Abstract
In recent years there has been an increasing interest of the scientific community on exosome research, with particular emphasis on the mechanisms by which tumor-derived exosomes can promote tumor growth. Particularly, exosome-mediated immune-escape is under deep investigation and still represents a quite controversial issue. Tumor-derived exosomes are carriers of information able to reprogram functions of immune target cells, influencing their development, maturation, and antitumor activities. They deliver proteins similar to those of the parent cancer cells, but also genetic messages like genomic DNA, mRNA, and microRNAs (miRNAs) that ultimately share the so called “tumor microenvironment” in a pro-tumoral fashion. The content of tumor-derived exosomes could be implicated in several signaling pathways operating in the tumor microenvironment, providing a further modality of dys-regulation of antitumor immunity. The aim of this review is to provide a state-of-the-art highlight of to the most recent discoveries in the field of interaction between tumor-derived exosomic miRNAs and the cells of immune system.
Keywords: Tumor-derived exosomes, microRNAs, Cancer, Immune system, Microenvironment
Introduction
Membraneous vesicles (MV) were first isolated by Taylor et al. in 1983, from the supernatants of human cancer cell lines, and were described to express several molecular markers also present in the parental cancer cell plasma membranes [1–3]. While biochemical and structural characterization of cancer-derived MV was on-going, other investigators discovered vescicular structures originated during reticulocytes maturation, which were named “exosomes” [4]. Nowadays exosomes, including tumor-derived exosomes (TEX), are classified as a class of small (30–150 nm) membrane-bound extracellular vesicles (EVs), heterogeneous in size, present in all body fluids, made and delivered by many (if not all) cells [5–8]. Cellular secretion of exosomes seems to occur spontaneously at a physiological rate, and their biogenesis differs from that of other extracellular vesicles (EVs), since it involves the intra-cellular endosomal compartment [9]. TEX can be discriminated from exosomes derived from normal cells because of their specific molecular signature both in terms of cargo and as surface markers [9,10]. Intriguingly, it has been shown that TEX circulating in the plasma of cancer patients express several soluble factors involved in tumor immune-escape, such as FasL, TRAIL, PD-L1, IL-10, TGF-β1, prostaglandin E2, CD39, and D73 [11,12]. Not only immunosuppressive molecules are representative of TEX content, but also immune stimulators like tumor-associated antigens (TAAs), and MCH components. This double role of TEX in shaping the immune response is source of controversy as to their biological functions [13,14]. The type of cargo and the tumor microenvironment (TME), defined as the surrounding cellular environment entangled around the cancer cells, seem to play a key role in determining which one of the two functions TEX could ultimately carry out. Overall, taking into account the active participation of TME in establishing immunosuppressive conditions, and the increased number of TEX with tumor stage and progression, the hypothesis that TEX promote immune-escape mechanisms appears more likely [5,15]. Such statement suffers of the limitations implied in all generalizations. It is indeed quite possible that the overall effect of TEX in cancer biology is different for different types of cancers. In addition to proteins and lipids, TEX deliver also genetic messages such as DNA, RNA, and microRNAs (miRNAs) all of which can be shuttled inter-cellularily as a form of cell-cell communications [16]. Valadi et al., provided the first evidence that miRNAs and messenger RNAs (mRNAs) could be functionally transferred from one cell to another through exosomes [17]. Since this seminal discovery, miRNAs have emerged as critical players in shaping the inflammatory TME. They are the most studied non-coding RNAs, and form a large family (in humans about 2600 unique mature miRNAs) of short (19–24 nucleotides), highly conserved, single-strand RNAs with important roles in a variety of biological and pathological processes such as cell differentiation, proliferation, death, and stress response [18,19]. Because of their high tissue- and cell-specificity, and their rigorously controlled expression based on developmental stage, they affect essentially every aspect of cell biology through a fine control gene expression by recognizing a 2–7 nucleotides long target sequence, called “seed-region”, which can be found into the 3′-UTR [20], 5′-UTR [21], or coding region [22] of target mRNAs. The complementarity miRNA:mRNA can lead to mRNA degradation (imperfect complementarity) or, more frequently, will prevent mRNA translation, typically resulting in down-regulation of the encoded protein [23]. Some miRNAs may also up-regulate translation of their specific mRNA target [24]. Recently, Melo et al. described how breast-cancer TEX are able to include precursor miRNAs (pre-miRNAs) complexed with Dicer, TRBP and AGO2 proteins displaying a cell-independent capacity to process pre-miRNAs into mature form, providing the first report of a cell-autonomous process occurring in exosomes when secreted into the extracellular space [25]. To date the information about how TEX, TME and miRNAs interact each other in the modulation of the immune system is still very scarce, especially regarding TEX-mediated communication between cancer cells and immune cells. The relevant impact that the understanding of this relationship could have on the development of new therapeutic and diagnostic strategies highlights the need for further studies aimed at clarifying this interaction. The aim of this review is to give a critical state-of-the-art perspective on the current knowledge of TEX mediated cancer-immune cell interactions.
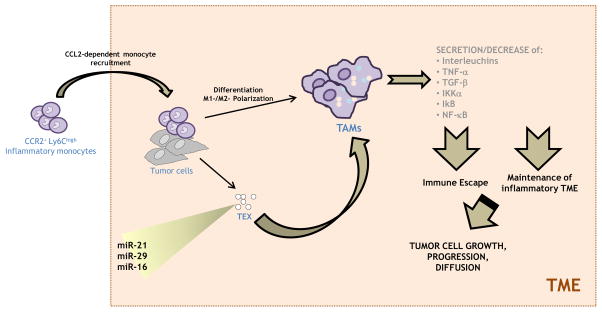
Cancer to Macrophages
Tumor-associated macrophages (TAMs) are currently the most widely studied inflammatory cell component of TME. They principally originate from bone marrow monocytes, but they have also been shown to derive from circulating and splenic monocytes as wells [26]. CCR2+Ly6Chigh inflammatory monocytes, representing the main TAM precursor, are recruited by TME cells into the tumor tissue, in a chemokine (C-C motif) ligand 2 (CCL2)-dependent manner, as demonstrated by Qian et al. in tumor-bearing mice in which inhibition of CCL2–CCR2 signaling blocked the enrollment of inflammatory monocytes, inhibited metastasis, and prolonged the survival [27,28]. Within the TME, TAMs can either undergo M1-, or M2-polarization: while M1 cells produce pro-inflammatory, antitumor cytokines such as inteleukin-12 (IL-12) and inteleukin-23 (IL-23) and are characteristic of early stage and regressing tumors [29], M2 macrophages are immunosuppressive, are involved in tissue repair and are frequently abundant in advanced cancers where they promote cancer progression and metastasis [30]. TAMs could be activated by TEXs, but in this case they show a different cytokine profile than the one induced by lipopolysaccharide (LPS) or inteleukin-4 (IL-4) [31]. Subsequent to exosome stimulation, macrophages exhibited reduced levels of metallopeptidase inhibitor 1 (TIMP1), interferon gamma (IFNγ), inteleukin-16 (IL-16) and increased levels of inteleukin-8 (IL-8), macrophage inflammatory protein-2 (MIP2), interleukin-1 receptor antagonist (IL-1RA), and CCL2 which were closely associated with tumor invasion and metastasis. MiRNA-mediated regulation of TAMs is one of the currently most investigated topics in the field of TAM-associated carcinogenesis. In 2012, Fabbri et al. identified a new mechanism of communication between TAMs and cancer cells via miRNAs. They demonstrated that miR-21 and miR-29a, secreted into exosomes by non-small cell lung cancer (NSCLC) cells, were recruited in the TME by TAMs and bind to their Toll-like receptor 8 (TLR8), triggering the NF-κB pathway and the secretion of inteleukin-6 (IL-6) and Tumor necrosis factor -α (TNF-α). The release of these cytokines promotes an inflammatory TME that favors cancer growth and dissemination [32]. Interestingly, in neuroblastoma, TEX miR-21 is able to up-regulate miR-155 in TAMs, following its binding with TLR8. MiR-155 is then secreted in TAM-derived exosomes and transferred back to neuroblastoma cells, where it inhibits Telomeric Repeat Binding Factor 1 (TERF1), a telomerase inhibitor whose down-regulation increases Cisplatin resistance [33]. Jang et al. showed that epigallocatechin gallate (EGCG), a molecule with well known anti-tumor effects, suppressed tumor growth in 4T1 murine breast cancer cell line in association with decreased TAM and M2 macrophage infiltration. In particular, in EGCG-treated mice they observed a lower expression of chemokine for monocytes (CSF-1 and CCL-2) in tumor cells, while TAM cytokines switched from M2- to M1-like phenotype as evidenced by decreased IL-6 and Transforming growth factor-β (TGF-β) and increased TNF-α. Ex vivo incubation of TAMs with EGCG-treated 4T1 exosomes drove the increase of IL-6 and TGF-β, decrease of TNF-α, IκB kinase α (IKKα) suppression and concomitant IκB increase, ex vivo incubation of isolated tumor cells with EGCG inhibited the CSF-1 and CCL-2 expression. Moreover, they found that EGCG could up-regulate miR-16 both in 4T1 cells and in their exosomes. Treatment of TAM and cancer cells with exosomes from 4T1 cells treated with EGCG or which underwent miR-16 knocking-down, can restore the observed effects on chemokines, cytokines, and Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway, suggesting a novel mechanism by which EGCG exerts anti-tumor activity via regulation of TAM in tumor microenvironment. This could be carried out through EGCG up-regulation of miR-16 in tumor cells, and its subsequent transfer to TAM via exosomes [34].
By targeting the 3′UTR of multiple genes including rho-dependent kinase-2 (ROCK2), miR-511-3p acts as a key regulator of CD206+ TAM tumorigenic actions [35]. While during macrophage differentiation CUE domain containing 2 (CUEDC2) expression is significantly upregulated, CUEDC2 deficiency results in triggering pro-inflammatory cytokine which could be correlated to both colitis and colon carcinogenesis. The level of CUEDC2 in TAMs is downregulated by miR-324-5p, which in turn is upregulated by IL-4. Notably, CUEDC2 expression is almost undetectable in macrophages in human colon cancer, and this decreased CUEDC2 expression is associated with high levels of interleukin-4 and miR-324-5p [36]. Sonda et al. showed that miR-142-3p downregulation enhanced macrophage M2-differentiation with immunosuppressive functions in tumor. Indeed, miR-142-3p is able to downregulate and repress glycoprotein 130 (gp130), the common subunit of IL-6 cytokine receptor family, and CCAAT-enhancer-binding protein β (C/EBPβ), a transcription factor, whose LAP* isoform is induced by tumor-released cytokines signaling through gp130. Both these molecules are critical for generating pro-tumoral M2 macrophages through regulation of TGF-β signals.
Both in vitro and in vivo, miR-142-3p forced expression impaired macrophage differentiation. Mice constitutively expressing miR-142-3p in the bone marrow confirmed a marked increase in survival subsequent to immunotherapy with tumor-specific T lymphocytes. This study demonstrated how miR-142-3p could modify the TME and favor antitumor immunity, also stressing the feasibility of altering tumor-induced macrophage differentiation as a potent tool to improve the efficacy of cancer immunotherapy [37].
MiRNAs involved in the line of communication cancer-macrophages are shown in Figure 1.
Figure 1. miRNAs involved in the line of communication cancer-macrophages.
MiRNAs released by tumor via exosomes can interact with tumor-associated macrophages in order to maintain a TME inflammatory status which in turn promote cancer growth, progression and diffusion.
Cancer to Dendritic Cells
Dendritic cells (DCs) are antigen-presenting cells whose main function is to process the antigen material and present it on the cell surface to T cells, acting like messengers between the innate and the adaptive immune systems. They express a broad range of TLRs and cytokines, which take an active part in activating the immune response [38]. It has been demonstrated that in DCs-based immunotherapy, the expression of TLR4 has an important anticancer effect [39]. Nevertheless, the release of TEX in the TME can inhibit both differentiation and maturation of DCs, which can change their role from effective antigen presenting cells into negative modulators of immune responses [40]. In bone marrow, TEX can prevent the differentiation of myeloid precursor into DCs giving rise to a cluster of immature myeloid-derived suppressor cells (MDSCs) able to help tumor progression [41]. An in vitro study based on a culture system of mouse DCs demonstrated that this interference occurred via induction of IL-6 [42]. Several studies focused on the protein content of TEX to explain their inhibitory role [42–44], but only a few also took miRNAs into consideration. Based on the evidence that TLR4 was a target of miR-203, and the fact that miR-203, overexpressed in pancreatic adenocarcinoma compared to normal pancreatic tissue, may be involved in the progression of pancreatic cancer [45,46], Zhou et al. investigated the influence of pancreatic TEX miR-203 on TLR4 and downstream cytokines. They showed that exposure of DCs to these exosomes downregulated the expression of TLR4 and decreased the expression of related cytokines such as TNF-α, necessary for the maturation of DCs, and IL-12, beneficial for Th1 differentiation, enhancing cellular immunity. Tumor-derived exosomes interfere with DCs via miR-203, in fact exosomal miR-203 is likely to regulate and control the expression of TLR4 and the production of TNF-α and IL-12 by DCs, therefore contributing to their dysfunction [47]. Ding et al. explored how miR-212-3p delivered to DCs by pancreatic cancer-derived exosomes downregulated regulatory factor X-associated protein (RFXAP), decreasing MHC II expression and inducing immune tolerance of DCs [48]. After showing that pancreatic cancer (PC)-derived exosomes (PEs) can induce immune tolerance by downregulating the expression of TLR4 in dendritic cells (DCs) via miR-203, Que et al. tried to verify if PC-derived exosomal miRNAs could modulate the anti-tumor activity of DCs/cytokine-induced killer cells (CIKs). In particular they analyzed the potential role of miRNA-depleted exosomes in activating DCs/CIKs. PEs were extracted from PANC-1 cell line supernatants, then ruptured or ultrafiltrated (UELs). DCs co-cultured with CIKs after stimulation with lipopolysaccharide (LPS), PEs, and UEL. UELs-stimulated DCs/CIKs showed a higher killing rate compared to a LPS and PEs stimulation, suggesting that miRNA-depleted exosome proteins may be promising agonists for activating DC/CIKs against pancreatic cancer [49].
MiRNAs involved in the line of communication cancer-DCs are shown in Table 1.
Table 1.
miRNAs involved in the line of communication cancer-DCs
Cancer to Lymphocytes
MiRNAs also play a role in the biology of NK cells and T lymphocytes. Natural killer (NK) are a sub-population of T cells with a role as tumor cell killer, based on their production of antitumor cytokines, including IL-4, IFN-γ, FasL, IL-13, and perforin [50]. However, their efficiency is abrogated by exposure to TGF-β. Donatelli et al. showed that this inhibition was a consequence of TGF-β induction of miR-183, which targeted and repressed DNAX activating protein 12 kDa (DAP12), a signal adaptor for lytic function in NK cells, becoming a key factor in TGF-β–mediated immunosuppression [51]. More recently, Berchem et al. focused their attention on intratumoral hypoxia, being an integral component of all solid tumors, demonstrating for the first time the existence of a relationship between hypoxic tumor derived microvesicles (TD-MVs) and NK-mediated citotoxicity. Using several tumor models they showed that microvescicles derived from hypoxic tumor cells were qualitatively different, and can inhibit NK cell function much more if compared to normoxic ones. Indeed, hypoxic TD-MVs package two immunosuppressive factors that can contribute to NK cell cytotoxicity dysregulation, in vitro and in vivo. TD-MVs are able to determine a decrease of the activating natural-killer group 2 member D (NKG2D) receptor expression on the NK surface, which is mediated by the transfer of TGF-β1 from TD-MVs to NK cells. Next, miRNA profiling revealed the presence of high levels of miR-210 and miR-23a in hypoxic TD-MVs. MiR-210 controls the antigen specific immune response and has been frequently reported as the master regulator of tumor hypoxic responses [52], while miR-23a directly targets the expression of CD107a acting as an additional immunosuppressive factor [53]. This study highlights the existence of a novel way to induce immune suppression mediated by hypoxic TD-MVs further improving the knowledge of the immunosuppressive mechanisms prevailing in the hypoxic tumor TME. Tang et al. investigated the role of miR-92a in the development of tolerant natural killer T (NKT) cells, in glioma. They reported that in U87 cells primary glioma, abundant IL-6+ IL-10+ NKT cells were detected, due to the expression of miR-92a by glioma cells. The expression of the antitumor molecules, including perforin, FasL, and IFN-γ, appeared significantly attenuated if compared with control. The IL-6+ IL-10+ NKT cells showed less potential in inducing apoptosis in glioma cells, but showed the ability to suppress cytotoxic CD8+ T cells [54]. An increase in the population of CD4+CD25highFoxp3+ regulatory T cells (Tregs), a subset of CD4+ T cells able to maintain self-tolerance and modulate immune responses, is among the mechanisms developed by cancer cells to mediate immune evasion [55,56]. It has been demonstrated that during carcinogenesis Tregs are increased [57], and their depletion holds promises as an anti-cancer therapy [58,59]. Yin et al. investigated the mechanisms underlying the above observation. They found that miR-214 was abundantly secreted into microvescicles to recipient T-cells, various types of human cancers and mouse tumor models. Tumor-derived miR-214 efficiently decreased phosphatase and tensin homolog (PTEN), and promoted Treg expansion in targeted mouse peripheral CD4+ T cells. Tregs induced by cancer cell-secreted miR-214 produced higher levels of IL-10 and promoted tumor growth in nude mice in addition to enhance immune suppression and tumor implantation/growth in mice as demonstrated by the block of Treg expansion and tumor growth when microvescicles containing anti-miR-214 antisense oligonucleotides were transfer to mice implanted with tumor [60]. Breast cancer stem-like cells (BCSCs) are highly metastatic and resistant to therapy because of their resistance to tumor-infiltrating NK cells cytotoxicity. This is due to downregulation in BCSCs of MHC class I polypeptide-related sequence A/B (MICA/MICB), two ligands for the stimulatory NK cell receptor NKG2D, mediated by aberrantly expression of oncogenic miR-20a. All-trans retinoic acid, an agent inducing breast cancer cell differentiation, restored the miR-20a-MICA/MICB axis and sensitized BCSC to NK cell citotoxicity, therefore reducing immune escape-associated BCSCs metastasis [61]. This correlation mir-20a/MICA/MICB/NK was showed also in human ovarian cancer tissue where this miRNA enhances long-term cellular proliferation and invasion capabilities. MiR-20a binds directly to the MICA/B mRNA 3′UTR region, resulting in its degradation and reducing its protein levels on the plasma membrane thus leading to tumor cell evasion of NK-mediated killing. The treatment with anti-miR-20a enhanced NKG2D-mediated killing of cancer cells both in vitro and in vivo [62]. Similarly, miR-10b downregulates MICB expression with comparable effects on NK cells and tumor proliferation [63]. Together these findings provide evidence that cancer cells can actively shape immune cells functions through delivery of exosomes containing specific miRNAs.
MiRNAs involved in the line of communication cancer-lymphocytes are shown in Table 2.
Table 2.
miRNAs involved in the line of communication cancer-lymphocytes
Summary and Future Directions
The TME is emerging as a focal component in cancer biology and resistance to treatments. In TME, defined as the tumor surrounding cellular environment, each components communicates with and affects the behavior of cancer cells, leading to a constant balancing between pro- and anti-tumor phenotype. One of the main ways of communication between these cells is mediated by EVs. The most extensively studied EVs are exosomes (30–100 nm), in particular TEX that are currently accepted as mediators of carcinogenesis [64], and can functionally transfer their content to the recipient cells [16]. Immune-evasion represents one of the hallmarks of cancer success growth in an organism, and several studies have highlighted the role of TEX as a winning strategy exploited by cancer cells to evade immune-surveillance [65]. TEX contain not only proteins and lipids, but also genetic messages like genomic DNA, mRNA, and miRNAs. Increasing evidence points towards a central role of exosomal miRNAs in the dialogue between cancer cells and immune cells, and the outcome of cancer-immune system interactions is orchestrated by this inter-cellular shuttling of these small non-coding RNAs. Certainly this observation does not exclude a role for other tumor-derived exosome cargo molecules. Actually, a systematic analysis of the effects of proteins versus mRNAs versus miRNAs versus long non-coding RNAs versus DNA is missing and only such a systematic approach will address the issue of the relative contribution of any of these macromolecules in immune-escape scenarios. A further layer of complexity is represented by the complexity of the TME and the fact that shuttling of exosomal cargo constantly occurs among cancer cells and the several different cell populations of the TME. Moreover, the stoichiometry of these interactions is completely unknown. For instance, it is unknown how different concentrations of TEX miRNAs will affect targeting of mRNAs in surrounding immune-cells, therefore affecting the overall balance between immune-stimulation and immune-escape.
Future efforts will have to address these questions, especially to promote our translational endeavors towards the development of new therapies that impair TEX-mediated immune-escape and re-establish a healthier and reactive anti-tumoral immune response.
Acknowledgments
Dr. Fabbri is a St. Baldrick Foundation’s Scholar and is supported by a Hyundai Hope on Wheels grant, a William Lawrence & Blanche Hughes Foundation grant, a Jean Perkins Foundation grant, the Nautica Malibu Triathlon Funds, a STOP Cancer grant, the Hugh and Audy Lou Colvin Foundation grant, an Alex’s Lemonade Stand Foundation grant, and the award number P30CA014089 from the National Cancer Institute.
Footnotes
Authors’ contributions
Both authors were involved in drafting the manuscript, revising it for intellectual content, and reading and approving the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taylor DD, Chou IN, Black PH. Isolation of plasma membrane fragments from cultured murine melanoma cells. Biochem Biophys Res Commun. 1983;113:470–476. doi: 10.1016/0006-291x(83)91749-7. [DOI] [PubMed] [Google Scholar]
- 2.Taylor DD, Taylor CG, Jiang CG, Black PH. Characterization of plasma membrane shedding from murine melanoma cells. Int J Cancer. 1988;41:629–635. doi: 10.1002/ijc.2910410425. [DOI] [PubMed] [Google Scholar]
- 3.Taylor DD, Gercel-Taylor C, Lyons KS, Stanson J, Whiteside TL. T-cell apoptosis and suppression of T-cell receptor/CD3-zeta by Fas ligand-containing membrane vesicles shed from ovarian tumors. Clin Cancer Res. 2003;9:5113–5119. [PubMed] [Google Scholar]
- 4.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 5.Keller S, Ridinger J, Rupp A-K, Janssen JWG, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poliakov A, Spilman M, Dokland T, Amling CL, Mobley JA. Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. Prostate. 2009;69:159–67. doi: 10.1002/pros.20860. [DOI] [PubMed] [Google Scholar]
- 7.Sharma S, Gillespie BM, Palanisamy V, Gimzewski JK. Quantitative Nano-structural and Single Molecule Force Spectroscopy bio-molecular analysis of human saliva derived exosomes. Langmuir. 2011;27:14394–14400. doi: 10.1021/la2038763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skriner K, Adolph K, Jungblut PR, Burmester GR. Association of citrullinated proteins with synovial exosomes. Arthritis Rheum. 2006;54:3809–3814. doi: 10.1002/art.22276. [DOI] [PubMed] [Google Scholar]
- 9.Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Wieckowski E, Whiteside TL. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol Res. 2006;36:247–254. doi: 10.1385/IR:36:1:247. [DOI] [PubMed] [Google Scholar]
- 11.Boyiadzis M, Whiteside TL. Information transfer by exosomes: A new frontier in hematologic malignancies. Blood Rev. 2015;29:281–290. doi: 10.1016/j.blre.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Bobrie A, Thery C. Exosomes and communication between tumours and the immune system: are all exosomes equal? Biochem Soc Trans. 2013;41:263–267. doi: 10.1042/BST20120245. [DOI] [PubMed] [Google Scholar]
- 13.Gabrielsson S, Scheynius A. Exosomes in immunity and cancer--friends or foes? Semin Cancer Biol. 2014;28:1–2. doi: 10.1016/j.semcancer.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11:1010–1020. [PubMed] [Google Scholar]
- 16.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.KA, G-JS miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014 Jan;42(Database issue):D68–73. doi: 10.1093/nar/gkt1181. Epub 2013 Nov 25. (n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.BP L, Burge CB, CB B, DP B. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005 Jan 14;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. (n.d.) [DOI] [PubMed] [Google Scholar]
- 21.L JR, Y TA, Steitz JA, S JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007 Jun 5;104(23):9667–72. doi: 10.1073/pnas.0703820104. Epub 2007 May 29. (n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.JJ F, A L-M, Coller HA, HA C. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008 Sep 30;105(39):14879–84. doi: 10.1073/pnas.0803230105. Epub 2008 Sep 23. (n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.AV The functions of animal microRNAs. Nature. 2004 Sep 16;431(7006):350–5. doi: 10.1038/nature02871. (n.d.) [DOI] [PubMed] [Google Scholar]
- 24.VS, TY, Steitz JA, JA S. Switching from repression to activation: microRNAs can up-regulate translation. Sci. Dec 21;318(5858):1931–4. doi: 10.1126/science.1149460. Epub 2007 Nov 29. (n.d.) [DOI] [PubMed] [Google Scholar]
- 25.Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortez-Retamozo V, Etzrodt M, Newton A, Rauch PJ, Chudnovskiy A, Berger C, Ryan RJH, Iwamoto Y, Marinelli B, Gorbatov R, Forghani R, Novobrantseva TI, Koteliansky V, Figueiredo J-L, Chen JW, Anderson DG, Nahrendorf M, Swirski FK, Weissleder R, Pittet MJ. Origins of tumor-associated macrophages and neutrophils. Proc Natl Acad Sci. 2012;109:2491–2496. doi: 10.1073/pnas.1113744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su S, Liu Q, Chen J, Chen J, Chen F, He C, Huang D, Wu W, Lin L, Huang W, Zhang J, Cui X, Zheng F, Li H, Yao H, Su F, Song E. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25:605–620. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 28.Qian B-Z, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. http://dx.doi.org/10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marelli G, Allavena P, Erreni M. Tumor-associated macrophages, multi-tasking cells in the cancer landscape. Cancer Res Front. 2015;1:149–161. doi: 10.17980/2015.149. [DOI] [Google Scholar]
- 30.Colvin EK. Tumor-associated macrophages contribute to tumor progression in ovarian cancer. Front Oncol. 2014;4:137. doi: 10.3389/fonc.2014.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marton A, Vizler C, Kusz E, Temesfoi V, Szathmary Z, Nagy K, Szegletes Z, Varo G, Siklos L, Katona RL, Tubak V, Howard OMZ, Duda E, Minarovits J, Nagy K, Buzas K. Melanoma cell-derived exosomes alter macrophage and dendritic cell functions in vitro. Immunol Lett. 2012;148:34–38. doi: 10.1016/j.imlet.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–6. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, Kennedy R, Ivan C, Zhang X, Vannini I, Fanini F, Amadori D, Calin GA, Hadjidaniel M, Shimada H, Jong A, Seeger RC, Asgharzadeh S, Goldkorn A, Fabbri M. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang J-Y, Lee J-K, Jeon Y-K, Kim C-W. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer. 2013;13:421. doi: 10.1186/1471-2407-13-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Squadrito ML, Pucci F, Magri L, Moi D, Gilfillan GD, Ranghetti A, Casazza A, Mazzone M, Lyle R, Naldini L, De Palma M. miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell Rep. 2012;1:141–154. doi: 10.1016/j.celrep.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Wang S-X, Mu R, Luo X, Liu Z-S, Liang B, Zhuo H-L, Hao X-P, Wang Q, Fang D-F, Bai Z-F, Wang Q-Y, Wang H-M, Jin B-F, Gong W-L, Zhou T, Zhang X-M, Xia Q, Li T. Dysregulation of the miR-324-5p-CUEDC2 axis leads to macrophage dysfunction and is associated with colon cancer. Cell Rep. 2014;7:1982–1993. doi: 10.1016/j.celrep.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Sonda N, Simonato F, Peranzoni E, Calì B, Bortoluzzi S, Bisognin A, Wang E, Marincola FM, Naldini L, Gentner B, Trautwein C, Sackett SD, Zanovello P, Molon B, Bronte V. miR-142-3p Prevents Macrophage Differentiation during Cancer-Induced Myelopoiesis. Immunity. 2013;38:1236–1249. doi: 10.1016/j.immuni.2013.06.004. http://dx.doi.org/10.1016/j.immuni.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto M, Oshikawa T, Tano T, Ahmed SU, Kan S, Sasai A, Akashi S, Miyake K, Moriya Y, Ryoma Y, Saito M, Sato M. Mechanism of anticancer host response induced by OK-432, a streptococcal preparation, mediated by phagocytosis and Toll-like receptor 4 signaling. J Immunother. 2006;29:78–86. doi: 10.1097/01.cji.0000192106.32206.30. [DOI] [PubMed] [Google Scholar]
- 40.Gabrilovich D. Mechanisms and functional significance of tumour-induced dendritic-cell defects. Nat Rev Immunol. 2004;4:941–952. doi: 10.1038/nri1498. [DOI] [PubMed] [Google Scholar]
- 41.Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32:19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Xiang X, Zhuang X, Zhang S, Liu C, Cheng Z, Michalek S, Grizzle W, Zhang H-G. Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells. Am J Pathol. 2010;176:2490–2499. doi: 10.2353/ajpath.2010.090777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu S, Liu C, Su K, Wang J, Liu Y, Zhang L, Li C, Cong Y, Kimberly R, Grizzle WE, Falkson C, Zhang H-G. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J Immunol. 2007;178:6867–6875. doi: 10.4049/jimmunol.178.11.6867. [DOI] [PubMed] [Google Scholar]
- 44.Valenti R, Huber V, Filipazzi P, Pilla L, Sovena G, Villa A, Corbelli A, Fais S, Parmiani G, Rivoltini L. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-beta-mediated suppressive activity on T lymphocytes. Cancer Res. 2006;66:9290–9298. doi: 10.1158/0008-5472.CAN-06-1819. [DOI] [PubMed] [Google Scholar]
- 45.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 46.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 47.Zhou M, Chen J, Zhou L, Chen W, Ding G, Cao L. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cell Immunol. 2014;292:65–69. doi: 10.1016/j.cellimm.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Ding G, Zhou L, Qian Y, Fu M, Chen J, Chen J, Xiang J, Wu Z, Jiang G, Cao L. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget. 2015;6:29877–29888. doi: 10.18632/oncotarget.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Que L, Ri-sheng, Cheng Lin, Guo-ping Ding, Zheng-rong Wu, Cao Increasing the immune activity of exosomes: the effect of miRNA-depleted exosome proteins on activating dendritic cell/cytokine-induced killer cells against pancreatic cancer. J Zhejiang Univ B. 2016;17:352–360. doi: 10.1631/jzus.B1500305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berzins SP, Ritchie DS. Natural killer T cells: drivers or passengers in preventing human disease? Nat Rev Immunol. 2014;14:640–646. doi: 10.1038/nri3725. [DOI] [PubMed] [Google Scholar]
- 51.Donatelli SS, Zhou J-M, Gilvary DL, Eksioglu EA, Chen X, Cress WD, Haura EB, Schabath MB, Coppola D, Wei S, Djeu JY. TGF-β–inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci U S A. 2014;111:4203–4208. doi: 10.1073/pnas.1319269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noman MZ, Buart S, Romero P, Ketari S, Janji B, Mari B, Mami-Chouaib F, Chouaib S. Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res. 2012;72:4629–4641. doi: 10.1158/0008-5472.CAN-12-1383. [DOI] [PubMed] [Google Scholar]
- 53.Berchem G, Noman MZ, Bosseler M, Paggetti J, Baconnais S, Le cam E, Nanbakhsh A, Moussay E, Mami-Chouaib F, Janji B, Chouaib S. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-β and miR23a transfer. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2015.1062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang B, Wu W, Wei X, Li Y, Ren G, Fan W. Activation of Glioma Cells Generates Immune Tolerant NKT Cells. J Biol Chem. 2014;289:34595–34600. doi: 10.1074/jbc.M114.614503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakaguchi S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 56.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 57.Fu J, Xu D, Liu Z, Shi M, Zhao P, Fu B, Zhang Z, Yang H, Zhang H, Zhou C, Yao J, Jin L, Wang H, Yang Y, Fu Y-X, Wang F-S. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology. 2007;132:2328–2339. doi: 10.1053/j.gastro.2007.03.102. [DOI] [PubMed] [Google Scholar]
- 58.Quezada SA, Peggs KS, Simpson TR, Shen Y, Littman DR, Allison JP. Limited tumor infiltration by activated T effector cells restricts the therapeutic activity of regulatory T cell depletion against established melanoma. J Exp Med. 2008;205:2125–2138. doi: 10.1084/jem.20080099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Q, Bucher C, Munger ME, Highfill SL, Tolar J, Munn DH, Levine BL, Riddle M, June CH, Vallera DA, Weigel BJ, Blazar BR. Depletion of endogenous tumor-associated regulatory T cells improves the efficacy of adoptive cytotoxic T-cell immunotherapy in murine acute myeloid leukemia. Blood. 2009;114:3793–3802. doi: 10.1182/blood-2009-03-208181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin Y, Cai X, Chen X, Liang H, Zhang Y, Li J, Wang Z, Chen X, Zhang W, Yokoyama S, Wang C, Li L, Li L, Hou D, Dong L, Xu T, Hiroi T, Yang F, Ji H, Zhang J, Zen K, Zhang C-Y. Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Res. 2014;24:1164–1180. doi: 10.1038/cr.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang B, Wang Q, Wang Z, Jiang J, Yu S-C, Ping Y-F, Yang J, Xu S-L, Ye X-Z, Xu C, Yang L, Qian C, Wang JM, Cui Y-H, Zhang X, Bian X-W. Metastatic Consequences of Immune Escape from NK Cell Cytotoxicity by Human Breast Cancer Stem Cells. Cancer Res. 2014;74:5746–5757. doi: 10.1158/0008-5472.CAN-13-2563. http://cancerres.aacrjournals.org/content/74/20/5746.abstract. [DOI] [PubMed] [Google Scholar]
- 62.Xie J, Liu M, Li Y, Nie Y, Mi Q, Zhao S. Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell Mol Immunol. 2014;11:495–502. doi: 10.1038/cmi.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsukerman P, Stern-Ginossar N, Gur C, Glasner A, Nachmani D, Bauman Y, Yamin R, Vitenshtein A, Stanietsky N, Bar-Mag T, Lankry D, Mandelboim O. MiR-10b downregulates the stress-induced cell surface molecule MICB, a critical ligand for cancer cell recognition by natural killer cells. Cancer Res. 2012;72:5463–5472. doi: 10.1158/0008-5472.CAN-11-2671. [DOI] [PubMed] [Google Scholar]
- 64.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whiteside TL. Tricks tumors use to escape from immune control. Oral Oncol. 2009;45:e119–23. doi: 10.1016/j.oraloncology.2009.03.006. [DOI] [PubMed] [Google Scholar]