Abstract
Ovarian cancer is a major cause of cancer mortality among women, largely due to late diagnosis of advanced metastatic disease. More extensive molecular analysis of metastatic ovarian cancer is needed to identify post-translational modifications of proteins, especially glycosylation that is particularly associated with metastatic disease to better understand the metastatic process and identify potential therapeutic targets. Glycoproteins in ascites fluid were enriched by affinity binding to lectins (ConA or WGA) and other affinity matrices. Separate glycomic, proteomic, and glycopeptide analyses were performed. Relative abundances of different N-glycan groups and proteins were identified from ascites fluids and a serum control. Levels of biomarkers CA125, MUC1, and fibronectin were also monitored in OC ascites samples by Western blot analysis. N-Glycan analysis of ascites fluids showed the presence of large, highly fucosylated and sialylated complex and hybrid glycans, some of which were not observed in normal serum. OC ascites glycoproteins, haptoglobin, fibronectin, lumican, fibulin, hemopexin, ceruloplasmin, alpha-1-antitrypsin, and alpha-1-antichymotrypsin were more abundant in OC ascites or not present in serum control samples. Further glycopeptide analysis of OC ascites identified N-and O-glycans in clusterin, hemopexin, and fibulin glycopeptides, some of which are unusual and may be important in OC metastasis.
Keywords: ovarian cancer, malignant ascites, glycoproteomics, N-glycans, proteomics, glycopeptides, mass spectrometry
Graphical abstract
INTRODUCTION
Ovarian cancer (OC) is a major cause of cancer mortality in women.1 Often, by the time OC is diagnosed, it has already metastasized, leaving the patient with few treatment options and little chance of survival. Although considerable attention is being paid to biomarkers of early-stage disease,2 it is also important for us to conduct more extensive molecular analyses of metastatic, late-stage epithelial OC to identify glycoproteomic biomarkers in addition to the well-established OC biomarker CA125.3,4 Information about metastatic glycan-associated biomarkers could potentially serve as prognostic and predictive biomarkers to help with diagnosis and treatment decisions as well as identify additional therapeutic targets.
The objective of this current study is to perform glycomic, proteomic, and glycopeptide analyses on malignant ascites obtained from women with metastatic OC. Ascites is produced when malignant tumor cells escape from the primary tumor site into the surrounding abdominal cavity, where tumor cells find a rich, nourishing environment, enabling them to grow and proliferate. Eventually these tumor cells can enter the circulation and metastasize to other organ sites. Because metastasis is the major cause of mortality in OC, identification of glycosylated proteins produced by or in response to the metastatic tumor cells helps us to better understand the metastatic process and gives us insight into how to interfere with the growth and metastasis of OC tumor cells. OC ascites is a complex sample consisting largely of fluid and a variety of different cell types, mostly immune cells (lymphocytes, macrophages, dendritic cells), red blood cells (RBCs), and fibroblasts with varying numbers of tumor cells (few to hundreds of millions cells per sample). The fluid is separated from the cells by centrifugation. Ascites fluids contain valuable molecular information specific to each individual patient that uniquely reflects their age, gender, genetics, and different lifestyle influences from diet and environmental, in addition to having metastatic disease.5,6
For this study, concurrent N-glycan and proteomic analyses were first performed on OC metastatic ascites fluids and a normal serum control. Ascites fluids were obtained from women diagnosed with metastatic OC. Glycosylated proteins were enriched through affinity binding to three immobilized lectins: concanavalin A (ConA) that preferentially binds neutral and high-mannose-type glycans, wheat germ agglutinin (WGA) that tends to bind N-acetyl glucosamine (GlcNAc) and sialylated glycans, and Aleuria aurantia lectin (AAL), which selectively binds fucosylated glycosylated proteins. Western blot (WB) analyses of CA125, fibronectin, and MUC1 were included in this study. CA125 is still the most reliable biomarker for OC, while fibronectin and MUC1 have been previously reported as potential biomarkers of OC.7–11 Fibronectin is a medium abundant plasma/serum protein, more easily detected by proteomic methods, whereas CA125 and MUC1 are present in much lower amounts. Analysis of these glycoproteins helps us to determine the overall enrichment of these glycoproteins by ConA or WGA lectin binding.
N-Glycan analysis was conducted using analytical glycomic methods designed to maximize the numbers of glycans identified.12–15 Proteomic analysis of lectin-bound samples was performed using conventional trypsin digestion, followed by peptide mass fingerprinting.16 Although proteomic analysis after lectin binding still showed mostly abundant plasma/serum proteins, an increase in certain types of N-linked glycans was identified. These N-glycans are not usually identified in plasma or serum samples and may be indicative of cancer tumor antigens important to the metastasis of tumor cells. These intriguing results show the value of performing concurrent glycomic and proteomic analyses of ascites fluids and how lectin affinity chromatography can be used to enrich for potentially important glycans. Further glycopeptide analysis was performed to identify site occupancy and heterogeneity of attached glycans to specific proteins. These studies will eventually help us to identify specific glycoproteins and their glycosylation as a means of identifying better candidate glycoproteomic biomarkers of metastatic OC.
EXPERIMENTAL SECTION
Acquisition and Processing of Ascites Samples
Samples of metastatic OC ascites were obtained from the Cancer Center Biorepository, as part of the UCDCC #183 clinical study of OC (Dr. Gary Leiserowitz) with patient consent utilizing an institutionally approved IRB protocol. The ascites specimens were deidentified (coded) so that the identity of the individual donating the samples could not be identified by the research personnel. Ascites specimens (ranging in volume from 10 mL to 1.5 L) were transferred to sterile centrifuge tubes or bottles and centrifuged at 300–450g (1200–1400 rpm, Beckman GPR centrifuge) for 10 min to pellet cells (malignant, immune, and other cells) in the sample. Supernatant (fluid) was removed, aliquoted, and frozen for further molecular analyses (−80 °C).
Reagents
PNGase F was acquired from New England Biolabs (Ipswich, MA). Graphitized carbon cartridges came from Grace Alltech (Deerfield, IL). Trizma and gelatin (fish scale) came from Sigma-Aldrich (St. Louis, MO), sodium dodecyl sulfate from BioRad (Hercules, CA), and 30% acrylamide/bis acrylamide and SafeBlue colloidal Coomassie blue stain from National Diagnostic (Charlotte, NC). All other reagents, solvents, and buffers used in this project were of analytical grade.
Lectin Binding of Ascites Fluid
Proteins in ascites were bound to and eluted from immobilized ConA and WGA lectins according to the manufacturer instruction (Thermo-Fisher Scientific, Pierce Division, Rockford, IL). Eluted samples were concentrated (Amicon, EMD Millipore, Billerica, MA, MW 10 000 cutoff). Samples of the lectin-bound proteins were analyzed by 7.5% SDS-PAGE, followed by carbohydrate staining (ProQ Emerald, Invitrogen, Carlsbad, CA) and then protein staining (Safe-Blue, National Diagnostics, Charlotte, NC). Duplicate samples were run and electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon, EMD Millipore, Billerica, MA), blocked with 5% gelatin (Sigma, St. Louis, MO), and probed for CA125 (Zymed, now Invitrogen, San Diego, CA; this CA125 antibody is no longer available). The blots were sequentially stripped and probed for MUC1, MUC2, and MUC3 (Geneway, San Diego CA) and fibronectin (Neomarker, Fremont, CA).
Glycomic Analysis of Ascites Samples: Release and Processing of Glycans
N-Glycans were released and processed as previously described with slight modifications.17 Aliquots (50 μL) of ascites fluid or control serum (Sigma) were added to an equal volume of 200 mM ammonium bicarbonate (Sigma-Aldrich, St. Louis, MO) solution with 10 mM dithiothreitol (DTT) (Promega, Madison, WI) and heat-denatured. Peptide-N-glycosidase F (PNGaseF) was added (2 μL, equivalent to 1000 NEB units or 15 IUB mU, New England Biolabs, Ipswich, MA). N-Glycan release was performed in a microwave oven (20 W for 10 min) (CEM, Matthews, NC). Deglycosylated proteins were precipitated by adding 400 μL of ice-cold ethanol, followed by incubation at −80 °C for 1 h and then centrifugation (14 000g, 20 min, Eppendorf centrifuge). Supernatants containing the released N-glycans were transferred to new microcentrifuge tubes and dried in vacuo. Solid-phase extraction of glycans was performed on a Gilson GX-274 ASPEC liquid handler equipped with a cartridge rack. Using the liquid handler, cartridges (Carbograph, Alltech Associates, Deerfield, IL) containing porous graphitized carbon (150 mg/cartridge) were first washed with nanopure water and then conditioned with 6 mL of 0.1% trifluoroacetic acid (TFA) in 80% acetonitrile (ACN)/H2O (v/v), followed by a final rinse of water. Glycan solutions were applied to the cartridges and subsequently washed with 12 mL of nanopure (E-Pure, Barnstead) water at a flow rate of 1 mL/min for desalting. Glycans were eluted with 4.5 mL of 10% ACN/H2O (v/v), 20% ACN/H2O (v/v), and 40% ACN/H2O (v/v) with 0.05% TFA and dried in a centrifugal evaporator apparatus. Each sample was reconstituted in 50 μL of nanopure water prior to MS analysis.
nanoHPLC-chip-TOF or qTOF MS Analysis
Glycan samples (1 μL) were analyzed using an Agilent 6200 Series HPLC–Chip/TOF MS system equipped with a microwell-plate autosampler (maintained at 20 °C), capillary sample loading pump, nanopump, HPLC–chip interface and the Agilent 6210 TOF LC/MS, or an Agilent 6520 nanoLC-Chip-Q-TOF equipped with an Agilent 1200 series nano-LC system.12,18 The HPLC–Chip (Glycan Chip) consisted of a 40 nL enrichment column and a 43 × 0.075 mm2 inner diameter (i.d.) analytical column, both with graphitized carbon (5 μm) as stationary phase. Upon injection, the sample was loaded onto the enrichment column and subsequently separated on the analytical column using a gradient of 3% ACN with 0.5% formic acid (FA) (solvent A) to 90% ACN with 0.5% FA (solvent B). The mass spectrometers were operated in positive mode.
Data Processing
Processing of the glycan data was performed using Masshunter qualitative analysis (version B.06.00, Agilent) and Microsoft Excel 2013 as described in Hua et al.19 with modifications. Data were loaded into Masshunter qualitative analysis, and glycan features were identified and integrated using the Molecular Feature Extractor algorithm. Signals above a signal-to-noise (S/N) threshold of 5.0 were first considered, after which the signals were deconvoluted using a tolerance of 0.0025 m/z ± 10 ppm. Resulting deconvoluted masses were subsequently annotated using a retrosynthetic theoretical glycan library,20 with a 15 ppm mass error allowance.
Protein Identification by Tandem Mass Spectrometry (LC–MS/MS)
Samples of ascites fluid and lectin-bound (WGA- and ConA-bound) were submitted to the UC Davis Proteomic Core for protein identification using trypsin digestion, followed by tandem mass spectrometry (MS/MS). In brief, proteins were reduced and alkylated according to previously described procedure21 and digested with sequencing-grade trypsin per the manufacturer’s recommendations (Promega, Madison, WI).
Liquid chromatography tandem mass spectrometry (LC–MS/MS) protein identification was performed using an Agilent 6330 XCT ion trap mass spectrometer connected to an Agilent HPLC-Chip system (Santa Clara, CA). Peptides were loaded on to an Agilent 5 cm C18 peptide chip and separated using a 15 min gradient of 2–80% buffer B (Buffer A = 0.1% FA, Buffer B = 95% ACN 0.1% FA) at 200 nL/min. The top 10 ions in each MS scan were subjected to automatic low-energy collision-induced dissociation (CID).
Database Searching: Tandem mass spectra were extracted by Bioworks version 3.2. Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using X! Tandem (The GPM, thegpm.org; version TORNADO (2010.01.01.4)). X! Tandem was set up to search the Uniprot complete human database (August 2010, 44 512 entries), plus 110 nonhuman contaminant protein sequences from the common “Repository of Adventitious Proteins” database (http://www.thegpm.org/crap/), plus an equal number of reverse sequences, assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 0.40 Da and a parent ion tolerance of 1.8 Da. Iodoacetamide derivatization of cysteine was specified in X! Tandem, as a fixed modification. Deamidation of asparagine, oxidation of methionine, sulfonation of methionine, tryptophan oxidation to formylkynurenin of tryptophan, and acetylation of the N-terminus were specified in X! Tandem as variable modifications.
Criteria for Protein Identification
Scaffold (version Scaffold_3_00_02, Proteome Software, Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at >95.0% probability, as specified by the Peptide Prophet algorithm.22 Protein identifications were accepted if they could be established at >90.0% probability and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm.23 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Decoy False Discovery Rate
For the ConA and WGA ascites fractions: 106 proteins were identified at 50.0%, 1.0% Decoy FDR and 2592 Spectra at 50.0% Minimum, 0.04% Decoy FDR
Sample Preparation for Glycopeptide Analyses
Ascites fluid samples and serum controls were prepared using affinity chromatography with hemoglobin-agarose (H-agarose, Sigma H8756) to enrich for heme-iron binding proteins such as hemopexin and haptoglobin. After elution from H-agarose, ascites samples from different patients were pooled (five different ascites fluids: asc1, asc412, asc436, asc526, and asc528), and half of the sample was desialylated using acid hydrolysis by treatment with 0.1 M FA for 1 h at 80 °C, followed by lyophilization (Sample #2). The same procedure was performed on a serum control sample (Sample #1). Desialylation can improve the overall detection of glycosylation in glycopeptides where heavy sialylation can sometimes interfere with comprehensive glycan analysis of glycopeptides. The desialylated samples (Samples #1 and #2) and the remaining H-bound serum control sample (Sample #3) and H-bound pooled ascites fluid sample (Sample #4) were trypsin-digested as previously described. Also analyzed were samples prepared using AAL-agarose to enrich for fucose-containing glycosylated proteins (Sample #5) as well as OC ascites fluid that was depleted of HSA and IgG (Agilent HSA/IgG MARS column) (Sample #6). These six samples (desialylated and native OC ascites and serum control, AAL-bound and HSA/IgG-depleted OC ascites) were subjected to trypsin digestion before transport to the Proteomic Core at UC Davis.
Peptide Analysis Using an Orbitrap Q Exactive Plus MS with Tandem MS/MS
Liquid chromatography separation for peptide analysis was done using on an Easy nLC II (Thermo Fisher Scientific), a Proxeon nanospray source. The digested peptides were reconstituted in 2% ACN/0.1% TFA, and ~3 μg of each sample was loaded onto a 100 μm × 25 mm Magic C18 100 Å 5U reverse-phase trap, which desalted the samples before being separated using a 75 μm × 150 mm Magic C18 200 Å 3U reverse-phase column. Peptides were eluted with a gradient of 0.1% FA (A) and 100% ACN (B), flow rate of 300 nL/min. A 60 min gradient was run with 5–35% B over 50 min, 35–80% B over 3 min, 80% B for 1 min, 80–5% B over 1 min, and finally held at 5% B for 10 min. Each of the gradients was followed by a 1 h column wash.
Mass spectra were collected on an Orbitrap Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) in a data-dependent mode with one MS precursor scan, followed by 15 MS/MS scans. A dynamic exclusion of 15 s was used. MS spectra were acquired with a resolution of 70 000 and a target of 1 × 106 ions or a maximum injection time of 30 ms. MS/MS spectra were acquired with a resolution of 17 500 and a target of 5 × 104 ions or a maximum injection time of 50 ms. Peptide fragmentation was performed using higher energy collision dissociation (HCD) with a normalized collision energy (NCE) value of 27. Unassigned charge states as well as +1 and ions > +5 were excluded from MS/MS fragmentation. The mass spectrometry proteomics data have been deposited to the Proteome Xchange Consortium via the PRIDE24 partner repository with the data set identifier PXD003794. Byonic software v2.6 (Protein Metrics, San Carlos, CA) was used to analyze the peptide raw data for the presence of N- and O-linked glycans.
RESULTS
Lectin Binding Enriches Glycosylated Proteins and Removes Most Albumin
We first used concurrent glycomic and proteomic analyses of OC ascites fluids and a serum control before and after lectin binding utilizing two different immobilized lectins, ConA and WGA, to enrich for glycosylated proteins. OC ascites fluids were prepared by centrifugation to separate the fluid from cells. OC fluid samples were aliquoted and frozen at −80 °C and subsequently thawed for use in the lectin binding studies. Initial analysis of glycoproteins isolated by ConA or WGA lectin binding was performed by SDS-PAGE, followed by carbohydrate and then protein staining of the same gel, as shown in Figure 1A,B for one ascites sample (Asc1) (see Experimental Section for details). In the protein stained gel, albumin (MW 69 kDa) was clearly the most abundant protein in the flow-thru or unbound (UB) fractions from both ConA and WGA affinity bindings (Figure 1A, lanes 1 and 4), which did not stain with the carbohydrate stain because albumin is not glycosylated (Figure 1B, lanes 1 and 4). Albumin accounted for at least 80% of the total protein in the original OC ascites fluid (Figure 1A, lane 7, Asc), showing that lectin binding removed most of this abundant protein, thereby making it easier to analyze the remaining glycosylated proteins.
Figure 1.
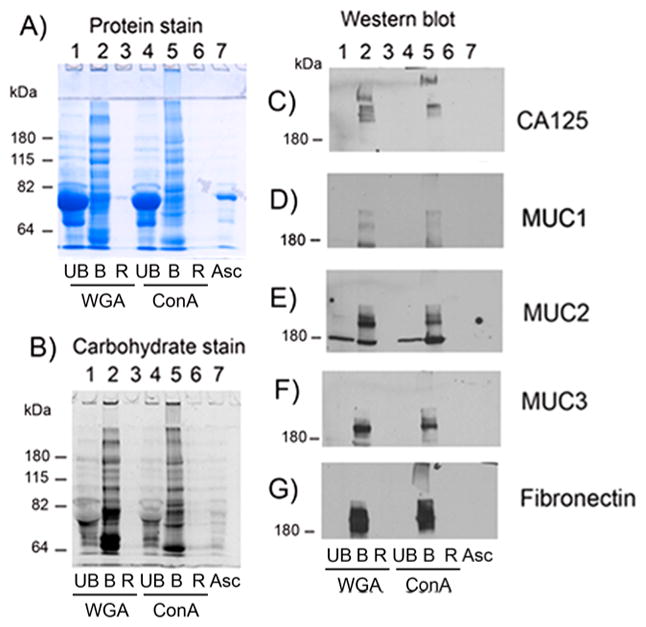
SDS-PAGE of WGA- and ConA-bound ascites (asc1) proteins with Western blot analysis for CA125, fibronectin, and mucins 1, 2, and 3. (A) Protein stain, (B) carbohydrate stain, (C) Western blot analysis of CA125, (D) MUC1, (E) MUC2, (F) MUC3, and (G) fibronectin. UB, unbound; B, bound; R, resin; WGA, wheat germ agglutinin; ConA, concanavalin A; asc, ascites.
Most glycosylated proteins in Asc1 fluid were bound and eluted from ConA- and WGA-immobilized lectins, as shown by staining for carbohydrate (Figure 1B, lanes 2 and 5) and protein (Figure 1A, lanes 2 and 5) of the same gel. Very little protein remained bound to the immobilized resin (R) (Figure 1A,B, lanes 3 and 6) showing the efficiency of the glycoprotein binding to the immobilized lectins. Bound WGA and ConA proteins (Figure 1A, lanes 2 and 5) were highly glycosylated large proteins (Figure 1B, lanes 2 and 5), some of which were unable to enter the SDS-PAGE stacking gel (Figure 1A,B, lanes 2 and 5). These large glycoproteins may have molecular weights as high as several million daltons. Differences in protein- and carbohydrate-stained glycoproteins were observed between those bound to ConA when compared with those bound to WGA (Figure 1A,B, lane 2 compared with lane 5), indicating specificity of each lectin for certain glycoproteins.
WB analysis was performed on a duplicate set of samples for Asc1 (fluid and ConA- and WGA-bound samples) probing for CA125, fibronectin, MUC1, MUC2, and MUC3. Monitoring the presence of these proteins enabled us to determine if these particular proteins could be enriched by lectin binding. CA125 was detected in the WGA- and ConA-bound fractions (Figure 1C, lanes 2 and 5) as extremely large heterogeneous molecular weight proteins with several protein bands detected, some of which did not even enter the stacking gel in the ConA-bound sample. CA125 was not detected in the original ascites fluid (Asc) likely due to its being below the level of detection in this assay. CA125 was also not detected in the unbound lectin fractions (Figure 1C, lanes 1 and 4), showing that it was completely bound to the lectins. Also completely bound to the lectins were MUC1, MUC2, MUC3, and fibronectin (Figure 1D–G, lanes 2 and 5).
Glycomic Analysis of Lectin Bound Proteins
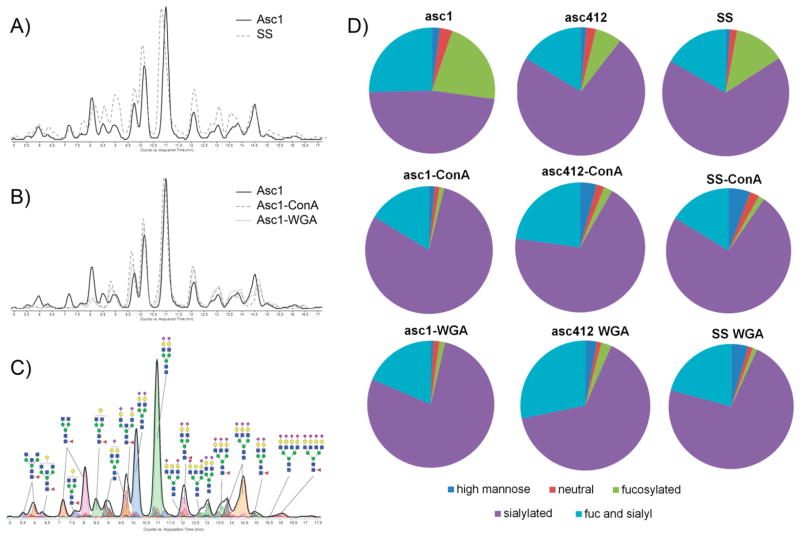
N-Glycan analysis of two OC ascites fluids (Asc1 and Asc412) and their corresponding ConA- and WGA-bound fractions was performed to produce N-glycan profiles for each ascites sample. The idea behind total N-glycomic profiling of these samples was to identify the relative amounts of different N-glycans present in the original ascites fluids compared with each corresponding lectin-bound sample. N-Glycans were isolated from two different ascites fluids (Asc1 and Asc412) as well as from a normal serum control (SS) and each of their lectin-(ConA or WGA) bound fractions. Global N-glycan analysis was performed (see Experimental Section for details). After lyophilization, the samples were reconstituted and subjected to nano-LC Chip/TOF MS or nano-LC Chip/qTOF MS.12,15,25 Examples of the total ion chromatograms of one of the ascites sample (Asc1) (Figure 2A, solid line) compared with that of the serum control (Figure 2A, dashed line) shows differences in the glycan profile. Chromatograms of N-glycans isolated from proteins in the original ascites fluid (Asc1, solid line) compared with those isolated from proteins bound to ConA (dashed lines) and WGA lectins (dotted lines) (Figure 2B) also appear to show some distinct glycan changes in the chromatographs, especially in the early eluted glycans (Figure 2B). Identification of N-glycans was based on the elution time and mass of the glycans using a theoretical database.15,20,26,27 A chromatogram, annotated with N-glycan structures from the ascites fluid, is shown in Figure 2C, which shows the more neutral glycans eluting early while the more negatively charged sialylated glycans eluted later. Pie charts comparing N-glycans from Asc1 and Asc412 fluids along with their ConA- and WGA-bound proteins (Figure 2D) as well as a serum control for comparison show the percentages for each type of glycan (high mannose, neutral, sialylated, fucosylated, and sialylated). ConA-and WGA-bound proteins of all samples (Asc1, Asc412, and SS) showed a much lower percentage of fucosylated glycans but an increase in the percentage of sialylated only and fucosylated-sialylated glycans. These overall glycan changes are important because they will have functional consequences to the proteins to which they are attached in terms of recognition, binding to other proteins, stability, and protein functional activities.
Figure 2.
Nano-LC Chip/TOF MS analysis of N-glycans released from proteins present in OC ascites fluid and ConA- and WGA-bound ascites. (A) Comparison of glycan chromatographs between a serum control and ascites fluid (solid line represents ascites fluid, dotted line represents serum control). (B) Comparison of chromatographs of N-glycans isolated from ascites and ConA- and WGA-bound proteins (solid line represents ascites fluid, dashed line represents ConA, and dotted line represents WGA glycans). (C) Annotated N-glycan profile of the ascites fluid analyzed by nano-LC Chip/TOF MS. (D) Pie charts of the different types (classes) of N-glycans obtained from OC ascites fluid and ConA- and WGA-bound proteins.
Glycomic Results of Ascites and ConA- and WGA-Bound Proteins
Further detailed analyses of the N-glycans from OC ascites fluid samples and a serum control were conducted to show the relative abundance and types of glycans by composition and structure as determined by nano-LC Chip/TOF MS. Composition of the N-glycans identified is presented in the following manner Hex_HexNAc_fucose_sialic acid using the H (Hex), N (HexNAc), F (fucose), and S (sialic acid) configuration. Because it is not possible to distinguish different hexoses (mannose, galactose, glucose) by mass spectrometry, hexose content (H) representing all mannoses, galactoses, and glucose, is presented first; thenm the N-acetyl hexosamine content (N-acetyl galactosamine or GlcNAc) is presented in the second position (N = HexNAc), followed by the fucose content (F= fucose) and finally the sialic acid content (S = sialic acid).
N-Glycan results are presented for a serum control (SS) and two ascites fluid samples, Asc1 and Asc412. In this study the serum control (SS) was also subjected to lectin binding with ConA and WGA. N-Glycan data results will be presented in bar graphs comparing glycan results from the three starting samples with glycan results from their respective ConA- and WGA-bound protein fractions. N-Glycan results with possible structures are shown in Supplemental Table S1. Data results are organized to show related modifications of glycan compositions such as the addition of fucose and sialic acid to core and antennary structures.
Abundant H5N4 N-Glycans in Ascites Fluids
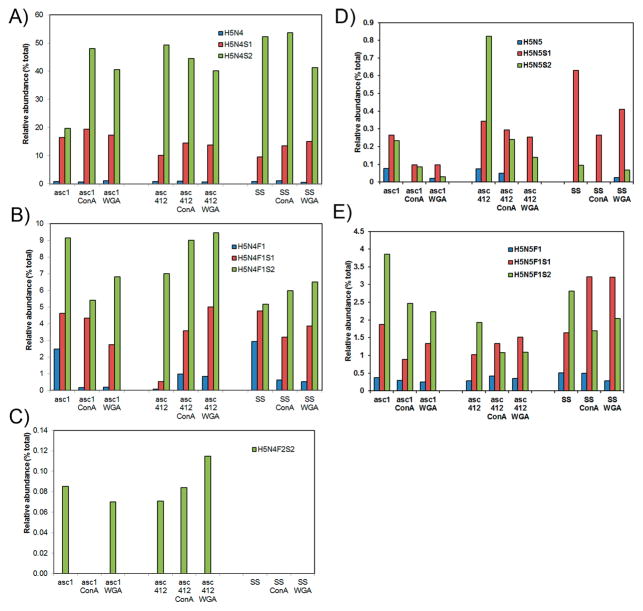
The most abundant N-glycan species in all samples were biantennary H5N4 isoforms, especially H5N4F0S2 and H5N4F0S1 (Figure 3A,B, structures in Supplemental Table S1). These glycans accounted for 51–72% of all glycans in the original samples, ~78% in all ConA samples, and ~68% in all WGA samples. Biantennary doubly sialylated H5N4S2 had the highest glycan content in all samples that ranged from 20% in Asc1 to 40–53% in the rest of the samples (Figure 3A, third bar in each series). This glycan is likely attached to the most abundant glycosylated proteins in ascites and SS samples. The second most prominent glycan in all samples was the monosialylated biantennary H5N4S1 species (10–18%) (Figure 3A, second bar in each series). The fucosylated versions of these glycans are shown in Figure 3B. Fucosylated doubly sialylated H5N4F1S2 (5–8%)(Figure 3B, third bar) was the most abundant of the fucosylated H5N4 glycoforms in all samples and particularly high in Asc1, followed by fucosylated, monosialylated H5N4F1S1 (2–4%) (Figure 3B, second bar). A separate graph was included for the bifucosylated, doubly sialylated H5N4F2S2 isoform (Figure 3C) to show this glycan present in ascites samples but not in the SS samples. This glycoform was present in proteins bound to WGA for both ascites fluids but only to ConA in Asc412.
Figure 3.
Comparative analysis of N-glycans obtained from two ascites samples Asc1 and Asc412 and a serum control sample (SS). (A) Abundant N-glycans H5N4, H5N4S1, and H5N4S2 N-glycans before and after lectin (ConA or WGA) binding in Asc1 (left), Asc412 (middle), and SS (right); (B) H5N4F1, H5N4F1S1, and H5N4F1S2 N-glycans; (C) H5N4F2S2 N-glycan; (D) H5N5, H5N5S1, and H5N5S2 N-glycans; and (E) H5N5F1, H5N5F1S1, and H5N5F1S2 N-glycans. H represents hexose; N represents N-acetyl glucosamine or GlcNAc; F represents fucose; S represents sialic acid.
Also highly present in all samples were the complex/hybrid possibly bisected H5N5-type glycans with sialylated glycans presented in Figure 3D and fucosylated, sialylated glycans (H5N5S1 and H5N5S2) presented in Figure 3E (putative structures in Supplemental Table S1). Profiles of this glycan group show clear differences in the relative abundance of these glycans in the three starting samples (Asc1, Asc412, SS) with more doubly sialylated H5N5S2 in Asc412 (Figure 3D, third bar, middle section) than in Asc1 (Figure 3D, third bar, left section) and really low in SS samples (Figure 3D, third bar, right section). SS samples contained mostly H5N5S1 (Figure 3D, first bar, right section). Two fucosylated glycans, mono-and doubly sialylated H5N5F1S1 and H5N5F1S2, were relatively abundant (1 to 3.7%) in all samples (Figure 3E, second and third bars in each series). Interestingly, Asc1-ConA and Asc1-WGA samples appeared to have higher content of the doubly sialylated form than Asc412-ConA and Asc412-WGA samples, likely reflecting the glycoprotein content of these samples. In contrast, SS-ConA and SS-WGA samples had a high content of monosialylated fucosylated H5N5F1S1 (up to 3% of total glycans). Bisected fucosylated and doubly sialylated N-glycan H5N5F1S2 have been described in blood samples from women with late-stage metastatic OC.28
Glycan Groups H3N4, H4N4, and H4N5 in OC Ascites
Some glycan groups had one glycan that was predominate and mostly present in the original samples with little or none present in the lectin-bound samples (Supplemental Figure S1A–C). This was the situation with glycan groups H3N4, H4N5, and H4N5, which are likely bi- and triantennary complex glycans containing fucosylation and some sialylation (Supplemental Table S1). The fucosylated form, H3N4F1, H4N4F1, and H4N5F1 were the most abundant glycans in each group, all likely having core fucosylation (Supplemental Figure S1A–C). Each predominant glycan ranged in relative abundance from ~11% for H3N4F1 in Asc1 to ~2% for Asc412 and SS (Supplemental Figure S1A) and 3% H4N4F1 in Asc1, 2% in Asc412, and 5% in SS original samples (Supplemental Figure S1B). There was ~2% H4N5F1 in Asc1, 0.5% in Asc412, and 1.2% in SS (Supplemental Figure S1C). H3N4F1 and H4N5F1 were particularly high in Asc1, whereas H4N4F1 was highest in SS. Interestingly, none of these glycans were enriched by lectin binding. Very little sialylation was observed in these glycans, although sialylated H4N4S1 (Supplemental Figure S1B) was present in ConA-bound ascites samples (Asc1-ConA and Asc412-ConA) but not detected in SS-ConA. Small amounts of this glycan were detected in WGA-bound ascites and SS samples (Supplemental Figure S1B). Fucosylated, sialylated H4N5F1S1 was present in all samples except for SS-ConA, with more present in Asc1 samples and in Asc1-ConA and Asc412-ConA samples (Supplemental Figure S1C).
High Mannose N-Glycans in Serum and OC Ascites Fluid Samples
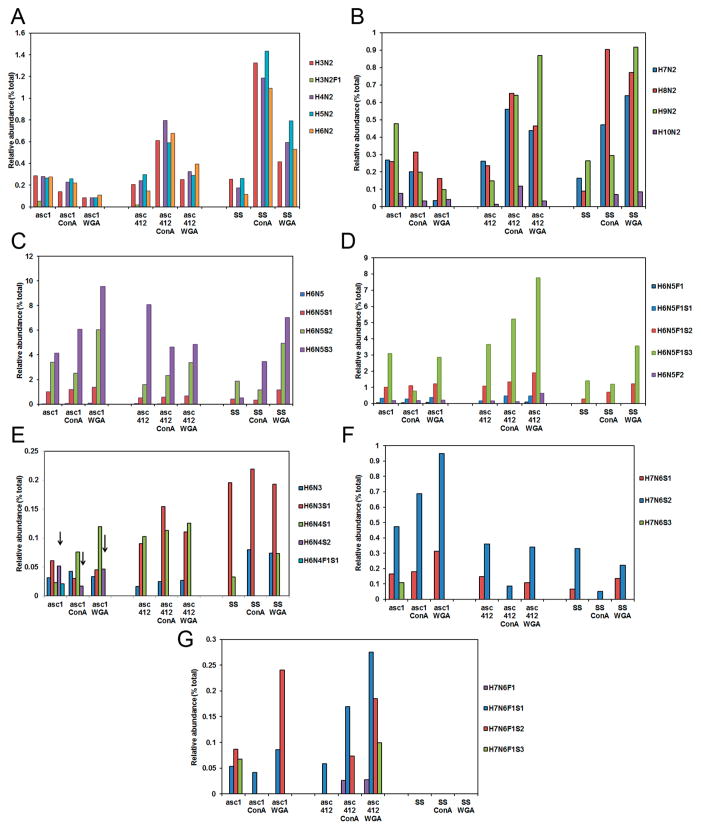
Differences in the amount of high mannose containing N-glycans in serum of women with OC have been previously reported;29,30 therefore, we were particularly interested in examining high mannose N-glycans in our OC ascites samples. High mannose glycans in Asc1 (Figure 4 left section), Asc412 (middle section), and serum (SS) (right section) with their ConA- and WGA-bound samples are shown starting with the shorter mannose glycans H3N2 (Man3) to H6N2 (Man6) in Figure 4A and with larger high-mannose-containing glycans H7N2 (Man7) to H10N2 (Man9+hex) in Figure 4B. Unexpectedly, there was considerably less relative abundance of mannose-containing N-glycans (H3N2 to H6N2) in Asc1 than in Asc412 or SS samples. Lectin binding enriched for mannose-containing N-glycans was observed in Asc412 and SS samples but not in Asc1 samples (Figure 4A,B).
Figure 4.
N-Glycans in Asc1, Asc412, and SS before and after lectin binding. (A) High mannose glycans that range from H3N2 to H6N2; (B) high mannose that range from H7N2 to H10N2; (C) N-glycan groups H6N5, H6N5S1, H6N5S2, and H6N5S3 (without fucosylation); (D) N-glycan groups H6N5, H6N5S1, H6N5S2, H6N5S3 with one fucose, and H6N5F2; (E) N-glycans H6N3, H6N3S1, H6N4S1, H6N4S2, and H6N4F1S1; (F) N-glycans H7N6S1, H7N6S2, and H7N6S3 (no fucosylation); and (G) N-glycans H7N6F1S1, H7N6F1S2, and H7N6F1S3 (with fucosylation).
Glycans H3N2 to H6N2 were particularly high in ConA samples from Asc412 and control serum (SS) (Figure 4A). Asc412-ConA had about half the relative content of these mannose-containing glycans as SS-ConA and about three times more content than Asc1-ConA (Figure 4A). There appeared to be 50% less H3N2, H4N2, H6N2, and H6N2 in proteins bound to WGA for all samples when compared with ConA-bound samples. A small amount of fucosylated H3N2F1 was only detected in Asc1 and Asc412, but none was detected in any other samples.
Interestingly, larger high mannose glycans H7N2, H8N2, and H9N2 were found to be greatly enriched by both ConA and WGA lectin binding in the Asc412 and SS samples (Figure 4B, middle and right sections), whereas the same high mannose glycans in Asc1 samples (Figure 4B, left section) did not have enriched binding to ConA or to WGA lectins (Figure 3B, left section), indicating that proteins containing these mannose glycans in Asc1 did not bind well to these lectins. High mannose glycans have been reported in one study to be elevated in blood of OC patients28 but in another OC blood study29 were reduced, so these intriguing high mannose results in ascites fluids will need further investigation, especially into their protein sources.
Glycan Group H6N5 Forms in OC Ascites
Next to be shown are the H6N5 group of glycans (Figure 4C,D). This group of glycans has a variety of different structures that can be complex or hybrid and possibly bisected, thus showing the heterogeneity in this group of glycans. Up to three sialic acids attached to this glycan group have been described for vitronectin,31 alpha-1-antitrypsin (A1AT),32 and serotransferrin,33 and the doubly fucosylation form has been described in epidermal growth factor receptor.34 An examination of the relative abundance of the unsialylated and sialylated forms of this glycan in these samples is shown in Figure 4C, while the corresponding fucosylated forms of these glycans are shown in Figure 4D. Little or no H6N5 was detected in any sample. The predominant glycans in all samples were trisialylated H6N5S3 (Figure 4C) and fucosylated trisialylated H6N5F1S3 (Figure 4D), followed by doubly sialylated (H6N5S2) (Figure 4C) and then the monosialylated glycoform (H6N5S1) (Figure 4C), also with fucosylation (H6N5F1S2 and H6N5F1S1) (Figure 4D), especially in the ascites samples. The relative abundance of these glycoforms ranged from <1% in SS up to 9% in the Asc1-WGA sample for H6N5S2 (Figure 4C, left) and above 7% for H6N5F1S3 in the Asc412-WGA sample (Figure 4D, middle). Proteins containing H6N5F1S3 appeared to bind especially well to lectins in Asc412-ConA and Asc412-WGA (Figure 4D, middle section, third bar). A small amount of bifucosylated H6N5F2 (Figure 4D, fifth bar in series) was present in ascites samples and not in SS samples.
N-Glycan Isoforms from H6N3, H6N4, and H7N6 Species Are Enriched in Ascites Fluids Showing Increased Fucosylation and Sialylation
N-Glycan H6N3 glycoform (Figure 4E, first bar in series) was detected in all samples with the exception of SS (Figure 4E, first bar in series). Although it was not identified in the SS starting sample, it was enriched in SS-ConA and SS-WGA (Figure 4E, right section), showing enrichment of proteins containing this glycan by lectin binding. H6N3 is a hybrid glycan, meaning that it contains both high mannose and complex glycan structures. The monosialylated glycoform, H6N3S1 (Figure 4E, second bar in series), was a predominant glycan, especially in SS samples (Figure 4E, right section). It was also highly present in Asc412 samples (Figure 4E, middle section) but not in Asc1 samples (Figure 4E, left section).
H6N4 was not detected in any samples, but the monosialylated form H6N4S1 was detected in all but not in SS-ConA (Figure 4E, third bar in series). H6N4S1 is also a hybrid-type glycan and possibly bisected (see Supplemental Table S1 for possible structures). This glycan was particularly enriched by lectin binding in Asc412 (Figure 4E, middle section). Interestingly, the doubly sialylated form H6N4S2 was only detected in Asc1 samples (Figure 4E, left section, see arrows) and not in any Asc412 or SS samples. This glycan was enriched by WGA binding in Asc1 samples. A small amount of fucosylated, monosialylated H6N4F1S1 was only detected in Asc1 (Figure 4, left section, fifth bar in series). H6N4F1S1 and H6N4S1 are glycan structures that have been identified in human milk sIgA.35 The doubly sialylated form H6N4S2 has been identified in ascites fluid and urine of a female with infantile sialidosis.36
Next examined were H7N6 glycoforms. Monosialylated H7N6S1 was present in all but Asc412-ConA and SS-ConA samples (Figure 4F, first bar in series). Doubly sialylated H7N6S2 was present in all samples and especially present in Asc1 samples, particularly in Asc1-WGA (Figure 4F, left section, second bar in series). It was less abundant in Asc412 and SS samples with only a small amount detected in Asc412-ConA and SS-ConA (Figure 4F, middle and right sections). Trisialylated H7N6S3 was only detected in Asc1 with none detected in lectin-bound Asc1-ConA or Asc1-WGA (Figure 4F, third bar). It was not detected in any of the Asc412 or SS samples.
The fucosylated forms of these glycans are shown in Figure 4G. Fucosylated H7N6F1 was detected only in Asc412-ConA and Asc412-WGA (Figure 4G, first bar in series, middle section) and not in any Asc1 (Figure 7C, left section) or SS samples (Figure 4G, right section). Fucosylated, monosialylated H7N6F1S1 (Figure 4G, second bar in series) was detected in both ascites samples, especially in Asc412-ConA and Asc412-WGA (Figure 4G), but none was detected in SS samples (Figure 4G, right section). Fucosylated, doubly sialylated H7N6F1S2 (Figure 4G, third bar in series) was highly present in Asc1, Asc1-WGA, Asc412-ConA, and Asc412-WGA but not in Asc1-ConA or Asc412. It was not detected in any SS samples (Figure 4G, right section). Finally, fucosylated, trisialylated H7N6F1S3 was detected only in Asc1 and Asc412-WGA (Figure 4G, fourth bar, left and middle sections). An explanation of why there were such differences in the presence of the H7N6 glycoforms in ascites samples that were not detected in SS samples remains to be determined.
Figure 7.
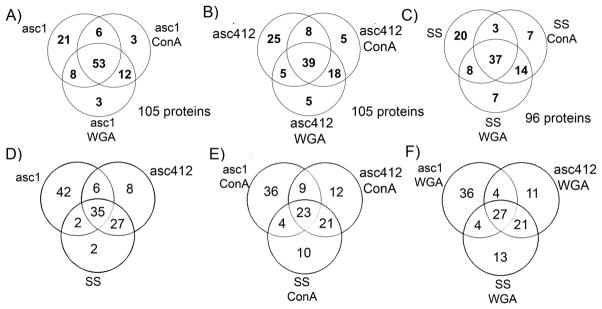
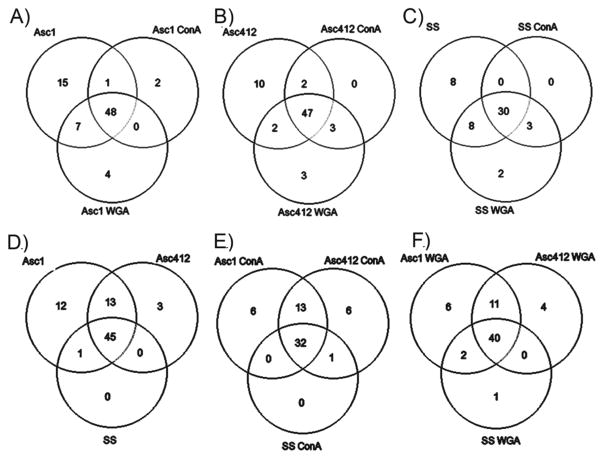
Venn diagrams comparing ascites (asc1 and asc142) and SS proteins. (A) asc1, asc1- ConA, and asc1-WGA; (B) asc142, asc142-ConA, and asc142-WGA; (C) SS, SS-ConA, and SS-WGA; (D) asc142 and SS; (E) asc142-ConA and SS-ConA; and (F) asc142-WGA and SS-WGA.
Venn Diagrams Comparing Glycans in SS, Asc1, and Asc412 Samples Show Common and Different N-Glycans in These Samples
There were 77 different glycans detected in Asc1 samples, 67 in Asc412, and 51 in SS samples (see Supplemental Table S2). Venn diagrams showing the overlap and differences in glycans comparing the lectin-bound samples with each original sample (Asc1, Asc412, SS) are shown in Figure 5A–C (Supplemental Table S2). A comparison is also made between N-glycans in the original samples comparing glycans in Asc1, Asc412, and SS (Figure 5D), followed by a comparison of the ConA-bound samples (Figure 5E) and finally a comparison of the WGA-bound samples (Figure 5F) (Supplemental Table S3).
Figure 5.
Venn diagrams comparing N-glycans from SS, Asc1, and Asc142. (A) Comparing original fluids with lectin-bound samples. (B) Comparing SS, Asc1, and Asc412 before lectin binding, with ConA binding and with WGA binding.
Venn diagrams show that the majority of the glycans identified (48 glycans or 62% for Asc1, 47 glycans or 70% for Asc412, and 30 glycans or 59% for SS) were common to each original and its corresponding lectin-bound sample (ConA and WGA). Unexpectedly, there were many glycans (15 glycans for Asc1, 10 glycans for Asc412, and 8 glycans for SS) identified only in the original samples and not detected in the lectin-bound samples. Most glycans were common to both ConA and WGA samples for each starting material (Asc1, Asc412, and SS), with a few glycans unique to the ConA samples (2 in Asc1-ConA) and to WGA samples (4 in Asc1-WGA, 3 in Asc412-WGA, and 2 in SS-WGA). (Glycans are identified in Supplemental Table S3.)
The more interesting comparisons are shown in the lower Venn diagrams (Figure 5D–F) (Supplemental Table S3) that directly compare N-glycans from Asc1, Asc412, and SS, separated by type of sample (original or ConA- or WGA-bound). In this comparison, 45 glycans (61%) were common to all original samples, 32 glycans (55%) were common in all ConA-bound samples, and 40 glycans (63%) were common in all WGA-bound samples. All glycans in SS were detected in Asc1 and Asc412, with one exception being a single glycan (H5N3) that was common to Asc1 and SS but not identified in Asc412. In contrast, Asc1 had 12 unique glycans and Asc412 had 3 unique glycans, with 13 glycans common between them that were not identified in SS.
The ConA- and WGA-bound compared samples gave a similar pattern with most of the SS glycans identified in all samples (32 glycans for ConA and 40 glycans for WGA). There were 6 unique glycans detected in Asc1-ConA and 6 in Asc412-ConA, with 13 glycans common to the ascites ConA-bound samples. For the WGA-bound samples, 6 were unique to Asc1-WGA, 4 to Asc412-WGA, and 1 to SS-WGA. The overlap between Asc1-WGA and Asc412-WGA was 11 glycans. (Glycan comparisons are shown in Supplemental Table S3.). Of the N-glycans that were present in both ascites, including their lectin-bound samples (cancer) and not in SS (control) samples, four N-glycans, H6N5, H6N5F1S1, H6N5F2S3, and H7N6F1S1, were identified, which could possibly be specific to OC ascites fluids and to metastatic cancer.
Proteomic Analysis of ConA- and WGA-Bound Ascites Proteins
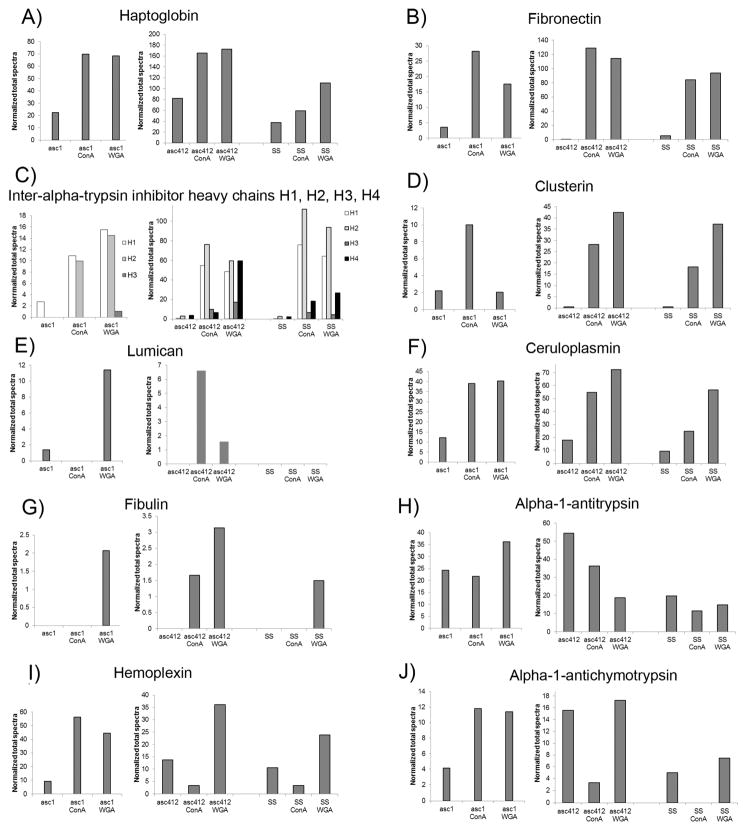
Total proteomic analyses were carried out on the ascites and SS samples to identify the proteins present in theses samples (see Experimental Section). Proteomic analyses of Asc1 samples (Asc1, Asc1-ConA, and Asc1-WGA) were performed earlier from the Asc412 and SS samples (Asc412, Asc412-ConA, Asc412-WGA, SS, SS-ConA, and SS-WGA), so the results are presented in separate Supplemental Tables (Supplemental Table S4 for Asc1 samples and Supplemental Table S5 for Asc412 and SS samples). One hundred and six proteins were identified in the Asc1 samples (Supplemental Table S4) and 114 proteins were identified in Asc412 and SS samples (Supplemental Table S5). Proteomic results are displayed for specific proteins: haptoglobin, fibronectin, interalpha-trypsin inhibitor heavy chains H1, H2, and H3, clusterin, lumican, ceruloplasmin, fibulin, alpha-1-antitrypsin, hemopexin, and alpha-1-antichymotrypsin in a series of bar graphs that compare results for each sample analyzed (Figure 6A–J). Because proteomic analysis of Asc1 samples was conducted separate from those of Asc412 and SS, which were analyzed together, the Asc1 results are shown in separate bar graphs from those of Asc412 and SS (determined using Scaffold v4). Proteins used for the figures were selected because all have previous reports in the literature connecting them to OC (haptoglobin, fibronectin, interalpha-trypsin inhibitor, clusterin, ceruloplasmin, alpha-1-antitrypsin, and hemopexin)37 or were enriched in the OC ascites samples (lumican, fibulin, alpha-1-antichymotrypsin).
Figure 6.
Proteins identified in ascites fluids and SS before and after lectin (ConA and WGA) lectin binding. (A) Haptoglobin, (B) fibronecctin, (C) interalpha-trypsin inhibitor heavy chains H1, H2, H3, and H4, (D) clusterin, (E) lumican, (F) ceruloplasmin, (G) fibulin, (H) alpha-1-antitrypsin, (I) hemopexin, and (J) alpha-1-antichymotrypsin.
Haptoglobin was identified in all samples and enriched two to three times with lectin binding (Figure 6A). Asc1 samples are shown in the left section. Asc412 in middle section. and SS in right section of each graph. There appeared to be less haptoglobin in SS samples.
Interestingly, fibronectin was one of the more abundant proteins detected in all samples and particularly enriched by lectin binding (Figure 6B). Fibronectin was enriched 5–8 times in Asc1 with lectin binding, 192–216 fold in Asc412, and 15–17 fold in SS samples.
Interalpha-trypsin inhibitors consist of three or four heavy chains (interalpha-trypsin inhibitor heavy chains: H1, H2, H3, and H4) that combine with one light chain (AMBP or SPINT2) to function together as a covalent protein–glycosaminoglycan–protein complex to act as a protease inhibitor as part of an inflammatory response.38 Interalphatrypsin inhibitors (ITIH) proteins, especially ITIH4 and ITIH1, have been previously reported in OC.37,39,40 The interalpha-trypsin inhibitor heavy chains H1, H2, and H3 were detected in Asc1, Asc412, and SS samples, but ITH4 was not detected in Asc1 samples (Figure 6C). ITIH1 was enriched 4-fold in asc1-ConA and 6-fold in Asc1-WGA, 91-fold in Asc412-ConA and 81-fold in Asc412-WGA. In comparison, ITIH1 was greatly enriched 138-fold in SS-ConA and 116-fold in SS-WGA. ITIH2 was not detected in the starting sample Asc1 but was highly present in lectin-bound Asc1-ConA and Asc1-WGA. It was enriched 25-fold in Asc412-ConA, 20-fold in Asc412-WGA, and similar to ITIH1, it was greatly enriched 41-fold in SS-ConA and 34-fold in SS-WGA. ITIH3 was not detected in Asc1 or Asc1-ConA, but a small amount of ITIH3 was detected in Asc1-WGA (Figure 6C, left section). There was also little ITIH3 in Asc412 and SS samples. As previously mentioned, ITIH4 was not detected in Asc1 samples but was enriched 2-fold in Asc412-ConA and 17-fold in Asc412-WGA (Figure 6C, middle). ITIH4 was enriched 8-fold in SS-ConA and 12-fold in SS-WGA. There appears to be lectin-binding differences of ITIH proteins, with SS ITIH proteins binding better to lectin than ascites proteins.
Clusterin (Figure 6D) and hemopexin (Figure 6I) are also proteins reported in OC samples.41 In ascites fluid samples, clusterin was enriched 5-fold by ConA binding in Asc1 but not by WGA binding, whereas in Asc412-ConA, clusterin was enriched 47-fold and 71-fold in Asc412-WGA. SS-Con enriched clusterin 33-fold and 67-fold by SS-WGA. Hemopexin was enriched in Asc1-ConA by 6-fold and in Asc1-WGA by 5-fold, but clusterin in Asc412 did not bind to ConA but was enriched (3-fold) by WGA. ConA also did not enrich hemopexin, but WGA did enrich hemopexin in SS samples. These results show variability between clusterin and hemopexin in the ascites fluid samples. For these proteins, Asc412 samples were more similar to SS samples, showing the need to further assess more individual patient cases.
Alpha-1-antichymotrypsin (Figure 6J) showed a similar pattern to hemopexin for these samples with strong binding to Asc1-ConA and Asc1-WGA, little binding to ConA for Asc412 and SS samples, and good binding to WGA. There appeared to be less alpha-1-antichymotrypsin in SS samples. Alpha-1-antitrypsin (Figure 6H) also showed less amounts in SS samples, with little or no enrichment with lectin binding. Ceruloplasmin showed enrichment by lectin binding to all samples, especially to Asc1-ConA and Asc412-ConA, but not to SS-ConA. WGA bound ceruloplasmin in all samples.
Finally, lumican (Figure 6E) and fibulin (Figure 6G) were more highly detected in ascites samples. Lumican was detected in Asc1, Asc1-WGA, Asc412-ConA, and Asc412-WGA and not in any SS samples. Ishiwata et al. reported that lumican could inhibit HEK cell attachment and growth and inhibit activation of integrin pathways,42 while Yamamoto et al. determined that secreted lumican could stimulate growth and inhibit invasion of human pancreatic cancer.43
Fibulin was detected in Asc1-WGA but not in Asc1-ConA. It was detected in Asc412-ConA and Asc412-WGA with some detected in SS-WGA. Fibulin is of interest because fibulin-1 is an extracellular matrix (ECM), fibronectin -binding protein that can be secreted by human OC cells and regulated by estrogen.44
Not shown are proteomic results that determined attractin was detected only in Asc1 WGA and not identified in any Asc412 or in SS samples; afamin and heparin cofactor were identified in Asc1 ConA and WGA samples but not in Asc412 or SS samples; and zinc-alpha-2-glycoprotein was present in Asc1, not binding to ConA, and weakly binding to WGA. These results are shown in Supplemental Table S4 for Asc1 samples and Supplemental Table S5 for Asc412 and SS samples. Modulator of apoptosis 1 and dynein heavy chain 1 were identified in Asc412 ConA but not in any Asc1 or SS samples. Collagen (alpha-1) and C-reactive protein were detected in Asc412 and bound well to ConA and WGA (Supplemental Table S5). Collagen was detected in Asc1 but did not appear to bind to the lectins. C-reactive protein was detected in Asc412 but not detected in asc1 samples. Ficolin 3 and X-ray radiation resistance-associated protein 1 were identified in Asc412 WGA and not in any Asc1 or SS samples.
Venn Diagrams Comparing the Proteins Identified in Ascites and Serum Control Samples Show the Common and Different Proteins
Venn diagrams were constructed to show a comparison of proteins identified in the Asc1 Asc412 and SS samples, especially to show the common and different proteins identified in each sample. The results show that although the majority of proteins were common to the lectin-bound and original sample (50.5% for Asc1, 37.1% for Asc412, and 38.5% for SS) (Figure 7A–C, Supplemental Table 6A), there were considerable numbers of proteins identified in each sample that were not common to all samples, specific to each source (Asc1, Asc412, or SS) and not bound to lectins. In Asc1 there were 21 proteins or 20%, in Asc412 there were 25 proteins or 23.8%, and in SS there were 20 proteins or 20.8% not bound to lectins. More overlap was observed in proteins identified bound to lectins (12 proteins or 11.4% for Asc1 samples, 8 proteins or 17.1% for Asc412, and 14 proteins or 14.6% for SS samples), suggesting that the majority of glycosylated proteins in each sample were bound to both lectins.
Comparing the samples in each category (original sample or ConA- or WGA-bound) with each other (Figure 7D–F, Supplemental Table 6B) shows a slightly different picture. There were less common proteins (35 for the original samples, 23 for the ConA-bound samples, and 27 for the WGA-bound sample) and more unique proteins in the ascites samples in each category, especially for Asc1 (42 for Asc1, 36 for Asc1-ConA, and 36 proteins for Asc1-WGA). There was more similarity between Asc412 and SS (27 for original samples, 21 for ConA, and 21 for WGA) than between Asc1 and SS (2 original samples and 4 ConA- and 4 WGA-bound proteins). Unexpectedly there were some SS-ConA (10 proteins) and SS-WGA proteins (13 proteins) that were not detected in the ascites fluids. A list of these proteins for Figure 7D–F is provided in Supplemental Table S6B. These protein profiles show the heterogeneity in the ascites samples as well as the similarity between proteins in ascites and the serum control.
When comparing the protein profiles, three proteins, collagen alpha-1, leucine-rich alpha-2-glycoprotein, and lumican, were identified in all ascites samples (including lectin-bound samples) but not in SS samples. Forty-two proteins were uniquely identified in Asc1, 8 in Asc412, and 2 in SS. Proteins in Asc1 that bound to both lectins were N-acetylmuramoyl-L-alanine amidase, heparin cofactor 2, afamin, zinc-alpha-2-glycoprotein, and complement factor I, whereas haptoglobin-related protein bound to only to ConA and not to WGA. Asc1 proteins binding to WGA but not to ConA were angiotensinogen, complement component C6, attractin, and corticosteroid-binding globulin. There were fewer unique proteins in Asc412 when compared with Asc1 or in SS samples. C-reactive protein in Asc412 bound to both lectins, whereas dynein heavy chain 1 (axonemal) and modulator of apoptosis 1 only bound to ConA in Asc412 ascites samples.
Glycopeptide- and Peptide-Site-Specific Glycan Analysis of Ascites Proteins
Further glycopeptide- and glycan-site-specific analysis was conducted on OC ascites protein samples using (1) hemoglobin affinity chromatography (H-agarose) designed to enrich for iron-binding proteins such as hemopexin and haptoglobin (Samples #1–4) ; (2) AAL lectin affinity chromatography to enrich for fucose containing glycosylated proteins (Samples #5); and (3) IgG and human serum albumin depletion to remove the most abundant serum/plasma proteins, thus reducing the complexity of the sample to be analyzed (Sample #6). A serum control was also subjected to H-agarose affinity chromatography. Samples (ascites and serum control) obtained from hemoglobin affinity binding were divided in half, and one-half were subjected to desialylation (desialylated serum control was in Sample #1, and desialylated ascites were in Sample #2) to determine if the removal of sialic acids would improve the overall glycopeptide analysis. The other half of the samples from serum control and ascites were separately analyzed (serum control in Sample #3 and ascites in Sample #4). All protein samples were alkylated and trypsin-digested before proteomic analysis (see Experimental Section). We report here the glycopeptide results for three of the glycosylated proteins identified above: clusterin, fibulin, and hemopexin. Interestingly, clusterin and fibulin were not detected in the serum control samples but only in the OC ascites samples.
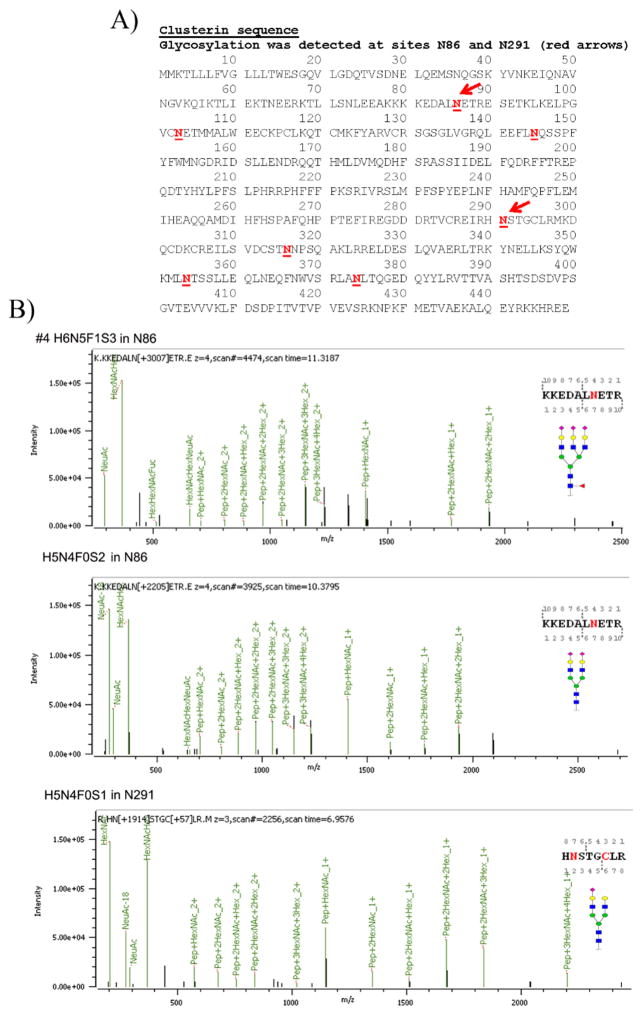
Glycosylation in clusterin has been reported at N86, N103, N145, N291, N317, N354, and N374 sites in isoform 1 (Figure 8A, red underlined bold type), but we only identified N-glycans in sites N86 and N291 in clusterin in OC ascites samples #2, #4, and #6 (Table 1). No clusterin peptides were detected in samples #1 and #3 (serum control samples). Clusterin peptides were detected in #5 (AAL-bound ascites), but none were glycosylated. N-Glycans in clusterin sites N86 and N291 were largely bi- and tri-antennary N-glycans (H5N4F0S0 and H6N5F0S0) (Table 1) with some fucosylated H6N5F1S0 (27%) detected in desialylated ascites sample #2. In the H-bound, sialylated OC ascites sample (sample #4), N-glycans were largely doubly sialylated biantennary (H5N4F0S2) and triply sialylated triantennary (H6N5F0S3) glycans located in sites N86 and N291. Examples of MS/MS spectra for the N-glycans in N86 of sample #4 are shown in Figure 8B. A tetra-antennary H7N6F0S0 was weakly detected in clusterin site N291 (6%) in the H-bound, desialylated OC ascites sample #2 (Table 1). We were unable to detect glycosylation in the other reported sites likely because glycopeptides containing these sites were not detected in our analysis. The amount of clusterin, although enriched by the H binding, was still low, making it difficult to get better coverage. Sample #5 did not produce any clusterin glycopeptides, and sequence coverage was low (19%).
Figure 8.
(A) Clusterin sequence with N-glycosylation sites identified by bold, red, underlined type. Red arrows identify the most prevalent sites identified in serum and OC ascites samples. (B) Tandem MS/MS of two OC ascites clusterin glycopeptides with glycan and glycan site identified in each glycopeptide.
Table 1.
Glycan Sites and Heterogeneity of Glycans Attached to Glycopeptides Obtained from Clusterin Present in OC Ascites Fluids
| Sample #2 - desialyiated | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| PID | Site | Sequence | Glycans | Score | structure | composition | # peptides | total# glycopeptides in site | % heterogeneity |
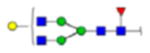
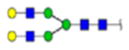
| 2 | 684133 | N86 | K.KKEDALN[+2133.77171]ETR.E | HexNac(5)Hex(6)Fuc(1)NeuAc(0) | 161.8 |
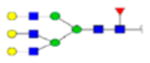
|
H6N5F1S0 | 3 | 27% | |
| 5 | 684333 | N86 | K.KKEDALN[+2133.77171]ETR.E | HexNac(5)Hex(6)Fuc(0)NeuAc(0) | 213.9 |
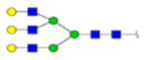
|
H6N5F0S0 | 3 | 27% | |
| 12 | 650092 | N86 | K.KKEDALN[+1622.58161]ETR.E | HexNac(4)Hex(5)Fuc(0)NeuAc(0) | 235.7 |
|
H5N4F0S0 | 5 | 11 | 45% |
| 24 | 709068 | N291 | R.TVC[+57.02146]REIRHN[+1606.58669]STGC[+57.02146]LR.M | HexNac(4)Hex(4)Fuc(1)NeuAc(0) | 129.2 |
|
H4N4F1S0 | 2 | 13% | |
| 26 | 683653 | N291 | R.TVC[+57.02146]REIRHN[+1460.52878]STGC[+57.02146]LR. M | HexNac(4)Hex(4)Fuc(0)NeuAc(0) | 128.8 |
|
H4N4F0S0 | 2 | 13% | |
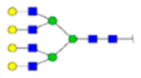
| 1 | 683013 | N291 | R.HN[+2352.84600]STGC[+57.02 146]LR.M | HexNac(6)Hex(7)Fuc(0)NeuAc(0) | 83.1 |
|
H7N6F0S0 | 1 | 6% | |
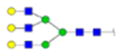
| 8 | 649370 | N291 | R.HN[+1987.71380]STGC[+57.02 146]LR.M | HexNac(5)Hex(6)Fuc(0)NeuAc(0) | 238.4 |
|
H6N5F0S0 | 3 | 19% | |
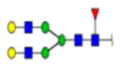
| 11 | 603402 | N291 | R.HN[+1768.63952]STGC[+57.02 146]LR.M | HexNac(4)Hex(5)Fuc(1)NeuAc(0) | 227.7 |
|
H5N4F1S-0 | 1 | 6% | |
| 18 | 603002 | N291 | R.HN[+1622.58161]STGC[+57.02 146]LR.M | HexNac(4)Hex(5)Fuc(0)NeuAc(0) | 463.2 |
|
H5N4F0S0 | 7 | 16 | 44% |
| Sample #4 - same as #2 except not desialylated | |||||||||
| 1 | 715191 | N86 | K.KKEDALN[+3007.05796]ETR.E | HexNac(5)Hex(6)Fuc(1)NeuAc(3) | 30 |
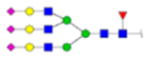
|
H6N5F1S3 | 2 | 40% | |
| 3 | 715231 | N86 | K.KKEDALN[+2861.00005]ETR.E | HexNac(5)Hex(6)Fuc(0)NeuAc(3) | 30 |
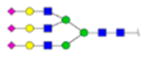
|
H6N5F0S0 | 1 | 20% | |
| 5 | 641812 | N86 | K.KKEDALN[+2204.77244]ETR.E | HexNac(4)Hex(5)Fuc(0)NeuAc(2) | 124.2 |
|
H5N4F0S2 | 2 | 5 | 40% |
| 4 | 607891 | N291 | R.HN[+2278.80922]STGC[+57.02 146]LR.M | HexNac(5)Hex(6)Fuc(0)NeuAc(1) | 104.1 |
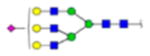
|
H6N5F0S1 | 1 | 17% | |
| 7 | 608131 | N291 | R.HN[+1913.67702]STGC[+57.02 146]LR.M | HexNac(4)Hex(5)Fuc(0)NeuAc(1) | 347.6 |
|
H5N4F0S1 | 5 | 6 | 83% |
| Sample #6 - HSA/IgG depleted sample | |||||||||
| 1 | 413677 | N291 | R.EIRHN[+2204.77244]STGC[+57.02146]LR.M | HexNac(4)Hex(5)Fuc(0)NeuAc(2) | 132.9 |
|
H5N4F0S2 | 2 | 100% | |
| 3 | 363539 | N291 | R.HN[+2204.77244]ST GC[+57.02146]LR.M | HexNac(4)Hex(5)Fuc(0)NeuAc(2) | 349.2 |
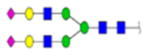
|
H5N4F0S2 | 3 | 5 | 100% |
Clusterin with the most heterogeneity and different types of N-glycans was identified in sample #2 (H-bound desialylated), while peptides from sample #6 (IgG/HSA depleted) had only one type glycan (H5N4F0S2) in a single site, N291 (Table 1). The cellular sources of these clusterin molecules are presently unknown but could originate from stromal cells and not from a liver source. Clusterin reportedly has a chaperon role in the ECM that may bind misfolded proteins, which, in turn, may interfere with improper protein–protein interactions, thus preventing aggregation and keeping proteins in solutions.45,46
Fibulin peptides were identified in OC ascites samples #2, #4, #5, and #6 (Supplemental Figure S2) and not in serum control samples #1 and #3. Surprisingly, glycopeptides from fibulin were identified in only two samples, #2 (H-agarose-bound, desialylated) and #5, (AAL-bound sample) (Supplemental Figures S2 and S3). No glycopeptides were detected in #4 (H-agarose-bound, sialylated proteins) or in #6 (IgG/HSA depleted), likely due to low coverage (Supplemental Figure S2). Coverage was less for fibulin in #2 (23%), #4 (21%), and #5 (7.7%) (Supplemental Figure S2) compared with clusterin in samples #2 (41%), #4 (35%), and #5 (19%), suggesting that there was less fibulin than clusterin in many of these samples, which is consistent with our previous proteomic analysis of ascites and serum control, where there was generally more clusterin detected than fibulin (Figure 6).
O-Linked glycosylation (H2N2F0S2) was detected attached to two peptides at site T56 in fibulin-1 in sample #2 (Fibulin sequence in Supplemental Figure S3A). N-Linked glycans were detected in fibulin-1 present in lectin-bound AAL sample (sample #5) at N539 (Supplemental Figure S3B). The types of N-glycan attached to site N539 of fibulin-1 from sample #5 were unusual and interesting and were H7N5F0S2, H6N4F0S2, and H6N4F0S1 (three glycopeptides) (Supplemental Figure 3B). These putative glycan compositions will need further verification at each specific glycopeptide site. Fibulins are secreted, extracellular proteins that may moderate cell morphology, growth, adhesion, and motility, which are all possible functions that could help promote tumor growth and assist the tumor cells in ascites to metastasize.47 The fact fibulin was not detected in the serum control samples analyzed (#1 and #3) was not surprising because ECM proteins would not be expected to be present in serum in high amounts but closer to the source of tumor growth. The contribution of glycosylation to fibulin function is still unknown. Fibulin-1 has epidermal-growth-factor-like repeats in its domain II and may be involved with aggregating proteoglycans in the ECM,48 which could help contribute to metastasis of tumor cells.
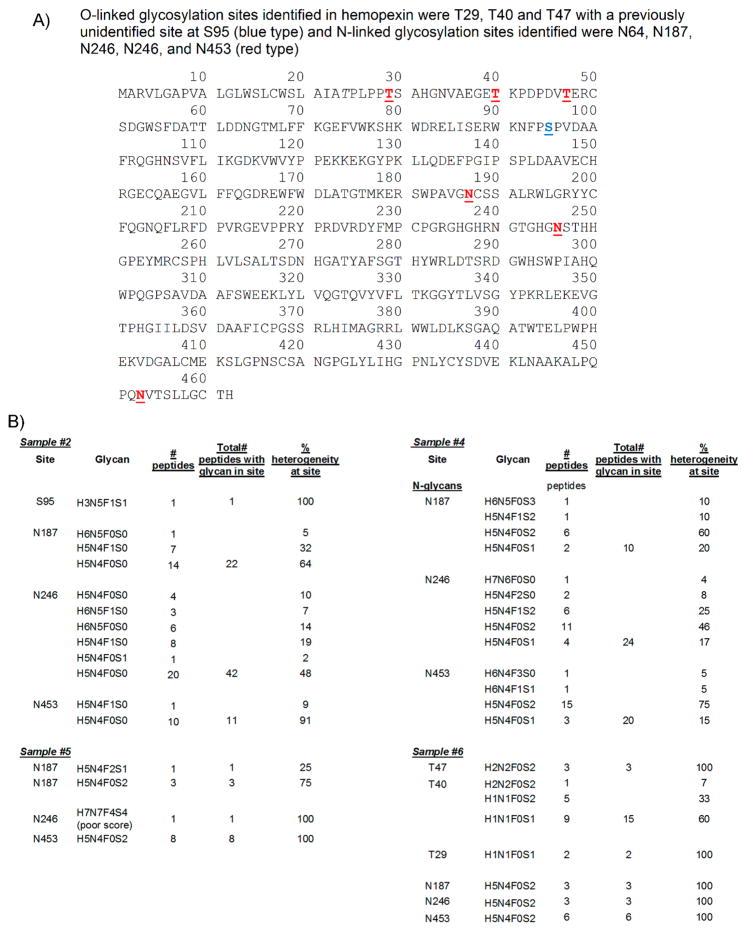
Hemopexin was another glycosylated protein identified in the ascites and serum control samples analyzed. Hemopexin peptides were detected in serum control samples #1 and #3 with poor coverage (27% for #1 and 5.6% in #3). Surprisingly, no glycopeptides were detected in these samples (Supplemental Table S9). In contrast, highly glycosylated hemopexin was detected in all OC ascites samples (#2, #4, #5, and #6) with good coverage (70, 66, 59, and 72%, respectively), suggesting that H-affinity binding selects for glycosylated hemopexin. Because sample #2 was desialylated, little or no sialylated glycans were detected, as expected. O-Linked glycans were detected only in hemopexin from OC ascites sample #2 (desialyated H-agarose-bound) and sample #6 (IgG/HSA depleted) (Figure 9A,B). An unusual O-linked glycan H3N5F1S1 was identified attached to site S95 from sample #2 (Figure 9B). In sample #6, O-linked glycans were identified attached to T29, T40, and T47 with H1N1F0S1 (two peptides) identified in T29, three different O-glycans, H2N2F0S2 (one peptide), H1N1F0S2 (five peptides), and H1N1F0S1 (nine peptides) were identified attached to site T40, and H2N2F0S2 (three peptides) was identified attached to site T47 (Figure 9B).
Figure 9.
Hemopexin sequence, with glycan sites, number of glycopeptides identified, and % heterogeneity at each site in hemopexin present in OC ascites. (A) Hemopexin sequence. (B) Hemopexin glycan sites, composition, # peptides, and % heterogeneity in OC ascites samples #2, #4, #5, and #6.
Three N-linked sites (N187, N240 and N453) were identified in OC ascites hemopexin present in all four OC ascites samples (#2, #4, #5, #6) (Figure 9A,B). Little glycosylated hemopexin (only three glycopeptides) was identified in #5 (AAL-bound) (Figure 9B), suggesting that most glycosylated hemopexin are heavily sialylated. The predominant N-glycan in hemopexin from samples #5 and #6 was H5N4F0S2 (Figure 9B), an abundant doubly sialylated biantennary N-glycan, which was attached to all three N-linked sites. In sample #2, a complex glycan H6N5F0S0 was detected attached to N187 in hemopexin, but the predominant N-glycans in N187 were fucosylated biantennary H5N4F1S0 (32%) and nonfucosylated biantennary H5N4F0S0 (64%) N-glycan (Figure 9B). In site N240 of sample #2, more fucosylated H6N5F1S0 (7%) and nonfucosylated H6N5F0S0 (14%) were detected, but fucosylated H5N4F1S0 (20%) and nonfucosylated H5N4F0S0 (47.6%) were still the more predominant glycans (Figure 9B). In site N453, 9% of glycopeptides were fucosylated biantennary H5N4F1S0, with by far the most predominant glycan in this site being the nonfucosylated biantennary H5N4F0S0 N-glycans (90%) (Figure 9B).
In OC ascites sample #4, no O-glycans were identified in hemopexin. All N-glycans identified in site N187 were sialylated with 10% H6N5F0S3, 10% H5N4F1S2, 60% H5N4F0S2, and 20% H5N4F0S1 identified in this site. In site N240, an unusual N-glycan H7N5F0S0 (4%) was identified, but the most predominant N-glycans were biantennary doubly fucosylated H5N4F2S0 (8%) and monofucosylated doubly sialylated H5N4F1S2 (25%) but mostly doubly sialylated biantennary H5N4F0S2 (45.8%) with a small percentage of monosialylated biantennary H5N4F0S1 (17%) (Figure 9B). In site N453, the predominant glycan was doubly sialylated H5N4F0S2 (75%), with a small percentage of monosialylated H4N4F0S1 (15%). Traces of two other unusual N-glycans were identified in this site, H6N4F3S0 and H6N4F1S1 (5% each), but the scores for these glycans were poor.
Hemopexin was not an abundant protein in sample #5, the AAL-bound sample. Site N187 had only four glycopeptides identified, with H5N4F0S2 (75%) being the most predominant (Figure 9B). N453 had eight peptides identified, all with the doubly sialylated biantennary N-glycan H5N4F0S2 attached. There was slight evidence of a rare glycan, H7N7F2S4, in site N246 with a weak score (score = 30).
Surprisingly, sample #6 had the most evidence of O-glycans in hemopexin. Highly sialylated O-glycans were detected in T29, T40, and T47 (Figure 9B), with N-glycans detected in N187, N246, and N453, all of which were doubly sialylated biantennary H5N4F0S2 (Figure 9B).
Glycoproteomic analysis of hemopexin, present in six differently OC ascites samples (Samples #1–6), showed that ascites samples prepared in different ways can make a considerable difference in the subsequent glycan analysis. The number and type of glycans detected at each site varied considerably depending on how the sample was prepared.
DISCUSSION
Performing proteomic and glycomic analyses of ascites fluid can help us to better understand the complexity of OC glycoproteomics and how the diversity of glycosylation might contribute to the metastatic process of OC. A top-down approach was used on glycoproteins present in human OC ascites fluid aided by lectin binding. Separate proteomic and glycomic analyses were conducted on two ascites fluid samples; then, glycopeptide analysis was performed on affinity-bound ascites and serum samples to identify the proteomic and glycomic components in the original and lectin- and affinity-bound samples. The objective of this research study was to apply glycomic, proteomic, and glycoproteomic methods to help identify potential metastatic glycomic biomarkers that can then be related back to the metastatic tumor cells and their ascites microenvironment, especially because changes in glycosylation accompany tumorigenesis and metastasis.49–51 Glycosylation is implicated in the tumor cell microenvironment and survival and is predicted to have a critical role in the ability of tumor cells to intravasate into the circulation and metastasize to other organ sites.52,53 The type and heterogeneity of glycosylation directly impacts protein function, as shown by Taniguchi and colleagues, who showed that core fucosylation and bisecting N-glycans can regulate receptor recognition, adhesion, and motility.54 We find evidence that these types of glycosylation are relatively more abundant in the ascites fluid when compared with a normal serum control.
In this manuscript, we show results comparing N-glycans and proteins acquired from two OC ascites fluid samples and have compared these results with a well-known, standardized control sample, which was a commercially available normal human serum sample. We used a commercial source of pooled serum as a control in this study because it was already being used as a standard for technical (sample processing) and analytical mass spectrometry methods in our prior clinical studies.55 We have extensively characterized the N-glycome of serum proteins in these samples as normal glycosylated serum/plasma proteins in our databases.15,56 Our future studies will utilize the methods/technology developed in this study toward clinical larger studies of OC patient ascites and sera matched with suitable controls.
OC ascites fluid is a rich source of cancer glycan biomarkers. The amount of sample obtained from a single individual can range from a few milliliters to over a liter, whereas the amount of blood we can get from an individual is usually no more than 4 to 5 mL. Importantly, tumor cells thrive and proliferate in ascites, so the cancer tumor antigens are expected to be enriched compared with circulating blood. Similar to serum/plasma samples, the most abundant protein in ascites is serum albumin, which in plasma is 60% (approximately 500–800 μM/L).57 Because serum albumin is not glycosylated and does not bind to immobilized lectins, the use of lectins eliminates this abundant protein from our glycomic and proteomic analyses and greatly improves our ability to detect lower abundant glycosylated proteins. IgG, the second most abundant protein in serum and plasma,57 is glycosylated and so was expected to bind to lectins, but surprisingly it was not the second most abundant protein in ascites fluids (Supplemental Table S2). IgG has N-linked glycan sites present in the constant region of the heavy chain as well as other N-linked glycan sites in the variable light chain.57,58 N-Glycans from in the Fc constant and variable regions of IgG are released by PNGase F,58 so these glycans are likely to be highly present in our glycan analysis. OC ascites fluid IgG and IgA site-specific glycosylation in ascites fluids will be separately reported (to be published).
WB analyses of CA125, fibronectin, MUC1, MUC2, and MUC3, all heavily glycosylated proteins considered to be biomarkers of OC, were included in this study to follow their enrichment with lectin binding. WB analysis of CA125 initially proved to be challenging because not all commercial antibodies were able to recognize the high-molecular-weight glycosylated species we knew to be present in these samples. CA125 contains both N- and O-linked glycosylation, which causes native CA125 to range in size from 250 kDa to several million daltons).59 Fibronectin in the lectin-bound fractions (Figure 1G, lanes 2 and 5) also showed heterogeneity in size that ranged from 180 kDa to ~250 kDa, with some very large high-molecular-weight proteins indicated in the ConA-bound fraction (Figure 1G, lane 5). There is also the possibility that dimers and multimers in fibronectin cause it to run as a larger protein of ~480 kDa on a reduced SDS-PAGE gel.60 MUC1, 2, and 3, like CA125, are highly O-linked glycosylated with similar molecular sizes, ~250 kDa. These mucin proteins were not detected in the original ascites fluid (Asc1), likely because their abundance was too low, so lectin binding enriched these proteins, making their detection by WB analysis possible. Multiple protein isoforms and glycan heterogeneity largely due to how glycans are synthesized and attached to proteins is are major challenges to correctly analyzing metastatic cancer glycoproteins. Only fibronectin was detected in our direct proteomic analysis of ascites fluid. CA125 and MUC1, 2, and 3 were not detected, which shows the limitation of our ability to detect these very low abundant proteins.
Glycomic Analysis of OC Ascites Fluids by Lectin Binding
Liquid chromatography greatly improves our ability to detect glycosylation changes.50 We previously used MALDI FT ICR MS for our glycomic analysis, which was extremely sensitive, but lacked the separation we now have with nano-LC Chip/TOF and nano-LC Chip/qTOF MS.12 We are able to identify more glycans using nano-LC with Chip/TOF MS.12 The patterns of N-glycan binding to ConA and WGA lectins for a normal serum control (SS) and ascites fluid samples obtained from two individual OC cases were very informative. H5N4S1 and H5N4S2 and the core-fucosylated versions H5N4F1S1 and H5N4F1S2 were the most abundant glycans in all samples, including the lectin-bound fractions. These glycans are known to be present in alpha-2-macroglobulin, transferrin, IgG, antitrypsin, AGP, IgM, and IgA,15 all proteins that were highly present in our samples (Supplemental Tables S4 and S5).
Another abundant group of glycans were those based on H5N5 with high amounts of core-fucosylated mono- and disialylated H5N5F1S1 and H5N5F1S2 present in SS and Asc1 samples and less so in Asc412 samples. H5N5 is predominately a bisected-type glycan. Synthesis of bisected-type glycans is dependent on N-acetylglucosaminyltransferase (GnT) III (MGAT3). Bisected-type glycans have been reported to increase in cancer, and increased expression of bisected glycans and MGAT3 in OC tissues has been reported.61
High-mannose-type N-glycans are of particular interest because they have been reported as glycomic biomarkers for a number of different cancer types including OC,29 CA125 from OVCAR, an OC cell line,59 prostate cancer,62 and breast cancer.63 In our glycomics analysis, most high mannose glycoforms were in the SS serum control and to a much lesser extent in Asc1 samples with Man3 (H3N2) to Man6 (H6N2) high in SS samples, especially bound to ConA. Man7 (H7N2), Man8 (H8N2), and Man9 (H9N2) were high in SS and Asc412 samples but not Asc1 samples. As previously mentioned, high mannose glycans are highly present in apolipoprotein B-10064 and complement C3.65 High-mannose-type glycans H7N2, H8N2, and H9N2 have also been identified in IgA, IgM alpha-2-macrogloblin, and AGP.15 The reason for the reduction in high-mannose-type glycans in Asc1 remains to be investigated. Fourteen different complement factors or subcomponents were identified in the samples analyzed (Supplemental Tables S4 and S5). Complement C3, C4A, and C4B were the most prevalent complement factors identified in all samples. Complement C1r, 9, and H were also detected in all samples and appeared to bind well to lectins, whereas Complement factor 5 did not appear to bind well to lectins (Supplemental Tables S4 and S5). All complement proteins are glycosylated with largely core-fucosylated N-glycans.66 High-mannose N-glycans are reported to be highly present in complement C3.65 Complement C3 was determined in our study to be one of the most abundant proteins in Asc1, making this protein a possible source of high mannose glycans detected in this fluid. Interestingly, highly glycosylated complement C3 was not detected in samples #1, #2, #3, #4, or #5 but was detected in sample #6, the HSA/IgG-depleted OC ascites. It was highly glycosylated with highmannose-type glycans (H5N2, H6N2, H7N2, H8N2, N9N2, and H10N2).
Fucosylated glycans H3N4F1, H4N4F1, and H4N5F1, which are glycans that are typically found on IgG, were not detected in samples that bound to lectins (Supplemental Figure S1A–C), suggesting that fucosylation did not assist with ConA or WGA lectin binding. In contrast, glycans that were fucosylated and sialylated H5N5F1S1 and H5N5F1S2 (Figure 3E) and H6N5F1S2, H6N5F1S3, and H7N6F1S2 (Figure 4G) were detected in lectin-bound samples, showing the affinity of lectins (ConA and WGA) for proteins with these types of N-glycans.
Proteins Identified in OC Ascites
The majority of proteins identified in Asc1, Asc412, and SS control were highly abundant proteins, most of which were among the most commonly identified proteins in plasma.57 Unexpectedly, fibronectin, ceruloplasmim, and hemopexin were in higher relative quantities in OC ascites (Asc1 and Asc412) than usually measured in human plasma57 and in our serum control (SS). These results show proteomic differences between normal serum/plasma proteins and ascites fluids.
Previous proteomic analysis of ascites fluid was performed by Kuk et al.,67 but their focus was on smaller molecular weight proteins of <30 kDa to avoid having to include high abundant proteins such as albumin. Of the 52 proteins identified in their study, clusterin precursor and isoform C of fibulin-1 precursor were identified, which were also identified in our study.
Lectin binding enriched most glycosylated proteins and removed many unglycosylated proteins. Albumin, apolipoproteins A-1, A4, E, and C–III, vitamin D-binding protein, and gelsolin from Asc1 did not bind to lectins (Supplemental Table S4). Certain glycosylated proteins such as pigment epithelium-derived factor from asc1 also did not appear to bind to ConA or WGA lectins. Serotransferrin, which is glycosylated, bound to WGA but not to ConA lectin in Asc1 and did not bind to either lectin in Asc412 and SS samples (Supplemental Table S5). Apolipoprotein B-100, complement C4-A, and alpha 1-acid glycoprotein appeared to bind better to WGA than to ConA in Asc1. In contrast, serotransferrin, lumican, afamin, and attractin from Asc1 bound better to ConA (Supplemental Table S4). These differences in binding to lectins demonstrate the variability in glycosylation in these proteins from different sources that may have biological and functional consequences in tumor metastasis. Glycosylation of serotransferrin was examined in the six samples analyzed by proteomic methods. Although among the most abundant proteins in sample #6 (HSA/IgG -depleted OC ascites), very little glycosylation was detected (four peptides, two different sites, weakly detected with O-glycans and two with the common N-glycan H5N4F0S2, out of 513 total peptides with 81.66% coverage), which likely explains the poor binding of serotransferrin to ConA and WGA lectins. Vitamin D binding protein was also detected in OC ascites sample #6 with no glycosylation confirming why this protein would not bind to lectins.
CA125 and MUC1,2,3 were detected by WB analysis but not detected by our proteomic analysis, showing that our present proteomic analysis was not sensitive enough to detect these very low abundant proteins. Better enrichment methods are needed to improve our ability to isolate and perform glycomic analysis on these individual proteins.
Specific Glycosylation of Proteins
Further glycomic analysis of some of these proteins was performed to determine the heterogeneity and site identification of glycans of clusterin, fibulin, and hemopexin in OC ascites fluid samples. This type of analysis has recently been reported by Chandler et al. for haptoglobin68 and ITIH4.69 N-Glycans attached to specific haptoglobin sites showed the heterogeneity of glycans identified in each site.68 The majority of glycans described were sialylated with some possessing core and others with outer arm antennary fucose attached to bi- and triantennary glycoforms.
Glycans H6N5, H6N5F1S1, H6N5F2S3, and H7N6F1S1 were N-glycans that we identified as being present in ascites fluids but less abundant or missing in the serum control samples. Interestingly, H6N5 is a glycan structure synthesized through GlcNAc transferase-V (GNT-V, also known as MGATV), which catalyzes the α1–6 branching of N-glycans. An important aspect of this type of glycan structure is that it could lead to expression of poly-N-acetyllactosamines that, in turn, can get fucosylated and sialylated, which are structures potentially recognized by galectins and selectins. These glycan structures can play an important role in carcinogenesis as well as immune responses.70 The H6N5 glycans were identified in peptides from clusterin but not in OC ascites peptides from fibulin or in hemopexin.
CONCLUSIONS
The approach taken in this study was to obtain glycomic and proteomic profiles of proteins present in OC ascites fluids enriched by differential binding to ConA and WGA lectins while removing most of the largely abundant serum albumin and increasing the presence of less abundant proteins. These less abundant proteins might harbor much of the abnormal glycosylation potentially related to OC tumor metastasis. Performing the proteomic analysis in concert with the glycomic analysis enables us to identify potential protein sources of the abnormal glycomic profiles we identified. The challenge still remains to identify the actual protein sources of the abnormal glycosylation, which we are actively pursuing. OC ascites fluids provide us with a rich source of material to develop the necessary glycoproteomic analytical methods needed to accomplish this task. Further glycoproteomic investigation could also help us to better understand the metastatic process as well as how a system response to the growth of OC tumor cells as well as how they might be able to evade and circumvent immune surveillance. Biological context of the glycosylated protein is important. Separation of protein populations produced different glycopeptide results for the same protein, which suggests a functional connotation and purpose for each glycosylation. Moreover, isolation of protein complexes with biologically relevant samples is likely to produce more meaningful results. We suspect that gathering information from the same sample processed in different manners will enable us to piece together vital data necessary to help us solve the complex question of how tumor cells are able to metastasize and escape immune surveillance. Importantly, we showed that how the samples were prepared could significantly impact the subsequent glycoproteomic analysis. Another important aspect of these glycoproteomic studies of OC ascites fluids is the presence and glycosylation of ECM components, such as clusterin, fibulin, and hemopexin, that may be important for OC tumor growth, survival, and metastasis. What unites many of the OC ascites proteins identified in this study is that most of them are ECM proteins with potentially important functions impacting metastasis of OC tumor cells (chaperons, adhesion, protein interactions, and communication).
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Hai Chi Pham and Lauren Dimapasoc to this research project. Protein identifications of the lectin-bound fractions from the ascites fluid were performed by the UC Davis Proteomic Core (Brett Phinney, Director, and Diana Tran, Junior Specialist). Funding was provided by the Ovarian Cancer Research Foundation award to G.S.L., NIH R01 GM049077 to C.B.L., and private donations to G.L. and the UC Davis Cancer Center for ovarian cancer research projects. We also acknowledge the support of the Ovarian Cancer Education and Research Network (OCERN).
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b00548.
Supplemental Figure S1. Additional glycan groups comparing samples. Supplemental Figure S2. Fibulin sequence coverage and modifications. Supplemental Figure S3. Fibulin sequence with glycosylation sites and glycopeptides with putative glycan structures. Supplemental Table S1. N-glycan analysis of the ascites fluid, ConA- and WGA-bound proteins with structures. Supplemental Table S2. A comparison of N-glycans in samples. Supplemental Table S3. Data used for glycan Venn diagrams. Supplemental Table S4: Proteomic data for Asc1, Asc1-ConA, Asc1-WGA. Supplemental Table S5: Proteomic data for Asc412, Asc412-ConA, Asc412-WGA, SS, SS-ConA, SS-WGA. Supplemental Tables S6A and S6B. Proteomic data used for Venn diagrams comparing proteins from ascites samples and control. Supplemental Table S7. Clusterin glycosylation sites, glycan composition, % occupancy, no. peptides, and total glycan containing peptides. Supplemental Table S8. Type of N-glycans attached to clusterin isolated from OC ascites fluid. Supplemental Table S9. All hemopexin peptides from OC ascites samples #1–6 with glycan information. (PDF)
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. Ca-Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi E, Ueda Y, Matsuzaki S, Yokoyama T, Kimura T, Yoshino K, Fujita M, Kimura T, Enomoto T. Biomarkers for screening, diagnosis, and monitoring of ovarian cancer. Cancer Epidemiol, Biomarkers Prev. 2012;21(11):1902–12. doi: 10.1158/1055-9965.EPI-12-0646. [DOI] [PubMed] [Google Scholar]
- 3.Felder M, Kapur A, Gonzalez-Bosquet J, Horibata S, Heintz J, Albrecht R, Fass L, Kaur J, Hu K, Shojaei H, Whelan RJ, Patankar MS. MUC16 (CA125): tumor biomarker to cancer therapy, a work in progress. Mol Cancer. 2014;13:129. doi: 10.1186/1476-4598-13-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu KH, Skates S, Hernandez MA, Bedi D, Bevers T, Leeds L, Moore R, Granai C, Harris S, Newland W, Adeyinka O, Geffen J, Deavers MT, Sun CC, Horick N, Fritsche H, Bast RC., Jr A 2-stage ovarian cancer screening strategy using the Risk of Ovarian Cancer Algorithm (ROCA) identifies early-stage incident cancers and demonstrates high positive predictive value. Cancer. 2013;119(19):3454–61. doi: 10.1002/cncr.28183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed N, Stenvers KL. Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front Oncol. 2013;3:256. doi: 10.3389/fonc.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puiffe ML, Le Page C, Filali-Mouhim A, Zietarska M, Ouellet V, Tonin PN, Chevrette M, Provencher DM, Mes-Masson AM. Characterization of ovarian cancer ascites on cell invasion, proliferation, spheroid formation, and gene expression in an in vitro model of epithelial ovarian cancer. Neoplasia. 2007;9(10):820–9. doi: 10.1593/neo.07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward BG, McGuckin MA, Hurst TG, Khoo SK. Expression of multiple tumour markers in serum from patients with ovarian carcinoma and healthy women. Aust N Z J Obstet Gynaecol. 1989;29(3 Pt 2):340–5. doi: 10.1111/j.1479-828x.1989.tb01759.x. [DOI] [PubMed] [Google Scholar]
- 8.Devine PL, McGuckin MA, Ward BG. Circulating mucins as tumor markers in ovarian cancer (review) Anticancer Res. 1992;12(3):709–17. [PubMed] [Google Scholar]
- 9.Jacobs IJ, Menon U. Progress and challenges in screening for early detection of ovarian cancer. Mol Cell Proteomics. 2004;3(4):355–66. doi: 10.1074/mcp.R400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Budiu RA, Mantia-Smaldone G, Elishaev E, Chu T, Thaller J, McCabe K, Lenzner D, Edwards RP, Vlad AM. Soluble MUC1 and serum MUC1-specific antibodies are potential prognostic biomarkers for platinum-resistant ovarian cancer. Cancer Immunol Immunother. 2011;60(7):975–84. doi: 10.1007/s00262-011-1010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demeter A, Sziller I, Csapo Z, Olah J, Keszler G, Jeney A, Papp Z, Staub M. Molecular prognostic markers in recurrent and in non-recurrent epithelial ovarian cancer. Anticancer Res. 2005;25(4):2885–9. [PubMed] [Google Scholar]
- 12.Chu CS, Ninonuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, Killeen K, Miyamoto S, Grimm R, Lebrilla CB. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9(7):1939–51. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B, An HJ, Kirmiz C, Lebrilla CB, Lam KS, Miyamoto S. Glycoproteomic analyses of ovarian cancer cell lines and sera from ovarian cancer patients show distinct glycosylation changes in individual proteins. J Proteome Res. 2008;7(9):3776–88. doi: 10.1021/pr800297u. [DOI] [PubMed] [Google Scholar]
- 14.Froehlich JW, Barboza M, Chu C, Lerno LA, Jr, Clowers BH, Zivkovic AM, German JB, Lebrilla CB. Nano-LC-MS/MS of glycopeptides produced by nonspecific proteolysis enables rapid and extensive site-specific glycosylation determination. Anal Chem. 2011;83(14):5541–7. doi: 10.1021/ac2003888. [DOI] [PubMed] [Google Scholar]
- 15.Aldredge D, An HJ, Tang N, Waddell K, Lebrilla CB. Annotation of a serum N-glycan library for rapid identification of structures. J Proteome Res. 2012;11(3):1958–68. doi: 10.1021/pr2011439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shevchenko A, Chernushevich I, Ens W, Standing KG, Thomson B, Wilm M, Mann M. Rapid ‘de novo’ peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun Mass Spectrom. 1997;11(9):1015–24. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1015::AID-RCM958>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 17.Kronewitter SR, de Leoz ML, Peacock KS, McBride KR, An HJ, Miyamoto S, Leiserowitz GS, Lebrilla CB. Human serum processing and analysis methods for rapid and reproducible N-glycan mass profiling. J Proteome Res. 2010;9(10):4952–9. doi: 10.1021/pr100202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song T, Ozcan S, Becker A, Lebrilla CB. In-depth method for the characterization of glycosylation in manufactured recombinant monoclonal antibody drugs. Anal Chem. 2014;86(12):5661–6. doi: 10.1021/ac501102t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst. 2011;136(18):3663–71. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kronewitter SR, De Leoz ML, Strum JS, An HJ, Dimapasoc LM, Guerrero A, Miyamoto S, Lebrilla CB, Leiserowitz GS. The glycolyzer: automated glycan annotation software for high performance mass spectrometry and its application to ovarian cancer glycan biomarker discovery. Proteomics. 2012;12(15–16):2523–38. doi: 10.1002/pmic.201100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hjernø K. Protein Identification by Peptide Mass Fingerprinting. Mass Spectrometry Data Analysis in Proteomics. 2006;367:61–75. doi: 10.1385/1-59745-275-0:61. [DOI] [PubMed] [Google Scholar]
- 22.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 23.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 24.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–56. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ninonuevo M, An H, Yin H, Killeen K, Grimm R, Ward R, German B, Lebrilla C. Nanoliquid chromatography-mass spectrometry of oligosaccharides employing graphitized carbon chromatography on microchip with a high-accuracy mass analyzer. Electrophoresis. 2005;26(19):3641–9. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- 26.Kronewitter SR, An HJ, de Leoz ML, Lebrilla CB, Miyamoto S, Leiserowitz GS. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics. 2009;9(11):2986–94. doi: 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu S, Salcedo J, Tang N, Waddell K, Grimm R, German JB, Lebrilla CB. Employment of tandem mass spectrometry for the accurate and specific identification of oligosaccharide structures. Anal Chem. 2012;84(17):7456–62. doi: 10.1021/ac301398h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alley WR, Jr, Vasseur JA, Goetz JA, Svoboda M, Mann BF, Matei DE, Menning N, Hussein A, Mechref Y, Novotny MV. N-linked glycan structures and their expressions change in the blood sera of ovarian cancer patients. J Proteome Res. 2012;11(4):2282–300. doi: 10.1021/pr201070k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biskup K, Braicu EI, Sehouli J, Fotopoulou C, Tauber R, Berger M, Blanchard V. Serum glycome profiling: a biomarker for diagnosis of ovarian cancer. J Proteome Res. 2013;12(9):4056–63. doi: 10.1021/pr400405x. [DOI] [PubMed] [Google Scholar]
- 30.An HJ, Miyamoto S, Lancaster KS, Kirmiz C, Li B, Lam KS, Leiserowitz GS, Lebrilla CB. Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. J Proteome Res. 2006;5(7):1626–35. doi: 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa H, Yoneda A, Seno N, Hayashi M, Ishizuka I, Hase S, Matsumoto I. Structures of the N-linked oligosaccharides on human plasma vitronectin. Eur J Biochem. 1995;230(3):994–1000. doi: 10.1111/j.1432-1033.1995.tb20647.x. [DOI] [PubMed] [Google Scholar]
- 32.Mega T, Lujan E, Yoshida A. Studies on the oligosaccharide chains of human alpha 1-protease inhibitor. II. Structure of oligosaccharides. J Biol Chem. 1980;255(9):4057–61. [PubMed] [Google Scholar]
- 33.Fu D, van Halbeek H. N-glycosylation site mapping of human serotransferrin by serial lectin affinity chromatography, fast atom bombardment-mass spectrometry, and 1H nuclear magnetic resonance spectroscopy. Anal Biochem. 1992;206(1):53–63. doi: 10.1016/s0003-2697(05)80010-7. [DOI] [PubMed] [Google Scholar]
- 34.Stroop CJ, Weber W, Gerwig GJ, Nimtz M, Kamerling JP, Vliegenthart JF. Characterization of the carbohydrate chains of the secreted form of the human epidermal growth factor receptor. Glycobiology. 2000;10(9):901–17. doi: 10.1093/glycob/10.9.901. [DOI] [PubMed] [Google Scholar]
- 35.Pierce-Cretel A, Pamblanco M, Strecker G, Montreuil J, Spik G, Dorland L, Van Halbeek H, Vliegenthart JF. Primary structure of the N-glycosidically linked sialoglycans of secretory immunoglobulins A from human milk. Eur J Biochem. 1982;125(2):383–8. doi: 10.1111/j.1432-1033.1982.tb06694.x. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura Y, Takahashi Y, Yamaguchi S, Omiya S, Orii T, Yara A, Gushiken M. Severe infantile sialidosis–the characteristics of oligosaccharides isolated from the urine and the abdominal ascites. Tohoku J Exp Med. 1992;166(4):407–15. doi: 10.1620/tjem.166.407. [DOI] [PubMed] [Google Scholar]
- 37.Kim JH, Kim YW, Kim IW, Park DC, Kim YW, Lee KH, Jang CK, Ahn WS. Identification of candidate biomarkers using the Experion automated electrophoresis system in serum samples from ovarian cancer patients. Int J Oncol. 2013;42(4):1257–62. doi: 10.3892/ijo.2013.1803. [DOI] [PubMed] [Google Scholar]
- 38.Zhuo L, Hascall VC, Kimata K. Inter-alpha-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J Biol Chem. 2004;279(37):38079–82. doi: 10.1074/jbc.R300039200. [DOI] [PubMed] [Google Scholar]
- 39.Clarke CH, Yip C, Badgwell D, Fung ET, Coombes KR, Zhang Z, Lu KH, Bast RC., Jr Proteomic biomarkers apolipoprotein A1, truncated transthyretin and connective tissue activating protein III enhance the sensitivity of CA125 for detecting early stage epithelial ovarian cancer. Gynecol Oncol. 2011;122(3):548–53. doi: 10.1016/j.ygyno.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin B, White JT, Wu J, Lele S, Old LJ, Hood L, Odunsi K. Deep depletion of abundant serum proteins reveals low-abundant proteins as potential biomarkers for human ovarian cancer. Proteomics: Clin Appl. 2009;3(7):853–861. doi: 10.1002/prca.200800141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Xie X, Nie S, Buckanovich RJ, Lubman DM. Altered expression of sialylated glycoproteins in ovarian cancer sera using lectin-based ELISA assay and quantitative glycoproteomics analysis. J Proteome Res. 2013;12(7):3342–52. doi: 10.1021/pr400169n. [DOI] [PubMed] [Google Scholar]
- 42.Ishiwata T, Yamamoto T, Kawahara K, Kawamoto Y, Matsuda Y, Ishiwata S, Naito Z. Enhanced expression of lumican inhibited the attachment and growth of human embryonic kidney 293 cells. Exp Mol Pathol. 2010;88(3):363–70. doi: 10.1016/j.yexmp.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto T, Matsuda Y, Kawahara K, Ishiwata T, Naito Z. Secreted 70 kDa lumican stimulates growth and inhibits invasion of human pancreatic cancer. Cancer Lett. 2012;320(1):31–9. doi: 10.1016/j.canlet.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 44.Clinton GM, Rougeot C, Derancourt J, Roger P, Defrenne A, Godyna S, Argraves WS, Rochefort H. Estrogens increase the expression of fibulin-1, an extracellular matrix protein secreted by human ovarian cancer cells. Proc Natl Acad Sci U S A. 1996;93(1):316–20. doi: 10.1073/pnas.93.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyatt A, Yerbury J, Poon S, Dabbs R, Wilson M. The Chaperone Action of Clusterin and Its Putative Role in Quality Control of Extracellular Protein Folding. Adv Cancer Res. 2009;104:89–114. doi: 10.1016/S0065-230X(09)04006-8. [DOI] [PubMed] [Google Scholar]
- 46.Pucci S, Mazzarelli P, Nucci C, Ricci F, Spagnoli LG. CLU “In and Out”: Looking for a Link. In: Bettuzzi S, Pucci S, editors. Clusterin, Part B. Vol. 105. Elsevier; Amsterdam: 2009. pp. 93–113. [DOI] [PubMed] [Google Scholar]
- 47.Gallagher WM, Currid CA, Whelan LC. Fibulins and cancer: friend or foe? Trends Mol Med. 2005;11(7):336–40. doi: 10.1016/j.molmed.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Aspberg A, Adam S, Kostka G, Timpl R, Heinegard D. Fibulin-1 is a ligand for the C-type lectin domains of aggrecan and versican. J Biol Chem. 1999;274(29):20444–9. doi: 10.1074/jbc.274.29.20444. [DOI] [PubMed] [Google Scholar]
- 49.Varki A, Freeze HH, Manzi AE. Overview of glycoconjugate analysis. Curr Protoc Protein Sci. 2009:1–8. doi: 10.1002/0471140864.ps1201s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim Biophys Acta, Gen Subj. 2012;1820(9):1347–53. doi: 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 51.Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc Natl Acad Sci U S A. 2002;99(16):10231–3. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schultz MJ, Swindall AF, Bellis SL. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 2012;31(3–4):501–18. doi: 10.1007/s10555-012-9359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin Chem. 2010;56(2):223–36. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi M, Kuroki Y, Ohtsubo K, Taniguchi N. Core fucose and bisecting GlcNAc, the direct modifiers of the N-glycan core: their functions and target proteins. Carbohydr Res. 2009;344(12):1387–90. doi: 10.1016/j.carres.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 55.Ruhaak LR, Taylor SL, Miyamoto S, Kelly K, Leiserowitz GS, Gandara D, Lebrilla CB, Kim K. Chip-based nLC-TOF-MS is a highly stable technology for large-scale high-throughput analyses. Anal Bioanal Chem. 2013;405(14):4953–8. doi: 10.1007/s00216-013-6908-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song T, Aldredge D, Lebrilla CB. A Method for In-Depth Structural Annotation of Human Serum Glycans That Yields Biological Variations. Anal Chem. 2015;87(15):7754–62. doi: 10.1021/acs.analchem.5b01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hortin GL, Sviridov D, Anderson NL. High-abundance polypeptides of the human plasma proteome comprising the top 4 logs of polypeptide abundance. Clin Chem. 2008;54(10):1608–16. doi: 10.1373/clinchem.2008.108175. [DOI] [PubMed] [Google Scholar]
- 58.Mimura Y, Ashton PR, Takahashi N, Harvey DJ, Jefferis R. Contrasting glycosylation profiles between Fab and Fc of a human IgG protein studied by electrospray ionization mass spectrometry. J Immunol Methods. 2007;326(1–2):116–26. doi: 10.1016/j.jim.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 59.Kui Wong N, Easton RL, Panico M, Sutton-Smith M, Morrison JC, Lattanzio FA, Morris HR, Clark GF, Dell A, Patankar MS. Characterization of the oligosaccharides associated with the human ovarian tumor marker CA125. J Biol Chem. 2003;278(31):28619–34. doi: 10.1074/jbc.M302741200. [DOI] [PubMed] [Google Scholar]
- 60.Sakai K, Fujii T, Hayashi T. Cell-free formation of disulfide-bonded multimer from isolated plasma fibronectin in the presence of a low concentration of SH reagent under a physiological condition. Matrix Biol. 1994;14(3):415–21. doi: 10.1093/oxfordjournals.jbchem.a124353. [DOI] [PubMed] [Google Scholar]
- 61.Abbott KL, Nairn AV, Hall EM, Horton MB, McDonald JF, Moremen KW, Dinulescu DM, Pierce M. Focused glycomic analysis of the N-linked glycan biosynthetic pathway in ovarian cancer. Proteomics. 2008;8(16):3210–20. doi: 10.1002/pmic.200800157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Leoz ML, An HJ, Kronewitter S, Kim J, Beecroft S, Vinall R, Miyamoto S, de Vere White R, Lam KS, Lebrilla C. Glycomic approach for potential biomarkers on prostate cancer: profiling of N-linked glycans in human sera and pRNS cell lines. Dis Markers. 2008;25(4–5):243–58. doi: 10.1155/2008/515318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Leoz ML, Young LJ, An HJ, Kronewitter SR, Kim J, Miyamoto S, Borowsky AD, Chew HK, Lebrilla CB. Highmannose glycans are elevated during breast cancer progression. Mol Cell Proteomics. 2011;10(1):M110 002717. doi: 10.1074/mcp.M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harazono A, Kawasaki N, Kawanishi T, Hayakawa T. Site-specific glycosylation analysis of human apolipoprotein B100 using LC/ESI MS/MS. Glycobiology. 2004;15(5):447–62. doi: 10.1093/glycob/cwi033. [DOI] [PubMed] [Google Scholar]
- 65.Crispin MD, Ritchie GE, Critchley AJ, Morgan BP, Wilson IA, Dwek RA, Sim RB, Rudd PM. Monoglucosylated glycans in the secreted human complement component C3: implications for protein biosynthesis and structure. FEBS Lett. 2004;566(1–3):270–4. doi: 10.1016/j.febslet.2004.04.045. [DOI] [PubMed] [Google Scholar]
- 66.Ritchie GE, Moffatt BE, Sim RB, Morgan BP, Dwek RA, Rudd PM. Glycosylation and the complement system. Chem Rev. 2002;102(2):305–20–19. doi: 10.1021/cr990294a. [DOI] [PubMed] [Google Scholar]
- 67.Kuk C, Kulasingam V, Gunawardana CG, Smith CR, Batruch I, Diamandis EP. Mining the ovarian cancer ascites proteome for potential ovarian cancer biomarkers. Mol Cell Proteomics. 2009;8(4):661–9. doi: 10.1074/mcp.M800313-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chandler KB, Pompach P, Goldman R, Edwards N. Exploring site-specific N-glycosylation microheterogeneity of haptoglobin using glycopeptide CID tandem mass spectra and glycan database search. J Proteome Res. 2013;12(8):3652–66. doi: 10.1021/pr400196s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chandler KB, Brnakova Z, Sanda M, Wang S, Stalnaker SH, Bridger R, Zhao P, Wells L, Edwards NJ, Goldman R. Site-specific glycan microheterogeneity of inter-alpha-trypsin inhibitor heavy chain H4. J Proteome Res. 2014;13(7):3314–29. doi: 10.1021/pr500394z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horst AK, Wagener C. Bitter sweetness of complexity. Top Curr Chem. 2008;288:1–15. doi: 10.1007/128_2008_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.