Abstract
The lack of information on bacteriophages of Clostridium difficile prompted this study. Three of 56 clinical C. difficile isolates yielded double-stranded DNA phages φC2, φC5, φC6, and φC8 upon induction. Superinfection and DNA analyses revealed relatedness between the phages, while partial sequencing of φC2 showed nucleotide homology to the sequenced C. difficile strain CD630.
Clostridium difficile has risen from relative obscurity 20 years ago to be an important hospital pathogen recognized as the cause of a wide spectrum of enteric diseases, including C. difficile-associated diarrhea (CDAD) (28). Almost all antimicrobials have now been implicated as being able to incite CDAD, including the two agents most commonly used for treatment, vancomycin and metronidazole (12). Thus, alternative nonantibiotic treatment modalities have been sought. There has been renewed interest in phage therapy (35) with the recent success of whole-phage and purified-phage components used in treating infections (7) or as antimicrobial agents (18, 31). Although phages of the Clostridium species have been described previously (22), there have been a limited number of studies of C. difficile phages (20, 27). Some studies deal with their use in strain typing (10, 32), but extensive studies of the molecular biology of C. difficile phages are lacking. The aim of this study, therefore, was to isolate and characterize bacteriophages specific for C. difficile as a preliminary step to assessing their potential as novel therapeutic agents.
Fifty-six C. difficile strains were isolated from patients with CDAD in Sir Charles Gairdner Hospital, Perth, Western Australia, Australia, by previously described methods (29). Clostridium perfringens (13 isolates), Clostridium septicum (2 isolates), Staphylococcus aureus (10 isolates), Lactobacillus spp. (10 isolates), and Bacillus subtilis ATCC 6633 were obtained from the culture collection of The Western Australian Centre for Pathology and Medical Research. All cultures were incubated in an anaerobic chamber (80% N2, 10% CO2, and 10% H2; model Mark III; Don Whitley Scientific Limited) at 37°C, except B. subtilis ATCC 6633. Stationary-phase (16- to 18-h) and log-phase (2.5- to 3-h) cultures of bacteria were prepared in brain heart infusion broth. MRS medium was also used for Lactobacillus spp. Anaerobic plaque assays were done on anaerobe basal agar (ABA) and overlay agar (0.74% [wt/vol] ABA, 0.01 M CaCl2, 0.4 M MgCl2). Rogosa agar was also used for Lactobacillus spp., and aerobic plaque assays of S. aureus and B. subtilis were done on nutrient agar.
C. difficile phages were not detected in environmental samples.
Eight samples of soil and animal feces were collected from areas around and away from Sir Charles Gairdner Hospital; untreated sewage was collected from a treatment plant on five different occasions. Soil and fecal samples were mixed with 2 volumes of phosphate-buffered saline; these and the sewage samples were centrifuged at 17,700 × g for 60 min at 4°C and filtered through 0.22-μm-pore-size filters (Acrodisc). Three different methods, i.e., direct assay (2), specific enrichment (2), and host adsorption (26), were used in attempts to detect virulent phages from environmental sources. Samples were assayed for plaques by spotting 10 μl of sample onto 1.5 ml of overlay agar seeded with 600 μl of indicator culture. Sixteen of the 56 C. difficile strains were used as indicators. Despite the use of multiple methods and samples, no virulent phages specific to C. difficile were isolated. Various factors that may influence phage isolation (time of adsorption incubation, volume of the host used for adsorption, temperature of samples, and indicator strains used) were changed several times in an attempt to optimize each isolation technique based on theoretical considerations. C. difficile is a strict anaerobe that exists as spores in the environment (3). Virulent phages require their host to be in an active state of growth for infection and multiplication to occur. Cell surface structures such as pili, which serve as receptors for phages, are absent in a spore. This may explain the absence or rarity of C. difficile-specific virulent phages in the environment that we sampled. A lysogenic cycle is presumably convenient for phages of spore-forming anaerobes, as prophages are not limited by the availability of metabolically active anaerobic cells in an often hostile environment (34).
Four temperate phages were isolated, and lysogens of the phages were unstable.
Three (5.4%) of 56 C. difficile strains (producers) induced with 3 μg of mitomycin C ml−1 (Sigma) as previously described (32) were lysogenic (Table 1). Phages φC2 and φC5 were propagated in stationary cultures of CD062, while φC6 and φC8 were propagated in log-phase cultures of CD60 and CD843, respectively, on agar from a single plaque (20, 32). Phage was extracted by mixing the overlay agar and phage buffer (0.15 M NaCl, 10 mM Tris [pH 6.5], 10 mM MgSO4, 1 mM CaCl2) in the ratio 1:3 and then filtering the supernatant (0.8- and 0.2-μm-pore-size filters; Acrodisc) to obtain crude phage suspensions.
TABLE 1.
General properties of four C. difficile phages
| Property | Result for phage:
|
|||
|---|---|---|---|---|
| φC2 | φC5 | φC6 | φC8 | |
| Induced host strain | CD242 | CD578 | CD371 | CD371 |
| Propagating strain | CD062 | CD062 | CD60 | CD843 |
| Plaque appearance, diam (mm) | Clear, 1 | Clear, 1 | Turbid, 0.5 | Clear, 1.5 |
| Particle morphology | ||||
| Width of particle head (nm)a | 64.8 ± 3.4 | 57.9 ± 6.9 | 69.6 ± 2 | 59.8 ± 3.7 |
| Length of tail (nm)a | 147.7 ± 46.9 | 118.3 ± 9.6 | 337.4 ± 9.8 | 139.6 ± 22.3 |
| Sheathg | + | + | − | + |
| Morphotypeb | A1 | A1 | B1 | A1 |
| Single-step growth curve | ||||
| Burst sizec | 5 | 7 | 19 | 33 |
| Latent period (min)d | 32 | 36 | 118 | 90 |
| MOIe | 0.0018 | 0.0019 | 0.035 | 0.007 |
| Estimated genome size (kb)f | 43.3 ± 3.6 | 45.9 ± 3.8 | 36.3 ± 1.2 | 54.5 ± 3.8 |
Means ± standard deviations, 4 to 10 complete or defective phages were measured.
Morphotype A1 belonged to Myoviridae, and B1 belonged to Siphoviridae.
Average yield of virus particles per infected host cell.
Minimum length of time from host cell adsorption to lysis.
The multiplicity of infection (MOI) is the ratio of adsorbed phage particles to bacteria.
Estimated by adding up fragment sizes of phage DNA digested with HindIII. Means ± SD standard deviations of sizes of bands in three profiles were determined.
+, sheath present; −, sheath absent.
Lysogens are bacterial cells harboring prophage(s) that may be induced to produce phage particles (19). In order to isolate lysogens of the four phages, hosts were infected with phage and assayed for plaques. The centers of clear or turbid plaques were picked and cultured anaerobically on blood agar for 48 h. Single colonies were tested for prophage by plaque assay with the uninfected strain following induction with 3 μg of mitomycin C ml−1. The stability of lysogenic status after storage at −70°C was also determined by colony blotting (16) with digoxigenin (DIG)-labeled phage probes as described below. Of the 50 lysogens isolated in this manner, 72% spontaneously lost their prophage after storage, and no plaques were detected postinduction. A total of 14 lysogens, i.e., 3 of φC2, 5 of φC6, and 6 of φC8, remained stable. No stable lysogens of φC5 were isolated. Reasons for the apparent high rates of prophage instability among lysogens were not determined; however, this may be a consequence of host stress resulting in phage induction (33).
φC2, φC5, and φC8 have similar primary characteristics.
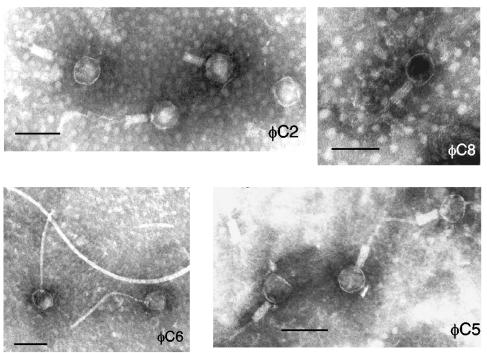
Particle morphology was observed by electron microscopy. Crude phage preparations (4.5 ml) were centrifuged at 30,000 ×g for 2 h at 4°C, and the pellet was resuspended in 50 to 100 μl of phage buffer and applied to carbon-coated Formvar grids (300 mesh Supergrids; SPI Supplies). The grids were stained for 2 min with 1% (wt/vol) aqueous Alcian blue dissolved in highly pure water, briefly rinsed with water, and blotted dry. Grids were negatively stained with 2% (wt/vol) sodium silicotungstate (pH 6.8) and air dried before they were viewed under a Phillips 401 transmission electron microscope operating at 80 kV. φC2, φC5, and φC8 particles (Myoviridae) were morphologically similar, while φC6 particles (Siphoviridae) differed in that they had long, flexuous, striated tails (Fig. 1; Table 1). Other primary properties determined include single-step growth curves, calculated as described previously (2), except that all manipulations were performed anaerobically. φC2 and φC5 had short latent periods and small burst sizes, while φC6 and φC8 had longer latent periods and larger burst sizes (Table 1). Overall, burst sizes were smaller than for temperate phages of other bacterial species (6, 15, 38); possibly, optimal growth conditions were not achieved (1). To determine phage host ranges, serial dilutions of crude phage preparations (≥109 or ≥106 PFU per ml−1 for φC6) were assayed for plaques on stationary- and log-phase bacterial cultures. The host ranges of each phage against C. difficile were different, although some overlap was evident between φC2 and φC5 (3 of 56 isolates) and between φC6 and φC8 (14 of 56 isolates), suggesting the recognition of common phage receptors for each pair. φC8 had the broadest host range, lysing 26 out of 56 C. difficile isolates (46%), followed by φC6 (43%), φC5 (20%), and φC2 (16%). The phages were not active against the C. perfringens, C. septicum, S. aureus, B. subtilis, and Lactobacillus species isolates tested.
FIG. 1.
Phages φC2, φC8, and φC5 possess contractile sheaths and rigid tails, while φC6 particles have long, flexible tails. Bar, 100 nm.
φC6 is distantly related to φC2, φC5, and φC8, which are similar.
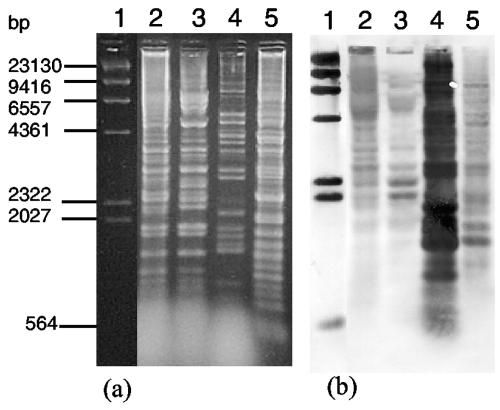
Phage relationships were investigated by restriction analyses, superinfection experiments, and Southern hybridizations. Crude phage suspensions were concentrated and purified through a preformed CsCl density gradient before DNA extraction (30). Phage DNA was purified (Wizard Clean-up system; Promega) and digested according to the manufacturer's instructions (Promega). Banding patterns of φC2 and φC5 generated by HindIII and XbaI double digestion had the majority of bands in common (Fig. 2a). These digests were cross-hybridized under stringent conditions with φC5 and φC6 DNA probes to determine the degree of genomic similarity. Digested DNA was transferred onto a membrane (Amersham Pharmacia Biotech) (30), fixed by heating in a microwave at 700 W for 2.5 min (5), and hybridized according to the manufacturer's instructions (DIG High Prime DNA labeling and detection starter kit I; Roche) to DIG-labeled phage probes prepared using purified whole-phage DNA. Southern hybridizations were repeated at least three times. Of the 20 hybridized bands for φC8 (Fig. 2b, lane 5), approximately 25% were similar in molecular weight to φC6 bands, compared to the restriction profile of φC6 (Fig. 2a, lane 4). Of the 12 and 11 hybridized bands for φC2 and φC5 (Fig. 2b, lanes 2 and 3), respectively, approximately 12.5 and 16%, respectively, were similar in molecular weight to φC6 bands. These results indicated moderate homology between φC6 and φC8 and low homology to φC2 and φC5. Conversely, cross-hybridization of the φC5 probe occurred most in φC2 and φC8 and least in φC6 (data not shown), suggesting high sequence homology between φC2 and φC5, some homology between them and φC8, and little homology to φC6. Immunity of lysogens to phage superinfection was tested as previously described (37). Strains lysogenic for φC2 and φC5 were immune to superinfection by φC2 and φC5 but were susceptible to φC6 and φC8. Lysogens of φC6 and φC8 were susceptible to the other three phages. Thus, φC2 and φC5 were related and used similar repressors, while φC6 and φC8 were either unrelated to the other phages or had different operator sequences. φC6 is probably most distantly related to the other phages; however, its genetic sequence similarity to φC8 could have resulted from horizontal genetic transfer or recombination with φC8 while coinfecting CD371. The method for determination of cohesive ends was described previously (23). Restriction enzymes for 1 μg of DNA were PvuII or XbaI for φC2, XbaI or HindIII for φC6, and BamHI for λ, a positive control. Cohesive ends were not detected in φC2, as there was no difference between the restriction profiles of untreated and ligase-treated DNA, while cohesive ends were detected in φC6.
FIG. 2.
Restriction digests and Southern hybridization of phage DNA. (a) HindIII and XbaI double digest of phage DNA. Lanes: 1, λ, HindIII DIG-labeled marker (Roche); 2, φC2; 3, φC5; 4, φC6; 5, φC8. (b) Southern hybridization of gel shown in panel a with DIG-labeled φC6 genomic probe (25 ng ml−1).
φC2 integrates into the host chromosome.
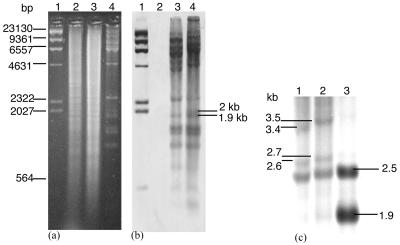
Of the four phages, φC2 was the most robust, reproducing in moderately high titers. It was therefore chosen to determine whether its prophage existed extrachromosomally or chromosomally. In order to isolate C. difficile DNA, an overnight broth culture (10 ml) was concentrated 10-fold in Tris-EDTA buffer (pH 8). Cells were treated with 2 μg of lysozyme ml−1 at 37°C for 30 min followed by 1% (vol/vol) sodium dodecyl sulfate, 40 mM EDTA, and 500 μg of proteinase K ml−1 at 55°C for 1 to 2 h and 100 μg of RNase A ml−1 at 37°C for 30 min. DNA was extracted twice with equal volumes of phenol-chloroform-isoamyl alcohol (25:24:1) followed by chloroform, precipitated with 0.3 M sodium acetate (pH 5.2) and ethanol, dissolved in 50 μl of Tris-EDTA buffer (pH 8), and purified (Wizard Clean-up system; Promega). Phage and bacterial DNA were digested with HindIII, separated in agarose, transferred to a membrane, and hybridized to the φC2 probe as described above. Undigested chromosomal DNA of lysogen CD839C2 hybridized to the φC2 probe, while the parental strain CD839 did not, indicating probable chromosomal integration of φC2 (data not shown). Further investigation of the prophage integration site through comparing hybridization patterns of HindIII-digested phage and lysogen DNA revealed a 1.9- to 2-kb band for φC2 that was absent for CD839C2 (Fig. 3b). The 1.9- and 2-kb HindIII fragments were extracted from agarose (QIAEX II; QIAGEN) and DIG labeled (Roche). Use of the 1.9-kb fragment as a probe for hybridization to DNA of lysogens resulted in the appearance of new 2.6- and 3.4-kb bands for CD839C2 (Fig. 3c, lane 1) and 2.7- and 3.5-kb bands for CD843C2 (Fig. 3c, lane 2), while use of the 2-kb fragment did not result in the detection of new bands. The 1.9-kb band was not detected in the lysogens, suggesting that an attP site was within the fragment. It is likely that prophage integration occurred at the same site in both lysogens, despite the 1-kb difference between the new bands, which may have been due to protein contamination. The probe also bound to a 2.5-kb band present for both phage and lysogen DNA, possibly due to gene duplication or incomplete digestion of φC2.
FIG. 3.
Determining the site of chromosomal prophage integration through Southern hybridization of DIG-labeled φC2 to DNA of lysogen(s). (a) Agarose gel of HindIII-digested DNA. Lanes: 1, λ HindIII DIG-labeled marker (Roche); 2, CD839; 3, CD839C2; 4, φC2. (b) A Southern blot of the gel shown in panel a with a φC2 genome probe (25 ng ml−1) revealed differences between the banding patterns of CD839C2 and φC2 in the 1.9- to 2-kb region. (c) Southern hybridization of the 1.9-kb HindIII fragment (2.5 ng ml−1) to DNA of lysogens CD839C2 and CD843C4 resulted in new bands. Lanes: 1, CD839C2; 2, CD843C2; 3, φC2.
Cloning and sequence analysis of part of the φC2 genome reveals homology to a variety of organisms.
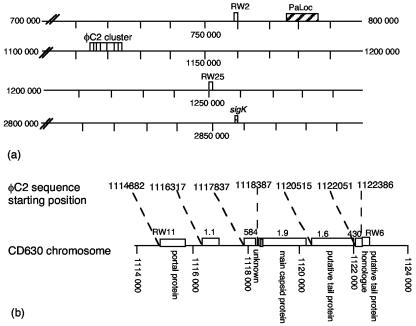
In total, 9 (i.e., sequences 1.9, 1.6, 1.1, 584, 430, RW2, RW6, RW11, and RW25) (Fig. 4a) of 16 φC2 HindIII fragments were cloned (30). Fragments cloned into pUC18 were transformed into Escherichia coli XL1-Blue {recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 lac [F′ proAB lacIqZM15:: Tn10(Tetr)]}. Plasmids were extracted (mini plasmid kit; QIAGEN) and sequenced (ABI 377 or ABI 3700 DNA sequencer; Applied Biosystems) and searched against databases using FASTA (24) and BLAST (4). φC2 sequences have the GenBank accession numbers AY522333 to AY522341. All translated sequences were homologous to putative gram-positive phage structural proteins, except for sequences 584, RW2, and RW25. For example, sequence 1.9 had 26% identity over 292 amino acids to a Lactobacillus delbrueckii phage main capsid protein (E value of 6e−21). Sequences 1.6, 430, and RW6 had 33% identity (181 amino acids; E value, 1e−19), 36% identity (105 amino acids; E value, 3e−09), and 31% identity (108 amino acids; E value, 1e−05), respectively, to B. subtilis PBSX putative phage tail proteins, while RW11 was homologous to a B. subtilis phage portal protein. It is not unusual for a phage genome to have diverse origins, and there is now overwhelming evidence that phages exchange genes in a modular fashion through the different species of hosts that they infect (8, 11, 14). Nucleotide homology between φC2 sequences and the recently sequenced C. difficile strain CD630 was found in three regions on CD630 (Fig. 4a), with seven of nine sequences clustered in nucleotide positions 1114882 to 1122754 (Fig. 4b). Given the relative order of sequences on CD630, it is possible that sequences 1.6, 430, and RW6 encoded phage tail proteins. The position of sequence RW11 relative to sequences 1.1, 584, 1.9, 1.6, 430, and RW6 in CD630 (Fig. 4b) is consistent with the locations of portal proteins relative to those of capsid and tail proteins in other phages, such as Lactobacillus lactis phages bIL285 and bIL286 (9) and Streptococcus thermophilus phage Sfi27 (11). RW2 and RW25 were located more than one phage genome length (i.e., >47 kb) from the main cluster, indicating that φC2 is not identical to the prophage in CD630. Furthermore, the putative attP in sequence 1.9 was intact on CD630, indicating that the prophage did not arrive in CD630 through site-specific integration at this attP. Although a putative attP sequence consisting of a central palindromic sequence flanked by peripheral imperfect repeats was located in sequence 1.9, further evidence is required to substantiate its function. The proximity of the C. difficile pathogenicity locus (PaLoc) to RW2 may indicate that it was once part of a prophage and may be interesting for discussions on the origins of the PaLoc, which are not known. The sigK gene, reported to contain a phage-like element (13), was not in close proximity to φC2 sequences (Fig. 4a).
FIG. 4.
φC2 prophage nucleotide positions in C. difficile CD630 genome. (a) φC2 sequences (blank boxes) were located between nucleotide positions 765221 and 765669, 1114882 and 1122754, and 1254421 and 1254907. Of the nine sequences, seven were clustered, and six of the seven showed protein homology to phage structural proteins. RW2 and RW25 were located 349,183 and 131,667 bases, respectively, from the cluster. The PaLoc (larger striped box) is shown located between nucleotide positions 786149 and 795379, approximately 20 and 319 kb from RW2 and RW11, respectively. sigK (smaller striped box) was not in close proximity to phage sequences. (b) The φC2 cluster on CD630 with the proposed attP in sequence 1.9 is shown in grey.
Finding φC2-like sequences in the genome of CD630 suggests that functional or cryptic prophages may be prevalent in C. difficile. There is amino acid sequence homology between tcdE in the PaLoc of C. difficile and phage holins (36), suggesting a possible association between the PaLoc and phage. Further characterization of C. difficile phages will enable a greater understanding of the genetics of these phages and elucidate their role, if any, in the pathogenicity of C. difficile. Although φC2 may not be suitable as a therapeutic agent for people with CDAD because of its temperate nature, it may have potential as a transducing phage. Its development should add to the small but growing number of integrative cloning vectors available for use with C. difficile (13, 17, 21, 25) and form the basis of a genetic tool useful in studies of clostridia.
Nucleotide sequence accession numbers.
Nucleotide sequence accession numbers AY522333 to AY522341 were allocated to the C2 sequences deposited in GenBank.
Acknowledgments
We thank Brian Mee and Manfred Beilharz for valuable advice on phage and molecular biology and Marina Silich-Carrara and Paul Caterina for their help with the transmission electron microscope.
REFERENCES
- 1.Abedon, S. T., T. D. Herschler, and D. Stopar. 2001. Bacteriophage latent-period evolution as a response to resource availability. Appl. Environ. Microbiol. 67:4233-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, M. 1959. Bacteriophages. Interscience Publishers, Inc., New York, N.Y.
- 3.Al Saif, N., and J. S. Brazier. 1996. The distribution of Clostridium difficile in the environment of South Wales. J. Med. Microbiol. 45:133-137. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angeletti, B., E. Battiloro, E. Pascale, and E. D'Ambrosio. 1995. Southern and Northern blot fixing by microwave oven. Nucleic Acids Res. 23:879-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashelford, K. E., J. C. Fry, M. J. Bailey, A. R. Jeffries, and M. J. Day. 1999. Characterization of six bacteriophages of Serratia liquefaciens CP6 isolated from the sugar beet phytosphere. Appl. Environ. Microbiol. 65:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas, B., S. Adhya, P. Washart, B. Paul, A. N. Trostel, B. Powell, R. Carlton, and C. R. Merril. 2002. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 70:204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brussow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 9.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dei, R. 1989. Observations on phage-typing of Clostridium difficile: preliminary evaluation of a phage panel. Eur. J. Epidemiol. 5:351-354. [DOI] [PubMed] [Google Scholar]
- 11.Desiere, F., S. Lucchini, and H. Brussow. 1999. Comparative sequence analysis of the DNA packaging, head, and tail morphogenesis modules in the temperate cos-site Streptococcus thermophilus bacteriophage Sfi21. Virology 260:244-253. [DOI] [PubMed] [Google Scholar]
- 12.Fekety, R., J. Silva, B. Buggy, and H. G. Deery. 1984. Treatment of antibiotic-associated colitis with vancomycin. J. Antimicrob. Chemother. 14(Suppl. D):97-102. [DOI] [PubMed] [Google Scholar]
- 13.Haraldsen, J. D., and A. L. Sonenshein. 2003. Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol. Microbiol. 48:811-821. [DOI] [PubMed] [Google Scholar]
- 14.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendry, G. S., and P. C. Fitz-James. 1974. Characteristics of φT, the temperate bacteriophage carried by Bacillus megaterium 899a. J. Virol. 13:494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karcher, S. J. 1995. Molecular biology: a project approach. Academic Press, San Diego, Calif.
- 17.Liyanage, H., S. Kashket, M. Young, and E. R. Kashket. 2001. Clostridium beijerinckii and Clostridium difficile detoxify methylglyoxal by a novel mechanism involving glycerol dehydrogenase. Appl. Environ. Microbiol. 67:2004-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeffler, J. M., D. Nelson, and V. A. Fischetti. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170-2172. [DOI] [PubMed] [Google Scholar]
- 19.Lwoff, A. 1953. Lysogeny. Bacteriol. Rev. 17:269-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahony, D. E., P. D. Bell, and K. B. Easterbrook. 1985. Two bacteriophages of Clostridium difficile. J. Clin. Microbiol. 21:251-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullany, P., M. Wilks, L. Puckey, and S. Tabaqchali. 1994. Gene cloning in Clostridium difficile using Tn916 as a shuttle conjugative transposon. Plasmid 31:320-323. [DOI] [PubMed] [Google Scholar]
- 22.Ogata, S., and M. Hongo. 1979. Bacteriophages of the genus Clostridium. Adv. Appl. Microbiol. 25:241-273. [DOI] [PubMed] [Google Scholar]
- 23.Pajunen, M., S. Kiljunen, and M. Skurnik. 2000. Bacteriophage φYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182:5114-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purdy, D., T. A. O'Keeffe, M. Elmore, M. Herbert, A. McLeod, M. Bokori-Brown, A. Ostrowski, and N. P. Minton. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46:439-452. [DOI] [PubMed] [Google Scholar]
- 26.Purdy, R. N., B. N. Dancer, M. J. Day, and D. J. Stickler. 1984. A novel technique for the enumeration of bacteriophage from water. FEMS Microbiol. Lett. 21:89-92. [Google Scholar]
- 27.Ramesh, V., J. A. Fralick, and R. D. Rolfe. 1999. Prevention of Clostridium difficile-induced ileocecitis with bacteriophage. Anaerobe 5:69-78. [Google Scholar]
- 28.Riley, T. V. 1998. Clostridium difficile: a pathogen of the nineties. Eur. J. Clin. Microbiol. Infect. Dis. 17:137-141. [DOI] [PubMed] [Google Scholar]
- 29.Riley, T. V., G. L. O'Neill, R. A. Bowman, and C. L. Golledge. 1994. Clostridium difficile-associated diarrhoea: epidemiological data from Western Australia. Epidemiol. Infect. 113:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schuch, R., D. Nelson, and V. A. Fischetti. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884-889. [DOI] [PubMed] [Google Scholar]
- 32.Sell, T. L., D. R. Schaberg, and F. R. Fekety. 1983. Bacteriophage and bacteriocin typing scheme for Clostridium difficile. J. Clin. Microbiol. 17:1148-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder, L., and W. Champness. 1997. Molecular genetics of bacteria. ASM Press, Washington, D.C.
- 34.Stewart, F. M., and B. R. Levin. 1984. The population biology of bacterial viruses: why be temperate. Theor. Popul. Biol. 26:93-117. [DOI] [PubMed] [Google Scholar]
- 35.Summers, W. C. 2001. Bacteriophage therapy. Annu. Rev. Microbiol. 55:437-451. [DOI] [PubMed] [Google Scholar]
- 36.Tan, K. S., B. Y. Wee, and K. P. Song. 2001. Evidence for holin function of tcdE gene in the pathogenicity of Clostridium difficile. J. Med. Microbiol. 50:613-619. [DOI] [PubMed] [Google Scholar]
- 37.Williamson, S. J., M. R. McLaughlin, and J. H. Paul. 2001. Interaction of the φHSIC virus with its host: lysogeny or pseudolysogeny? Appl. Environ. Microbiol. 67:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon, S. S., R. Barrangou-Poueys, F. Breidt, Jr., T. R. Klaenhammer, and H. P. Fleming. 2002. Isolation and characterization of bacteriophages from fermenting sauerkraut. Appl. Environ. Microbiol. 68:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]