Abstract
Listeria monocytogenes can be isolated from a range of food products and may cause food-borne outbreaks or sporadic cases of listeriosis. L. monocytogenes is divided into three genetic lineages and 13 serotypes. Strains of three serotypes (1/2a, 1/2b, and 4b) are associated with most human cases of listeriosis. Of these, strains of serotypes 1/2b and 4b belong to lineage 1, whereas strains of serotype 1/2a and many other strains isolated from foods belong to lineage 2. L. monocytogenes is isolated from foods by selective enrichment procedures and from patients by nonselective methods. The aim of the present study was to investigate if the selective enrichment procedure results in a true representation of the subtypes of L. monocytogenes present in a sample. Eight L. monocytogenes strains (four lineage 1 strains and four lineage 2 strains) and one Listeria innocua strain grew with identical growth rates in the nonselective medium brain heart infusion (BHI), but differed in their growth rate in the selective medium University of Vermont medium I (UVM I). When coinoculated in UVM I, some strains completely outgrew other strains. This outcome was dependent on the lineage of L. monocytogenes rather than the individual growth rate of the strains. When inoculated at identical cell densities in UVM I, L. innocua outcompeted L. monocytogenes lineage 1 strains but not lineage 2 strains. In addition, lineage 2 L. monocytogenes strains outcompeted lineage 1 L. monocytogenes strains in all combinations tested, indicating a bias in strains selected by the enrichment procedures. Bias also occurred when coinoculating two lineage 2 or lineage 1 strains; however, it did not appear to correlate with origin (clinical versus food). Identical coinoculation experiments in BHI suggested that the selective compounds in UVM I and II influenced this bias. The results of the present study demonstrate that the selective procedures used for isolation of L. monocytogenes may not allow a true representation of the types present in foods. Our results could have a significant impact on epidemiological studies, as lineage 1 strains, which are often isolated from clinical cases of listeriosis, may be suppressed during enrichment by other L. monocytogenes lineages present in a food sample.
Listeria monocytogenes is a food-borne pathogenic bacterium which can cause outbreaks or sporadic cases of listeriosis. The incidence of listeriosis is low; however, the bacterium is very important for food safety, since listeriosis is associated with very high mortality, approximately 25%, in susceptible individuals (9). It is crucial for implementation of control measures in the food industry that the food sources causing listeriosis be identified at points in the food chain where contamination occurs and that potential differences in virulence among different subtypes be understood. To be able to perform such analyses, subtyping of L. monocytogenes is crucial and is primarily based on serotyping or molecular methods (35). L. monocytogenes is divided into 13 known serovars, of which especially three, 4b, 1/2a, and 1/2b, are isolated from cases of listeriosis (20). Nucleotide variation in three virulence genes (actA, inlA, hly) grouped strains of L. monocytogenes into three groups (lineage 1, 2, and 3) (28, 36). The majority of clinical strains were found within lineage 1, which covers serotypes 4b and 1/2b, whereas serotype 1/2a belongs to lineage 2 (21). No clinical isolates were found within lineage 3 (36).
Listeria monocytogenes is commonly isolated from many different types of foods, including ready-to-eat products. Listeriosis is primarily associated with such products as they are not subjected to thermal treatment before consumption and an extended refrigerated shelf life may allow growth of the microorganism. The bacterium is often isolated from food-processing environments, where it may persist and cause continuous contamination of food products (11, 31). Recently, it has been suggested that biofilm formation by L. monocytogenes and, hence, potentially its ability to persist may differ among strains of different lineages (1).
Detection of L. monocytogenes from foods and food processing environments can be difficult because the bacterium in contaminated foods is normally found in very low numbers in a heterogeneous microflora. Hence, as is common practice for a number of food-borne pathogenic bacteria, enrichment steps are included in the analyses, and detection is often limited by the performance of the enrichment broth. Several studies have evaluated the performance of different isolation methods for their ability to detect low levels of L. monocytogenes, as well as injured cells (4, 25, 32, 33). One of the most commonly used enrichment broths is the University of Vermont medium (UVM), which contains nalidixic acid (suppresses gram-negative bacteria) and acriflavin (suppresses gram-positive bacteria) as selective supplements. Ideally, such enrichment procedures and selective medium should allow only the target organisms to proliferate. Also, since L. monocytogenes is often isolated from foods as part of epidemiological investigations, the procedures for enrichment and detection should ensure that all subtypes present in a sample be represented after enrichment. However, when sampling from foods, Loncarevic et al. (17) found more L. monocytogenes clones by direct plating compared to isolation following enrichment procedures.
Other Listeria species such as Listeria innocua are often found in foods, and since L. innocua may have a faster growth rate than L. monocytogenes in UVM and other enrichment broths, L. innocua may overgrow L. monocytogenes, which then escapes detection (6, 18, 26). Such observations also raise the question of whether the enrichment procedures allow equal growth of all subtypes of L. monocytogenes. If not, the enrichment procedure may be biased in selecting specific strains and subsequent epidemiological studies may be seriously hampered. Gracieux et al. (13) concluded that virulent L. monocytogenes strains reached significantly higher cell counts on selective agar media such as PALCAM, Oxford, Rapid L. mono (RLM), and ALOA Listeria agar than did nonvirulent strains. However, their study did not address any biases of these enrichment procedures.
Any systematic bias in the enrichment procedure vis-a-vis subtypes will clearly have a major impact on epidemiological studies. This is true in cases where food sources causing outbreaks are traced as well as studies of contamination of food-processing environments. In clinical investigations, L. monocytogenes will typically be isolated from blood or cerebrospinal fluid samples either directly on nonselective plates or following enrichment in nonselective broths. Obviously, the clinical isolates which are compared to food isolates have been isolated following a very different procedure. A number of studies have attempted to determine the links between specific ready-to-eat products and listeriosis by comparing large collections of L. monocytogenes isolated from foods with strains from clinical cases. Subtyping large collections of isolates by combinations of different molecular subtyping methods (pulsed-field gel electrophoresis, ribotyping, and randomly amplified polymorphic DNA) have not revealed any links between specific foods and disease (12, 19).
Based on the above, the purpose of the present study was to determine if one of the most common enrichment procedures for L. monocytogenes is biased in terms of differentially selecting specific subgroups. Also, the competition between L. innocua and L. monocytogenes during enrichment in UVM was investigated.
MATERIALS AND METHODS
Bacterial strains and storage conditions.
Eight strains of L. monocytogenes and one strain of L. innocua were used in the experiments (Table 1). The strains were chosen to allow lineages 1 and 2 to be represented by both food isolates and clinical strains. Six of the eight strains and the L. innocua strain were chosen as representing the major clusters described by Fonnesbech Vogel et al. (10). Cultures were stored at −80°C and streaked on brain heart infusion (BHI) plates (Oxoid CM225, Oxoid Ltd. Basingstoke, Hampshire, United Kingdom) and incubated for 24 h at 37°C before further culturing.
TABLE 1.
Listeria strains used in this study
| Species | Serogroup | Lineage | Strain | Origin | Reference |
|---|---|---|---|---|---|
| L. monocytogenes | 1/2a | 2 | La22 | Salmon smokehouse | 11 |
| 1/2a | 2 | HU4239 | Clinical case | 36 | |
| 1/2a | 2 | V5a | Salmon smokehouse | 11 | |
| 1/2a | 2 | C1-056 | Clinical case | 36 | |
| 4b | 1 | V518a | Salmon smokehouse | 11 | |
| 4b | 1 | 4542 | Clinical case | 11 | |
| 4b | 1 | C1-109 | Clinical case | 36 | |
| 1/2b | 1 | 7418 | Sausage | 11 | |
| L. innocua | R255a | Salmon smokehouse | 11 |
Growth rate of individual Listeria strains in UVM and BHI.
To determine if any possible selection in the selective University of Vermont medium (UVM) (Oxoid CM863 supplemented with SR0142 or SR0143) enrichment medium is caused by different growth rates, the growth rate and maximum cell density of all strains were determined in UVM I. One colony from each strain was inoculated into 10 ml of BHI broth and incubated for 24 h at 37°C. The cultures were diluted in BHI to a cell density of approximately 108 CFU/ml. Cell densities were adjusted based on optical density at 450 nm and standard curves relating colony counts to optical density were constructed for each strain.
The bacterial cultures were inoculated in 250 ml of UVM I (Oxoid CM863) at an initial level of 40 CFU/ml. Initial cell densities were confirmed by spread plating of the inoculation culture on BHI agar (incubated for 24 h at 37°C). Growth in UVM I at 30°C for 24 h was followed by sampling every 4 h and determining cell densities by spread plating on BHI (incubated for 24 h at 37°C); 100 μl of 24-h UVM I culture was transferred to 9.9 ml of UVM II and incubated for 24 h at 30°C, after which cell densities were determined by plate counts on BHI (incubated for 24 h at 37°C). Growth rates were determined from the exponential part of the growth curve (typically 102 to 107 CFU/ml) and comparisons of growth rates were done with Student's t test. Also, the maximum cell densities after growth for 24 h at 30°C in BHI were determined for all strains following an inoculation of approximately 40 CFU/ml.
Competition between Listeria strains in UVM I and II and BHI.
All strains were tested in selected combinations, allowing mixtures of L. monocytogenes and L. innocua as well as mixtures of different serotypes or lineages of L. monocytogenes from different sources to be studied. Cell densities of the precultures, grown as monocultures, were adjusted, reaching an initial cell density of 40 CFU/ml. After incubation of UVM I at 30°C for 24 h, 0.1 ml of the UVM I culture was transferred to 9.9 ml of UVM II (Oxoid CM863) and incubated for a further 24 h at 30°C. The density of L. monocytogenes and L. innocua in mixed cultures was determined by spread plating on Rapid L. mono (RLM) (Bio-Rad, Ivry-sur-Seine, France), where L. monocytogenes appears as blue colonies and L. innocua as white colonies.
To ensure that the selective plating medium itself did not bias the counting, these mixed cultures were also spread plated onto blood agar plates (incubated for 24 h at 37°C), where L. monocytogenes colonies appeared with zones of hemolysis whereas L. innocua was nonhemolytic. Cell densities of mixtures of two L. monocytogenes strains were determined by spread plating on BHI-agar, and to determine the ratio of the two strains, 34 to 40 colonies were isolated from the BHI plates and each was inoculated in 5 ml of BHI for 24 h at 37°C. Discrimination between the two L. monocytogenes, strains were done with serotyping and/or randomly amplified polymorphic DNA typing of the individual isolates (see below). Competition experiments within BHI were conducted as for UVM I.
Serotyping and lineage determination.
Serotyping was performed with commercial O-antigen Listeria antisera one and four (Bacto Listeria O antiserum 1 223001 and Bacto Listeria O antiserum 4 223011; Difco Laboratories). Colonies were grown in BHI broth for 24 h, and 1.5 ml of culture was boiled for 1 h. The samples were centrifuged at approximately 9,500 × g for 2 min, and the supernatant was removed. The pellets were resuspended in the remaining liquid. One drop of the resuspended organisms was mixed with one drop of antiserum on a glass plate and the plate was gently rocked for 1 min. Positive reaction was seen as coagulation in the sample. All samples were tested with both antisera one and four. The differentiation between serotypes a and b was done by Health Canada. The division in lineage 1 and 2 strains was derived from Fonnesbech Vogel et al. (10).
Randomly amplified polymorphic DNA typing.
DNA purification and randomly amplified polymorphic DNA (RAPD) were performed as previously described (11). Isolates were grown for 24 h in BHI at 37°C and DNA was purified with Dynabeads Dynal direct system 1 (Dynal A/S & Nordic, Oslo, Norway). Randomly amplified polymorphic DNA (RAPD) amplification was carried out with Ready-To-Go RAPD analysis beads (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) and primer HLWL85 (5′-ACAACTGCTC; DNA Technology, Aarhus, Denmark). The PCR was performed as follows: after denaturing for 2 min at 95°C, the next 10 cycles took place at 1 min of denaturing at 95°C, followed by annealing at 45 to 36°C, reducing the temperature with 1 degree Celsius for each cycle, and followed by 2 min at 72°C. The last 30 cycles used denaturing at 95°C for 1 min followed by annealing at 35°C for 1 min and extension at 72°C for 2 min followed by 10 min at 72°C. Nucleotide bands were separated in 2% agarose run at 90 V for 4 h and were visualized by staining with ethidium bromide.
Interaction between Listeria strains.
The possible antagonistic interaction between Listeria strains was determined to evaluate if the differential selection of strains was caused by inhibitory activities. Outgrown cultures as well as sterile filtered supernatants of outgrown cultures from all the Listeria spp. grown in BHI or UVM I were tested in an agar well-diffusion bioassay (23) against all of the Listeria spp. used in this study. One colony from each Listeria spp. was taken from a BHI plate and transferred to 10 ml of BHI broth or 10 ml of UVM I and incubated at 30°C for 24 h. The cell density was adjusted to approximately 107 CFU/ml; 500 μl of culture was inoculated in 50 ml of BHI broth (kept at 46°C) with 0.1% Tween 80, and the culture was immediately poured into plates. Wells (7 mm) were punched in the solidified agar and 50 μl of sterile supernatant or outgrown cultures was added to the wells. The plates were incubated at 30°C for 24 h, and any inhibition was seen as inhibition zones around the wells. Bacteriocin-producing Carnobacterium piscicola (23) was used as a positive control.
RESULTS
Individual growth.
All Listeria species reached almost identical cell densities (5 × 109 to 7 × 109 CFU/ml) when grown in BHI broth (Table 2). In contrast, UVM I limited growth and cell densities were 2 to 3 log10 units lower in UVM I than BHI broth after 24 h. Two clinical isolates (C1-056 and C1-109) were especially sensitive to the selective principles. Subsequent growth in UVM II allowed cultures to reach 108 to 109 CFU/ml (Table 2). Based on these results, the ability of the strains to grow on the selective substrate PALCAM was tested. Identical counts were found when comparing counts on PALCAM and BHI from outgrown cultures of all Listeria strains in BHI (data not shown).
TABLE 2.
Growth of six Listeria monocytogenes strains and one Listeria innocua strain in brain heart infusion broth and in UVM I and II
| Species | Strain | Log cell density after 24 h (CFU/ml)
|
Generation time (h) in UVM Ic | ||
|---|---|---|---|---|---|
| BHIa | UVM Ia | UVM IIb | |||
| L. monocytogenes | La22 | 9.70 ± 0.15 | 7.59 ± 0.27 | 8.98 ± 0.19 | 1.06 ± 0.05a,b |
| Hu4239 | 9.76 ± 0.12 | 7.64 ± 0.20 | 8.62 ± 0.29 | 1.09 ± 0.03a,b | |
| V5a | 9.81 ± 0.05 | 7.21 ± 0.14 | 9.11 ± 0.04 | 1.18 ± 0.09b,c,d | |
| C1-056 | 9.80 ± 0.04 | 6.72 ± 0.71 | 8.97 ± 0.06 | 1.28 ± 0.15c,d,e | |
| V518a | 9.77 ± 0.10 | 7.78 ± 0.21 | 9.05 ± 0.01 | 1.05 ± 0.02a | |
| C1-109 | 9.80 ± 0.21 | 6.22 ± 0.38 | 7.74 ± 1.79 | 1.31 ± 0.09c,e | |
| 4542 | 9.55 ± 0.15 | 7.04 ± 0.01 | 8.63 ± 0.03 | 1.18 ± 0.02c,d | |
| 7418 | 9.42 ± 0.06 | 7.30 ± 0.04 | 8.46 ± 0.05 | 1.16 ± 0.01c,d | |
| L. innocua | R255a | 9.75 ± 0.04 | 7.32 ± 0.81 | 8.98 ± 0.03 | 1.15 ± 0.11a,c,d,e |
After growth at 30°C for 24 h at an initial cell density of 40 cells/ml.
When inoculating 100 μl from UVM I into 9.9 ml of UVM II, followed by growth at 30°C for 24 h.
Growth rates with the same roman letter are not statistically significantly different (Student's t test at 5% level).
L. innocua versus L. monocytogenes in UVM I and UVM II.
Adjusting inoculation levels in cocultures based on absorbance measurements was very accurate and the inoculum levels of the two bacterial strains were almost identical (Table 3), with an average of 35 CFU/ml (data not shown). When L. innocua was coinoculated with L. monocytogenes lineage 2 strains, variation in the ratios of L. monocytogenes after 24 h of incubation was found, but both species were easily isolated after both UVM I and UVM II culturing. However, L. monocytogenes lineage 1 strains were outcompeted by L. innocua as only 2 to 6% of the colonies emerging after UVM I selective enrichment were L. monocytogenes and virtually no L. monocytogenes could be detected after UVM II selection (Table 3). Counts of hemolytic and nonhemolytic colonies on blood-agar plates were identical to counts of blue and white colonies on RLM plates (data not shown), indicating that the selective plates did not bias the counts.
TABLE 3.
Proportion of Listeria monocytogenes and Listeria innocua after coculturing in UVM I and UVM II at 30°C for 24 ha
| Lineage | Strains
|
No. of independent trials | Inoculation level (%)b
|
Distributionc (%)
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| UVM I
|
UVM II
|
||||||||
| A | B | A | B | A | B | A | B | ||
| 2 | La22 | R255a | 3 | 45 | 55 | 20 | 80 | 19 | 81 |
| 50 | 50 | 35 | 65 | 41 | 59 | ||||
| 47 | 53 | 50 | 50 | 68 | 32 | ||||
| 2 | V5a | R255a | 3 | 50 | 50 | 9 | 91 | 10 | 90 |
| 54 | 46 | 14 | 86 | 27 | 73 | ||||
| 49 | 51 | 77 | 23 | 72 | 28 | ||||
| 49 | 51 | 72 | 28 | 51 | 49 | ||||
| 2 | C1-056 | R255a | 1 | 50* | 50* | 95 | 5 | 72 | 28 |
| 50* | 50* | 89 | 11 | 74 | 26 | ||||
| 2 | Hu4239 | R255a | 1 | 51* | 49* | 25 | 75 | 51 | 49 |
| 51* | 49* | 27 | 73 | 50 | 50 | ||||
| 1 | V518a | R255a | 3 | 49 | 51 | 6 | 94 | 1 | 99 |
| 42 | 58 | 4 | 96 | 0.3 | 99.7 | ||||
| 47 | 53 | ND | ND | 1 | 99 | ||||
| 1 | C1-109 | R255a | 1 | 55* | 45* | 3 | 97 | 0 | 100 |
| 55* | 45* | 2 | 98 | 0 | 100 | ||||
A, L. monocytogenes; B, L. innocua.
*, inoculations done in duplicate.
Distribution determined by plate counts on Rapid L. mono (RLM). ND, not done
L. monocytogenes versus L. monocytogenes in UVM I and UVM II.
The very systematic difference in lineage competition ability prompted us to systematically investigate how different L. monocytogenes lineages behave in UVM I and UVM II. When lineage 2 and lineage 1 strains were combined, lineage 2 strains outcompeted the lineage 1 strains in all combinations. We isolated a total of 1,208 colonies from combinations of lineage 1 and lineage 2 strains after enrichment in UVM I (32 combinations with approximately 34 to 40 colonies per combination [Table 4 ]), and 987 colonies (82%) were lineage 2 and 221 colonies (18%) were lineage 1 (data not shown). After enrichment in UVM II, a total of 1,200 colonies were tested (Table 4), and 1,160 colonies (97%) were lineage 2 and only 40 colonies (3%) belonged to lineage 1 (data not shown). However, within lineage 1, the strains belonging to serotype 4b (strains C1-109, V518a, and 4542) seemed to be more affected by the lineage 2 competition than serotype 1/2b (strain 7418).
TABLE 4.
Proportion of two strains of Listeria monocytogenes after coculturing in UVM I and UVM II at 30°C for 24 ha
| Lineage combination | Strain
|
No. of independent trials | Inoculation level (%)
|
Distribution (%)
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| UVM I
|
UVM II
|
||||||||
| A | B | A | B | A | B | A | B | ||
| 2 vs. 1 | La22 | V518a | 5 | 47 | 53 | 64 | 36 | 100 | 0 |
| 44 | 56 | >99 | <1 | 100 | 0 | ||||
| 50 | 50 | ND | ND | 100 | 0 | ||||
| 48 | 52 | 100 | 0 | 97 | 3 | ||||
| 46* | 54 | 87 | 13 | ND | ND | ||||
| 46 | 54* | 95 | 5 | ND | ND | ||||
| La22 | 4542 | 1 | 53 | 47* | 89 | 11 | 100 | 0 | |
| 53* | 47* | 95 | 5 | 100 | 0 | ||||
| La22 | 7418 | 1 | 39* | 61* | 56 | 44 | 100 | 0 | |
| 39* | 61* | 64 | 36 | 83 | 17 | ||||
| V5a | 7418 | 1 | 55* | 45 | 55 | 45 | 81 | 19 | |
| 55* | 45* | 38 | 62 | 93 | 7 | ||||
| V5a | V518a | 2 | 48 | 52 | 91 | 9 | 100 | 0 | |
| 42 | 58 | 96 | 4 | 100 | 0 | ||||
| V5a | C1-109 | 2 | 54 | 46 | 100 | 0 | 100 | 0 | |
| 46 | 54 | 100 | 0 | 100 | 0 | ||||
| V5a | 4542 | 1 | 53* | 47* | 80 | 20 | 98 | 2 | |
| 53* | 47* | 75 | 25 | 100 | 0 | ||||
| C1-056 | C1-109 | 3 | 44 | 56 | ND | ND | 100 | 0 | |
| 59 | 41 | 100 | 0 | 100 | 0 | ||||
| 54 | 46 | 97 | 3 | 100 | 0 | ||||
| C1-056 | 4542 | 1 | 55* | 45 | 100 | 0 | 100 | 0 | |
| 55* | 45* | 100 | 0 | 100 | 0 | ||||
| C1-056 | 7418 | 1 | 58* | 42* | 67 | 33 | 95 | 5 | |
| 58* | 42* | 58 | 42 | 88 | 12 | ||||
| Hu4239 | 7418 | 1 | 42* | 58* | 60 | 40 | 93 | 7 | |
| 42* | 58* | 50 | 50 | 85 | 15 | ||||
| Hu4239 | V518a | 2 | 47 | 53 | 48 | 52 | 96 | 4 | |
| 44 | 56 | 91 | 9 | 90 | 10 | ||||
| Hu4239 | C1-109 | 2 | 52 | 48 | 96 | 4 | 100 | 0 | |
| 45 | 55 | 100 | 0 | 100 | 0 | ||||
| Hu4239 | 4542 | 1 | 52* | 48* | 93 | 7 | 100 | 0 | |
| 52* | 48* | 90 | 10 | 97 | 3 | ||||
| 2 vs. 2 | La22 | V5a | 3 | 40 | 60 | 80 | 20 | 75 | 25 |
| 48 | 52 | 63 | 37 | 48 | 52 | ||||
| 56 | 44 | 87 | 13 | ND | ND | ||||
| La22 | C1-056 | 2 | 52 | 48 | 76 | 24 | 100 | 0 | |
| 49 | 51 | 88 | 12 | 91 | 9 | ||||
| La22 | Hu4239 | 4 | 47 | 53 | 100 | 0 | 100 | 0 | |
| 52 | 48 | 73 | 27 | 92 | 8 | ||||
| 60 | 40 | 100 | 0 | 86 | 14 | ||||
| 53 | 47 | 51 | 49 | 80 | 20 | ||||
| 1 vs. 1 | V518a | C1-109 | 2 | 49 | 51 | 76 | 24 | 100 | 0 |
| 56 | 44 | 85 | 15 | 97 | 3 | ||||
| V518a | 4542 | 1 | 51* | 49* | 45 | 55 | 45 | 55 | |
| 51* | 49* | 40 | 60 | 31 | 69 | ||||
| 7418 | 4542 | 1 | 52* | 48* | 76 | 24 | 50 | 50 | |
| 52* | 48* | 72 | 28 | 70 | 30 | ||||
| 7418 | V518a | 1 | 47* | 53* | 68 | 32 | 75 | 25 | |
| 47* | 53* | 67 | 33 | 65 | 35 | ||||
See Table 3, footnotes a and b. ND, not done.
When two lineage 2 strains were coinoculated, both appeared in most cases after UVM I and UVM II selection, albeit in various proportions. A total of 576 colonies were tested from combinations between two lineage 2 strains (data not shown). Strain La22 (lineage 2) appeared to be a “strong” isolate and accounted for 80 to 100% of the colonies after growth in UVM I and UVM II with the other lineage 2 strains (Hu4239 and C1-056). When the four lineage 1 strains were tested against one another, only one combination (V518a against C1-109) resulted in complete dominance by one strain (V518a). This dominance could be explained by differences in generation time, 1.05 h (V518a) versus 1.31 h (C1-109) (Table 2).
Listeria versus Listeria in BHI.
To test if the selective pressure in UVM caused the difference in bias between lineage 1 and lineage 2 strains, a few combinations of Listeria strains were tested as cocultures in BHI (Table 5). A higher proportion of L. monocytogenes lineage 1 appeared when grown with L. innocua in BHI than when grown in UVM I and II. The proportions of lineage 2 strains were similar after growth in BHI as when grown in UVM I. When testing strain La22 (lineage 2) against strain V518a (lineage 1) in BHI, V518a appeared in higher proportions compared to the competition in UVM I. These results indicate that the selective compounds in UVM could be important for the bias during enrichment.
TABLE 5.
Proportion of Listeria monocytogenes and Listeria innocua after coculturing in BHI broth at 30°C for 24 h
| Lineage | Strain
|
No. of independent trials | Inoculation level (%)a
|
Distribution in BHIb (%)
|
|||
|---|---|---|---|---|---|---|---|
| A | B | A | B | A | B | ||
| 2 | La22 | R255a | 1 | 50 | 50 | 49 | 51 |
| 50 | 50 | 46 | 54 | ||||
| 2 | C1-056 | R255a | 1 | 50 | 50 | 92 | 8 |
| 50 | 50 | 89 | 11 | ||||
| 1 | V518a | R255a | 2 | 48 | 52 | 97 | 3 |
| 48 | 52 | 98 | 2 | ||||
| 56 | 44 | 99 | 1 | ||||
| 56 | 44 | 98 | 2 | ||||
| 1 | C1-109 | R255a | 1 | 55 | 45 | 50 | 50 |
| 55 | 45 | 49 | 51 | ||||
| 2 vs. 1 | La22 | V518a | 1 | 46 | 54 | 73 | 28 |
| 46 | 54 | 75 | 25 | ||||
Combinations inoculated in duplicate.
Distribution was determined by performing plate counts on RLM.
Growth experiment with lineage 2 (La22) versus lineage 1 (V518a) in UVM I.
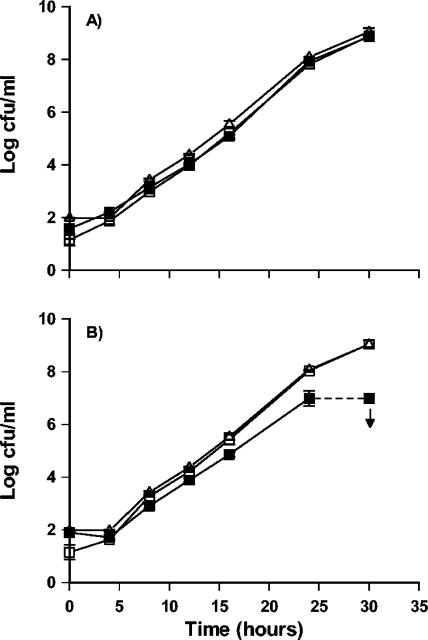
When grown as single cultures, strain La22 and strain V518a grew with identical growth rates in UVM I (Fig. 1A). To determine at what point during the selective enrichment the cultures interacted, samples were withdrawn every 4 h, and 80 colonies were isolated from each sampling point and subtyped to differentiate between the two strains. The growth rates of strains La22 and V518a were similar until the cell density reached approximately 106 CFU/ml (Fig. 1B). After 24 h, the combination consisted of 91% La22 and 9% V518a, and after 30 h, no colonies of V518a could be isolated. Hence, interaction occurred between stain La22 and strain V518a only at high cell densities.
FIG. 1.
A) Growth in UVM I of Listeria monocytogenes strain La 22 (□, lineage 2) and strain V518a (▪, lineage 1) as single cultures andthe two strains grown in coculture. B) Growth in UVM I of L. monocytogenes strain La22 (□) and strain V518a (▪) detected in the coculture by serotyping of 80 colonies and of a combination of the two strains (▵).
Bioassay.
The interaction between strain La22 (lineage 2) and strain V518a (lineage 1) which was seen in the growth experiment together with the overall bias between lineage 1 and lineage 2 during enrichment in UVM indicate that members of lineage 2 strains and L. innocua could have produced inhibitory compounds. To test if such compounds were present, supernatants and cultures from both BHI and UVM I broth of all Listeria species were tested in a well diffusion assay against all Listeria species. No strains were inhibitory to any of the other Listeria strains in this assay (data not shown).
DISCUSSION
The Listeria selective procedures used during the enrichment of food and environmental samples are believed to allow detection of most (80 to 95%) samples containing Listeria species (31). However, the present study demonstrates that one of the common selective enrichment procedures (University of Vermont medium) used for the detection of L. monocytogenes in foods may not result in an equal representation of the subtypes of L. monocytogenes present in a food sample. Even though our data are based on a limited number of strains, a consistent pattern was found indicating that the method favors L. monocytogenes lineage 2 strains at the expense of lineage 1 strains. This bias is very unfortunate, since two of the three most common serotypes (4b and 1/2b) associated with human cases of listeriosis are lineage 1 strains (16, 20). Hence, epidemiological investigations where clinical strains, which are typically isolated by nonselective procedures, are compared to strains isolated from foods may have difficulties providing links between food sources and disease (12, 19). Several studies have noted that although serotype 4b is often involved in listeriosis, it is not found as commonly in foods or the environment as other serotypes (9, 16). Furthermore, Loncarevic et al. (17) found that direct plating yielded more L. monocytogenes clones than the enrichment procedure when sampling from foods. Our data indicate that this discrepancy could be caused by bias in the selection procedure used when sampling from food products and the environment.
Growth in nonselective medium was almost identical for the nine Listeria strains studied, and maximum cell densities of almost 1010 CFU/ml were reached. In agreement with this, Gracieux et al. (13) found that the growth of 40 L. monocytogenes strains in a nonselective medium (TSA) was similar. In contrast, the Listeria strains were differently affected by the selective agents present in UVM I, and growth of all strains was inhibited compared to their growth in BHI in agreement with other studies (5, 18).
Both L. monocytogenes and L. innocua are commonly isolated from and appear to grow equally well in foods (8, 14, 26, 29). Concern has been raised that L. innocua could outgrow L. monocytogenes during the detection enrichment procedures and hence mask the presence of the pathogen (6, 26). Some studies have indicated that L. innocua can grow faster in Listeria selective medium (18), whereas other studies (5) have not been able to demonstrate any differences in growth in selective medium. Our study clearly demonstrates that the presence of L. innocua may indeed mask the presence of L. monocytogenes; however, the selective advantage of L. innocua was only apparent when coinoculated with L. monocytogenes lineage 1 strains. L. monocytogenes lineage 2 strains grew as well as L. innocua strains during enrichment (Table 3). Most studies finding a dominance of L. innocua have used L. monocytogenes serotype 4b, which is a lineage 1 strain (6, 26).
The bias in strain selection when a lineage 1 and a lineage 2 L. monocytogenes strain were cocultured could not be explained by differences in growth rate in UVM I (Table 2). One may ask if a bias also occurs in food products in the presence of different L. monocytogenes strains. This seems not to be the case, since Porto et al. (27) inoculated five different L. monocytogenes strains (both lineage 1 and 2 strains) onto frankfurters, and all strains were detected after 28 days, although the percentages of lineage 2 strains were higher than those of lineage 1 strains.
The outcome postenrichment when two lineage 2 or two lineage 1 strains were cocultured could, in one case of lineage 2 (La22 versus C1-056) and one case of lineage 1 (V518a versus C1-109), be explained by significant differences in the individual growth rates of the strains (P < 0.05 and P < 0.01, respectively). In other cases (La22 versus Hu4239 or V5a), there were no significant differences in the individual growth rate yet the same strain dominated in all cases even though different percentage levels were found.
When comparing individual growth with growth in combination of strain La22 (lineage 2) and V518a (lineage 1) (Fig. 1), it appeared that La22 inhibited the growth of strain V518a in the coculture when a cell density of 106 cell/ml was reached. When grown as monocultures, the growth of the two organisms was identical.
Changing the coculture experiment to the nonselective medium BHI shifted the bias for combinations between L. monocytogenes lineage 1 strains and L. innocua, but no shift was seen for combinations with L. monocytogenes lineage 2 strains and L. innocua. Additionally, strain V518a reached a higher percentage after growth in combination with strain La22 in BHI compared to the average level obtained after enrichment in UVM. Hence, the selective principles of UVM I and II (acriflavin and nalidixic acid) may contribute to the inhibition of one Listeria spp. strain by another during enrichment.
Nalidixic acid in UVM is used to suppress the growth of gram-negative bacteria and Bacillus spp. but has no effect on the growth of L. monocytogenes (15). Acriflavine is used in UVM to suppress non-Listeria gram-positive bacteria, but Jacobsen (15) found that L. monocytogenes strains were also inhibited by acriflavine and that different L. monocytogenes strains varied in their sensitivity to acriflavine. Acriflavine is used as the only supplement to suppress gram-positive bacteria in other Listeria selective enrichments medium such as BAMS, BCM, Fraser, and LRBS (32). Hence, the bias during enrichment in UVM of L. monocytogenes lineage 1 when present together with lineage 2 presented in this study is likely to occur when using other enrichment media.
Many gram-positive bacteria such as lactic acid bacteria produce bacteriocins that are inhibitory against L. monocytogenes (22, 30), and it has been shown that L. innocua can produce a bacteriocin(s) which inhibits L. monocytogenes (37). We did not, however, find that bacteriocin production (or other extracellular factors) caused the differential selection during growth in UVM I broth. Bacteriocin-negative lactic acid bacteria may be inhibitory to both lineage 1 and 2 L. monocytogenes strains, and it has been suggested that this is partly due to nutrient competition and that an interaction of this type could also take place between two L. monocytogenes strains (2, 24).
Our study indicates that the lineages of L. monocytogenes (28, 36), which are also reflected in the serotyping of the strains (21), could be physiologically different. Although large strain-to-strain variations are seen in all studies, some differences between lineages have been detected. Buncic et al. (3) found that serotype 1/2a (lineage 2) appeared to tolerate bacteriocins better than serotype 4b (lineage 1) and that lineage 2 strains appeared to attach better to surfaces than lineage 1 strains (1). Other studies have not been able to demonstrate a systematic, lineage-dependent difference between L. monocytogenes strains either in heat resistance (7) or in the ability to grow in sodium chloride (34). A shotgun DNA microarray was used to analyze 44 L. monocytogenes strains, and this revealed that 47 genes were absent in lineage 1 strains compared to lineage 2 strains (38). Some of the genes found in lineage 2 strains and not in lineage 1 strains are involved in stress response, transport of small molecules, or synthesis of cell wall molecules. One may speculate that while lineage 1 strains when growing as monocultures in UVM medium may cope with the stressful conditions and have a sufficient supply of nutrients such as amino acids, they cannot cope when cocultured with a more stress-tolerant lineage which is perhaps capable of more efficient uptake.
Listeriosis is a serious food-borne disease, and to implement appropriate control measures, one needs to understand the ecology of the virulent types as well as their niches, distribution, and transmission. To facilitate such investigations and assess the true distribution of serotypes and lineages, there is a need for the development of methods allowing all species and subtypes of Listeria to be detected.
Acknowledgments
We thank Martin Wiedmann for providing L. monocytogenes strains for the study, Tina Nørgaard and Anemone Bundvad for technical assistance, and Jeffrey Farber for serotyping of the strains. Constructive comments from three reviewers were most helpful.
The study was financed by the Danish Ministry for Food, Agriculture and Fisheries (grant no. 93s-24F4-Å02-00036).
REFERENCES
- 1.Borucki, M. K., J. D. Peppin, D. White, F. Loge, and D. R. Call. 2003. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 69:7336-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchanan, R. L., and L. K. Bagi. 1997. Microbial competition: Effect of culture conditions on the suppression of Listeria monocytogenes Scott A by Carnobacterium piscicola. J. Food Prot. 60:254-261. [DOI] [PubMed] [Google Scholar]
- 3.Buncic, S., S. M. Avery, J. Rocourt, and M. Dimitrijevic. 2001. Can food-related environmental factors induce different behaviour in two key serovars, 4b and 1/2a, of Listeria monocytogenes? Int. J. Food Microbiol. 65:201-212. [DOI] [PubMed] [Google Scholar]
- 4.Capita, R., C. Alonso-Calleja, M. Prieto, M. Garcia-Fernandez, and B. Moreno. 2001. Comparison of PALCAM and modified Oxford plating media for isolation of Listeria species in poultry meat following UVM II or Fraser secondary enrichment broths. Food Microbiol. 18:555-563. [Google Scholar]
- 5.Cornu, M., M. Kalmokoff, and J. P. Flandrois. 2002. Modelling the competitive growth of Listeria monocytogenes and Listeria innocua in enrichment broths. Int. J. Food Microbiol. 73:261-274. [DOI] [PubMed] [Google Scholar]
- 6.Curiale, M. S., and C. Lewus. 1994. Detection of Listeria monocytogenes in samples containing Listeria innocua. J. Food Prot. 57:1048-1051. [DOI] [PubMed] [Google Scholar]
- 7.De Jesus, A. J., and R. C. Whiting. 2003. Thermal inactivation, growth, and survival studies of Listeria monocytogenes strains belonging to three distinct genotypic lineages. J. Food Prot. 66:1611-1617. [DOI] [PubMed] [Google Scholar]
- 8.Duffy, G., D. Walsh, J. J. Sheridan, C. M. Logue, D. Harrington, I. S. Blair, and D. A. McDowell. 2001. Comparison of selective and non-selective enrichment media in the detection of Listeria monocytogenes from meat containing Listeria innocua. J. Appl. Microbiol. 90:994-999. [DOI] [PubMed] [Google Scholar]
- 9.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonnesbech Vogel, B., V. Fussing, B. Ojeniyi, L. Gram, and P. Ahrens. 2004. High-resolution genotyping of Listeria monocytogenes by fluorescent amplified-fragment length polymorphism analysis as compared to typing by PFGE, RAPD, ribotyping and PCR-RFLP. J. Food Prot. 67:1656-1665. [DOI] [PubMed] [Google Scholar]
- 11.Fonnesbech Vogel, B., H. H. Huss, B. Ojeniyi, P. Ahrens, and L. Gram. 2001. Elucidation of Listeria monocytogenes contamination routes in cold-smoked salmon processing plants detected by DNA-based typing methods. Appl. Environ. Microbiol. 67:2586-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilot, P., A. Genicot, and P. Andre. 1996. Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J. Clin. Microbiol. 34:1007-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gracieux, P., S. M. Roche, P. Pardon, and P. Velge. 2003. Hypovirulent Listeria monocytogenes strains are less frequently recovered than virulent strains on PALCAM and Rapid′ L. mono media. Int. J. Food Microbiol. 83:133-145. [DOI] [PubMed] [Google Scholar]
- 14.Gudbjörnsdttir, B., M.-L. Suihko, P. Gustavsson, G. Thorkelsson, S. Salo, A.-M. Sjöberg, O. Nichlasen, and S. Bredholt. 2004. The incidence of Listeria monocytogenes in meat, poultry and seafood plants in the Nordic countries. Food Microbiol. 21:217-225. [DOI] [PubMed] [Google Scholar]
- 15.Jacobsen, C. N. 1999. The influence of commonly used selective agents on the growth of Listeria monocytogenes. Int. J. Food Microbiol. 50:221-226. [Google Scholar]
- 16.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 17.Loncarevic, S., W. Tham, and M. L. Danielsson-Tham. 1996. The clones of Listeria monocytogenes detected in food depend on the method used. Lett. Appl. Microbiol. 22:381-384. [DOI] [PubMed] [Google Scholar]
- 18.Macdonald, F., and A. D. Sutherland. 1994. Important differences between the generation times of Listeria monocytogenes and Listeria innocua in two Listeria enrichment broths. J. Dairy Res. 61:433-436. [DOI] [PubMed] [Google Scholar]
- 19.Martinez, I., L. M. Rorvik, V. Brox, J. Lassen, M. Seppola, L. Gram, and B. Fonnesbech Vogel. 2003. Genetic variability among isolates of Listeria monocytogenes from food products, clinical samples and processing environments, estimated by RAPD typing. Int. J. Food Microbiol. 84:285-297. [DOI] [PubMed] [Google Scholar]
- 20.Mclauchlin, J. 1990. Distribution of serovars of Listeria monocytogenes isolated from different categories of patients with listeriosis. Eur. J. Clin. Microbiol. 9:210-213. [DOI] [PubMed] [Google Scholar]
- 21.Nadon, C. A., D. L. Woodward, C. Young, E. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nes, I. F., and H. Hole. 2000. Class II antimicrobial peptides from lactic acid bacteria. Biopolymers 55:50-61. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson, L., L. Gram, and H. H. Huss. 1999. Growth control of Listeria monocytogenes on cold-smoked salmon using a competitive lactic acid bacteria flora. J. Food Prot. 62:336-342. [DOI] [PubMed] [Google Scholar]
- 24.Nilsson, L., Y. Y. Ng, J. N. Christiansen, B. L. Jorgensen, D. Grotinum, and L. Gram. 2004. The contribution of bacteriocin to inhibition of Listeria monocytogenes by Carnobacterium piscicola strains in cold-smoked salmon systems. J. Appl. Microbiol. 96:133-143. [DOI] [PubMed] [Google Scholar]
- 25.Patel, J. R., and L. R. Beuchat. 1995. Evaluation of enrichment broths for their ability to recover heat-injured Listeria monocytogenes. J. Appl. Bacteriol. 78:366-372. [DOI] [PubMed] [Google Scholar]
- 26.Petran, R. L., and K. M. J. Swanson. 1993. Simultaneous growth of Listeria monocytogenes and Listeria Innocua. J. Food Prot. 56:616-618. [DOI] [PubMed] [Google Scholar]
- 27.Porto, A. C. S., L. Wonderling, J. E. Call, and J. B. Luchansky. 2003. Use of pulsed-field gel electrophoresis to monitor a five-strain mixture of Listeria monocytogenes in frankfurter packages. J. Food Prot. 66:1465-1468. [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 29.Rawles, D. D., G. Flick, A. Diallo, and R. Croonenberghs. 1995. Growth of mixed cultures of Listeria monocytogenes and Listeria innocua in blue-crab (Callinectes-sapidus) meat. J. Food Prot. 58:1268-1270. [DOI] [PubMed] [Google Scholar]
- 30.Ross, R. P., S. Morgan, and C. Hill. 2002. Preservation and fermentation: past, present and future. Int. J. Food Microbiol. 79:3-16. [DOI] [PubMed] [Google Scholar]
- 31.Ryser, E. T., S. M. Arimi, M. M. C. Bunduki, and C. W. Donnelly. 1996. Recovery of different Listeria ribotypes from naturally contaminated, raw refrigerated meat and poultry products with two primary enrichment media. Appl. Environ. Microbiol. 62:1781-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silk, T. M., T. M. T. Roth, and C. W. Donnelly. 2002. Comparison of growth kinetics for healthy and heat-injured Listeria monocytogenes in eight enrichment broths. J. Food Prot. 65:1333-1337. [DOI] [PubMed] [Google Scholar]
- 33.Suh, J. H., and S. J. Knabel. 2001. Comparison of different enrichment broths and background flora for detection of heat-injured Listeria monocytogenes in whole milk. J. Food Prot. 64:30-36. [DOI] [PubMed] [Google Scholar]
- 34.Vialette, M., A. Pinon, E. Chasseignaux, and M. Lange. 2003. Growths kinetics comparison of clinical and seafood Listeria monocytogenes isolates in acid and osmotic environment. Int. J. Food Microbiol. 82:121-131. [DOI] [PubMed] [Google Scholar]
- 35.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524-531. [PubMed] [Google Scholar]
- 36.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoyama, E., S. Maruyama, and Y. Katsube. 1998. Production of bacteriocin-like-substance by Listeria innocua against Listeria monocytogenes. Int. J. Food Microbiol. 40:133-137. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, C. M., M. Zhang, J. L. Ju, J. Nietfeldt, J. Wise, P. M. Terry, M. Olson, S. D. Kachman, M. Wiedmann, M. Samadpour, and A. K. Benson. 2003. Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: Identification of segments unique to lineage II populations. J. Bacteriol. 185:5573-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]