolysosome and the resulting molecules are recycled to the cytoplasm.
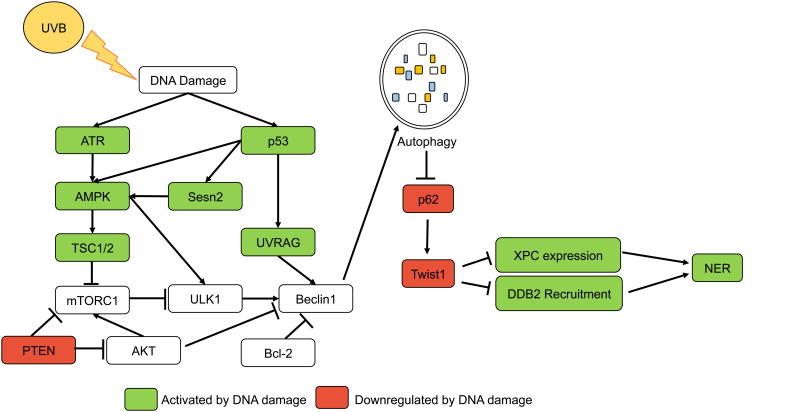
Figure 2. Autophagy in UVB response.
UVB radiation causes direct DNA damage in the form of photoproducts, and autophagy is induced by UVB to promote photoproduct repair. UVB directly induces autophagy activators AMPK, UVRAG and p53. p53, upon stabilization by UVB, activates transcription of AMPK, Sesn2, TSC2, and UVRAG to activate autophagy. ATR is also induced by UVB-induced DNA damage and can activate AMPK signaling. Sesn2 interacts with AMPK, TSC1, and TSC2 to activate autophagy in response to genotoxic stress. Activation of autophagy by these factors leads to the degradation of p62 and thus the decrease in Twist1 stability, which is required for XPC upregulation following UVB, and recruitment of DDB2 to sites of UVB-induced DNA damage. Conversely, autophagy activator PTEN is inhibited in response to UVB and this impairs NER by downregulating XPC.
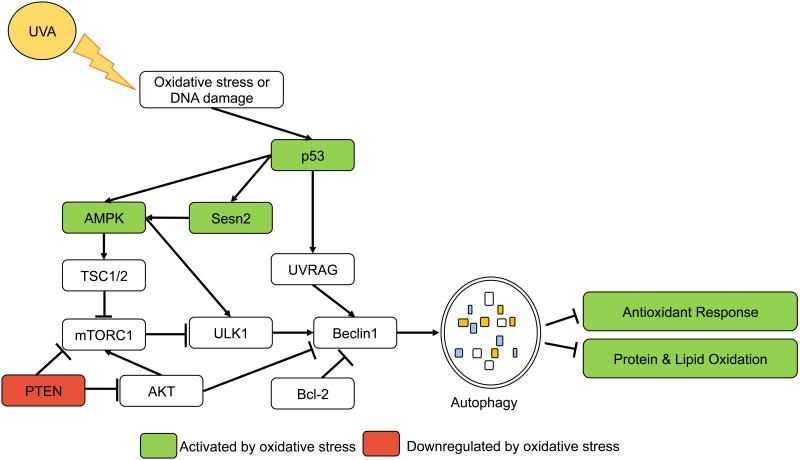
Figure 3. Autophagy in UVA response.
UVA-induced ROS production stimulates autophagy, and autophagy regulates the oxidative stress response following UVA. UVA stabilizes p53, which induces transcription of autophagy activators AMPK and Sesn2. Conversely, UVA suppresses PTEN expression, which may negatively impact autophagy induction following UVA. Autophagy clears oxidized proteins and lipids following UVA exposure, but suppresses Nrf2-mediated antioxidant response following UVA.
Oxidative stress induced by the formation of reactive oxygen species (ROS) similarly induces autophagy15 (Figure 3). ROS are highly reactive molecules, including radicals (singlet oxygen) and non-radicals such as hydrogen peroxide (H2O2). Low basal levels of ROS act as signaling molecules in healthy cells, but elevated ROS levels cause oxidative damage to proteins, lipids, and DNA, as is seen in the response to UVA16. Autophagy provides protection against oxidative stress by clearing the damaged proteins, lipids, and DNA, and restoring metabolic homeostasis17. The antioxidant response triggered by nuclear factor (erythroid-derived 2)-like 2 (Nrf2) is linked to autophagy through a feedback loop with adaptor protein p62. p62 competes with Nrf2 to bind to Kelch-like ECH-associated protein 1 (KEAP1)18,19, preventing KEAP1-mediated degradation of Nrf2. Increasing p62 levels, therefore, leads to the stabilization of Nrf2, and the degradation of KEAP1 by autophagy20. Nrf2 in turn binds to the antioxidant response element (ARE) within the p62 promoter to activate p62 transcription18.
Another autophagy regulator is the microphthalmia-associated transcription factor (MITF) family member transcription factor EB (TFEB)21–23. TFEB is a basic helix-loop-helix leucine zipper transcription factor, which binds to the Coordinated Lysosomal Expression and Regulation (CLEAR) binding site found in the promoter of many autophagy and lysosomal genes23. TFEB overexpression enhances transcription of an autophagy and lysosomal gene program21, as well as the degradation of autophagy substrates, mitochondria, and lipid droplets24. TFEB has been reported to activate the transcription of an autophagy program in response to starvation21.
TFEB activation is thought to be regulated primarily through phosphorylation. Under nutrient-rich conditions, TFEB is primarily cytosolic and inactive25. Upon nutrient deprivation, TFEB rapidly translocates to the nucleus and is activated to induce transcription of autophagy genes25. Phosphorylation at two sites, Ser211 and Ser142, determine the localization and activity of TFEB26. Ser211, when phosphorylated, is a docking site for chaperone 14-3-3, and this interaction retains TFEB in the cytoplasm25,27. Furthermore, to maintain its cytosolic localization and inactivation, TFEB is phosphorylated at Ser211 by mammalian target of rapamycin complex 1 (mTORC1), a known negative regulator of autophagy 27.
mTORC1 is a key regulator of cell growth, proliferation, protein synthesis, and autophagy28 (Figure 1). mTORC1 consists of the core components mLST8, serine/threonine kinase mTOR, and adaptor protein Raptor. mTORC1 is regulated in response to cellular stress by 5′-AMP-activated protein kinase (AMPK)28 (Figure 1). AMPK and its activator LKB1 sense reductions in cellular ATP and induce autophagy to replenish ATP stores. AMPK activation leads to the phosphorylation and activation of tuberous sclerosis complexes 1 and 2 (TSC1/2). TSC2 is a GTPase-activating protein which acts on small G-protein Rheb to inhibit mTORC1, and mTORC1 inhibition by TSC2 induces autophagy28 (Figure 1).
mTORC1 is also regulated by the phosphatidylinositol-3 kinase (PI3K)-protein kinase B (AKT) pathway, and its negative regulator phosphatase and tensin homolog (PTEN). PI3K phosphorylates PIP2 to form signaling intermediate PIP3, while PTEN dephosphorylates PIP3. PIP3 activates AKT, which in turn activates mTOR and phosphorylates Beclin1 to inhibit autophagy29,30. PTEN has been shown to negatively regulate mTOR to induce autophagy in a variety of cell types31,32, and is activated by ATM in response to DNA damage to induce autophagy33.
Autophagy Initiation
The mTORC1/AMPK pathway regulates the initiation of autophagy (Figure 1). In nutrient rich conditions, mTORC1 binds, phosphorylates, and inactivates Unc-51-like kinase 1 (ULK1) and Atg1334. AMPK can also bind and phosphorylate ULK135, blocking inhibition by mTOR and activating ULK136. ULK1 activation by AMPK or by mTOR inhibition allows ULK1 to phosphorylate Atg13 and focal adhesion kinase family-interacting protein of 200 kD (FIP200)34. ULK1, Atg13, and FIP200 form a scaffold (called the ULK1 complex)34, which localizes to the burgeoning phagophore and promotes the recruitment of other proteins essential for autophagy.
The ULK1 complex activates essential autophagy gene Beclin137,38, via ULK1-mediated phosphorylation39 (Figure 1). Beclin1, which is bound to antiapoptotic protein Bcl-2 in normal conditions, dissociates40 and binds to UVRAG38. UVRAG promotes the formation of the Beclin1 complex, consisting of core components Beclin1, Vps34, and p15041. The Beclin1 complex can interact with other proteins to induce phagophore nucleation in response to a variety of stressors42.
Elongation
Upon initiation of phagophore nucleation by Beclin1 complex formation, two sequential ubiquitin-like conjugation systems are induced to facilitate phagophore elongation43 (Figure 1). In the first, ubiquitin-like Atg12 is activated by the E1-like activating enzyme Atg7. Atg7 transfers Atg12 to the E2 conjugating enzyme Atg10, and Atg12 is irreversibly attached to Atg5. The Atg12-Atg5 conjugate binds to Atg16 and attaches to the phagophore43. In the second system, LC3 is then processed by cysteine protease Atg4 to allow activation by Atg7. Atg7 transfers the activated LC3 (LC3-I) to E2 enzyme Atg3. Finally, a phosphatidylethanolamine (PE) lipid is attached to LC3-I by Atg12-Atg5 conjugate to form LC3-II. LC3-II is linked to the phagophore membrane by the attached PE. The amount of LC3-II ultimately controls the size of the autophagosome, and the amount of cargo included for degradation43.
Degradation
LC3-II facilitates selective autophagy of long-lived proteins, protein aggregates, or damaged organelles by forming interactions with adaptor proteins (Figure 1). Selective autophagy adaptors, such as p62/SQSTM1, bind to cargo for degradation, and to LC3-II on the autophagosome membrane through the LC3-interacting region (LIR)44. Upon autophagosome-lysosome fusion, both the adaptor and cargo are degraded. The products of autophagy are recycled to the cytosol to maintain essential cellular processes after starvation or stress.
Autophagy in UV-Induced DNA Damage Response
UV-Induced DNA Photodamage
UVB radiation is efficiently absorbed by DNA within the epidermis5 and damages DNA directly to form photoproducts. The most common UVB-induced photoproducts are cyclopyrimidine dimers (CPDs) and, to a lesser extent, pyrimidine-(6-4)-pyrimidone photoproducts (6-4PPs)45. Of these, 6-4PPs are bulkier, but more efficiently repaired than CPDs46–48. As a result of this ineffective repair as well as the abundance of damage, CPDs are responsible for most (∼80%) UVB-induced mutations49,50.
The ability of UVA to induce photoproduct formation, and the extent to which this contributes to skin damage, remains controversial. UVA causes the formation of far fewer photoproducts than UVB51,52, and was previously thought to be harmless. However, multiple studies have reported UVA-induced CPD formation, and in many cases, CPDs were the predominant lesion formed by UVA51,53–56. UVA does not cause significant formation of 6-4PPs51,53,54,57. UVA-induced CPDs are predominantly T-T dimers58, and these dimers persist longer than UVB-induced CPDs51. The abundance of UVA-induced CPDs, and ineffective repair of these lesions may therefore form a significant source of UVA-induced skin damage56.
UV radiation-induced DNA photodamage activates DNA damage repair, cell cycle arrest, and apoptotic pathways. Cell cycle arrest allows time for recognition and repair of photoproducts, but if DNA damage is extensive and irreparable, apoptosis is initiated59. In surviving cells, proliferation is induced to replace dying cells and maintain tissue homeostasis60. Disrupting signaling through these pathways can lead to cancerous expansion of damaged cells.
One of the key signaling mediators is p53, a transcription factor that plays a key role in balancing the pathways activated in response to DNA photodamage (Figure 2). UV-induced photoproduct formation leads to the stabilization of p5361. p53 then activates transcription of a complex program of cell cycle inhibitors, DNA damage response genes62,63, and apoptotic genes64,65. p53 induces the transcription of cell cycle inhibitor p21 to arrest the cell cycle at the G1 phase following UV exposure66, and p53-mediated DNA damage response gene induction is required for the repair of UV-induced DNA damage62. The decision to induce pro-apoptotic signals by p53 is context dependent, and can be impacted by p53 expression prior to UV exposure67, skin cell layer68,69, length of time since UV exposure67, and cell type64.
DNA Damage Repair
UV-induced photoproducts are repaired by either transcription-coupled (TC-) or global genome (GG-) nucleotide excision repair (NER)70–73. TC-NER allows the rapid recognition and repair of damage in transcriptionally active genes, while GG-NER repairs damage throughout the genome independent of gene transcription. TC-NER begins when RNA polymerase stalls at distorted DNA. Initiation of GG-NER requires recognition of distorted DNA by xeroderma pigmentosum complementation group C (XPC) or damage specific DNA binding protein 2 (DDB2, also called XPE)70.Transcription of XPC and DDB2/XPE is induced by p53 following UVB irradiation62.
Upon recognition of DNA damage, both NER subpathways converge on a common repair pathway70. Transcription factor II H (TFIIH), a dimer of helicases XPB and XPD, is recruited to the damage site and unwinds DNA around the damage site. XPA and replication protein A (RPA) bind to the damaged strand and undamaged strand, respectively. This binding allows the recruitment of endonucleases XPG and XPF-ERCC1. XPG and XPF-ERCC1 then excise the damaged DNA to create a single strand of DNA complementary to the damage site. Replication factor C (RFC) loads proliferating cell nuclear antigen (PCNA) onto the DNA strand and a DNA polymerase synthesizes a strand complementary to the damage site. Finally, DNA ligase seals the nicks to complete NER70.
Cell cycle arrest following UV exposure is critical to provide ample time for DNA damage repair, and to prevent proliferation of damaged cells. UV-induced DNA damage activates the sensors ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) to trigger cell cycle arrest via p53 stabilization65 (Figure 2). ATR recognizes single strand DNA breaks caused by UV, including RPA-bound ssDNA formed during NER. ATR then phosphorylates checkpoint kinase 1 (Chk1) to activate checkpoints at the G1, S, and G2/M phases74. ATM, which recognizes double strand breaks (DSBs), phosphorylates Chk2 to delay the cell cycle75. XPC and DDB2 have been shown to facilitate the recruitment of ATM and ATR to sites of DNA damage and promote the activation of cell cycle arrest pathways76.
Regulation of autophagy by UV-induced DNA damage response
UVB exposure directly and rapidly activates AMPK9, UVRAG77, and p5378 to activate autophagy (Figure 2). Upon activation by UV, p53 induces transcription of autophagy activators AMPK, TSC2, Sestrin 1, and Sestrin 279–81. Sestrin 1 and 2 have been shown to interact with AMPK, TSC1, and TSC2 to inhibit mTOR signaling in response to genotoxic stress80.
Regulation of UV-induced DNA damage response by autophagy
Genotoxic stress is a trigger for autophagy, and repair of UV-induced DNA damage is regulated by autophagy. Knockdown of key autophagy genes AMPK, Atg5, Atg7, Atg12, and Atg14 impairs the repair of UVB-induced DNA damage82,83. Recent work has identified multiple pathways through which autophagy regulates UV-induced DNA damage repair.
First, we have shown that autophagy positively regulates the recognition of DNA damage by XPC and DDB2. Knockdown of essential autophagy gene Atg5 decreases XPC transcription following UVB radiation by a Twist1-dependent mechanism83. Autophagy deficiency leads to the accumulation of Twist1, which in turn activates transcriptional repressor complex E2F4-RBL2 through AKT signaling. The E2F4-RBL2 complex represses XPC transcription in autophagy-defective cells, leading to an accumulation of DNA damage83.
Recognition of UV-induced DNA damage by DDB2 is also dependent on autophagy. Autophagy deficiency impairs DDB2 recruitment to CPD sites following UVB exposure83 (Figure 2). This defect was found to be caused by Twist1 binding to and inhibiting p300, a key factor in DDB2 recruitment83. We have previously shown that Twist1 is stabilized by p62 in autophagy-deficient conditions84, and autophagy-mediated degradation of Twist1 can facilitate DDB2 recruitment and CPD repair83. Collectively, this work suggests a tumor suppressive role for autophagy in the promotion of DNA damage repair.
UVB rapidly induces autophagy through activation of AMPK to regulate DNA damage repair9,82 (Figure 2). AMPK activation by UVB increases XPC protein levels and increases CPD repair. Knockdown of AMPK reduces CPD repair following UVB, but has no effect on 6-4PP repair82. This work further links autophagy to the positive regulation of UVB-induced DNA damage repair.
Autophagy activator UVRAG was initially identified in a genetic screen as a protein able to partially complement the UV sensitivity of xeroderma pigmentosum (XP) cells defective in GG-NER85. Until recently, however the function of UVRAG in DNA damage response was unknown. UVRAG has recently been shown to be essential for both autophagy and GG-NER in response to UV86. UVRAG was found to facilitate the recruitment of DDB1 and DDB2 to sites of UV-induced DNA damage by binding DDB1. Knockdown of UVRAG inhibits transfer of damaged DNA from DDB1 to XPC during NER initiation. However, UVRAG activates autophagy and GG-NER independently86, suggesting UVRAG may act as a signaling hub for concurrent activation of DNA damage repair and autophagy (Figure 2).
Conversely, autophagy activator PTEN is inhibited by Sestrin 287 and in response to UVB88 (Figure 2). PTEN downregulation impairs GG-NER by downregulating XPC transcription89. It is unclear, however, whether downregulation of autophagy has a significant role in PTEN-regulated DNA damage repair in response to UVB.
Autophagy in Oxidative Stress Response
UV-Induced Oxidative Damage
The most established effect of UVA is causing oxidative damage to DNA, proteins, and lipids (Figure 3). UVA is absorbed by cellular photosensitizers, which transfer energy in either type I or type II photosensitization reactions90. In type I reactions, energy is transferred directly to DNA through free radical formation, resulting in oxidative modifications. In type II reactions, photosensitizers transfer energy to molecular oxygen, creating singlet oxygen91. Singlet oxygen reacts preferentially with guanine and can cause a number of oxidative DNA modifications. Of these, 8-oxo-2′-deoxyguanosine (8-oxo-dG) is the most common oxidative lesion caused by UVA55. 8-oxo-dG can be repaired through base excision repair by 8-oxo-guanine glycosylase (OGG1)-mediated excision92. However, defective repair of 8-oxo-dG lesions following UVA exposure can cause GC→TA transversion55. UVB can also cause ROS production and oxidative damage93, although this effect is likely secondary to the direct DNA damage caused by UVB.
UVA-induced oxidative stress damages lipids and proteins in addition to DNA. UVA induces the oxidation of phospholipids in keratinocytes10,94, and these oxidized phospholipids form adducts with proteins95,96. Accumulation of oxidized phospholipid-protein adducts is found in a number of degenerative diseases and also in photoaged skin93.
Accumulation of proteins with oxidative modifications is also seen in the dermis after UVA exposure93, likely due to reduced expression of antioxidant enzymes93,97. Recently, UVA has been shown to target OGG1 for oxidation98, and in doing so, compromises BER of oxidized DNA98. Furthermore, UVA has been shown to cause oxidative damage to single-strand DNA binding factor RPA, impairing NER16,99. This work suggests there are combinatorial effects of UVA and UVB in inducing and perpetuating DNA damage.
ROS generation in response to UVA triggers the antioxidant response pathway, beginning with the stabilization of antioxidant response factor Nrf2100. ROS formation triggers the dissociation of Nrf2 from KEAP1, stabilizing Nrf2 and allowing its nuclear translocation101. Nuclear Nrf2 activates an antioxidant response program through binding to antioxidant response elements in gene promoters101. Nrf2 also activates the transcription of genes involved in DNA damage response, including 8-oxo-dG-excising enzyme OGG1102,103.
Regulation of Autophagy by UV-Induced Oxidative Stress
The mechanisms by which UV-induced oxidative stress regulates autophagy have only recently been clarified. UVA induces autophagy10 (Figure 3), and treatment with the singlet oxygen quencher NaN3 impairs the induction of autophagy by UVA10. Similarly, antioxidants block the induction of autophagy following UVB exposure104. Collectively, this work suggests that UV-induced ROS production leads to the activation of autophagy.
UVA exposure leads to an increased number of oxidized phospholipids, oxysterols, and cholesterols in keratinocytes10,105. One oxidized lipid formed by UVA exposure, 25-hydroxycholesterol (25-OH), is sufficient to induce autophagy in keratinocytes105. This further implicates UV-induced oxidative damage in the activation of autophagy (Figure 3).
UVA has been shown to regulate the transcription of a number of genes involved in autophagy (Figure 3). UVA induces transcription of autophagy adaptor protein p62 in skin fibroblasts106, as well as autophagy activators p53107,108 and Sestrin281. As described above, p53 and Sestrin 2 can induce autophagy through AMPK signaling. In contrast, UVA suppresses expression of the autophagy activator PTEN109. PTEN activates autophagy through the PI3K pathway; however, it remains to be seen whether UVA-mediated PTEN downregulation impairs autophagy in UVA response.
Regulation of UV-Induced Oxidative Stress Response by Autophagy
The function of autophagy is critical for oxidative stress response110. However, little work has been done to establish the role of autophagy in the response to UVA-induced oxidative stress. UVA-induced oxidative stress triggers autophagy to clear oxidized lipids and proteins10 (Figure 3). Autophagy deficiency leads to the accumulation of oxidized phospholipids and protein aggregates following UVA exposure10. Furthermore, autophagy deficiency increases the Nrf2-dependent antioxidant response in keratinocytes, even prior to UVA exposure10. We have found that in melanocytes the autophagy activator Sestrin 2 reduces Nrf2 levels upon induction by UVA, and increases UVA-induced ROS production81. Autophagy induction also serves to inhibit Nrf2 stabilization by degrading p6210. This work indicates that autophagy plays a complex role in UVA-induced oxidative stress response, by clearing oxidized proteins and lipids, while minimizing antioxidant response in different cell types.
Autophagy in UV-Induced Cell Proliferation and Apoptosis
UV radiation exposure can induce apoptosis and proliferation60, often within the same sun-damaged tissue111,112. In normal skin, compensatory hyperproliferation is induced to replace the cells cleared by apoptosis and maintain homeostasis. The mechanisms governing UV-induced proliferation and apoptosis are tightly regulated to prevent cancerous expansion of cells damaged by UV.
UVA and UVB induce apoptosis of epidermal cells by increasing p53 and Bax expression, while decreasing expression of Bcl-2112,113. Furthermore, UVA and UVB can induce apoptosis through p38 activation9,114,115. UVA has been reported to downregulate PTEN and upregulate AKT signaling to protect against apoptosis116. Conversely, UVB-induced UV radiation resistance associated gene (UVRAG) expression suppresses apoptosis by sequestering Bax away from the mitochondria, where it induces apoptosis117. As UVRAG is a p53 target gene14, this would suggest that UVRAG serves as a possible negative feedback loop in the regulation of UV-induced apoptosis.
Another essential mediator of both UV-induced proliferation and apoptosis is cyclooxygenase-2 (COX-2). Both UVA and UVB upregulate COX-2118–120, an inducible prostaglandin synthase which catalyzes the rate-limiting step in prostaglandin E2 (PGE2) synthesis. PGE2 signals through autocrine and paracrine mechanisms to promote cell proliferation121 and suppress apoptosis122. COX-2 is induced in the skin of hairless mice following UV radiation to increase proliferation123, and induction of COX-2 protects against UVB-induced apoptosis119,123,124.
In addition, the mTOR signaling pathway regulates cell growth and proliferation in response to stress. Sestrin 2, a stress-inducible protein that is activated following UVB exposure87, inhibits cell proliferation through the negative regulation of mTOR signaling80. Similarly, AMPK, an inhibitor of mTOR signaling, suppresses cell proliferation in response to UVB82.
UV exposure induces apoptosis and cell proliferation to maintain tissue homeostasis. Research suggests that autophagy plays key roles in the regulation of apoptosis and proliferation, but the contributions of autophagy are likely context-dependent. Knockdown of the autophagy activators AMPK9,81, Sestrin 287, Beclin1125, Atg59,125, and UVRAG126 sensitizes skin and skin cancer cells to UVB-induced apoptosis, suggesting a protective role of autophagy. However, other research suggests that autophagy or AMPK signaling promote UVB-induced apoptosis104,127.
The antiapoptotic Beclin1-binding protein Bcl-2 is downregulated following UVB exposure112, which may free Beclin1 to bind UVRAG and induce autophagy38. UVB-induced UVRAG expression suppresses apoptosis by sequestering Bax126, independent of its role in autophagy induction126. Furthermore, UVRAG is key to suppressing proliferation after UVB, independent of its function in autophagy activation38. As the domains of UVRAG required for activating autophagy, promoting proliferation, and suppressing apoptosis differ, it is possible that UVRAG can activate these processes simultaneously, therefore forming a critical hub in the response to UV radiation.
UVB activates the AMPK signaling pathway to induce autophagy9,82 and suppress apoptosis9,128. Autophagy induction by UVB limits p62 levels to prevent p62-mediated p38 activation and subsequent apoptosis9. UVB similarly induces autophagy activator Sestrin 2 to promote cell survival87. This work suggests that autophagy is activated by UVB to promote cell survival, and likely contributes to UVB-induced tumorigenesis.
Furthermore, we have previously shown that autophagy activator AMPK regulates expression of XPC, and consequently UVB-induced DNA damage repair82. AMPK-mediated DNA damage repair suppressed cell proliferation in response to UVB82, suggesting a key role for AMPK in the regulation of autophagy, proliferation, and apoptosis.
PTEN, which has been shown to negatively regulate autophagy, is involved in the regulation of cell proliferation and survival following UV exposure. Both UVA and UVB suppress PTEN expression to promote survival of keratinocytes109,116,129. Sestrin 2 promotes the suppression of PTEN and subsequent AKT activation to promote survival in response to UVB87. PTEN suppression by UVA was accompanied by upregulation of AKT signaling and an increase in proliferation130. PTEN is therefore central to the regulation of proliferation and apoptosis after UV, and has previously been linked to autophagy. It is unclear whether PTEN regulates UV-induced autophagy, however.
Autophagy in Photodamage-Induced Disease
UV and Skin Cancer
Approximately 90% of skin cancers are attributed to UV exposure131,132. Skin cancer is the most common form of cancer, with 3.5 million cases diagnosed each year in the United States alone133,134. Skin cancer causes more than 20,000 deaths in the US and treatment costs the US $8.1 billion annually135. Worldwide, incidence of skin cancer is rapidly rising each year 134,136, increasing the number of people for whom skin cancer will become a costly and potentially deadly disease.
Skin cancers are broadly classified into two major types by the cell type of origin: melanoma and non-melanoma skin cancer (NMSC). NMSC consists of two major types: squamous cell carcinoma (SCC) and basal cell carcinoma (BCC). BCC is the most common type of skin cancer, representing 80% of skin cancer cases137. BCC accounts for approximately 3,000 deaths each year in the US and can be disfiguring137. SCC represents accounts for approximately 16% of skin cancers137 and has potential to metastasize138. An estimated 9,000 Americans die each year of SCC138. Melanoma is the least common, but most aggressive form of skin cancer. Despite accounting for only 4% of skin cancer cases, melanoma is responsible for nearly half of all skin cancer deaths139.
UVA was initially believed to be non-tumorigenic due to its poor ability to cause direct DNA damage. This indirectly promoted the development of UVB-specific sunscreens140 and the use of high-powered UVA lamps in indoor tanning beds141. It has since been shown that UVA induces skin carcinogenesis in vivo142–144 and indoor tanning, even intermittently, significantly increases skin cancer risk145,146. ROS production is thought to be the primary mechanism by which UVA causes skin cancer. Melanocytes are particularly sensitive to UVA, as melanin can act as a photosensitizer for UVA and enhances ROS production following UVA exposure56,147–149. Consequently, UVA radiation is believed to play a major causal role in 67% of melanoma cases150.
UVB radiation has been thought to contribute mostly to NMSC formation by causing direct DNA photodamage. UVB signature TC→TT and CC→TT mutations are commonly found in the p53 gene of skin cancer patients151, suggesting the importance of p53-mediated DNA damage response in preventing skin carcinogenesis. Defects in UV-induced DNA damage repair greatly accelerate skin cancer development, as is seen in xeroderma pigmentosum (XP) patients. XP is an autosomal recessive disorder caused by defects in GG-NER, and XP patients have a 10,000-fold increased susceptibility to UV-induced NMSC152. XP patients develop NMSC 50 years younger than the general population.
Tumor Suppressive Role of Autophagy in Skin Cancer
Autophagy can be tumor suppressive or pro-tumorigenic, depending on context153. Autophagy acts as a tumor suppressor by promoting ROS clearance154, DNA repair13,83,155, and degrading oncogene p62156. Autophagy deficiency causes accumulation of targets bound for degradation, including p62106, promotes ROS generation15, and causes genomic instability12.
Degradation of adaptor protein p62 has been found to be an important tumor suppressive function of autophagy156. In autophagy-deficient conditions or upon transcriptional upregulation by UVA106, p62 accumulates and acts as a signaling hub by forming interactions with a number of pro-tumorigenic proteins. We have found that p62 binds and stabilizes Twist1, a transcription factor involved in epithelial-mesenchymal transition84. In doing so, p62 promotes proliferation and migration of skin cancer cells in vitro84. In a mouse model of SCC, the p62-Twist1 interaction promotes tumor growth and metastasis84.
p62 similarly activates NF-κB signaling in a feed-forward loop. The interaction between p62 and TRAF6157,158, as well as interactions with death domain serine/threonine kinase RIP159 and atypical protein kinases160, ultimately activates NF-κBsignaling. NF-κB signaling in turn upregulates p62 transcription161. NF-κB signaling is activated by p62 to promote Ras-mediated tumorigenesis158.
Oncogenic Role of Autophagy in Skin Cancer
Autophagy can facilitate tumor development by promoting cell survival in times of genotoxic, oxidative, or metabolic stress, and by providing the macromolecules necessary to sustain a high rate of proliferation153. We have proposed an oncogenic function for autophagy in the development of SCC, in which autophagy promotes survival of SCC cells with extensive DNA damage9. Future investigations will help understand the role of autophagy in the regulation of critical signaling pathways in tumorigenesis.
UV and Photoaging
Photoaging (also called extrinsic aging) is premature aging of the skin caused by environmental effects, primarily UV exposure. Photoaging differs from intrinsic (chronological) aging, which affects the skin in ways similar to other organs and can be superimposed on photoaging162. While intrinsically aged skin is thin and smooth, photoaging can cause a leathery thickening, sagging, and wrinkling of skin163,164. Histologically, photoaged skin is characterized by a loss of dermal collagen165, induction of matrix metalloproteinases (MMPs)166, and accumulation of elastin167.
Both UVA and UVB have been shown to cause photoaging by inducing ROS production and subsequent oxidative damage to DNA, lipids, and proteins167. UVA-induced alterations to skin lipid composition105 and phospholipid oxidation10 cause skin sagging characteristic of photoaging. Furthermore, UV exposure leads to the accumulation of oxidatively modified proteins and depletes antioxidant enzymes in photoaged skin93,168.
Autophagy in Photoaging
UVA-induced ROS generation leads to the oxidation of phospholipids, and subsequent formation of oxidized phospholiplid-protein adducts10. Autophagy induced by UVA degrades these adducts10 to prevent damage caused by aggregation of heavily oxidized protein adducts169. Aging-related decline in autophagic clearance170 leads to the accumulation of oxidized phospholipid-protein adducts171, and oxidized protein aggregates172. Adduct accumulation contributes to skin photoaging171.
Implications for Autophagy in UV-Induced Disease Prevention and Treatment
Autophagy modulators are of clinical interest for the treatment and prevention of skin cancer and many other diseases. However, the context-dependent and often opposing functions of autophagy make it difficult to predict response to autophagy modulators in the clinic. Here, we examine the current understanding of autophagy modulation in UV-induced skin cancer.
We have reported that AMPK is activated by UVB to induce autophagy9, promote DNA repair82, impair cell proliferation82, and inhibit apoptosis9,128 under stress conditions. AMPK activation is reduced in human skin cancer samples82, and therefore, targeting AMPK for activation in skin cancers could provide an opportunity to block tumor growth. Our data supports the use of AMPK activators metformin and AICAR to block the growth of UVB-induced skin tumors in vivo82.
Autophagy activator rapamycin reduces UV-induced skin tumor formation and progression83,173. Rapamycin treatment increases XPC levels through Twist1 downregulation83, and consequently reduces the number of UV-induced mutations in p53 in skin tumors173. Inhibition of autophagy by Spautin-1 increases Twist1, decreases XPC, and increases tumor growth induced by UVB83. Collectively, this work suggests that autophagy plays a tumor suppressive function in UVB response.
Conclusions
While much progress has been made in understanding the regulatory and functional role of autophagy in energy stress response, the role of autophagy in response to UVB and UVA radiation is only beginning to be understood. Autophagy regulates UV-induced apoptosis, DNA damage repair, oxidized lipid removal, and other oxidative damage response. The role of autophagy in UV response, skin cancer, and aging remains to be elucidated.
First, while autophagy likely prevents the accumulation of oxidative damage that leads to UV-induced photoaging, it remains unclear whether activation of autophagy would be effective at preventing photoaging. Moreover, there is currently no clear indication that autophagy modulation would be effective in the treatment or prevention of skin cancer. The contribution of autophagy to the promotion or suppression of tumor growth and metastasis is likely dependent on a multitude of factors, including cell type and disease state. Understanding the context-dependency of the oncogenic and tumor suppressive roles of autophagy in skin cancer requires further research into UV response and could provide clearer insight into autophagy modulation as a treatment option for skin cancer. These future findings will define the regulatory and functional role of autophagy in UV response and skin cancer and aging, and provide molecular basis for targeting autophagy to prevent and treat skin cancer and aging-associated diseases.
Acknowledgments
We apologize to those investigators whose work could not be directly referenced owing to space limitations. Work in the authors' laboratorywas supported by the NIH/NIEHS grant ES024373 and ES016936 (YYH), the American Cancer Society (ACS) grant RSG-13-078-01 (YYH), the University of Chicago Cancer Research Center (P30 CA014599), the CTSA (UL1 TR000430), and the University of Chicago Friends of Dermatology Endowment Fund. We thank Dr. Ann Motten for critical reading of the manuscript.
References
- 1.van Weelden H, de Gruijl FR, van der Putte SC, Toonstra J, van der Leun JC. The carcinogenic risks of modern tanning equipment: is UV-A safer than UV-B? Arch Dermatol Res. 1988;280(5):300–307. doi: 10.1007/BF00440604. http://www.ncbi.nlm.nih.gov/pubmed/3178287. [DOI] [PubMed] [Google Scholar]
- 2.de Gruijl FR, van der Leun JC. Environment and health: 3. Ozone depletion and ultraviolet radiation. CMAJ. 2000;163(7):851–855. http://www.ncbi.nlm.nih.gov/pubmed/11033716. [PMC free article] [PubMed] [Google Scholar]
- 3.D'Orazio J, Jarrett S, Amaro-Ortiz A, Scott T. UV radiation and the skin. Int J Mol Sci. 2013;14(6):12222–12248. doi: 10.3390/ijms140612222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruls WAG, Slaper H, can der Leun JC, Berrens L. Transmission of human epidermis and stratum corneum as a function of thickness in the ultraviolet and visible wavelengths. Photochem Photobiol. 1984;40(4):485–494. doi: 10.1111/j.1751-1097.1984.tb04622.x. [DOI] [PubMed] [Google Scholar]
- 5.Meinhardt M, Krebs R, Anders A, Heinrich U, Tronnier H. Wavelength-dependent penetration depths of ultraviolet radiation in human skin. J Biomed Opt. 2008;13(4):44030. doi: 10.1117/1.2957970. [DOI] [PubMed] [Google Scholar]
- 6.De Gruijl FR. Photocarcinogenesis: UVA vs. UVB Radiation. Ski Pharmacol Appl Ski Physiol. 2002;15:316–320. doi: 10.1159/000064535. http://www.ncbi.nlm.nih.gov/pubmed/12239425. [DOI] [PubMed] [Google Scholar]
- 7.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res Mol Mech Mutagen. 2005;571(1):19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 8.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiang L, Wu C, Ming M, Viollet B, He YY. Autophagy controls p38 activation to promote cell survival under genotoxic stress. J Biol Chem. 2013;288(3):1603–1611. doi: 10.1074/jbc.M112.415224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Zhang CF, Rossiter H, et al. Autophagy is induced by UVA and promotes removal of oxidized phospholipids and protein aggregates in epidermal keratinocytes. J Invest Dermatol. 2013;133(6):1629–1637. doi: 10.1038/jid.2013.26. [DOI] [PubMed] [Google Scholar]
- 11.Bess AS, Ryde IT, Hinton DE, Meyer JN. UVC-induced mitochondrial degradation via autophagy correlates with mtDNA damage removal in primary human fibroblasts. J Biochem Mol Toxicol. 2013;27(1):28–41. doi: 10.1002/jbt.21440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vessoni A, Filippi-Chiela E, Menck C, Lenz G. Autophagy and genomic integrity. Cell Death Differ. 2013;20103(10):1444–1454. doi: 10.1038/cdd.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathew R, Kongara S, Beaudoin B, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21(11):1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenzelmann Broz D, Spano Mello S, Bieging KT, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27(9):1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22(10):377–388. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mcadam E, Brem R, Karran P. DNA Damage and Repair Oxidative Stress–Induced Protein Damage Inhibits DNA Repair and Determines Mutation Risk and Therapeutic Efficacy. Mol Cancer Res. 14(7):612–622. doi: 10.1158/1541-7786.MCR-16-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen X, Wu J, Wang F, Liu B, Huang C, Wei Y. Deconvoluting the role of reactive oxygen species and autophagy in human diseases. Free Radic Biol Med. 2013;65:402–410. doi: 10.1016/j.freeradbiomed.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Jain A, Lamark T, Sjøttem E, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285(29):22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 20.Taguchi K, Fujikawa N, Komatsu M, et al. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 2012;109(34):13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Settembre C, Zoncu R, Medina DL, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sardiello M, Palmieri M, di Ronza A, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 23.Palmieri M, Impey S, Kang H, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20(19):3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 24.Settembre C, De Cegli R, Mansueto G, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15(6):647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8(6):903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roczniak-Ferguson A, Petit CS, Froehlich F, et al. The Transcription Factor TFEB Links mTORC1 Signaling to Transcriptional Control of Lysosome Homeostasis. Sci Signal. 2012;5(228):ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46(6):372–383. doi: 10.3109/07853890.2014.912836. [DOI] [PubMed] [Google Scholar]
- 30.Wang RC, Wei Y, An Z, et al. Akt-Mediated Regulation of Autophagy and Tumorigenesis Through Beclin 1 Phosphorylation. Science (80-) 2012;338(6109):956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning L, Guo-Chun Z, Sheng-Li A, et al. Inhibition of autophagy induced by PTEN loss promotes intrinsic breast cancer resistance to trastuzumab therapy. Tumour Biol. 2016;37(4):5445–5454. doi: 10.1007/s13277-015-4392-0. [DOI] [PubMed] [Google Scholar]
- 32.Errafiy R, Aguado C, Ghislat G, et al. PTEN increases autophagy and inhibits the ubiquitin-proteasome pathway in glioma cells independently of its lipid phosphatase activity. PLoS One. 2013;8(12):e83318. doi: 10.1371/journal.pone.0083318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen JH, Zhang P, Chen WD, et al. ATM-mediated PTEN phosphorylation promotes PTEN nuclear translocation and autophagy in response to DNA-damaging agents in cancer cells. Autophagy. 2015;11(2):239–252. doi: 10.1080/15548627.2015.1009767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mack HID, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8(8):1197–1214. doi: 10.4161/auto.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8(7):688–698. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 39.Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pattingre S, Tassa A, Qu X, et al. Bcl-2 Antiapoptotic Proteins Inhibit Beclin 1-Dependent Autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Maiuri MC, Criollo A, Kroemer G, et al. Crosstalk between apoptosis and autophagy within the Beclin 1 interactome. EMBO J. 2010;29(3):515–516. doi: 10.1038/emboj.2009.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36(12):2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 45.Matsunaga T, Hieda K, Nikaido O. Wavelength dependent formation of thymine dimers and (6-4)photoproducts in DNA by monochromatic ultraviolet light ranging from 150 to 365 nm. Photochem Photobiol. 1991;54(3):403–410. doi: 10.1111/j.1751-1097.1991.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 46.Yoon JH, Lee CS, O'Connor TR, Yasui A, Pfeifer GP. The DNA damage spectrum produced by simulated sunlight. J Mol Biol. 2000;299(3):681–693. doi: 10.1006/jmbi.2000.3771. [DOI] [PubMed] [Google Scholar]
- 47.Kusumoto R, Masutani C, Sugasawa K, et al. Diversity of the damage recognition step in the global genomic nucleotide excision repair in vitro. Mutat Res. 2001;485(3):219–227. doi: 10.1016/s0921-8777(00)00082-3. http://www.ncbi.nlm.nih.gov/pubmed/11267833. [DOI] [PubMed] [Google Scholar]
- 48.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40(2):359–369. doi: 10.1016/0092-8674(85)90150-3. http://www.ncbi.nlm.nih.gov/pubmed/3838150. [DOI] [PubMed] [Google Scholar]
- 49.Nakajima S, Lan L, Kanno SI, et al. UV Light-induced DNA Damage and Tolerance for the Survival of Nucleotide Excision Repair-deficient Human Cells. J Biol Chem. 2004;279(45):46674–46677. doi: 10.1074/jbc.M406070200. [DOI] [PubMed] [Google Scholar]
- 50.Budden T, Bowden NA. The role of altered nucleotide excision repair and UVB-induced DNA damage in melanomagenesis. Int J Mol Sci. 2013;14(1):1132–1151. doi: 10.3390/ijms14011132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mouret S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc Natl Acad Sci U S A. 2006;103(37):13765–13770. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kielbassa C, Roza L, Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18(4):811–816. doi: 10.1093/carcin/18.4.811. http://www.ncbi.nlm.nih.gov/pubmed/9111219. [DOI] [PubMed] [Google Scholar]
- 53.Douki T, Reynaud-Angelin A, Cadet J, Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42(30):9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- 54.Courdavault S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Larger yield of cyclobutane dimers than 8-oxo-7,8-dihydroguanine in the DNA of UVA-irradiated human skin cells. Mutat Res Mol Mech Mutagen. 2004;556(1):135–142. doi: 10.1016/j.mrfmmm.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 55.Douki T, Perdiz D, Grof P, et al. Oxidation of Guanine in Cellular DNA by Solar UV Radiation: Biological Role. Photochem Photobiol. 1999;70(2):184–1. [PubMed] [Google Scholar]
- 56.Premi S, Wallisch S, Mano CM, et al. Photochemistry. Chemiexcitation of melanin derivatives induces DNA photoproducts long after UV exposure. Science. 2015;347(6224):842–847. doi: 10.1126/science.1256022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rochette PJ, Therrien JP, Drouin R, et al. UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 2003;31(11):2786–2794. doi: 10.1093/nar/gkg402. http://www.ncbi.nlm.nih.gov/pubmed/12771205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikehata H, Kudo H, Masuda T, Ono T. UVA induces C->T transitions at methyl-CpG-associated dipyrimidine sites in mouse skin epidermis more frequently than UVB. Mutagenesis. 2003;18(6):511–519. doi: 10.1093/mutage/geg030. [DOI] [PubMed] [Google Scholar]
- 59.Dunkern TR, Fritz G, Kaina B. Ultraviolet light-induced DNA damage triggers apoptosis in nucleotide excision repair-deficient cells via Bcl-2 decline and caspase-3/-8 activation. Oncogene. 2001;20(42):6026–6038. doi: 10.1038/sj.onc.1204754. [DOI] [PubMed] [Google Scholar]
- 60.Courdavault S, Baudouin C, Sauvaigo S, et al. Unrepaired cyclobutane pyrimidine dimers do not prevent proliferation of UV-B--irradiated cultured human fibroblasts. Photochem Photobiol. 2004;79(2):145–151. doi: 10.1562/0031-8655(2004)079<0145:ucpddn>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 61.Blattner C, Tobiasch E, Litfen M, Rahmsdorf HJ, Herrlich P. DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene. 1999;18(9):1723–1732. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- 62.Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc Natl Acad Sci U S A. 2002;99(20):12985–12990. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berg RJ, Ruven HJ, Sands AT, de Gruijl FR, Mullenders LH. Defective global genome repair in XPC mice is associated with skin cancer susceptibility but not with sensitivity to UVB induced erythema and edema. J Invest Dermatol. 1998;110(4):405–409. doi: 10.1111/j.1523-1747.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- 64.Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22(56):9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- 65.Latonen L, Laiho M. Cellular UV damage responses—Functions of tumor suppressor p53. Biochim Biophys Acta - Rev Cancer. 2005;1755(2):71–89. doi: 10.1016/j.bbcan.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 66.Bissonnette N, Hunting DJ. p21-induced cycle arrest in G1 protects cells from apoptosis induced by UV-irradiation or RNA polymerase II blockage. Oncogene. 1998;16(26):3461–3469. doi: 10.1038/sj.onc.1201899. [DOI] [PubMed] [Google Scholar]
- 67.McKay BC, Chen F, Perumalswami CR, Zhang F, Ljungman M. The tumor suppressor p53 can both stimulate and inhibit ultraviolet light-induced apoptosis. Mol Biol Cell. 2000;11(8):2543–2551. doi: 10.1091/mbc.11.8.2543. http://www.ncbi.nlm.nih.gov/pubmed/10930452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qin JZ, Chaturvedi V, Denning MF, et al. Regulation of apoptosis by p53 in UV-irradiated human epidermis, psoriatic plaques and senescent keratinocytes. Oncogene. 2002;21(19):2991–3002. doi: 10.1038/sj.onc.1205404. [DOI] [PubMed] [Google Scholar]
- 69.Tron VA, Li G, Ho V, Trotter MJ. Ultraviolet radiation-induced p53 responses in the epidermis are differentiation-dependent. J Cutan Med Surg. 1999;3(5):280–283. doi: 10.1177/120347549900300511. http://www.ncbi.nlm.nih.gov/pubmed/10381953. [DOI] [PubMed] [Google Scholar]
- 70.Shah P, He YY. Molecular regulation of UV-induced DNA repair. Photochem Photobiol. 91(2):254–264. doi: 10.1111/php.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10(11):756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 72.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5(7):564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 73.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 74.Smith J, Tho L Mun, Xu N, Gillespie DA. Chapter 3 – The ATM–Chk2 and ATR–Chk1 Pathways in DNA Damage Signaling and Cancer. Advances in Cancer Research. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 75.Bartek J, Falck J, Lukas J. CHK2 kinase--a busy messenger. Nat Rev Mol Cell Biol. 2001;2(12):877–886. doi: 10.1038/35103059. [DOI] [PubMed] [Google Scholar]
- 76.Ray A, Milum K, Battu A, Wani G, Wani AA. NER initiation factors, DDB2 and XPC, regulate UV radiation response by recruiting ATR and ATM kinases to DNA damage sites. DNA Repair (Amst) 2013;12(4):273–283. doi: 10.1016/j.dnarep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang W, Ju J, Lee K, Nam K, Oh S, Shin I. Protein kinase B/Akt1 inhibits autophagy by down-regulating UVRAG expression. Exp Cell Res. 2013;319(3):122–133. doi: 10.1016/j.yexcr.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 78.Crighton D, Wilkinson S, O'Prey J, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126(1):121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 79.Feng Z, Hu W, de Stanchina E, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67(7):3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 80.Budanov AV, Karin M, Astrinidis A, et al. p53 Target Genes Sestrin1 and Sestrin2 Connect Genotoxic Stress and mTOR Signaling. Cell. 2008;134(3):451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao B, Shah P, Qiang L, He TC, Budanov A, He YY. Distinct Role of Sesn2 in Response to UVB-Induced DNA Damage and UVA-Induced Oxidative Stress in Melanocytes. Photochem Photobiol. 2016 doi: 10.1111/php.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu CL, Qiang L, Han W, Ming M, Viollet B, He YY. Role of AMPK in UVB-induced DNA damage repair and growth control. Oncogene. 2013;32(21):2682–2689. doi: 10.1038/onc.2012.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Qiang L, Zhao B, Shah P, Sample A, Yang S, He YY. Autophagy positively regulates DNA damage recognition by nucleotide excision repair. Autophagy. 2016;12(2):357–368. doi: 10.1080/15548627.2015.1110667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qiang L, Zhao B, Ming M, et al. Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc Natl Acad Sci U S A. 2014;111(25):9241–9246. doi: 10.1073/pnas.1322913111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Perelman B, Dafni N, Naiman T, et al. Molecular cloning of a novel human gene encoding a 63-kDa protein and its sublocalization within the 11q13 locus. Genomics. 1997;41(3):397–405. doi: 10.1006/geno.1997.4623. [DOI] [PubMed] [Google Scholar]
- 86.Yang Y, He S, Wang Q, et al. Autophagic UVRAG Promotes UV-Induced Photolesion Repair by Activation of the CRL4DDB2 E3 Ligase. Mol Cell. 2016;62(4):507–519. doi: 10.1016/j.molcel.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao B, Shah P, Budanov AV, et al. Sestrin2 protein positively regulates AKT enzyme signaling and survival in human squamous cell carcinoma and melanoma cells. J Biol Chem. 2014;289(52):35806–35814. doi: 10.1074/jbc.M114.595397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ming M, Han W, Maddox J, et al. UVB-induced ERK/AKT-dependent PTEN suppression promotes survival of epidermal keratinocytes. Oncogene. 2010;29(4):492–502. doi: 10.1038/onc.2009.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ming M, Feng L, Shea CR, et al. PTEN positively regulates UVB-induced DNA damage repair. Cancer Res. 2011;71(15):5287–5295. doi: 10.1158/0008-5472.CAN-10-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wondrak GT, Jacobson MK, Jacobson EL. Endogenous UVA-photosensitizers: mediators of skin photodamage and novel targets for skin photoprotection. Photochem Photobiol Sci. 2005;5(2):215–237. doi: 10.1039/b504573h. [DOI] [PubMed] [Google Scholar]
- 91.Baier J, Maisch T, Maier M, Engel E, Landthaler M, Bäumler W. Singlet Oxygen Generation by UVA Light Exposure of Endogenous Photosensitizers. Biophys J. 2006;91(4):1452–1459. doi: 10.1529/biophysj.106.082388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dahle J, Brunborg G, Svendsrud DH, Stokke T, Kvam E. Overexpression of human OGG1 in mammalian cells decreases ultraviolet A induced mutagenesis. Cancer Lett. 2008;267(1):18–25. doi: 10.1016/j.canlet.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 93.Sander CS, Chang H, Salzmann S, et al. Photoaging is associated with protein oxidation in human skin in vivo. J Invest Dermatol. 2002;118(4):618–625. doi: 10.1046/j.1523-1747.2002.01708.x. [DOI] [PubMed] [Google Scholar]
- 94.Gruber F, Bicker W, Oskolkova OV, Tschachler E, Bochkov VN. A simplified procedure for semi-targeted lipidomic analysis of oxidized phosphatidylcholines induced by UVA irradiation. J Lipid Res. 2012;53(6):1232–1242. doi: 10.1194/jlr.D025270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gugiu BG, Mesaros CA, Sun M, Gu X, Crabb JW, Salomon RG. Identification of oxidatively truncated ethanolamine phospholipids in retina and their generation from polyunsaturated phosphatidylethanolamines. Chem Res Toxicol. 2006;19(2):262–271. doi: 10.1021/tx050247f. [DOI] [PubMed] [Google Scholar]
- 96.Podrez EA, Poliakov E, Shen Z, et al. Identification of a novel family of oxidized phospholipids that serve as ligands for the macrophage scavenger receptor CD36. J Biol Chem. 2002;277(41):38503–38516. doi: 10.1074/jbc.M203318200. [DOI] [PubMed] [Google Scholar]
- 97.Punnonen K, Jansén CT, Puntala A, Ahotupa M. Effects of In Vitro UVA Irradiation and PUVA Treatment on Membrane Fatty Acids and Activities of Antioxidant Enzymes in Human Keratinocytes. J Invest Dermatol. 1991;96(2):255–259. doi: 10.1111/1523-1747.ep12462271. [DOI] [PubMed] [Google Scholar]
- 98.Gueranger Q, Li F, Peacock M, et al. Protein oxidation and DNA repair inhibition by 6-thioguanine and UVA radiation. J Invest Dermatol. 2014;134(5):1408–1417. doi: 10.1038/jid.2013.509. [DOI] [PubMed] [Google Scholar]
- 99.Guven M, Brem R, Macpherson P, Peacock M, Karran P. Oxidative Damage to RPA Limits the Nucleotide Excision Repair Capacity of Human Cells. J Invest Dermatol. 2015;135(11):2834–2841. doi: 10.1038/jid.2015.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hirota A, Kawachi Y, Itoh K, et al. Ultraviolet A irradiation induces NF-E2-related factor 2 activation in dermal fibroblasts: protective role in UVA-induced apoptosis. J Invest Dermatol. 2005;124(4):825–832. doi: 10.1111/j.0022-202X.2005.23670.x. [DOI] [PubMed] [Google Scholar]
- 101.Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1(1):45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh B, Chatterjee A, Ronghe AM, et al. Antioxidant-mediated up-regulation of OGG1 via NRF2 induction is associated with inhibition of oxidative DNA damage in estrogen-induced breast cancer. BMC Cancer. 2013;13(1):253. doi: 10.1186/1471-2407-13-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tian FF, Zhang FF, Lai XD, et al. Nrf2-mediated protection against UVA radiation in human skin keratinocytes. Biosci Trends. 2011;5(1):23–29. doi: 10.5582/bst.2011.v5.1.23. http://www.ncbi.nlm.nih.gov/pubmed/21422597. [DOI] [PubMed] [Google Scholar]
- 104.Cavinato M, Koziel R, Romani N, et al. UVB-Induced Senescence of Human Dermal Fibroblasts Involves Impairment of Proteasome and Enhanced Autophagic Activity. Journals Gerontol Ser A Biol Sci Med Sci. 2016 Aug; doi: 10.1093/gerona/glw150. glw150. [DOI] [PubMed] [Google Scholar]
- 105.Olivier E, Dutot M, Regazzetti A, et al. Lipid deregulation in UV irradiated skin cells: Role of 25-hydroxycholesterol in keratinocyte differentiation during photoaging. J Steroid Biochem Mol Biol. 2016 doi: 10.1016/j.jsbmb.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 106.Lamore SD, Wondrak GT. Autophagic-lysosomal dysregulation downstream of cathepsin B inactivation in human skin fibroblasts exposed to UVA. Photochem Photobiol Sci. 2012;11(1):163–172. doi: 10.1039/c1pp05131h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reelfs O, Xu YZ, Massey A, Karran P, Storey A. Thiothymidine plus low-dose UVA kills hyperproliferative human skin cells independently of their human papilloma virus status. Mol Cancer Ther. 2007;6(9):2487–2495. doi: 10.1158/1535-7163.MCT-07-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Suh YA, Post SM, Elizondo-Fraire AC, et al. Multiple stress signals activate mutant p53 in vivo. Cancer Res. 2011;71(23):7168–7175. doi: 10.1158/0008-5472.CAN-11-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao B, Ming M, He YY. Suppression of PTEN Transcription by UVA. J Biochem Mol Toxicol. 2013;27(2):184–191. doi: 10.1002/jbt.21451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Navarro-Yepes J, Burns M, Anandhan A, et al. Oxidative stress, redox signaling, and autophagy: cell death versus survival. Antioxid Redox Signal. 2014;21(1):66–85. doi: 10.1089/ars.2014.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Einspahr JG, Alberts DS, Wameke JA, et al. Relationship of p53 Mutations to Epidermal Cell Proliferation and Apoptosis in Human UV-Induced Skin Carcinogenesis. Neoplasia. 1999;1(5):468–475. doi: 10.1038/sj.neo.7900061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee JK, Kim JH, Nam KT, Lee SH. Molecular events associated with apoptosis and proliferation induced by ultraviolet-B radiation in the skin of hairless mice. J Dermatol Sci. 2003;32(3):171–179. doi: 10.1016/S0923-1811(03)00094-X. [DOI] [PubMed] [Google Scholar]
- 113.Santamaria AB, Davis DW, Nghiem DX, et al. p53 and Fas ligand are required for psoralen and UVA-induced apoptosis in mouse epidermal cells. Cell Death Differ. 2002;9(5):549–560. doi: 10.1038/sj/cdd/4401007. [DOI] [PubMed] [Google Scholar]
- 114.Li JL, Liu N, Chen XH, Sun M, Wang CB. Inhibition of UVA-induced apoptotic signaling pathway by polypeptide from Chlamys farreri in human HaCaT keratinocytes. Radiat Environ Biophys. 2007;46(3):263–268. doi: 10.1007/s00411-007-0112-5. [DOI] [PubMed] [Google Scholar]
- 115.Dickinson SE, Olson ER, Zhang J, et al. p38 MAP kinase plays a functional role in UVB-induced mouse skin carcinogenesis. Mol Carcinog. 2011;50(6):469–478. doi: 10.1002/mc.20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.He YY, Pi J, Huang JL, Diwan BA, Waalkes MP, Chignell CF. Chronic UVA irradiation of human HaCaT keratinocytes induces malignant transformation associated with acquired apoptotic resistance. Oncogene. 2006;25(26):3680–3688. doi: 10.1038/sj.onc.1209384. [DOI] [PubMed] [Google Scholar]
- 117.Yin X, Cao L, Peng Y, et al. A critical role for UVRAG in apoptosis. Autophagy. 2011;79:1242–1244. doi: 10.4161/auto.7.10.16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Higashi Y, Kanekura T, Kanzaki T. Enhanced expression of cyclooxygenase (COX)-2 in human skin epidermal cancer cells: evidence for growth suppression by inhibiting COX-2 expression. Int J Cancer. 2000;86(5):667–671. doi: 10.1002/(sici)1097-0215(20000601)86:5<667::aid-ijc10>3.0.co;2-y. http://www.ncbi.nlm.nih.gov/pubmed/10797288. [DOI] [PubMed] [Google Scholar]
- 119.Chun KS, Akunda JK, Langenbach R. Cyclooxygenase-2 inhibits UVB-induced apoptosis in mouse skin by activating the prostaglandin E2 receptors, EP2 and EP4. Cancer Res. 2007;67(5):2015–2021. doi: 10.1158/0008-5472.CAN-06-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bachelor MA, Silvers AL, Bowden GT. The role of p38 in UVA-induced cyclooxygenase-2 expression in the human keratinocyte cell line, HaCaT. Oncogene. 2002;21(46):7092–7099. doi: 10.1038/sj.onc.1205855. [DOI] [PubMed] [Google Scholar]
- 121.Cao Y, Prescott SM. Many actions of cyclooxygenase-2 in cellular dynamics and in cancer. J Cell Physiol. 2002;190(3):279–286. doi: 10.1002/jcp.10068. [DOI] [PubMed] [Google Scholar]
- 122.Fischer SM, Pavone A, Mikulec C, Langenbach R, Rundhaug JE. Cyclooxygenase-2 expression is critical for chronic UV-induced murine skin carcinogenesis. Mol Carcinog. 2007;46(5):363–371. doi: 10.1002/mc.20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tripp CS, Blomme EAG, Chinn KS, Hardy MM, LaCelle P, Pentland AP. Epidermal COX-2 Induction Following Ultraviolet Irradiation: Suggested Mechanism for the Role of COX-2 Inhibition in Photoprotection. J Invest Dermatol. 2003;121(4):853–861. doi: 10.1046/j.1523-1747.2003.12495.x. [DOI] [PubMed] [Google Scholar]
- 124.Tsujii M, DuBois RN. Alterations in cellular adhesion and apoptosis in epithelial cells overexpressing prostaglandin endoperoxide synthase 2. Cell. 1995;83(3):493–501. doi: 10.1016/0092-8674(95)90127-2. [DOI] [PubMed] [Google Scholar]
- 125.Chen LH, Chu PM, Lee YJ, et al. Targeting protective autophagy exacerbates UV-triggered apoptotic cell death. Int J Mol Sci. 2012;13(1):1209–1224. doi: 10.3390/ijms13011209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yin X, Cao L, Kang R, et al. UV irradiation resistance-associated gene suppresses apoptosis by interfering with BAX activation. EMBO Rep. 2011;12(7):727–734. doi: 10.1038/embor.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cao C, Lu S, Kivlin R, et al. AMP-activated protein kinase contributes to UV- and H2O2-induced apoptosis in human skin keratinocytes. J Biol Chem. 2008;283(43):28897–28908. doi: 10.1074/jbc.M804144200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 128.Zhao B, Qiang L, Joseph J, Kalyanaraman B, Viollet B, He YY. Mitochondrial dysfunction activates the AMPK signaling and autophagy to promote cell survival. Genes Dis. 2016;3(1):82–87. doi: 10.1016/j.gendis.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ming M, Shea CR, Feng L, Soltani K, He YY. UVA induces lesions resembling seborrheic keratoses in mice with keratinocyte-specific PTEN downregulation. J Invest Dermatol. 2011;131(7):1583–1586. doi: 10.1038/jid.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.He YY, Council SE, Feng L, Chignell CF. UVA-induced cell cycle progression is mediated by a disintegrin and metalloprotease/epidermal growth factor receptor/AKT/Cyclin D1 pathways in keratinocytes. Cancer Res. 2008;68(10):3752–3758. doi: 10.1158/0008-5472.CAN-07-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koh HK, Geller AC, Miller DR, Grossbart TA, Lew RA. Prevention and early detection strategies for melanoma and skin cancer - Current status. Arch Dermatol. 1996;132(4):436–443. [PubMed] [Google Scholar]
- 132.Parkin DM, Boyd L, Walker LC. 16. The fraction of cancer attributable to lifestyle and environmental factors in the UK in 2010. Br J Cancer. 2011;105(Suppl):S77–81. doi: 10.1038/bjc.2011.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.American Cancer Society. Cancer Facts & Figures 2015. 2015 doi: 10.1097/01.NNR.0000289503.22414.79. [DOI] [Google Scholar]
- 134.Rogers HW, Weinstock MA, Harris AR, et al. Incidence estimate of nonmelanoma skin cancer in the United States, 2006. Arch Dermatol. 2010;146(3):283–287. doi: 10.1001/archdermatol.2010.19. [DOI] [PubMed] [Google Scholar]
- 135.Guy GP, Machlin SR, Ekwueme DU, Yabroff KR. Prevalence and costs of skin cancer treatment in the U.S., 2002-2006 and 2007-2011. Am J Prev Med. 2015;48(2):183–187. doi: 10.1016/j.amepre.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.World Health Organization, Cancer Research UK. World cancer factsheet. World Heal Organ. 2014;4 doi: 10.1002/ijc.27711. [DOI] [Google Scholar]
- 137.Mohan SV, Chang ALS. Advanced Basal Cell Carcinoma: Epidemiology and Therapeutic Innovations. Curr Dermatol Rep. 2014;3:40–45. doi: 10.1007/s13671-014-0069-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Karia PS, Han J, Schmults CD. Cutaneous squamous cell carcinoma: Estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol. 2013;68(6):957–966. doi: 10.1016/j.jaad.2012.11.037. [DOI] [PubMed] [Google Scholar]
- 139.Skin Cancer Foundation. Skin Cancer Facts & Statistics. 2015 [Google Scholar]
- 140.Gasparro FP. Sunscreens, skin photobiology, and skin cancer: The need for UVA protection and evaluation of efficacy. Environ Health Perspect. 2000;108(SUPPL. 1):71–78. doi: 10.1289/ehp.00108s171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.U.S. Department of Health and Human Services. Report on Carcinogens. Natl Toxicol Progr. (Twelfth) 2011:53. [Google Scholar]
- 142.De Laat A, Van Der Leun JC, De Gruijl FR. Carcinogenesis induced by UVA (365-nm) radiation: The dose-time dependence of tumor formation in hairless mice. Carcinogenesis. 1997;18(5):1013–1020. doi: 10.1093/carcin/18.5.1013. [DOI] [PubMed] [Google Scholar]
- 143.Sterenborg HJCM, Leun JC. Tumorigenesis by a long wavelength UV-A source. Photochem Photobiol. 1990;51(3):325–330. doi: 10.1111/j.1751-1097.1990.tb01718.x. [DOI] [PubMed] [Google Scholar]
- 144.Kelfkens G, de Gruijl FR, van der Leun JC. Tumorigenesis by short-wave ultraviolet A: papillomas versus squamous cell carcinomas. Carcinogenesis. 1991;12(8):1377–1382. doi: 10.1093/carcin/12.8.1377. [DOI] [PubMed] [Google Scholar]
- 145.Lazovich D, Vogel RI, Berwick M, Weinstock MA, Anderson KE, Warshaw EM. Indoor tanning and risk of melanoma: A case-control study in a highly exposed population. Cancer Epidemiol Biomarkers Prev. 2010;19(6):1557–1568. doi: 10.1158/1055-9965.EPI-09-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang M, Qureshi AA, Geller AC, Frazier L, Hunter DJ, Han J. Use of tanning beds and incidence of skin cancer. J Clin Oncol. 2012;30(14):1588–1593. doi: 10.1200/JCO.2011.39.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kipp C, Young AR. The soluble eumelanin precursor 5,6-dihydroxyindole-2-carboxylic acid enhances oxidative damage in human keratinocyte DNA after UVA irradiation. Photochem Photobiol. 1999;70(2):191–198. http://www.ncbi.nlm.nih.gov/pubmed/10461458. [PubMed] [Google Scholar]
- 148.Kvam E, Tyrrell RM. The role of melanin in the induction of oxidative DNA base damage by ultraviolet A irradiation of DNA or melanoma cells. J Invest Dermatol. 1999;113(2):209–213. doi: 10.1046/j.1523-1747.1999.00653.x. [DOI] [PubMed] [Google Scholar]
- 149.Kvam E, Dahle J. Melanin synthesis may sensitize melanocytes to oxidative DNA damage by ultraviolet A radiation and protect melanocytes from direct DNA damage by ultraviolet B radiation. Pigment Cell Res. 2004;17(5):549–550. doi: 10.1111/j.1600-0749.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 150.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Besaratinia A, Synold TW, Chen HH, et al. DNA lesions induced by UV A1 and B radiation in human cells: comparative analyses in the overall genome and in the p53 tumor suppressor gene. Proc Natl Acad Sci U S A. 2005;102(29):10058–10063. doi: 10.1073/pnas.0502311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bradford PT, Goldstein AM, Tamura D, et al. Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. J Med Genet. 2011;48(3):168–176. doi: 10.1136/jmg.2010.083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Poillet-Perez L, Despouy G, Delage-Mourroux R, Boyer-Guittaut M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol. 2015;4:184–192. doi: 10.1016/j.redox.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Karantza-Wadsworth V, Patel S, Kravchuk O, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21(13):1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mathew R, Karp CM, Beaudoin B, et al. Autophagy Suppresses Tumorigenesis through Elimination of p62. Cell. 2009;137(6):1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-κB activation. J Biol Chem. 2010;285(3):2077–2089. doi: 10.1074/jbc.M109.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Duran A, Linares JF, Galvez AS, et al. The Signaling Adaptor p62 Is an Important NF-κB Mediator in Tumorigenesis. Cancer Cell. 2008;13(4):343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 159.Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J. The interaction of p62 with RIP links the atypical PKCs to NF-κB activation. EMBO J. 1999;18(11):3044–3053. doi: 10.1093/emboj/18.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19(7):1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ling J, Kang Y, Zhao R, et al. Kras G12D-Induced IKK2/β/NF-κB Activation by IL-1α and p62 Feedforward Loops Is Required for Development of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2012;21(1):105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rabe JH, Mamelak AJ, McElgunn PJS, Morison WL, Sauder DN. Photoaging: Mechanisms and repair. J Am Acad Dermatol. 2006;55(1):1–19. doi: 10.1016/j.jaad.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 163.Kambayashi H, Yamashita M, Odake Y, Takada K, Funasaka Y, Ichihashi M. Epidermal changes caused by chronic low-dose UV irradiation induce wrinkle formation in hairless mouse. J Dermatol Sci. 2001;27(Suppl 1):S19–25. doi: 10.1016/s0923-1811(01)00113-x. http://www.ncbi.nlm.nih.gov/pubmed/11514121. [DOI] [PubMed] [Google Scholar]
- 164.Moloney SJ, Edmonds SH, Giddens LD, Learn DB. The hairless mouse model of photoaging: evaluation of the relationship between dermal elastin, collagen, skin thickness and wrinkles. Photochem Photobiol. 1992;56(4):505–511. doi: 10.1111/j.1751-1097.1992.tb02194.x. http://www.ncbi.nlm.nih.gov/pubmed/1454880. [DOI] [PubMed] [Google Scholar]
- 165.Boisnic S, Branchet-Gumila MC, Le Charpentier Y, Segard C. Repair of UVA-induced elastic fiber and collagen damage by 0.05% retinaldehyde cream in an ex vivo human skin model. Dermatology. 1999:43–48. doi: 10.1159/000051378. doi:51378. [DOI] [PubMed] [Google Scholar]
- 166.Herrmann G, Wlaschek M, Lange TS, Prenzel K, Goerz G, Scharffetter-Kochanek K. UVA irradiation stimulates the synthesis of various matrix-metalloproteinases (MMPs) in cultured human fibroblasts. Exp Dermatol. 1993;2(2):92–97. doi: 10.1111/j.1600-0625.1993.tb00015.x. http://www.ncbi.nlm.nih.gov/pubmed/8156175. [DOI] [PubMed] [Google Scholar]
- 167.Scharffetter-Kochanek K, Brenneisen P, Wenk J, et al. Photoaging of the skin from phenotype to mechanisms. Exp Gerontol. 2000;35(3):307–316. doi: 10.1016/s0531-5565(00)00098-x. [DOI] [PubMed] [Google Scholar]
- 168.Petropoulos I, Conconi M, Wang X, et al. Increase of oxidatively modified protein is associated with a decrease of proteasome activity and content in aging epidermal cells. J Gerontol A Biol Sci Med Sci. 2000;55(5):B220–7. doi: 10.1093/gerona/55.5.b220. http://www.ncbi.nlm.nih.gov/pubmed/10819308. [DOI] [PubMed] [Google Scholar]
- 169.Dunlop RA, Brunk UT, Rodgers KJ. Oxidized proteins: mechanisms of removal and consequences of accumulation. IUBMB Life. 2009;61(5):522–527. doi: 10.1002/iub.189. [DOI] [PubMed] [Google Scholar]
- 170.Yamaguchi O, Otsu K. Role of autophagy in aging. J Cardiovasc Pharmacol. 2012;60(3):242–247. doi: 10.1097/FJC.0b013e31824cc31c. [DOI] [PubMed] [Google Scholar]
- 171.Widmer R, Ziaja I, Grune T. Protein oxidation and degradation during aging: role in skin aging and neurodegeneration. Free Radic Res. 2006;40(12):1259–1268. doi: 10.1080/10715760600911154. [DOI] [PubMed] [Google Scholar]
- 172.Grune T, Reinheckel T, Davies KJ. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997;11(7):526–534. http://www.ncbi.nlm.nih.gov/pubmed/9212076. [PubMed] [Google Scholar]
- 173.Voskamp P, Bodmann CA, Rebel HG, et al. Rapamycin impairs UV induction of mutant-p53 overexpressing cell clusters without affecting tumor onset. Int J Cancer. 2012;131(6):1267–1276. doi: 10.1002/ijc.27391. [DOI] [PubMed] [Google Scholar]