Abstract
Spores from a number of different Bacillus species are currently being used as human and animal probiotics, although their mechanisms of action remain poorly understood. Here we describe the isolation of 237 presumptive gut-associated Bacillus spp. isolates that were obtained by heat and ethanol treatment of fecal material from organically reared broilers followed by aerobic plating. Thirty-one representative isolates were characterized according to their morphological, physiological, and biochemical properties as well as partial 16S rRNA gene sequences and screening for the presence of plasmid DNA. The Bacillus species identified included B. subtilis, B. pumilus, B. licheniformis, B. clausii, B. megaterium, B. firmus, and species of the B. cereus group, whereas a number of our isolates could not be classified. Intrinsic properties of potential importance for survival in the gut that could be advantageous for spore-forming probiotics were further investigated for seven isolates belonging to five different species. All isolates sporulated efficiently in the laboratory, and the resulting spores were tolerant to simulated gastrointestinal tract conditions. They also exhibited antimicrobial activity against a broad spectrum of bacteria, including food spoilage and pathogenic organisms such as Bacillus spp., Clostridium perfringens, Staphylococcus aureus, and Listeria monocytogenes. Importantly, the isolates were susceptible to most of the antibiotics tested, arguing that they would not act as donors for resistance determinants if introduced in the form of probiotic preparations. Together, our results suggest that some of the sporeformers isolated in this study have the potential to persist in or transiently associate with the complex gut ecosystem.
The widespread use of antibiotics as therapeutic and prophylactic agents and as growth promoters in animal husbandry has led to a worldwide increase in antibiotic resistance and the emergence of untreatable multidrug-resistant strains of bacteria (4, 33). This has prompted the European Union to phase out the use of these compounds as animal feed additives by 2006 (47). The use of competitive exclusion agents and probiotic feed additives in the livestock industry is therefore attracting increased attention as a cost-effective alternative to controlling animal disease and improving breeding performance (54).
The genus Bacillus comprises a diverse collection of aerobic endospore-forming bacteria whose spores consist of several protective layers surrounding the nucleoid in the spore core (20, 51). This structural organization makes the spores extremely resistant to external physical and chemical insults and in part determines their exceptional longevity in the environment (20, 38). The resilient and ubiquitous nature of these bacteria results in considerable daily intake of these organisms by humans and animals. This ingestion, which is often in the form of spores, occurs primarily through contaminated food and water.
Bacillus spores are being used as human and animal probiotics despite the fact that studies now indicate extensive mislabeling of constituent Bacillus strains (13, 21). Several of these strains have additionally been shown to be multidrug resistant and to harbor toxin genes (10, 21). Therefore, it is becoming increasingly clear that a more rigorous selection process is required for Bacillus probiotic candidates. Our understanding of competitive exclusion and probiotic properties relates mainly to the lactic acid bacteria however, with comparatively very little known regarding Bacillus spp. (45, 59). Nevertheless, it is anticipated that the competitive exclusion of pathogens by Bacillus probiotics will result from one or more modes of action, including immune exclusion, competition for adhesion sites, and production of antimicrobial agents, such as bacteriocins (45).
Although recent evidence suggests that Bacillus spores do germinate in the gastrointestinal tract (8, 22), it remains unclear which form, cell, spores, or both, is actually responsible for the competitive exclusion and probiotic effects. Despite these current limitations, the administration of spores as feed additives as opposed to vegetative cells clearly distinguishes Bacillus probiotics from other bacterial probiotic formulations and offers a number of clear advantages. These include low cost of production, ease of preparation, resistance to production processes, and extended shelf-life over a wide range of temperatures.
The use of competitive exclusion agents in the poultry industry has been described extensively in the prevention of several diseases caused by different avian pathogens, including zoonotic bacteria (1, 24, 40). However, the relatively undefined composition of some products and the uncertainty regarding their exact mechanisms of action have raised several ethical issues. Recent studies have shown that spores of a laboratory strain of Bacillus subtilis decreased different aspects of colonization of young chicks by the avian colibacillosis agent Escherichia coli O78:K80, Salmonella enterica serotype Enteritidis, and Clostridium perfringens (30, 31). We reasoned however that animal probiotics should preferentially originate from the target animal microflora, as their use would be ethically more acceptable and potentially more effective than exogenous strains. Moreover, there is evidence that laboratory strains of B. subtilis differ significantly from wild-type isolates, including properties that could enhance their efficacy as probiotics (7, 26; Barbosa et al., unpublished data).
Thus, in light of these arguments, we aimed in this study to isolate and identify aerobic sporeformers from the gastrointestinal tract of broilers. Strains were tested for intrinsic properties that would be beneficial to their survival in the gut ecosystem, properties that could be desirable for competitive exclusion or probiotic agents.
MATERIALS AND METHODS
Preparation of fecal samples, bacterial isolation, and growth conditions.
Twenty pooled samples of freshly voided fecal material were collected from the floors of buildings housing organically reared Hubbard 257 broilers in two different English farms. Care was taken to avoid gross contamination with environmental material. Fecal aliquots were initially diluted 1:1 (wt:vol) in buffered peptone-water (Oxoid) and resuspended by vigorous vortexing until an evenly distributed suspension was obtained. Aerobic spore-forming isolates were then selected by heat (39) and/or ethanol treatment (27). For heat treatment, the suspension was further diluted 1:10 in buffered peptone-water and incubated at 65°C for 20 or 45 min. For ethanol treatment, the original fecal suspension was diluted 1:1 in ethanol (final concentration, 50% vol/vol) and incubated for 1 h at room temperature (27). Subsequent plating of 0.1-ml aliquots of appropriate 10-fold serial dilutions in buffered peptone-water (up to 10−5) was done aerobically on Difco nutrient agar or Luria-Bertani (LB) plates, both of which support germination. Although no quantification was attempted, a measurable number of colonies (more than 10) was routinely obtained on 10−2 to 10−3 dilution plates after 24 to 48 h of incubation at 37°C. Colonies representing different morphologies were picked at random and purified by restreaking on agar plates of the same media. Production of spores by growth of the purified isolates on Difco sporulation medium (DSM) (39) plates was confirmed by phase-contrast microscopy before storage of the isolates at −80°C in Difco heart-infusion broth (HIB) with 30% glycerol.
Bacillus isolates were routinely grown aerobically at 37°C in LB or DSM, unless otherwise stated. The laboratory strain B. subtilis MB24 (trpC2 metC3) (17) (Table 1) was used as a control in most of the experiments described throughout this work. Other bacterial strains used as controls in different experiments or as indicator strains in antimicrobial activity assays are listed in Table 1.
TABLE 1.
Bacterial strains used in this study
| Bacterial species | Strain | Origina |
|---|---|---|
| Bacillus | ||
| B. subtilis | MB24, laboratory stock | A. O. Henriques |
| B. clausii | DSM 8716 | DSMZ |
| DSM 2515 | DSMZ | |
| B. licheniformis | 9945A (BGSC 5A2) | BGSC |
| B. cereus | T (BGSC 6A1) | BGSC |
| B. megaterium | 899 (BGSC 7A1) | BGSC |
| B. sphaericus | ATCC 33203 (BGSC 13A5) | BGSC |
| ATCC 14577 (BGSC 13A6) | BGSC | |
| B. cereus var. vietnami | Subtyl | S. Cutting |
| B. cereus | Biosubtyl Da Lat | S. Cutting |
| B. pumilus | Biosubtyl Nha Trang | S. Cutting |
| Others | ||
| Escherichia coli | O78:K80 (EC34195) | M. Woodward |
| O157:H7 (NCTC 12900) | S. Cutting | |
| Salmonella enterica | Enteritidis (S1400) | M. Woodward |
| Salmonella bareilly | Laboratory stock | S. Cutting |
| Citrobacter rodentium | Laboratory stock | M. Woodward |
| Pseudomonas aeruginosa | NCTC 12903 | S. Cutting |
| Enterobacter aerogenes | NCTC 10006 | S. Cutting |
| Staphylococcus aureus | RN4220 (pVF5) | V. Perreten |
| RN2442 (pE194) | V. Perreten | |
| Enterococcus faecalis | ATCC 29212 (DSM 2570) | T. Crespo |
| Listeria innocua | Laboratory stock | T. Crespo |
| Listeria monocytogenes | Laboratory stock | T. Crespo |
| Clostridium perfringens | fD00385 | M. Woodward |
Bacterial strains were obtained from public collections (BGSC, Bacillus Genetic Stock Centre, Department of Biochemistry, Ohio State University, Columbus, Ohio; DSMZ, DSM Collection, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany; NCTC, National Collection of Type Cultures, Central Public Health Laboratory, Colindale, England) from our laboratory stocks (A. O. Henriques and M. J. Woodward) or kindly supplied by Simon Cutting (School of Biological Sciences, Royal Holloway University of London, London, England), Vincent Perreten (Institute of Veterinary Bacteriology, University of Berne); S. aureus RN4220(pVF5) (48) and RN2442(pE194) (25); and Teresa Crespo (Instituto de Biologia Experimental e Tecnológica, Oeiras, Portugal).
Catalase test, growth characteristics, and hemolysis.
The catalase activity of bacterial isolates was detected by resuspension of a colony in a 3% solution of hydrogen peroxide (Sigma).
The ability to grow under alkaline conditions was determined by monitoring the optical density at 600 nm of cultures grown in alkaline nutrient broth (Difco nutrient broth buffered at pH 9.7 with a 4.2% NaHCO3, 5.3% Na2CO3 solution; formula from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany). Growth at 50°C was assessed by spotting 5 μl of an overnight LB culture on fresh LB plates which were incubated at 50°C for 24 h. Growth under anaerobic conditions was assessed by incubating LB plates streaked with test and control strains in an Oxoid anaerobic jar in the presence of an Anaerogen sachet. Growth was monitored at 24 h, the sachet was replaced, and the plates were incubated for another 24 h. Clostridium perfringens isolate fD00385 (Table 1) was included as a control for the anaerobic conditions.
Hemolysis was determined on Columbia 5% sheep blood agar plates (bioMérieux SA, Marcy-l′Etoile, France), streaked with colonies from fresh LB plates. Readings were taken after incubation at 37°C for 24 h.
API 50 CHB system test.
The biochemical profile of test isolates was determined with the API 50 CHB strips following the manufacturer's instructions (bioMérieux). This test allows bacterial strains to be classified according to their ability to ferment 49 different carbohydrates. The results were analyzed with the APILAB Plus software (bioMérieux).
DNA extraction and purification.
Total genomic DNA was extracted from overnight LB cultures by a modification of the method of Pitcher et al. (50). Overnight cultures of Bacillus strains grown in LB medium were inoculated as a 5-μl spot on LB agar plates. The presence of plasmid DNA in natural isolates was screened from overnight LB cultures by the method of Anderson and McKay with the small-scale procedure (2). This method was chosen since it allows detection of large plasmids frequently undetected by other methods.
PCR amplification and DNA sequencing of 16S rRNA.
Universal eubacterial primers fD1 (5′-CAACAGAGTTTGATCCTGGCTCAG-3′) (E. coli positions 8 to 27) and rD1 (5′-GCTTAAGGAGGTGATCCAGCC-3′) (E. coli positions 1541 to 1525) described by Weisburg et al. (66) were used to amplify small-subunit rRNA (16S rRNA) gene sequences of the Bacillus isolates. PCR amplification was carried out with Taq polymerase (MBI Fermentas) with an annealing temperature of 52°C. Partial or complete sequencing of the approximately 1.5-kb amplified products purified with the QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany) was obtained with the rD1 or fD1 primer and internal primers designed from previously determined sequences.
Computer analysis of the 16S rRNA sequences was performed with the Sequence Match software package through the Ribosomal Database Project II (http://rdp.cme.msu.edu/html) or by comparison with sequences in the GenBank nonredundant nucleotide database with BLAST (http://www.ncbi.nlm.nih.gov).
Biofilm formation.
Formation of biofilm was tested as described by Robleto et al. (57) with modifications. Essentially, 2 to 3 ml of Sterlini-Mandelstam medium (16) was inoculated with an exponential-phase culture grown in the same medium to an optical density at 600 nm of ≈0.01. The test was carried out in 13-ml polypropylene tubes that were incubated for 24 h without agitation at 37°C. Controls consisted of an uninoculated tube and a tube inoculated with exponential-phase cells for 10 min before washing. Tubes were then rinsed with water before staining with a solution of 1% crystal violet for 15 min and subsequently washed to remove excess stain. A positive result was recorded as the presence of a ring of staining at the interface between air and medium.
Antibiotic susceptibility.
The susceptibility of the Bacillus isolates to ampicillin, tetracycline, chloramphenicol, gentamicin, ciprofloxacin, and erythromycin was determined in Oxoid Muller-Hinton (MH) agar plates with E-test strips (AB Biodisk, Solna, Sweden). The MIC of lincomycin was determined in MH broth by the microdilution test on a 96-well microtiter plate.
The primers used in the preparation of specific internal probes and for PCR screening of the erm(A), erm(B), and erm(C) genes were those described by Sutcliffe et al. (64). The primers for erm(D) were 5′-GGACAGCATTTGATGCAT-3′ (forward) and 5′-GGTGAGCGTTCCAACGGT-3′ (reverse) and for erm(F) were 5′-GCCAACAATGTTGTTGCT-3′ (forward) and 5′-CGAAATTGACCTGACCTG-3′ (reverse). The forward primer for ermG was described previously (62), and the reverse primer was the complement of the reverse primer described in this article. Primers for erm (34) were 5′-ACTTAGCCGTGGAGAACCTC-3′ (forward) and 5′-TTGGAACATGCCGAACCATC-3′ (reverse).
The templates for erm(A), (B), and (F) were pEM9592, pJIR229, and pVA831 (pBF4), respectively, cloned in Escherichia coli and for erm(D) it was pEC101 in B. subtilis, and they were kindly supplied by M. C. Roberts (University of Washington, Seattle). The erm(C) probe was amplified from Staphylococcus aureus RN2442-pE194, kindly supplied by Vincent Perreten (Institute of Veterinary Bacteriology, University of Berne, Berne, Switzerland), and the erm(G) probe was amplified from plasmid pGERM, a kind gift from Abigail Salyers (University of Illinois, Urbana).
Southern blotting and hybridization.
Southern blotting and hybridization with erm-specific [α-32P]dCTP-labeled probes of EcoRI-digested genomic DNAs were carried out essentially as described elsewhere (60) with the RadPrime DNA labeling system (Invitrogen) and a hybridization temperature of 60°C.
Spore preparation and determination of sporulation efficiency.
Sporulation efficiency was measured by determining the titer of heat-resistant cells versus the total (viable) cell counts in DSM cultures 24 h after inoculation (18). Spores of 24- to 48-h cultures in DSM (as indicated) were purified on Gastrografin gradients essentially as described elsewhere (18). After determining the spore titers, the aqueous samples were stored frozen at −80°C.
Electron microscopy.
Samples of Bacillus isolates to be studied by electron microscopy were prepared from cultures that had been grown for 24 h in DSM. Aliquots (5 ml) were harvested, fixed, dehydrated, and embedded essentially as described elsewhere (19). Microscopic analysis was carried out on a Jeol transmission electron microscope operated at 60 keV.
Acid and bile salt tolerance.
The tolerance of Bacillus spores to bile salts and simulated gastric conditions was assayed as described by Duc et al. (10) with modifications. Essentially, ≈108 to 109 spores ml−1 (48-h spores) were resuspended in an isotonic buffer (Bott and Wilson salts: 1.24% K2HPO4, 0.76% H2PO4, 0.1% trisodium citrate, 0.6% [NH4]2SO4, pH 6.7) containing 0.2% bile salts (50% sodium cholate, 50% sodium deoxycholate; Sigma Chemical Co Ltd.) or in 0.85% NaCl, pH 2, containing 1 mg ml−1 pepsin (Sigma) and incubated at 37°C with agitation. Aliquots were taken immediately and after 1 and 3 h (for bile tolerance) or after 30 min and 1 h (for acid tolerance). Appropriate dilutions, made in Bott and Wilson salts, were plated directly onto LB plates and CFU were determined after incubation at 37°C for 24 h (48 h for isolate 259). Control samples were set up in parallel where spores were resuspended in Bott and Wilson salts or 0.85% NaCl only.
Resistance of vegetative cells to bile salts and simulated gastric conditions was determined as described for spores by harvesting overnight LB cultures of Bacillus isolates and resuspending cells in fresh LB, LB supplemented with 0.2% bile salts, or LB acidified to pH 2 with concentrated HCl and supplemented with pepsin at 1 mg ml−1, respectively.
Antimicrobial activity screening assay.
Antimicrobial activity was assessed with a colony overlay assay adapted from the method described by Pugsley et al. (53) to screen a different set of microorganisms. Overnight cultures of Bacillus strains grown in LB medium were inoculated as a 5-μl spot on LB agar plates and incubated at 37°C for 24 h prior to killing of the cells by exposure to chloroform vapors for 30 min. Plate covers were replaced, and the plates were aerated for 20 min before overlaying with 0.7% LB or Difco brain heart infusion agar (according to the requirements of the indicator strains) that had been inoculated with an overnight culture of the indicator strain. Zones of inhibition around the spots at any of the incubation times, 5, 8, 24, and 48 h at 37°C (30°C for Enterococcus aerogenes), were scored as positive.
RESULTS
Isolation of fecal sporeformers.
Spore-forming bacteria were selected by heat or ethanol treatment of chicken fecal material collected from the floors of organically reared Hubbard 257 broiler houses in two different English farms. Although heat treatment is a standard procedure for spore selection, treatment with ethanol is also an effective method for the selective isolation of sporeformers that eliminates potential complications derived from variations in the level of spore heat resistance between species (27) (see also below). Treated samples were subsequently plated and incubated aerobically. A total of 237 colonies representing the many different colony morphologies observed and sources of samples were chosen randomly for further analysis. Spore production, induced by nutrient exhaustion of all 237 isolates grown on DSM plates, was confirmed by phase-contrast microscopy. All the isolates were catalase positive, a characteristic that differentiates Bacillus from the anaerobic spore-forming Clostridium spp.
Characterization of selected isolates.
Of the 237 isolates, 31 were selected for further analysis. This selection was based on macroscopic differences in colony morphologies and also on the collection of samples at different sites and from different animal groups. Characterization of biochemical properties was performed with the API 50 CHB system (Table 2). However, some isolates could not be classified with this method, for example, isolates 19, 208, 268, and 369, which were unable to utilize any of the carbohydrates present in the strip. We also noted that the API system failed to identify or provided false readings for isolates that did not belong to any of the 19 species for which the method was developed (Table 2, and see below).
TABLE 2.
Identification and characterisation of spore-forming isolates
| Isolate no. | API50 CHBa
|
16S rRNA sequence analysis
|
Growthc
|
Plasmidsd | Hemolysise | |||
|---|---|---|---|---|---|---|---|---|
| Taxon | % ID/T | Closest known speciesb | % ID | 50°C | Anaerobiosis | |||
| 3 | B. megaterium 2 | 99.9/0.96 | B. megaterium | 99 | − | − | + | α |
| 10 | NI | B. macroides, B. simplex | 99 | − | − | − | γ | |
| 16 | B. megaterium 2 | 99.2/0.85 | B. megaterium | 99 | − | − | + | α |
| 17 | B. pumilus | 99.9/0.85 | B. pumilus | 100 | + | − | (−) | β |
| 19 | NI | Bacillus sp. strain VAN35 | 99 | − | − | (−) | γ | |
| 37 | B. subtilis | 95.3/1.00 | B. mojavensis, B. subtilis | 99 | + | +/− | − | “β” |
| 52 | Paenibacillus alvei, B. cereus 2 | 50.6/0.75, 39.4/0.56 | B. cereus group | 97 | + | + | + | β |
| 53 | NI | B. macroides, B. simplex | 99 | − | − | − | γ | |
| 56 | B. licheniformis | 98.0/0.87 | B. licheniformis | 99 | + | + | − | “α” |
| 62 | Brevibacillus brevis | 73.6/0.85 | B. silvestris | 95 | − | − | − | γ |
| 197 | B. pumilus | 99.2/0.82 | B. pumilus | 99 | + | − | − | β |
| 200 | B. subtilis | 95.3/1.00 | B. subtilis | 100 | + | +/− | − | “β” |
| 208 | NI | Bacillus sp. strain VAN35 | 99 | − | − | − | γ | |
| 210 | B. subtilis | 95.3/1.00 | B. subtilis | 100 | + | +/− | + | “β” |
| 235 | B. cereus 2 | 80.6/0.70 | B. cereus group | 99 | − | + | + | β |
| 241 | B. cereus 2 | 83.8/0.97 | B. cereus group | 99 | − | + | + | β |
| 243 | B. cereus 1 | 40.2/1.00 | B. cereus group | 99 | − | + | + | β |
| B. anthracis | 34.4/0.97 | |||||||
| B. mycoides | 25.2/0.99 | |||||||
| 257 | B. amyloliquefaciens | 49.0/0.74 | B. subtilis, B. atrophaeus, B. amyloliquefaciens, B. licheniformis | 99 | + | − | − | β |
| B. subtilis | 32.3/0.78 | |||||||
| B. licheniformis | 15.3/0.77 | |||||||
| 259 | B. licheniformis | 99.6/0.60 | B. clausii | 100 | −* | − | − | γ |
| 263 | B. pumilus | 99.9/0.96 | B. pumilus | 99 | + | − | − | β |
| 268 | NI | Bacillus sp. strain VAN35 | 99 | − | − | + | γ | |
| 278 | B. subtilis | 95.3/1.00 | B. subtilis, B. mojavensis, B. atrophaeus, B. licheniformis | 99 | + | + | − | β |
| 285 | B. subtilis | 97.1/0.74 | B. subtilis, B. mojavensis, B. atrophaeus, B. licheniformis | 99 | + | + | + | “α” |
| 287 | NI | Bacillus sp. strain N6 | 99 | − | − | + | γ | |
| 306 | B. licheniformis | 90.5/0.37 | B. pumilus | 99 | + | − | − | β |
| 317 | B. firmus | 86.6/0.54 | B. firmus | 98 | − | − | − | γ |
| Brevibacillus laterosporum | 12.6/0.45 | |||||||
| 340 | B. licheniformis | 99.9/0.74 | B. sonorensis, B. licheniformis | 99 | + | + | + | “α” |
| 344 | B. pumilus | 99.9/0.83 | B. pumilus | 99 | + | − | − | β |
| 345 | B. licheniformis | 99.9/0.98 | B. licheniformis | 99 | + | + | + | V |
| 369 | NI | Bacillus sp. strain VAN35 | 99 | − | − | + | γ | |
| 370 | NI | B. clausii | 99 | −* | − | − | γ | |
NI, the APILAB Plus software was unable to find an acceptable identity (ID).
Closest known species relative with BLAST (0 to 100%); underlined, closest known species relative with the Ribosomal Database Project II; bold, isolates for which the full sequence of the 16S rRNA was determined; for all other isolates, analysis is based on partial sequences of 600 to 800 nt. B. cereus group is listed when similar sequence identities were found for two or more species of the B. cereus group.
*, positive after 48 h of incubation. +/−, limited growth in the form of tiny colonies.
Parentheses indicate that the presence of a large plasmid is not clear.
Weak signals are in quotation marks. With the exception of hemolysis (performed twice), testing for growth at 50°C, anaerobiosis, and the presence of plasmids were repeated at least three times. V, variable result.
To further assist in identification of the isolates, a ≈1.5-kb fragment of the 16S rRNA gene was amplified from the genomic DNA of the 31 isolates. The nucleotide sequence at the 5′ end of the amplified product of all but 2 of the 31 isolates was obtained with primer fD1. For isolates 259 and 370, primer fD1 repeatedly failed to provide any sequence, and therefore the 3′-end sequence of the 16S rRNA was obtained with primer rD1. Partial sequences of the 16S rRNA (≈600 to 800 nucleotides) were compared to those deposited in GenBank and the Ribosomal Database Project (Table 2). For isolates 3, 37, 56, 197, 200, 210, and 259, which were later characterized in more detail (see below), complete unidirectional sequences of the 16S rRNA were obtained and analyzed as previously described (see below and Table 2).
Although combined biochemical and genetic analysis allowed the identification of the 31 isolates to the genus level as Bacillus spp., the API results and 16S rRNA sequences were not always in complete agreement. While this made definitive species identification more difficult in some instances (Table 2), the majority of the isolates exhibited more than 97% rRNA sequence identity to database entries and could be associated with known Bacillus species (32). These included B. subtilis, B. pumilus, B. licheniformis, B. clausii, B. megaterium, B. firmus, and species of the B. cereus group.
On the other hand isolates 52 and 62, which displayed only 95% and 97% sequence identity to their closest relatives, B. silvestris and B. cereus group species, respectively, may represent new species. However, determination of full rRNA sequences and further characterization will be required for these isolates to test this suggestion. Isolates 19, 208, 268, and 369 showed 99% identity with a database entry corresponding to a Bacillus strain isolated from vanilla beans undergoing postharvest processing, Bacillus sp. VAN35 (accession number AF286486). However, this strain has not been identified to the species level. The same was observed for isolate 287, which presented 99% sequence identity to Bacillus sp. N6 (accession number AB043854). Although phylogenetically these isolates are related to Bacillus spp., all showed a distinctive biochemical profile (see above), consisting of a negative result for all the tests in the API strip.
Characterization of the poultry gut Bacillus isolates was further assisted by the determination of multiple morphological and physiological parameters. For example, all B. licheniformis, B. pumilus, and B. subtilis isolates grew at 50°C (Table 2), while only B. licheniformis strains and isolates of the B. cereus group grew under anaerobic conditions (Table 2). Nevertheless, consistent with the reported ability of B. subtilis to grow anaerobically (37), different growth levels (varying from moderate growth to limited growth in the form of tiny colonies) were also detected for a number of B. subtilis isolates under these conditions (Table 2). We also observed that isolates 259 and 370 identified by 16S rRNA sequence analysis as B. clausii along with the control strains B. clausii DSM 8716 and B. clausii DSM 2515 were able to grow under alkaline conditions in consonance with their identification as B. clausii (61). The laboratory strain B. subtilis MB24 failed to grow under these conditions.
Approximately 60% of the 31 Bacillus isolates presented some level of hemolysis on 5% sheep blood agar, including all four isolates related to the B. cereus group (Table 2).
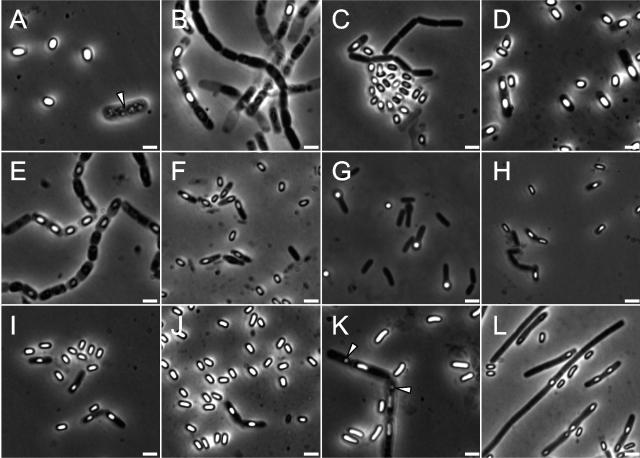
Microscopic examination of the isolates showed a diverse collection of rod-shaped bacteria producing endospores of different sizes and shapes, including those that caused swelling of the mother cell (Fig. 1). Some cells presented distinctive features such as the presence of one or multiple inclusions or vacuoles of undetermined nature (Fig. 1, arrowheads in panels A and K). In another example, and in agreement with observations reported for the B. clausii strain present in the probiotic Domuvar (21), B. clausii isolates 259 and 370 appeared to form long filaments (Fig. 1, panel L), which were septated, as revealed by staining with the membrane dye FM4-64 (Molecular Probes) (data not shown).
FIG. 1.
Morphological diversity of fecal sporeformers from poultry. Phase-contrast microscopy of poultry gut sporeformers grown in Difco sporulation medium (DSM). All microphotographs are at the same scale. Isolates represented in the different panels are as follows: A, 3; B, 10; C, 37; D, 52; E, 53; F, 56; G, 62; H, 197; I, 200; J, 210; K, 241; and L, 259. Scale bar, 2 μm. The presence of vacuoles or inclusions of undefined nature is indicated by an arrowhead in panels A and K.
Macroscopic analysis also identified distinctive traits for the diagnosis of some isolates. Isolate 241 was readily differentiated from the other B. cereus subgroup isolates as it released a dark brown pigment when grown on LB and DSM agar plates. In another case, the distant relationship of isolate 52 with the B. cereus subgroup revealed by 16S rRNA sequencing was strengthened by the small size of its colonies, which contrasted with the large flat colonies presented by the other three isolates of this subgroup. This was also supported by its inability to grow at 50°C (Table 2) (60, 63).
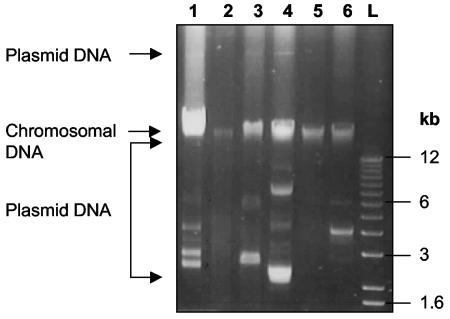
Of the 31 isolates analyzed, almost 50% contained detectable plasmid DNA, with some isolates harboring multiple plasmids (Fig. 2, Table 2). The electrophoretic mobility of these extrachromosomal elements varied greatly, with some strains containing large plasmid DNA molecules (Fig. 2) that migrated well above the chromosomal DNA band in agarose gels.
FIG. 2.
Plasmid profile of poultry fecal Bacillus isolates. Agarose gel electrophoresis of representative examples of plasmids isolated from poultry isolates by the method of Anderson and McKay with the small-scale procedure. Lanes: 1, 268; 2, 278; 3, 285; 4, 287; 5, 306; 6, 369; L, molecular size markers (the sizes of some of the bands are indicated for reference). The positions of chromosomal and plasmid DNAs are indicated by arrows on the left side of the figure.
Following this initial characterization, a group of seven isolates (B. megaterium 3, B. licheniformis 56, B. pumilus 197, B. clausii 259, and B. subtilis isolates 37, 200, and 210) was tested for properties that could be relevant for their survival or persistence in the animal gut ecosystem.
Biofilm formation.
Isolates that produced highly viscous colonies on LB plates also formed strong biofilms. This included isolates 37, 56, 200, and 210. Remarkably, for these isolates the film in the air-medium interface was strong enough to sustain the cultures when the tubes were inverted (not shown). Strains 3, 197, and laboratory strain MB24 grew without forming an attached ring, while isolate 259 was unable to grow in the experimental media and therefore could not be tested.
Antibiotic susceptibility analysis.
With rare exceptions, the isolates were sensitive to the antibiotics tested (Table 3). The exceptions included B. clausii 259 and B. licheniformis 56, which were both resistant to erythromycin and lincomycin. This suggested the involvement of an MLSB (macrolide-lincosamide-streptogramin B) resistance determinant (56). Exposure of both isolates to subinhibitory levels of erythromycin (0.4 μg ml−1) resulted in higher MICs, demonstrating the inducible nature of the resistance.
TABLE 3.
Susceptibility of selected Bacillus isolates to various antimicrobial agentsa
| Strain | MIC (μg ml−1)
|
||||||
|---|---|---|---|---|---|---|---|
| AM | EM | CL | TC | GM | CI | LC | |
| 3 | 0.064 | 0.032 | 0.75 | 0.032 | <0.016 | 0.032 | 125 |
| 37 | 0.023 | 0.047 | 1.5 | 0.064 | 0.047 | 0.023 | 31.25 |
| 56 | 0.25 | >256 | 12 | 0.094 | 0.19 | 0.023 | 250b |
| 197 | 0.023 | 0.032 | 6 | 0.094 | 0.064 | 0.047 | 7.8 |
| 200 | 0.023 | 0.064 | 2 | 3 | 0.094 | 0.047 | 62.5 |
| 210 | 0.023 | 0.064 | 1.5 | 0.064 | 0.064 | 0.032 | 31.25 |
| 259 | 0.19 | >256 | 4 | 0.064 | <0.016 | 0.38 | 2,500b |
MICs for ampicillin (AM), erythromycin (EM), chloramphenicol (CL), tetracycline (TC), gentamicin (GM), and ciprofloxacin (CI) were determined by the E-test method, while the MIC for lincomycin (LC) was determined by the microdilution method on a 96-well microtiter plate. The experiment was repeated three times, and representative results are shown.
MIC values after induction by subinhibitory concentrations of erythromycin (0.4 μg ml−1): 56, 2,500 μg ml−1 and 259, > 5,000 μg ml−1.
MLSB resistance is most commonly due to the presence of an erm gene, which encodes an rRNA methylase specific for adenine residues (56). In order to get a better understanding of the genetic basis for the resistance exhibited by these isolates, we used PCR and Southern blotting to screen for erm genes of classes A, B, C, D, F, and G. The results from these experiments identified a chromosomal erm(D) gene in B. licheniformis 56, but B. clausii 259 showed no significant similarity with any of the erm genes tested. PCR amplification with primers specific for erm(34), on the other hand, which was recently identified in the B. clausii probiotic Enterogermina and B. clausii reference strains (6), gave a product of the expected size, suggesting the presence of this gene. This was confirmed by sequencing of the amplified PCR product. DNA extractions from isolate 259 repeatedly failed to detect any plasmid DNA (Table 2), suggesting that the MLSB resistance of strain 259 was a consequence of the presence of erm(34) in the chromosome.
Sporulation properties.
We then calculated the sporulation efficiency of the Bacillus isolates, expressed as the percentage of heat-resistant cells (80°C, 20 min) present in 24-h DSM cultures. Although isolates 3, 37, 56, 197, 200, and 210 were able to grow well in DSM, B. clausii isolate 259 consistently showed an extended lag phase compared to most of the other isolates. However, this did not appear to affect the efficiency of sporulation. All isolates showed good sporulation efficiencies and showed spore titers in the order of 108 to 109 ml−1 (Table 4). Initially a low sporulation efficiency was observed for isolate 3, which did not correlate with the large number of spores observed by phase contrast microscopy of the 24-h DSM culture. However, this correlation was restored when the heat-resistant spore titers were determined at 65°C (Table 4).
TABLE 4.
Sporulation efficiencies of selected Bacillus isolatesa
| Isolate no. | No. of viable cells ml−1 | No. of spores ml−1 | Sporulation efficiency (%) |
|---|---|---|---|
| 3 (65°C) | 1.2 × 108 | 1.4 × 108 | 116.6 |
| 3 (80°C) | 1.2 × 108 | 2.9 × 106 | 2.4 |
| 37 | 5.8 × 108 | 5.8 × 108 | 100 |
| 56 | 6.3 × 108 | 2.8 × 108 | 44.4 |
| 197 | 2 × 109 | 1.8 × 109 | 90 |
| 200 | 8.4 × 108 | 9.1 × 108 | 108.3 |
| 210 | 7 × 108 | 6.9 × 108 | 98.6 |
| 259 | 1.1 × 108 | 1.9 × 108 | 172.7 |
| MB24 | 5.2 × 108 | 3 × 108 | 57.7 |
Sporulation was induced by exhaustion in DSM broth. At 24 h after inoculation, viable counts were measured before and after incubation at 80°C (and 65°C for 3) for 20 min prior to plating. Sporulation efficiency corresponds to the percentage of survivors. The experiment was performed three times, and a typical result is shown. Note that sporulation efficiencies slightly higher than 100% (see 3 and 200) are within the error of the method. However, this does not seem to be the case for isolate 259, which consistently gave sporulation efficiencies much higher than 100% (see text).
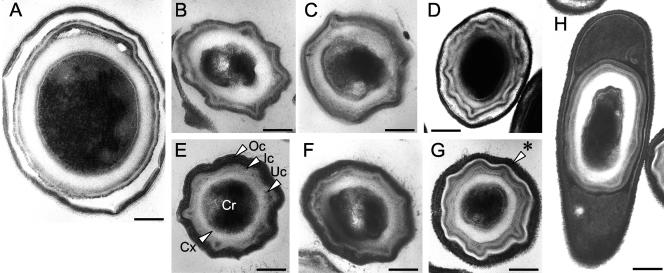
Spore ultrastructure.
All the seven poultry isolates herein examined conformed to the general structural pattern found to be common to all the Bacillus spores that have been examined so far (20) and that consists of a core compartment surrounded by a thick layer of cortex peptidoglycan, which in turn is covered by an inner and an outer coat layer (Fig. 3). The spores of the B. subtilis 37, 200, and 210 isolates were virtually indistinguishable from the spores of the laboratory strain MB24 (Fig. 3, panels B, E, and F). The structure of the B. megaterium 3 spore was also similar to that described in the literature for B. megaterium spores, with the “double” spore coat (3) (Fig. 3, panel A). However, one clear deviation was seen for spores of isolate 259. In these, the coat showed a thin electron-dense layer separating the inner coat from a more external lamellar layer (Fig. 3, panel G). In addition, the spores were associated with an external electron-dense layer with a porous appearance of unknown origin and undefined nature (Fig. 3, panels G and H).
FIG. 3.
Poultry fecal Bacillus spore morphological diversity. Thin-section electron micrographs of spores produced by selected gut Bacillus isolates, collected and processed from 24-h cultures in DSM. The following spore structures are indicated for a typical spore of B. subtilis (isolate 200, panel E): Cr, spore core; Cx, spore cortex; Uc, Ic, and Oc, under, inner, and outer spore coats, respectively. Other panels represent spores from the following isolates: A, 3; B, 37; C, 56; D, 197; F, 210; G, 259; and H, 259. An asterisk indicates the hair-like structures visible with spores of isolate 259. Scale bar: 0.1 μm in panels A to G; 0.2 μm in panel H.
While none of the spores examined presented any structure that could be described as an exosporium, spores of B. clausii 259 were very distinctive on account of the short hair-like structures protruding from the outermost coat layer (Fig. 3, arrowhead in panel G).
Acid and bile tolerance of Bacillus spores and cells.
Spores from isolates 3, 37, 56, 197, 200, 210, and 259 as well as those of the standard laboratory strain MB24 were tested for resistance to simulated gastric conditions and to bile salts (0.2%). Spores of all the isolates tested were similarly resistant to both simulated gastric conditions and to bile salts, as determined by less than one log difference in CFU at time zero and after exposure to both experimental conditions (data not shown).
Different studies have reported on the germination of Bacillus spores in the gastrointestinal tract, including the jejunum and ileum (8, 22, 35). For this reason we also assessed the survival rate of vegetative cells to exposure to both simulated gastric conditions and bile salts. In contrast to spores, vegetative cells of all isolates were very susceptible to both treatments. The results showed a decrease in cell viability of more than 98% within 5 min of exposure to the stress conditions (results not shown).
Antimicrobial activity of fecal Bacillus isolates.
An agar-spotting method was used to assess the production of antimicrobial compounds by the selected fecal Bacillus isolates from broiler chickens against a panel of gram-negative and gram-positive bacteria, which included food spoilage and pathogenic bacteria (Table 5). Although antimicrobial activity was detected for all the strains, there were considerable differences in their spectra and degrees of inhibition (Fig. 4, Table 5). Indeed, the antimicrobial activity appeared to be specific for a particular indicator and producer strain pair. A case in point relates to B. subtilis isolates 37, 200, and 210, each of which showed different spectra of activity. Strain 200, for example, presented the largest antibacterial spectrum, exhibiting inhibitory activity against several gram-positive species, such as Listeria monocytogenes, Listeria innocua, C. perfringens, Enterococcus faecalis, and Staphylococcus aureus, as well as different Bacillus species. Isolate 200 also produced a weak activity against the avian colibacillosis agent E. coli O78:K80. Importantly, several producer strains strongly inhibited C. perfringens (Table 5).
TABLE 5.
Antimicrobial activity of selected Bacillus isolatesa
| Indicator strains | Growth inhibition by strain:
|
||||||
|---|---|---|---|---|---|---|---|
| 3 | 37 | 56 | 197 | 200 | 210 | 259 | |
| E. coli O78:K80 | − | − | − | − | +/− | − | − |
| E. coli O157:H7 | − | − | − | − | − | − | − |
| S. enterica Serovar Enteritidis S1400 | − | − | − | − | − | − | − |
| S. bareilly | − | − | − | − | − | − | − |
| C. rodentium | − | − | − | − | − | − | − |
| P. aeruginosa | − | − | − | − | − | − | − |
| E. aerogenes | − | − | − | − | − | − | − |
| S. aureus RN4220(pVF5) | − | − | − | ++ | ++ | − | − |
| S. aureus RN2442(pE194) | − | +/− | − | ++ | +++ | +/− | − |
| E. faecalis ATCC 2570 | − | − | − | − | ++ | − | − |
| L. innocua | − | ++ | ++ | +/− | +++ | +++ | − |
| L. monocytogenes | − | + | + | − | ++ | ++ | − |
| C. perfringens fD00385 | − | +++ | + | ++ | +++++ | ++++ | v |
| B. megaterium 899 | + | +++ | +++ | ++ | +++++ | ++++ | + |
| B. cereus T | − | ++ | − | +/− | ++ | + | − |
| B. licheniformis 9945A | − | − | − | − | − | − | − |
| B. cereus var. vietnami (Subtyl) | − | ++ | +/− | − | +++ | + | − |
| B. cereus (Biosubtyl Da Lat) | − | ++ | ++ | +/− | +++ | + | − |
| B. pumilus (Biosubtyl Nha Trang) | − | − | + | − | − | − | − |
| B. subtilis MB24 | − | − | ++ | − | − | − | − |
| B. clausii DSM 8716 | − | − | +/− | − | − | − | − |
| B. sphaericus ATCC 33203 | − | + | ++ | ++ | + | − | − |
| B. sphaericus ATCC 14577 | − | + | ++ | + | + | − | − |
The different scores try to reflect different degrees of growth inhibition. +, clear halo of growth inhibition in at least one time point (results scored at 5, 8, 24, and 48 h of incubation); multiple +, increased activity as assessed visually by the increased diameter of the inhibition zone; −, no inhibition; +/−, reduction in growth intensity but not complete inhibition; v, results varied. All indicator strains were assayed at least twice.
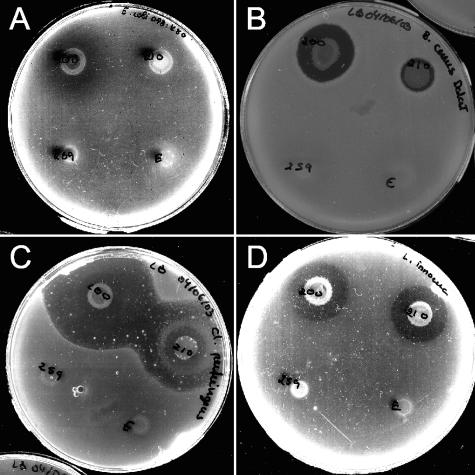
FIG. 4.
Representative examples of antimicrobial activity among poultry fecal Bacillus isolates. Antimicrobial activity was screened by a colony overlay assay where producing strains (clockwise starting on the bottom left: 259, 200, 210, and the B. clausii strain present in the probiotic Enterogermina, which was tested in parallel) grown as spots for 24 h on LB agar plates were overlaid with an indicator strain (A, E. coli O78:K80; B, B. cereus Biosubtyl Da Lat; C, C. perfringens fD00385; D, L. innocua).
DISCUSSION
Bacillus spp. have been described traditionally as aerobic saprophytic soil organisms. Several studies based on 16S rRNA analysis have failed to detect Bacillus species in the gastrointestinal tract of chicken, but this could be due in part to their presence in low numbers or as spores from which a representative sample of DNA would be difficult to extract (67 and references therein). Nevertheless, bacilli (12, 34) and Bacillaceae (29) have been detected in the cecal lumen and mucosa, respectively. The fact that we were able to isolate a large number of different Bacillus strains from broiler chicken feces suggests that a diverse collection of Bacillus species may be present in the poultry gut.
Bacillus species are frequently found in food (9, 14, 46), which could facilitate their entry into the different gut ecosystems. Earlier studies have reported on the frequent isolation of B. cereus from human feces (11, 65), and a recent study has reported the identification of B. subtilis, B. licheniformis, B. pumilus, and Bacillus fragilis in the cecum of rabbits and the role of sporulation by B. subtilis in the induction of gut-associated lymphoid tissue development (55). In parallel work, we also isolated aerobic sporeformers from the fecal material of sheep and cattle, which represent very different gut ecosystems (Barbosa et al., unpublished data). Together with some of the properties exhibited by the isolates described in this study, such as the resistance of their spores to simulated gastric conditions or to bile salts or biofilm formation (see below), and the suggestion that B. subtilis can germinate and sporulate in the gut (8, 22), this suggests that several Bacillus species may be well prepared for transit between the gut and the soil. Even if transient in the gut, detection of these organisms in all the fecal samples examined in our study suggests that their role in the intestinal microbial balance may have been underestimated.
As also found by others (14, 58), it became evident at an early point in this study that speciation of some Bacillus isolates was complex. However, we base our claim for diversity on the use of a range of different morphophysiological and biochemical parameters as well as genetic analysis to tentatively identify the isolates to the species level. For example, the API system only allows the identification of 19 of the best-defined and most common species and therefore failed to identify a number of our isolates. On the other hand, different species, such as those belonging to the B. cereus subgroup (B. cereus, B. anthracis, B. thuringiensis, and B. mycoides), may have essentially the same 16S rRNA sequences. This was the case for isolates 235, 241, and 243, where discrimination required both rRNA sequence data and information obtained from the API system. For these B. cereus isolates, we additionally noted that all three exhibited hemolytic activity, suggesting that none was B. anthracis (63).
Some isolates demonstrated properties that could be advantageous in gut ecosystems. In agreement with previous reports, all the isolates of B. licheniformis and of the B. cereus group were able to grow anaerobically (63). B. subtilis has been viewed as a strict aerobe, but recent work has shown that it can also grow anaerobically (37). Consistent with these reports, we observed that some B. subtilis isolates exhibited limited growth under the anaerobic conditions used and that anaerobic growth of B. subtilis strains 278 and 285 was considerably better than that observed for the other B. subtilis isolates, indicating strain-specific differences in the levels of tolerance to the particular anaerobic conditions used. Interestingly, it has also been shown that B. subtilis does not sporulate efficiently under anaerobic conditions (23), suggesting that its reported capacity to sporulate in the gut (22) may be strongly influenced by the particular niche occupied. Also, some isolates formed robust biofilms.
While the formation of biofilms by gut microorganisms is well recognized (43, 52), its relevance specifically to Bacillus strains in the gut has yet to be determined. Nevertheless, it is conceivable that biofilms on the surface of either the gastrointestinal tract or food particles could provide protection against some of the harsh physical and chemical stresses found in the gut and thereby promote their survival and persistence. Alternatively, but not exclusively, biofilms could play a role in the protection of the gut epithelium from adherence of pathogenic agents. Natural isolates of B. subtilis are able to form complex biofilms, which produce aerial structures or fruiting bodies, where spore formation preferentially occurs (7). In keeping with the suggestion that sporulation may occur in the gut (22, 55), it remains possible that biofilms serve as the preferred site of sporulation in this environment. A more detailed analysis of sporulation by these isolates appears pertinent.
We also note that all the selected isolates were capable of efficient sporulation under laboratory conditions, and their spores conformed to the general structural organization described for Bacillus species (20). However, the spores of B. clausii isolate 259 were considerably different from those described for the B. clausii probiotic Enterogermina (13) and included a hair-like layer at the spore edge. The presence of spore appendages in B. cereus has been implicated in their ability to adhere to surfaces (15, 28). The hair-like layer at the edge of the spores of isolate 259 is particularly intriguing due to the high similarity observed between the coat polypeptide composition of this isolate and Enterogermina, as assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown).
Spores of some Bacillus probiotic strains were recently shown to be susceptible to bile and simulated gastric conditions (10). Although vegetative cells of the broilers isolates were unable to survive the simulated gastrointestinal tract conditions, spores of all the different species tested, including those of B. megaterium 3 (which showed reduced heat resistance), were resistant to bile salts and acidic pH. It will be interesting to test whether this property correlates with the assembly of any of the spore structures seen. In any case, these findings suggest that the spores from the tested isolates could survive transit through the gastrointestinal tract's adverse environment. If so, their final outcome may then depend on the intestinal niche where spores persist and perhaps germinate (8, 22).
Bacillus species are being used as probiotics in both human and animals, and therefore properties such as the presence of extrachromosomal elements, resistance genes, or antimicrobial activity seemed pertinent to test. We found that plasmid DNA was common among poultry fecal Bacillus isolates, and their profiles also assisted in the discrimination of particular strains belonging to the same species. For example, the four isolates related to Bacillus sp. strain VAN 35 or the B. megaterium and B. licheniformis isolates could be differentiated on the basis of their distinctive plasmid profiles. Although the exact nature of the poultry Bacillus plasmids remains cryptic, in other Bacillus isolates the presence of plasmid DNA has frequently been associated with antibiotic resistance determinants and the production of toxins and antibiotic compounds (5, 36, 41). The occurrence of plasmid DNA could also be seen as an indicator of genetic stability and potential for genetic transfer in these species. This is important as gut ecosystems are thought to act as reservoirs of resistant bacteria and resistance genes that can be passed on to humans through the food chain. Given that the selected isolates were for the most part susceptible to the antibiotics tested, it is unlikely that these plasmids code for resistance determinants, but additional studies will be required to determine the exact nature of the extrachromosomal elements detected in this study.
In addition to B. clausii isolate 259, we found erm (34) in a second erythromycin-resistant poultry gut isolate, B. clausii strain 370, and also in the B. clausii strain present in the probiotic Domuvar (21) (data not shown). This suggests that the MLSB resistance of B. clausii isolates analyzed in this and previous studies (6) could be intrinsic to this bacterial species and occurs as a consequence of the presence of erm (34) in the chromosome.
Importantly, several Bacillus isolates demonstrated an antagonistic activity against a broad range of bacterial strains (44). Significantly in this context, the competitive exclusion of pathogens by probiotic strains, including the lactic acid bacteria, can be assisted by bacterial production of antimicrobial agents, such as bacteriocins (42). Also, in vitro, the anti-Helicobacter pylori activity of a probiotic strain of B. subtilis was shown to be dependent on the production of antibiotics (49). Of the target strains tested in our work, the inhibitory activity detected against L. monocytogenes and C. perfringens, a food-borne pathogen and the etiological agent of necrotic enteritis in poultry, respectively, is noteworthy. The nature of the antimicrobial activity of some of the poultry isolates is currently under study.
The extensive use of Bacillus species has not always been accompanied by rigorous scientific testing. The recent observation that several of these products contained mislabeled species, some of which were found to be multidrug resistant and/or produce toxins, raises additional questions regarding their use (10, 13, 21). Evidently, in order to establish the safety and effectiveness of probiotic products an initial unambiguous identification and characterization of the bacterial strain is essential. Recent studies have shown that a laboratory strain of B. subtilis was able to suppress poultry colonization by different avian pathogens (30, 31). Initially we hypothesized that indigenous isolates could potentially be safer and more effective.
The work described in this article aimed at testing whether a diverse collection of Bacillus species could be found in association with the gastrointestinal tract of chickens and additionally provided the first and necessary stage in the characterization of a selected group of strains in view of a potential application. Although we do not claim that any of the isolates herein described will have probiotic properties on the basis of the present investigation, the in vitro parameters tested suggest possible candidates for testing the safety and effectiveness of these organisms in suppressing poultry colonization by pathogens. This work is currently in progress.
Acknowledgments
This work was supported by a grant from the European Union 5th Framework (QLK5-CT-2001-01729) to A.O.H. and M.J.M.
We are grateful to bioMérieux, Portugal, for providing the APILAB Plus software.
REFERENCES
- 1.Abu-Ruwaida, A. S., M. Husseini, and I. M. Banat. 1995. Salmonella exclusion in broiler chicks by the competitive action of adult gut microflora. Microbios 83:59-69. [PubMed] [Google Scholar]
- 2.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson, A. I., and P. Fitz-James. 1976. Structure and morphogenesis of the bacterial spore coat. Bacteriol. Rev. 40:360-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbosa, T. M., and S. B. Levy. 2000. The impact of antibiotic use on resistance development and persistence. Drug. Resist. Updates 3:303-311. [DOI] [PubMed] [Google Scholar]
- 5.Bernhard, K., H. Schrempf, and W. Goebel. 1978. Bacteriocin and antibiotic resistance plasmids in Bacillus cereus and Bacillus subtilis. J. Bacteriol. 133:897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozdogan, B., S. Galopin, and R. Leclercq. 2004. Characterization of a new erm-related macrolide resistance gene present in probiotic strains of Bacillus clausii. Appl. Environ. Microbiol. 70:280-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casula, G., and S. M. Cutting. 2002. Bacillus probiotics: spore germination in the gastrointestinal tract. Appl. Environ. Microbiol. 68:2344-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damgaard, P. H., H. D. Larsen, B. M. Hansen, J. Bresciani, and K. Jorgensen. 1996. Enterotoxin-producing strains of Bacillus thuringiensis isolated from food. Lett. Appl. Microbiol. 23:146-150. [DOI] [PubMed] [Google Scholar]
- 10.Duc, L. H., H. A. Hong, T. M. Barbosa, A. O. Henriques, and S. M. Cutting. 2004. Characterisation of Bacillus probiotics available for human use. Appl. Environ. Microbiol. 70:2161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh, A. C. 1978. Prevalence of Bacillus cereus in the faeces of healthy adults. J. Hyg. (London) 80:233-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, P. M. Sabour, R. Wheatcroft, and S. Chen. 2002. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 208:1-7. [DOI] [PubMed] [Google Scholar]
- 13.Green, D. H., P. R. Wakeley, A. Page, A. Barnes, L. Baccigalupi, E. Ricca, and S. M. Cutting. 1999. Characterization of two Bacillus probiotics. Appl. Environ. Microbiol. 65:4288-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guinebretiere, M. H., O. Berge, P. Normand, C. Morris, F. Carlin, and C. Nguyen-The. 2001. Identification of bacteria in pasteurized zucchini purees stored at different temperatures and comparison with those found in other pasteurized vegetable purees. Appl. Environ. Microbiol. 67:4520-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hachisuka, Y., S. Kozuka, and M. Tsujikawa. 1984. Exosporia and appendages of spores of Bacillus species. Microbiol. Immunol. 28:619-624. [DOI] [PubMed] [Google Scholar]
- 16.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 17.Henriques, A. O., B. W. Beall, and C. P. Moran, Jr. 1997. CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henriques, A. O., B. W. Beall, K. Roland, and C. P. Moran, Jr. 1995. Characterization of cotJ, a sigma E-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J. Bacteriol. 177:3394-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriques, A. O., L. R. Melsen, and C. P. Moran, Jr. 1998. Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J. Bacteriol. 180:2285-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 21.Hoa, N. T., L. Baccigalupi, A. Huxham, A. Smertenko, P. H. Van, S. Ammendola, E. Ricca, and A. S. Cutting. 2000. Characterization of Bacillus species used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders. Appl. Environ. Microbiol. 66:5241-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoa, T. T., L. H. Duc, R. Isticato, L. Baccigalupi, E. Ricca, P. H. Van, and S. M. Cutting. 2001. Fate and dissemination of Bacillus subtilis spores in a murine model. Appl. Environ. Microbiol. 67:3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann, T., N. Frankenberg, M. Marino, and D. Jahn. 1998. Ammonification in Bacillus subtilis utilizing dissimilatory nitrite reductase is dependent on resDE. J. Bacteriol. 180:186-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hume, M. E., J. A. Byrd, L. H. Stanker, and R. L. Ziprin. 1998. Reduction of caecal Listeria monocytogenes in Leghorn chicks following treatment with a competitive exclusion culture (PREEMPT). Lett. Appl. Microbiol. 26:432-436. [DOI] [PubMed] [Google Scholar]
- 25.Iordanescu, S., M. Surdeanu, P. Della Latta, and R. Novick. 1978. Incompatibility and molecular relationships between small Staphylococcal plasmids carrying the same resistance marker. Plasmid 1:468-479. [DOI] [PubMed] [Google Scholar]
- 26.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 27.Koransky, J. R., S. D. Allen, and V. R. J. Dowell. 1978. Use of ethanol for selective isolation of spore-forming microorganisms. Appl. Environ. Microbiol. 35:762-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozuka, S., and K. Tochikubo. 1985. Properties and origin of filamentous appendages on spores of Bacillus cereus. Microbiol. Immunol. 29:21-37. [DOI] [PubMed] [Google Scholar]
- 29.Lan, P. T. N., H. Hayashi, M. Sakamoto, and B. Y. 2002. Phylogenetic analysis of cecal microbiota in chicken by the use of 16S rDNA clone libraries. Microbiol. Immunol. 16:371-382. [DOI] [PubMed] [Google Scholar]
- 30.La Ragione, R. M., G. Casula, S. M. Cutting, and M. J. Woodward. 2001. Bacillus subtilis spores competitively exclude Escherichia coli O78:K80 in poultry. Vet. Microbiol. 79:133-142. [DOI] [PubMed] [Google Scholar]
- 31.La Ragione, R. M., and M. J. Woodward. 2003. Competitive exclusion by Bacillus subtilis spores of Salmonella enterica serotype Enteritidis and Clostridium perfringens in young chickens. Vet. Microbiol. 94:245-256. [DOI] [PubMed] [Google Scholar]
- 32.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy, S. B. 2002. The antibiotic paradox. How the misuse of antibiotics destroys their curative powers, 2nd ed. Perseus Publishing, New York, N.Y.
- 34.Lu, J., U. Idris, B. Harmon, C. Hofacre, J. J. Maurer, and M. D. Lee. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazza, P. 1994. The use of Bacillus subtilis as an antidiarrhoeal microorganism. Boll. Chim. Farm. 133:3-18. [PubMed] [Google Scholar]
- 36.Monod, M., C. Denoya, and D. Dubnau. 1986. Sequence and properties of pIM13, a macrolide-lincosamide-streptogramin B resistance plasmid from Bacillus subtilis. J. Bacteriol. 167:138-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 38.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 40.Nisbet, D. J., G. I. Tellez, V. K. Lowry, R. C. Anderson, G. Garcia, G. Nava, M. H. Kogut, D. E. Corrier, and L. H. Stanker. 1998. Effect of a commercial competitive exclusion culture (Preempt) on mortality and horizontal transmission of Salmonella gallinarum in broiler chickens. Avian Dis. 42:651-656. [PubMed] [Google Scholar]
- 41.Okinaka, R. T., K. Cloud, O. Hampton, A. R. Hoffmaster, K. K. Hill, P. Keim, T. M. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouwehand, A., C. 1998. Antimicrobial components from lactic acid bacteria, p. 139-160. In W. A. Salminen (ed.), Lactic acid bacteria: microbiology and functional aspects, 2nd ed. Marcel Dekker Inc., New York, N.Y.
- 43.Palestrant, D., Z. E. Holzknecht, B. H. Collins, W. Parker, S. E. Miller, and R. R. Bollinger. 2004. Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct. Pathol. 28:23-27. [PubMed] [Google Scholar]
- 44.Parente, E., and A. Ricciardi. 1999. Production, recovery and purification of bacteriocins from lactic acid bacteria. Appl. Microbiol. Biotechnol. 52:628-638. [DOI] [PubMed] [Google Scholar]
- 45.Patterson, J. A., and K. M. Burkholder. 2003. Application of prebiotics and probiotics in poultry production. Poult. Sci. 82:627-631. [DOI] [PubMed] [Google Scholar]
- 46.Pepe, O., G. Blaiotta, G. Moschetti, T. Greco, and F. Villani. 2003. Rope-producing strains of Bacillus spp. from wheat bread and strategy for their control by lactic acid bacteria. Appl. Environ. Microbiol. 69:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perreten, V. 2003. Use of antimicrobials in food-producing animal in Switzerland and the European Union (EU). Mitt. Lebensm. Hyg. 94:155-163. [Google Scholar]
- 48.Perreten, V., N. Giampa, U. Schuler-Schmid, and M. Teuber. 1998. Antibiotic resistance genes in coagulase-negative staphylococci isolated from food. Syst. Appl. Microbiol. 21:113-120. [DOI] [PubMed] [Google Scholar]
- 49.Pinchuk, I. V., P. Bressollier, B. Verneuil, B. Fenet, I. B. Sorokulova, F. Megraud, and M. C. Urdaci. 2001. In vitro anti-Helicobacter pylori activity of the probiotic strain Bacillus subtilis 3 is due to secretion of antibiotics. Antimicrob. Agents Chemother. 45:3156-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 51.Priest, F. G. 1993. Systematics and ecology of Bacillus, p. 3-16. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 52.Probert, H. M., and G. R. Gibson. 2002. Bacterial biofilms in the human gastrointestinal tract. Curr. Issues Intest. Microbiol. 3:23-27. [PubMed] [Google Scholar]
- 53.Pugsley, A. P. 1985. Escherichia coli K12 strains for use in the identification and characterization of colicins. J. Gen. Microbiol. 131:369-376. [DOI] [PubMed] [Google Scholar]
- 54.Reuter, G. 2001. Probiotics—possibilities and limitations of their application in food, animal feed, and in pharmaceutical preparations for men and animals. Berl. Munch. Tierarztl. Wochenschr. 114:410-419. [PubMed] [Google Scholar]
- 55.Rhee, K. J., P. Sethupathi, A. Driks, D. K. Lanning, and K. L. Knight. 2004. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J. Immunol. 172:1118-1124. [DOI] [PubMed] [Google Scholar]
- 56.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robleto, E. A., I. Lopez-Hernandez, M. W. Silby, and S. B. Levy. 2003. Genetic analysis of the AdnA regulon in Pseudomonas fluorescens: nonessential role of flagella in adhesion to sand and biofilm formation. J. Bacteriol. 185:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowan, N. J., K. Deans, J. G. Anderson, C. G. Gemmell, I. S. Hunter, and T. Chaithong. 2001. Putative virulence factor expression by clinical and food isolates of Bacillus spp. after growth in reconstituted infant milk formulae. Appl. Environ. Microbiol. 67:3873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanders, M. E., L. Morelli, and T. A. Tompkins. 2003. Sporeformers as human probiotics: Bacillus, Sporolactobacillus, and Brevibacillus. Comprehensive Rev. Food Sci. Food Safety 2:101-110. [DOI] [PubMed] [Google Scholar]
- 60.Scott, K. P., T. M. Barbosa, K. J. Forbes, and H. J. Flint. 1997. High-frequency transfer of a naturally occurring chromosomal tetracycline resistance element in the ruminal anaerobe Butyrivibrio fibrisolvens. Appl. Environ. Microbiol. 63:3405-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Senesi, S., F. Celandroni, A. Tavanti, and E. Ghelardi. 2001. Molecular characterization and identification of Bacillus clausii strains marketed for use in oral bacteriotherapy. Appl. Environ. Microbiol. 67:834-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sneath, P. H. A. 1986. Endospore-forming gram-positive rods and cocci, p. 1104-1207. In N. S. M. P. H. A. Sneath, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md.
- 64.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turnbull, P. C., and J. M. Kramer. 1985. Intestinal carriage of Bacillus cereus: faecal isolation studies in three population groups. J. Hyg. (London) 95:629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu, X. Y., and R. D. Joergert. 2003. Composition of microbiota in content and mucus from ceca of broiler chickens as measured by fluorescent in situ hybridization with group-specific, 16S rRNA-targeted oligonucleotide probes. Poultry Sci. 82:1241-1249. [DOI] [PubMed] [Google Scholar]