Abstract
Iron-sulfur (Fe/S) cluster-containing proteins constitute one of the largest protein classes, with varied functions that include electron transport, regulation of gene expression, substrate binding and activation, and radical generation. Consequently, the biosynthetic machinery for Fe/S clusters is evolutionarily conserved, and mutations in a variety of putative intermediate Fe/S cluster scaffold proteins can cause disease states, including multiple mitochondrial dysfunctions syndrome (MMDS), sideroblastic anemia and mitochondrial encephalomyopathy. Herein, we have characterized the impact of defects occurring in the MMDS1 disease state that result from a point mutation (Gly208Cys) near the active site of NFU1, an iron-sulfur scaffold protein, via an in vitro investigation into the structural and functional consequences. Analysis of protein stability and oligomeric state demonstrates that the mutant increases the propensity to dimerize and perturbs the secondary structure composition. These changes appear to underlie the severely decreased ability of mutant NFU1 to accept an iron-sulfur cluster from physiologically relevant sources. Therefore, the point mutation on NFU1 impairs downstream cluster trafficking and results in the disease phenotype, because there does not appear to be an alternative in vivo reconstitution path, most likely due to greater protein oligomerization from a minor structural change.
Keywords: mitochondrial disease, iron-sulfur cluster, cluster exchange, protein stability, NFU1
Graphical abstract
Introduction
Mitochondria are complex eukaryotic organelles that serve as the site of aerobic cellular metabolism and energy production via oxidative phosphorylation. Furthermore, many important and diverse pathways for the production of essential biological cofactors are localized to the mitochondria [11–13]. Given the inherent structural and functional complexity of the mitochondria, combined with the necessary biosynthetic processes that occur there, mitochondrial defects can cause a wide variety of severe and generally untreatable disorders [11, 15]. Multiple mitochondrial dysfunctions syndrome (MMDS) constitutes a class of typically fatal diseases that result from the severe impairment of various metabolic pathways and energy production as a consequence of single nucleotide genetic mutations [16, 17]. Symptoms of MMDS include hypotonia, respiratory insufficiency, hyperglycinemia, encephalopathy, neurological regression, and failure to thrive, indicative of decreased functional actions of mitochondrial respiratory complexes [18–20]. Additional symptoms of MMDS are related to a deficiency of mitochondrial respiratory complexes and impaired function of lipoic acid dependent enzymes, such as pyruvate dehydrogenase (PDH) and protein H of the Glycine Cleavage System (GCS) [15, 16, 21]. Due to the severity and extent of symptoms described above, MMDS is typically fatal during perinatal stages [16]; however, some patients have lived until the age of two, at which point the symptoms culminated in death [17, 18, 20, 21].
Interestingly, all three types of MMDS so far identified are associated with genes that code for metalloproteins, specifically those involved in the biosynthesis of iron-sulfur clusters: IBA57, IscA2, BOLA3, or NFU1 [16, 17, 22–26]. Further inspection of the defects caused by these iron-sulfur cluster proteins has revealed a specific impairment of downstream [4Fe-4S]-cluster-containing proteins; by contrast, [2Fe-2S]-cluster-containing proteins and cytosolic iron-sulfur cluster proteins appear unaffected [15, 17, 22]. All four of the identified proteins have been implicated in iron-sulfur cluster biosynthetic pathways as mediators of cluster transfer and delivery. Iron-sulfur clusters are highly conserved inorganic prosthetic groups that are present across all kingdoms of life and play diverse roles in the cell, including electron transfer, regulation of gene expression, and disulfide reduction [27, 28].
Herein, we have focused on one of the aforementioned Fe/S proteins, NFU1. Human NFU1 consists of two domains, an N- and a C-terminal domain [4, 29–31], where the latter contains a highly conserved CXXC motif [31] that identifies this domain as the Nfu domain and suggests a role in iron-sulfur cluster binding, assembly, and transport, based on comparison to homologous proteins and the high level of sequence conservation [32–36]. The thermodynamic stability of NFU1 has been studied in depth to reveal that the isolated C-terminal domain of human NFU1 exhibits characteristics of a molten globular state [4, 30], while the isolated N-terminal domain demonstrates a highly ordered structure. However, when the two domains come together the overall protein maintains a relatively well-folded structure, suggesting a requirement for the unique N-terminal domain of the human protein to provide structural stabilization as well as serving a potential functional role [4, 30] that may include a protein oligomerization surface or a binding site for chaperones, such as Hsc20 [5, 37]. Structure-function characteristics of human NFU1 remain unclear, because this protein has been implicated in a variety of cellular roles. NFU1 is also known as the HIRA-interacting protein, where HIRA is the histone cell cycle regulation homologue A [38]. Furthermore, it exhibits thioredoxin-like activity in the apo form [29, 30], assembles and transfers a [2Fe-2S] cluster [1], assembles a [4Fe-4S] cluster [31, 39], and may transfer cluster to apo aconitase [5]. Homologs of human NFU1 have been implicated in similar roles, with [2Fe-2S] [33, 34, 36, 40] and [4Fe-4S] [32, 33, 35, 41] cluster transfer, suggesting that both functionalities are possible and potentially physiologically relevant. Most recently, NFU1 has been implicated in the disease phenotype of MMDS1 attributed to a c.622G>T missense mutation located in the gene encoding the protein NFU1 [17, 21]. This introduces a p.Gly208Cys missense mutation in the protein close to the iron-sulfur (Fe/S) cluster binding motif (Fig. 1) that alters the region around the cluster binding motif from GXCXXC to CXCXXC.
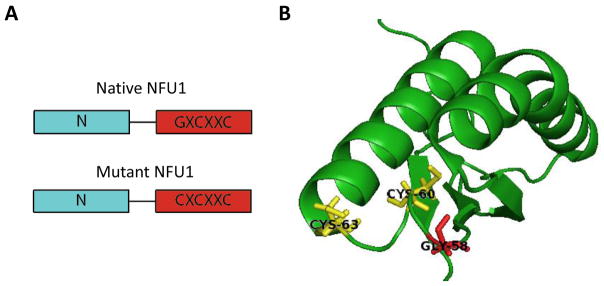
Figure 1.
(A) A representation of the two-domain composition of the native and mutant NFU1 proteins, with the N-terminal domain in blue and the C-terminal domain in red. NFU1 features the functional CXXC Fe/S cluster binding motif in its C-terminal domain. This pattern is altered in the mutant protein with the mutation of a nearby glycine residue at position 208 to cysteine, which gives a CXCXXC motif. (B) Solution NMR structure of human C-terminal domain of the NFU1 protein (PDB ID: 2M5O) with the cluster binding cysteines shown in yellow [5]. The glycine at position 208 in the full length protein (58 above), which is mutated to a cysteine in MMDS1, is colored red. Numbering is consistent with the C-terminal construct used in the structure determination.
In connection with the MMDS1 disease state, human NFU1 has been proposed to be involved in the maturation of the iron-sulfur clusters on the [4Fe-4S] target proteins lipoate synthase (LIAS) and succinate dehydrogenase (SDH) [17, 21, 39]. In patients with MMDS1, laboratory tests have demonstrated that the level of human NFU1 protein is not diminished [17, 21, 42], but the activity levels of LIAS and SDH are significantly impaired, while other [4Fe-4S] targets, such as aconitase, are relatively unaffected [17, 21, 42]. Despite the identification of the global cellular effect of the NFU1 mutation, there remains a lack of understanding of the influence of the mutation at the molecular level. For this reason, we have undertaken a biochemical approach to determine the impact of this mutation on the structure-function relationship of human NFU1.
Results
Given the drastic cellular consequences of the G208C mutation on NFU1, we hypothesized that there could be two major contributors to protein impairment. Since cellular studies have shown that the protein is present at normal levels [17, 21], the protein could be compromised in terms of its structural composition and recognition by partner proteins. Alternatively, it may be unable to perform functionally due to an inability to bind, receive or deliver an Fe/S cluster. By use of in vitro biochemical assays we set out to explore both possibilities.
Structural Alteration of G208C NFU1
G208C substitution does not significantly impact secondary structure
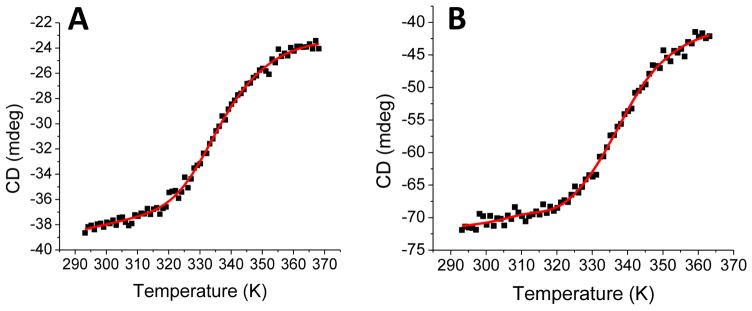
Much has been reported concerning the domain organization and protein stability of the recombinant native human NFU1 [4, 30], which provided a firm benchmark for study of the effect of the G208C substitution on human NFU1 with regard to structural and stability characterization, relative to the native protein. Secondary structure and stability was examined via variable temperature circular dichroism (VTCD) and comparison of the melting profiles and thermodynamic characteristics of the native human NFU1 and the G208C mutant. Data were fit to a one-state model to reduce error in the melting event parameters (Table 1) and yielded a satisfactory fit. Interestingly, the G208C NFU1 derivative shows a VTCD trace very similar to that of the native, with the mutant protein and the native exhibiting the same melting temperature within error, indicating that the two proteins are actually similar in their stabilities. Combining the VTCD data with an additional stability measurement, limited proteolysis (Fig. S2), we observe that the two proteins are both fairly equal in terms of stability, consistent with our other data. Both proteins exhibited reversible melting on VTCD (data not shown), indicating that the overall protein folds are relatively stable. Melting data were also fit to a two-state model (Fig. 2), due to the multidomain nature of the protein, which has been previously characterized [4]. However, the two-state model did not yield any interpretable improvements to the fit and was not considered further.
Table 1.
Variable temperature circular dichroism results. Native NFU1 and G208C NFU1 at 10 μM were subjected to melting from 20 to 95 °C at 0.4 °C per min. Table 1 shows the data fit to a one-state melting process using equation 1.
| Tm (°C) | ΔHV (kcal/mol) | |
|---|---|---|
| Native NFU1 | 65 ± 1 | 26 ± 2 |
| G208C NFU1 | 65 ± 2 | 31 ± 2 |
Figure 2.
VTCD traces for the melting of 10 μM native human NFU1 (A) and G208C NFU1 (B) in 40 mM phosphate, pH 7.4. Data were fit to equation 1 to obtain Tm and ΔHV, which are shown in Table 3. CD units of ellipticity (mdeg) were used directly without conversion to molar ellipticity because the van’t Hoff enthalpies are independent of such a factor [4].
Circular dichroism was also used to examine the overall characteristics of protein secondary structure by determining the percentages of secondary structural elements: α-helix, β-sheet and random coil. The values are summarized in Table 2 and demonstrate an overall increase in random coil and loss of secondary structural elements; however, the modest change in secondary structural elements has not impacted protein stability based on the one-state VTCD results (Fig. 2; Table 1) and limited proteolysis (Fig. S2).
Table 2.
Percentages of secondary structural elements measured by circular dichroism and analyzed using the analysis program CDSSTR [2] on the online Dichroweb server [8, 9].
| α-helix | β-sheet | Random Coil | |
|---|---|---|---|
| Native NFU1 | 47 % | 21 % | 33 % |
| G208C NFU1 | 37 % | 17 % | 46 % |
G208C substitution impacts tertiary structure by promoting dimerization
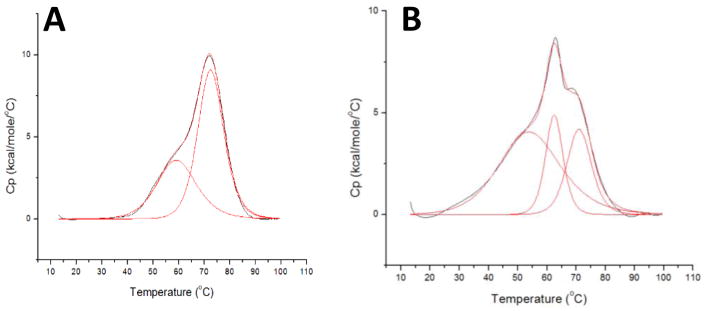
To clarify the results obtained via VTCD and extend the stability comparison to tertiary structural elements, Differential Scanning Calorimetry (DSC) was used to examine the thermodynamic properties of the NFU1 G208C mutant by determining the heat capacity of the protein in comparison to the native. The DSC profile of native human NFU1 (Fig. 3A) matches the parameters determined in prior literature [4] in that the protein follows a two-state melting process with Tm1 of 59.4 ± 0.20 °C and a second melting event at 72.6 ± 0.04 °C (Table 3). The lowest Tm value for native protein corresponds to the melting of the C-terminal domain and the higher melting temperature represents the melting of the well-folded N-terminal domain [4]. The DSC profile for the G208C derivative interestingly exhibited a three phase melting process (Fig. 3B): the first broad melting peak is indicative of the molten globular C-terminal domain with Tm1 of 49 ± 6 C, a second sharp peak not seen in the native melting profile with Tm2 of 64 ± 2 C and an additional shoulder from the structured N-terminal domain with Tm3 of 74 ± 4 C (Table 3). The similarity of melting temperatures and van’t Hoff enthalpies (within error) for the transitions in the substituted protein, relative to the native NFU1 (Table 4), confirm the comparable stabilities of each.
Figure 3.
Differential scanning calorimetry profiles for (A) 0.2 mM native human NFU1, (B) 0.3 mM G208C human NFU1. Both of the proteins were in 50 mM HEPES, 100 mM NaCl, and pH 7.4. The data were fit using Origin 7.0 to obtain Tm, ΔHcal, and ΔHV, all of which are listed in Tables 1
Table 3.
Melting temperatures as determined from fits to the DSC data. The DSC profiles used for fitting are shown in Figure 2.
| Tm1 (°C) | Tm2 (°C) | Tm3 (°C) | |
|---|---|---|---|
| Native NFU1 | 59.3 ± 0.2 | -- | 73 ± 3 |
| G208C NFU1 | 49 ± 6 | 64 ± 2 | 74 ± 4 |
Table 4.
Enthalpies of melting determined by DSC. The DSC profiles used for data fitting are shown in Figure 2.
| ΔHcal1 (kcal/mol) | ΔHV1 (kcal/mol) | ΔHcal2 (kcal/mol) | ΔHV2 (kcal/mol) | ΔHcal3 (kcal/mol) | ΔHV3 (kcal/mol) | |
|---|---|---|---|---|---|---|
| Native NFU1 | 73 ± 2 | 41 ± 2 | -- | -- | 122 ± 1 | 74 ± 5 |
| G208C NFU1 | 5.2 ± 0.4 | 33 ± 4 | 39 ± 2 | 50 ± 2 | 24 ± 4 | 74 ± 4 |
Previous DSC analysis of human NFU1 ascribed the two peaks in the melting trace as arising from C-terminal domain unfolding at low temperature, due to its molten globular nature, and N-terminal domain unfolding at higher temperature [4, 30]. For this reason, the presence of a third peak in the DSC trace for G208C NFU1 was quite surprising. An additional protein domain could not have been introduced by the point mutation, so we sought to understand the origin of the new peak by application of analytical ultracentrifugation (AUC) to examine oligomerization effects. AUC was completed for apo G208C NFU1 in the presence and absence of 1 mM TCEP to eliminate the potential for oligomerization due to disulfide bonds. In the absence of TCEP, a G208C NFU1 monomer was observed at 22.6 kDa (expected MW: 23.9 kDa) as 29% of the total observed species (Fig. 4, black). Dimer at 41.6 kDa accounted for 58%, and some higher order oligomer at 74.0 kDa, potentially corresponding to a tetrameric form, constituted 6% of the sample. However, in the presence of 1 mM TCEP (Fig. 4, red), monomer was still observed at 22.9 kDa, accounting for 30%. The percent of dimer at 41.9 kDa, was also practically unchanged at 60%. The largest change observed was for the higher order oligomeric species. A species at 83.5 kDa was observed as ~ 1.5% of the total species. The apo protein percentages contrast sharply with the AUC data collected for the native protein, which showed the monomer and dimer in approximately equal percentages [1], suggesting that the point mutation propagates the protein’s tendency to oligomerize, but not via disulfide bonding and could explain the additional melting peak present in the DSC trace. However, the apparent modest decrease in thermodynamic stability and increased propensity towards dimer formation does not alter the protein’s susceptibility to degradation in limited proteolysis (Fig. S2), reducing the likelihood that the point mutation initiates a major structural change of the protein. The mutant protein is not significantly less stable than the native form; however, the minor structural changes from secondary structure composition seem to promote a greater tendency to dimerize, and these perturbations may have large impacts on function and partner recognition.
Figure 4.
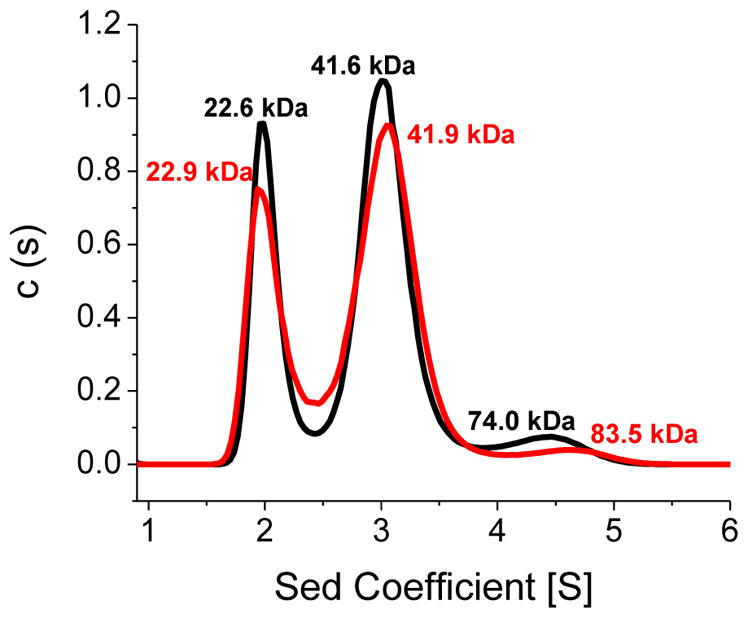
Analytical ultracentrifugation profiles for G208C NFU1. (A) Apo G208C was sedimented in the absence of TCEP (black) and in the presence of 1 mM TCEP (red). Sedimentation was monitored at 280 nm. The first peak of the black trace at 22.6 kDa accounts for 29% of the sample, the second peak at 41.6 kDa accounts for 58%, and the third peak at 74.0 kDa accounts for 6%. The first peak of the red trace at 22.9 kDa accounts for 30% of the sample, the second peak at 41.9 kDa accounts for 60%, and the third peak at 83.5 kDa accounts for 1.5%. The AUC results were fit to the Lamm equation [6, 7] using a continuous distribution model to obtain the peaks and molecular weights shown above.
Functional Impairment of G208C NFU1
G208C NFU1 can be reconstituted, but is primarily dimeric
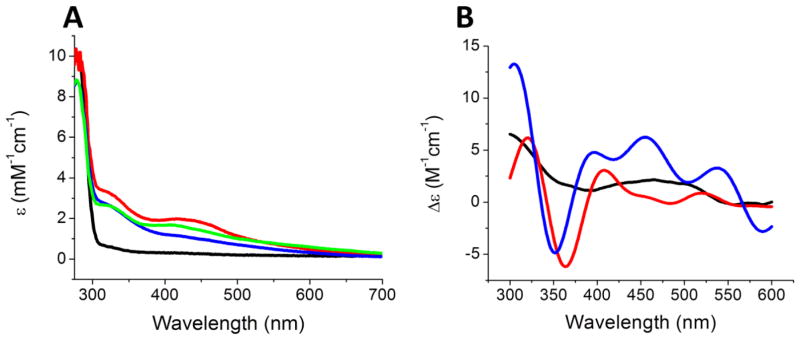
Following the thermodynamic and structural comparisons of G208C NFU1 with the native protein, we next examined the functional changes resulting from the mutation in order to link the oligomeric changes to the downstream effects. Similar to the native protein, G208C NFU1 could be reconstituted in vitro by use of Tm NifS and L-cysteine, and yielded a UV spectrum with typical iron-sulfur cluster charge-transfer bands at 330 and 420 nm (Fig. 5A, blue trace). However, in contrast to the native protein, which cannot be reconstituted with inorganic sulfide, G208C NFU1 was found to be readily reconstituted with ferric chloride and sodium sulfide to yield a native-like UV spectrum and a slightly more prominent 420 nm signature (Fig. 5A, green trace). Additionally, both reconstitution methods resulted in similar UV and CD spectra relative to native NFU1, although with a slight increase in intensity at certain wavelengths (Fig. 5). Inasmuch as native human NFU1 has been shown to bind a [2Fe-2S] cluster under enzymatic reconstitution conditions [1], the similarity in UV and CD spectra suggests that the G208C mutant is capable of binding the same type of cluster. Based on iron quantitation, reconstitution via sodium sulfide resulted in the highest maximum yield, with 0.86 [2Fe-2S] cluster/dimer, while reconstitution with L-cysteine gave 0.76 [2Fe-2S] cluster/dimer. However, both of these yields are in line with what has been typically achieved for the native protein, which yielded 0.75 [2Fe-2S] cluster/dimer [1]. Therefore, the G208C mutant is capable of binding an iron-sulfur cluster and in a similar environment to the native form. Protein oligomerization around the cluster was confirmed via AUC. Holo G208C was monitored at 420 nm to measure the cluster-bound form of the protein. The majority (65.6%) of the holo protein exists as a dimer with a minor tetrameric species (Fig. S3B), in contrast to the native protein, which demonstrated slightly more tetramer than dimer. For both the native and the mutant, no clear additional peaks were observed in the AUC trace [1, 5]. The remaining percentages correspond to protein aggregation and crashing out. The G208C mutation alters the ability of NFU1 to oligomerize in both the apo and holo forms relative to native protein. These data are confirmed by the AUC of the holo protein at 280 nm (Fig. S3A).
Figure 5.
UV (A) and CD (B) spectra following reconstitution of native and G208C NFU1. In both spectra, the black trace corresponds to apo G208C NFU1, and the red trace to native reconstituted holo NFU1. In (A), the blue trace is holo G208C NFU1 reconstituted with Tm NifS and L-cysteine, while the green trace is holo G208C NFU1 reconstituted with ferric chloride and sodium sulfide. In (B), the blue trace is holo reconstituted NFU1, since both reconstitution methods resulted in the same spectra.
G208C NFU1 cannot take up Fe/S clusters from relevant donor sources
Our previous analysis of native human NFU1 [1] has shown that the human protein was capable of receiving and transferring cluster from other Fe/S proteins, as well as from a physiologically relevant glutathione-bound iron-sulfur cluster complex [2Fe-2S](GS)4. This complex can be delivered to a number of iron-sulfur cluster proteins [43, 44] and is a viable substrate for the mitochondrial ABCB7 transporter [45, 46], suggesting a possible role for the [2Fe-2S](GS)4 complex as a component of the labile iron pool. Native human NFU1 can take up a [2Fe-2S] cluster from the complex with a second-order rate constant of 1930 M−1min−1 [1], as observed by circular dichroism (CD). However, G208C NFU1 is unable to take up a cluster from this complex, even with an 8-fold excess of the complex (data not shown).
Likewise, G208C NFU1 was unable to receive a Fe/S cluster from donor proteins. We have previously demonstrated that [2Fe-2S] cluster proteins are able to transfer an iron-sulfur cluster to native human NFU1 [10, 14]. Therefore, we have examined the ability of the G208C mutant protein to be reconstituted in the same way using CD spectroscopy. Holo reconstituted human IscU was added to apo G208C; however, no change in CD signal was observed that would represent the unique spectrum of holo G208C NFU1 (Fig. 5B), indicating that human IscU is incapable of transferring cluster into G208C NFU1 (data not shown). Because involvement by Hsc co-chaperones has been implicated in IscU-promoted cluster delivery to target proteins [16, 37, 47], we also examined the kinetics of transfer from holo human IscU to apo human NFU1 and G208C NFU1 in the presence of HSPA9, Hsc20, MgCl2 and ATP. With the chaperones, IscU was unable to transfer cluster in to either the native or the mutant (Fig. S4), even though transfer from IscU to the native protein can proceed without the chaperone system [14]. Similarly, we monitored the ability of S. pombe Isa1 to transfer a cluster into G208C NFU1, since that transfer was also observed for native NFU1 [10]; however, the appearance of the G208C NFU1 signature peaks was not observed, even over the course of 3 h (data not shown). Minor peak shifts were seen, but the overall signal mainly resembled Isa1, indicating possible formation of a heterodimeric complex. Nevertheless, no clear transfer was demonstrated from Isa1 to G208C NFU1. All of the donor sources for cluster that we examined were unable to reconstitute G208C NFU1 with an Fe/S cluster.
Chemically Reconstituted G208C NFU1 can still transfer Fe/S clusters to target proteins, albeit with perturbed second-order rate constants
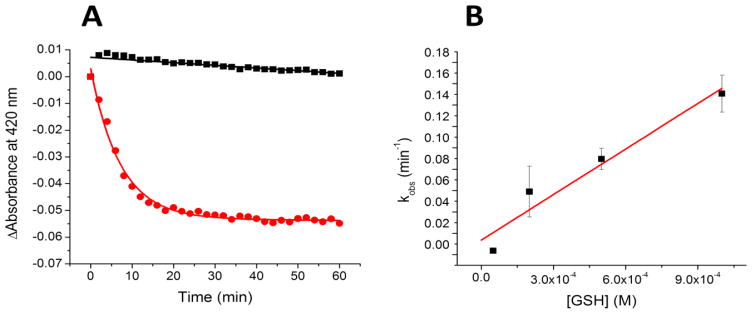
Our previous work with the native form of NFU1 has characterized aspects of NFU1 in Fe/S cluster delivery and trafficking. As such, the [2Fe-2S](GS)4 complex could be formed by cluster extraction from the [2Fe-2S] cluster-bound native NFU1 under conditions of excess glutathione (GSH), with a second-order rate constant of 130 ± 22 M−1min−1 [1]. Excess GSH is also able to extract the [2Fe-2S] cluster from holo G208C NFU1, and the decrease in absorbance at 420 nm was monitored over the course of an hour (Fig. 6A, red trace). The decrease in absorbance was not due to cluster breakdown on G208C NFU1, since a control in the absence of excess GSH (Fig. 6A, black trace) demonstrated very little change in absorbance. The change in absorbance data was determined at four different GSH concentrations and kobs was calculated from each concentration. The data were fit to a linear equation to determine the overall second-order rate constant for GSH extraction of 140 ± 20 M−1min−1 (Fig. 6B), which is in the same range as the second-rate constant found for the native protein, indicating that dimerization has not influenced the ability to transfer a cluster out of G208C NFU1 to this target. Extraction of the cluster by GSH to form the [2Fe-2S](GS)4 complex was confirmed by appearance of the cluster m/z peak of 1427.3 in ESI-MS, which corresponds to an adduct of the complex with one sodium [43, 46] (Fig. S5).
Figure 6.
GSH extraction of the [2Fe-2S] cluster from 10 μM reconstituted holo G208C human NFU1 to form the [2Fe-2S](GS)4 complex. The change in absorbance at 420 nm was monitored over the course of an hour and data were fit to an exponential to obtain the kobs. (A) Shows a representative trace of extraction by 1 mM GSH (red trace). The black trace shows a control of reconstituted holo G208C human NFU1 in the absence of GSH to demonstrate the rate of general cluster instability or breakdown. The concentration of GSH was varied, while keeping the concentration of G208C NFU1 constant to obtain a second-order rate constant. (B) The kobs data were plotted against the concentration of GSH and fit to a linear equation to determine an overall second-order rate constant of 140 ± 20 M−1min−1.
G208C NFU1 is also capable of transferring the [2Fe-2S] cluster to other apo protein targets. As found for native human NFU1, cluster transfer to ferredoxins 1 and 2 (Fdx1 and Fdx2) [1] and glutaredoxins 2 and 3 (Grx2 and Grx3) was observed [10]. Cluster transfer was monitored by use of CD spectroscopy because unique spectra are generated for each iron-sulfur cluster bound protein [48]. Although some these previous transfer reactions were not carried out in the preferred pseudo first-order kinetics range, as a result of limitations from detection sensitivity and protein solubility, we have developed a method to provide estimates for second-order rate constants in cluster transfer reactions using DynaFit software [3], similar to previously published literature [33, 49, 50]. We have shown in test cases that the apparent second-order rate constant obtained in this manner shows close agreement with the rate constant determined from more traditional concentration dependence measurements [1, 44]. Herein, we have used those same methods to determine apparent second-order rate constants for mutant NFU1 for comparison with the native transfer rates. Reconstituted holo G208C NFU1 was added to apo human Fdx1 and the increase in CD signal at 445 nm was monitored (Fig. 7A and 7B) and used to yield a second-order rate constant for the transfer of [2Fe-2S] cluster of 2600 ± 300 M−1min−1. Similarly, cluster transfer from G208C NFU1 to human Fdx2 yielded an overall second-order rate constant of 1200 ± 200 M−1min−1 (Fig. 7C and 7D).
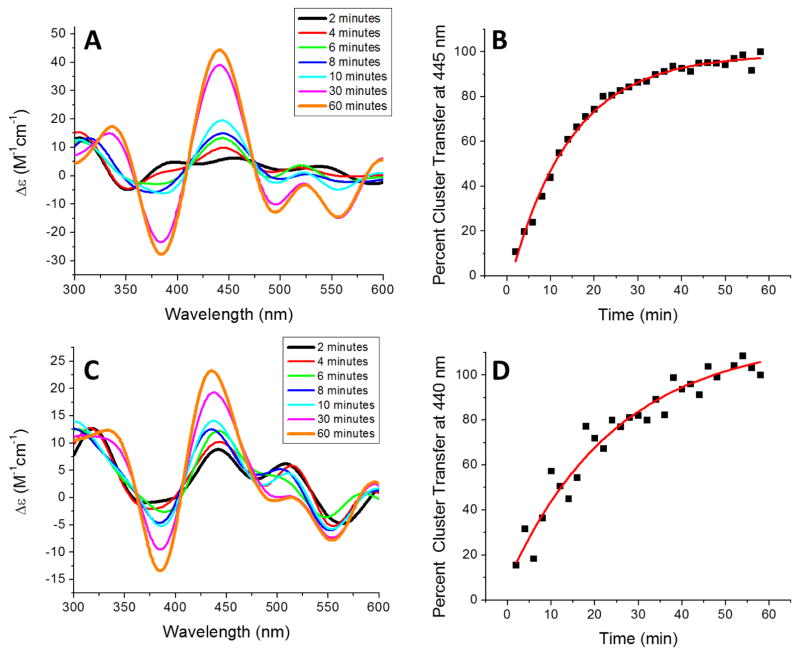
Figure 7.
Kinetics of [2Fe-2S] cluster transfer from holo reconstituted human G208C NFU1 to apo human ferredoxins. (A) Time course for cluster transfer to ferredoxin 1(Fdx1) monitored by CD in 50 mM HEPES, 100 mM NaCl, pH 7.5. Spectra were recorded every 2 min after the addition of holo NFU1, and converted to percent cluster transfer (B) to yield an apparent second-order rate constant from DynaFit of 2600 ± 300 M−1min−1 based on the concentration of the [2Fe-2S] cluster [3]. (C) Time course for cluster transfer from holo human NFU1 to apo human ferredoxin 2 (Fdx2) recorded by CD under the same conditions as for Fdx1. (D) The CD signal was again converted to the percentage of cluster transferred with time to yield an apparent second-order rate constant of 1200 ± 200 M−1min−1.
Cluster transfer kinetics for transfer to glutaredoxins was executed in the same manner as for the ferredoxins. Following addition of holo reconstituted G208C NFU1 to either apo Grx3 or apo Grx2, the change in CD signal was monitored. Since the glutaredoxins utilize glutathione as endogenous ligands, an excess of GSH was included in the reactions to be able to reconstitute a functional glutaredoxin protein. The [2Fe-2S](GS)4 complex is CD silent [43]. Given that no intermediate spectra were observed during the transfer from either native NFU1 or G208C NFU1 to glutaredoxins, the data is consistent with direct cluster transfer to the putative partner protein with no intermediate species formed by extraction of the cluster to form a transitory complex. In both cases, rapid transfer was observed such that by the end of 2 min the spectra already resembled those of the target holo proteins. For that reason, cluster transfer kinetics were monitored over 10 sec intervals, instead of 2 min intervals. Transfer to apo human Grx2 was also complete within the first 15 min (Fig. 8A and 8B), and based on the initial [2Fe-2S] cluster concentration of holo G208C NFU1, resulted in a large second-order rate constant of 22400 ± 5000 M−1min−1. Cluster transfer from holo G208C NFU1 to apo S. cerevisiae Grx3 (Fig. 8C and 8D) was complete within the first 15 min and yielded a second-order rate constant of 14500 ± 3500 M−1min−1.
Figure 8.
Kinetics of [2Fe-2S] cluster transfer from holo reconstituted human G208C NFU1 to apo glutaredoxins. (A) Time course for cluster transfer to apo human Grx2 monitored by CD in 50 mM HEPES, 100 mM NaCl, and pH 7.5 with 3 mM GSH. Spectra were recorded every 2 min after the addition of holo NFU1. However transfer was too rapid to monitor, so cluster transfer was monitored from 435–445 nm every 10 sec and converted to percent cluster transfer (B) to determine an apparent second-order rate constant using DynaFit of 22400 ± 5000 M−1min−1 based on the concentration of the [2Fe-2S] cluster [3]. (C) Time course for cluster transfer to apo S. cerevisiae Grx3 monitored by CD under identical conditions. Again, cluster transfer was too rapid to monitor, and so transfer was monitored from 450–460 nm every 10 sec and converted to percent cluster transfer (D) to determine an apparent second-order rate constant of 14500 ± 3500 M−1min−1.
G208C NFU1 was capable of delivering a [2Fe-2S] cluster to the same target proteins that have been examined for kinetic transfer from the native form. However, the rates of these transfer reactions are significantly different, especially when examining interactions with the glutaredoxin proteins.
Discussion
Previously we have examined and characterized the iron-sulfur cluster protein NFU1 in terms of its stability and iron-sulfur cluster binding and transfer capabilities [1, 29, 30] . Based on our findings from studies of the native human protein, we decided to investigate an important disease-causing mutation of NFU1. A genetic point mutation converts a glycine at position 208 of the amino acid sequence to a cysteine (G208C), resulting in the disease condition known as Multiple Mitochondrial Dysfunctional Syndrome 1 (MMDS1), where symptoms primarily result in death during the infant stage of life [17, 21]. Accordingly, to better determine the cause of the human disease phenotype and understand its molecular basis, we have characterized the stability and iron-sulfur cluster binding and transfer capabilities of the substituted G208C NFU1 protein. Given our two potential hypotheses for the disease phenotype (altered protein structure and recognition, or perturbation to Fe/S cluster uptake or transfer), we began with an examination of protein structure and stability, specifically in terms of secondary and tertiary structure characteristics. Initial analysis via VTCD demonstrated no significant change in secondary structure thermodynamics, suggesting that the native and mutant forms of NFU1 are relatively similar. However, modest changes were observed in secondary structure composition and DSC traces. G208C NFU1 exhibited an overall increase in random coil (Table 2), which does not demonstrate significant effects on stability. The major structural or stability impacts are reflected in the DSC trace, in conjunction with AUC, to show an increased tendency of the protein to oligomerize, specifically to the dimeric form.
The DSC trace of the mutant protein showed a new melting peak (Fig. 3A and 3B), relative to the recombinant native protein, which is consistent with a modest change in secondary structure that could perturb the dimerization interface. Examination by AUC showed that the dimer was present in the majority (Fig. 4), which contrasts with the more equal distribution of monomer and dimer for the native protein [1], suggesting the monomer-dimer equilibrium for apo to be skewed in the direction of dimer. The increased concentration of dimer could affect the DSC trace by altering the melting process. For the native protein, the unfolding of the dimer is concurrent with the melting of the molten globular C-terminal domain to yield a partially unfolded monomer [51, 52], as observed by the first transition in Figure 3A. Following this concerted melting process, the remainder of the protein melts to yield completely unfolded monomers, represented by the three-state mechanism shown in equation 1. However, G208C NFU1 exists primarily as a dimer, which shifts the unfolding process to exhibit a new and distinct melting peak in the DSC trace. The mechanism (equation 2) would now be reflective of a four-state melting process, in which the destabilized, molten globular C-terminal domain of the mutant protein begins to melt (the first transition of three), reducing dimer stability and causing the dimer to unfold in the second melting event (Fig. 2B) [51, 52]. Lastly, the N-terminal domain melts to complete the unfolding process, just as it had for the native.
| (1) |
| (2) |
Since the DSC and secondary structure data suggested a change in structure for the mutant protein, we submitted the protein sequence to Phyre2 [53] for structure prediction by homology modeling, which utilized the NMR structure of the C-terminal domain of mouse NFU1 (PDB ID: 1VEH) and the NMR structure of the C-terminal domain of human NFU1 (PDB ID: 2M5O) to generate a model for G208C NFU1. The resulting model predicts that the placement of the cysteines will change with the mutation (Fig. 9). The CXXC domain exists as a flexible loop, while the G208C NFU1 protein has an altered loop, which appears to lock the position of the C at residue 213 as the beginning of an α-helix (Fig. 9). The replacement of the flexible glycine [54], with a more rigid cysteine has altered the cluster binding loop such that the overall structure of the protein has changed, and could promote an interaction interface to support an increase in the presence of dimers, although how this change at the C-terminal domain might impact the overall protein is currently unknown.
Figure 9.
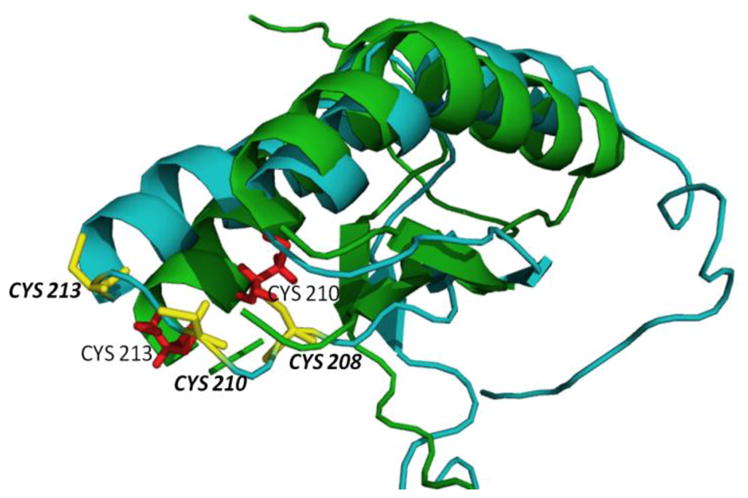
Alignment of the human C-terminal domain of the NFU1 protein (PDB ID: 2M5O) (green trace) and corresponding cysteines in red. The Phyre2 homology modeled structure generated for G208C NFU1 is shown in blue with the cysteines colored yellow.
Inasmuch as the investigation of the biochemical consequences of the G208C substitution demonstrate a structural change that promotes dimerization, but no major change in protein stability, we next examined how this mutation and its structural perturbations would impact function. Our studies of native NFU1 demonstrated a bound [2Fe-2S] cluster that can be transferred in and out of the protein by interactions with either other iron-sulfur cluster proteins or a physiologically relevant [2Fe-2S](GS)4 complex [1]. Similarly, the G208C NFU1 derivative was capable of undergoing reconstitution in vitro through the use of either sodium sulfide, or L-cysteine with Tm NifS as a sulfide source (Fig. 4), while native NFU1 can only be reconstituted using L-cysteine with Tm NifS; however, reconstitution yields in all cases were similar. Following reconstitution, both UV and CD spectra look analogous, indicating the presence of a similarly bound [2Fe-2S] cluster. The cluster on holo G208C NFU1 was stable over the course of an hour and remained bound to the protein even in the presence of 5 mM DTT (Fig. S6).
Although G208C NFU1 was capable of being reconstituted using in vitro methods, the protein was unable to accept cluster from other physiologically relevant sources, and most likely reflects deficiencies in reconstitution expected inside the cell. Also, in contrast to native protein, the G208C variant is unable to take up the [2Fe-2S](GS)4 complex [1]. Likewise, the mutant protein is unable to receive a [2Fe-2S] cluster from typical iron-sulfur scaffold proteins such as Isa1 and IscU (Table 5), which have been shown to deliver a cluster into native NFU1 [10, 14]. Current models for Fe/S cluster biogenesis indicate that transfer from IscU is promoted with assistance from heat shock chaperone proteins [16, 37, 47, 55]. We have found that transfer from holo IscU to apo native NFU1 or G208C NFU1 does not occur in the presence of the chaperones (Fig. S4), which is similar to transfer from holo human IscU to apo ferredoxin 2 (Fig. S7) or ferredoxin 1 [56], where transfer is unaffected in the case of the former, and inhibited for the latter, in the presence of chaperones. Inhibition most likely arises from stabilization of the cluster on holo IscU [56] that slows its ability to transfer the cluster. In the case of transfer to human NFU1 in the presence of chaperones, we postulate that no transfer is observed, at least on the time scale considered, due to possible steric interaction of NFU1 with the chaperone proteins [5, 57] in conjunction with the stabilization of cluster on IscU.
Table 5.
Apparent second-order rate constants determined using CD for [2Fe-2S] cluster transfer to and from G208C NFU1. Native transfer rates were previously determined using the same CD method [1, 10, 14].
| Rate Constant for Native NFU1 (M−1 min−1) |
Rate Constant for G208C NFU1 (M−1min−1) |
|
|---|---|---|
| Human NFU1 to human Fdx1 | 4700 ± 800 [1] | 2600 ± 300 |
| Human NFU1 to human Fdx2 | 3900 ± 1200 [1] | 1200 ± 200 |
| Human NFU1 to human Grx2 | 3740 ± 77 [10] | 22400 ± 5000 |
| Human NFU1 to S. cerevisiae Grx3 | 36200 ± 7700 [14] | 14500 ± 3500 |
| S. pombe Isa1 to human NFU1 | 6700 ± 1560 [10] | No transfer |
| [2Fe-2S](GS)4 to human NFU1 | 1930 ± 210 [1] | No transfer |
| Human IscU to human NFU1 | 4750 ± 8 [14] | No transfer |
| Human IscU to human NFU1 with HSPA9/Hsc20 and MgATP | No transfer | No transfer |
| Human NFU1 to GSH to form [2Fe- 2S](GS)4 complex | 130 ± 22 [1] | 140 ± 20 |
Heat shock chaperones are highly conserved across bacteria, yeast and humans, and serve crucial roles in Fe-S cluster biogenesis, mediating cluster delivery to key targets such as glutaredoxins and succinate dehydrogenase [37, 58]. As such, they have been studied in all of those organisms. Moreover, because the process of Fe-S cluster biosynthesis is highly conserved, the co-chaperone system is believed to play similar roles across various organisms, however, minor kinetic differences have been observed when making comparisons between organisms. For example, heat shock chaperones are observed to greatly increase the rate of transfer from IscU in Azotobacter vinelandii (22-fold to Fdx and 670-fold to Grx5) [50, 59] and on a more minor scale (5–10 fold) in E. coli [60, 61], but have actually been shown to slow a transfer reaction in humans [56]. The observed differences in rate enhancement across these organisms suggest that the chaperone systems could participate in distinct roles in support of Fe/S cluster biogenesis and/or trafficking, depending on the particular organism. In this case the human chaperone system appears to block IscU-mediated cluster delivery to NFU1 (Table 5), but serves no clear role in rate enhancement in subsequent delivery to Fdx’s (Table S1).
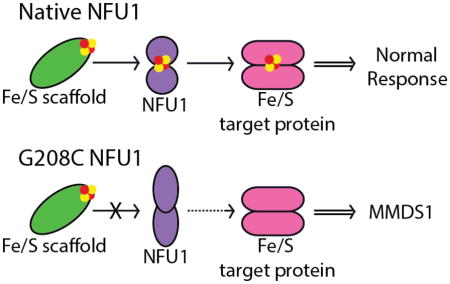
Since the mutant form of NFU1 cannot receive cluster from traditional sources, in contrast to the native protein (Table 5), this readily explains the detrimental phenotype and symptoms observed for those with the point mutation. Derivative G208C NFU1 not receive a cluster in vivo and would therefore be unable to transfer a cluster to downstream partners. The predicted change in structure (Fig. 9), suggesting that the CXXC loop in G208C NFU1 has been locked into a specific orientation, may align the cysteines in such a way that they no longer exhibit the conformational flexibility required for cluster uptake and reconstitution in the cell. Alternatively, the increased proportion of dimeric protein could sterically block the CXXC motif from cluster uptake. In either case, the protein cannot accept a pre-formed [2Fe-2S] cluster, since cluster assembly on G208C NFU1 is observed only under in vitro reconstitution conditions employing inorganic iron and sulfide. In addition to the possibility of impaired accessibility of the CXXC cluster binding motif, the replacement of a glycine with a cysteine near the cluster binding site could perturb cluster coordination, where the new cysteine now functions as a cluster ligand. This alternative is currently under investigation to determine if the CXXC motif is solely responsible for cluster coordination even with the added cysteine in the new CXCXXC sequence.
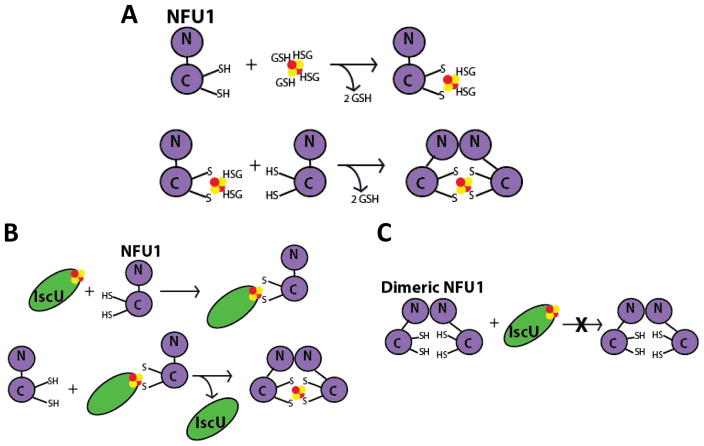
The inability of the mutant protein to receive cluster from relevant sources shows good correlation with the increased presence of dimer. In our examination of the native NFU1, cluster uptake from the [2Fe-2S](GS)4 complex was only possible when the protein was monomeric [1]. Kinetic assays for cluster uptake have revealed a model consistent with experimental data, where the cluster is taken up first by a monomer of NFU1, followed by formation of complete holo NFU1 when a second monomer is added to form the [2Fe-2S] cluster-bound dimer (Fig. 10). Cluster uptake into a preformed dimer did not occur and was not supported experimentally [1]. Since the mutant is primarily in the dimer form, the oligomeric state would prevent cluster uptake into G208C NFU1. A similar mechanism for cluster transfer from other holo proteins is most likely in place, where dimer formation is preventing G208C NFU1 from efficiently receiving cluster (Figure 10). A model, in which the monomeric form of NFU1 is the active species, is consistent with the emerging evidence for a functional NFU1-BolA complex involved in Fe/S cluster biogenesis [39, 62], due to the fact that NFU1 would need to be capable of forming cluster-bridged homo- and heterodimeric species.
Figure 10.
(A) A model for [2Fe-2S] cluster uptake by monomeric NFU1, represented by the N- and C-terminal domains, from the [2Fe-2S](GS)4 cluster complex to form an intermediate [2Fe-2S] species with two exogenous GSH ligands. A second monomeric NFU1 displaces the GSH molecules to form [2Fe-2S] dimeric NFU1. Adapted from [1]. (B) A model for [2Fe-2S] cluster uptake by monomeric NFU1, from a [2Fe-2S] cluster scaffold, such as IscU. Holo IscU interacts with monomeric NFU1 to form a transient heterodimeric complex. A second NFU1 monomeric displaces the now apo IscU to form the holo NFU1 dimer. In a case where NFU1 is a pre-formed dimer (C), such as in the G208C mutant, the dimeric NFU1 is unable to form the transient complex with holo IscU and therefore unable to receive the [2Fe-2S] cluster.
The structural change promoting dimer formation and the downstream consequences of this oligomerization could explain the dominant negative phenotype recently observed by introducing the MMDS1 mutation in yeast [39]. In utilizing yeast as a model system, Melber and colleagues introduced the MMDS1 mutation and found that co-expression of wild-type yeast Nfu1 could not rescue the effects [39]. Having a mutation that promotes protein oligomerization, thereby preventing cluster uptake and downstream transfer, readily explains this phenotype. Native human NFU1 has a natural tendency to dimerize [1] and the G208C mutation amplifies this characteristic. Therefore, it is plausible that a mixture of native and mutant Nfu1, such as in the co-expression system mentioned above, would also show high levels of dimer, in which the native protein could exist as both monomeric and dimeric states, the mutant would be primarily dimeric, and heterodimers could be formed between the native and the mutant monomeric proteins. With all of the dimers present, cluster uptake would be impaired and downstream cluster transfer perturbed, as we have shown here.
These kinetic limitations and alterations to the Fe/S biosynthesis pathway could generate the drastic consequences and the dominant negative phenotype. However, yeast is not the best model for human NFU1 concerning the MMDS1 condition. Yeast Nfu1 has been observed to behave differently from human NFU1 inasmuch as deletion of Nfu1 is not lethal [63], and negative effects are only exhibited when grown on specific media, which may be reflective of a conditional need for Nfu1 in yeast with a bypass mechanism in place to allow for survival [39]. Furthermore, introduction of the G208C equivalent mutation in yeast results in functional consequences that are not observed in patients, such as aconitase depletion [39]. Together, these differences may speak to potentially dissimilar functions across yeast and human Nfu-type proteins [39], indicating that these types of studies need to be addressed in an in vivo system that better replicates the MMDS1 disease condition in conjunction with in vitro studies, which are unique in being able to address issues at the molecular level, such as the dimerization effect.
While G208C NFU1 does not appear to have a mechanism of efficient in vivo reconstitution, the derivative did retain cluster transfer activity and was able to deliver clusters to both human ferredoxin 1 and 2 (Fig. 7), at a modest rate of approximately half of the second-order rate constant observed for the native protein (Table 5) [1]. A greater change in the transfer abilities of G208C NFU1 was noted for transfer of cluster to glutaredoxins (Fig. 8). The point mutation results in a changed preference for the glutaredoxins. Our previous work has indicated that [2Fe-2S] cluster from native NFU1 to apo Grx3 was a kinetic sink at 36000 M−1min−1, while transfer to Grx2 occurred, but at around the same level as other transfers from NFU1 (Table 5) [10, 14]. However, the mutant form of the protein now kinetically prefers cluster transfer to Grx2 by increasing the second-order rate constant seven-fold over transfer to Grx3, which decreased by three-fold. Although Grx2 is primarily present in the mitochondria [64], where Grx3 is cytosolic [65], the large change in second-order rate constants reflects a kinetically altered iron-sulfur cluster transfer pathway due to a single point mutation, which, again, could suggest why such severe phenotypes are observed, as there could be drastic downstream consequences for minor perturbations to the transfer pathway.
MMDS1 was only recently identified as a disease condition, with the first case studies emerging in 2011 [17, 21]; however, the number of patients diagnosed with MMDS1 due to the G208C NFU1 mutation has been growing since its initial disease classification. At least 22 patients have been diagnosed since 2011 [17–19, 21], but it is speculated that this is a highly underdiagnosed condition [19]. Patient case studies describing the same symptoms and disease characteristics, which were attributed to defects in the iron-sulfur cluster biosynthetic pathway, were first published in 2001; [66] but, in the absence of information provided by the human genome project, the exact cause of the disease remained unclear, suggesting the first subset of under-diagnosis. Furthermore, the disease symptoms resemble the majority of mitochondrial or energetic disorders and the patients often died of respiratory or mitochondrial failure, and so the exact cause of death had not been investigated in depth and the number of cases of MMDS1 is likely to be much higher [19]. Since the disease has only been examined recently, it is also possible that the majority of downstream target proteins for NFU1 that result in the severity of the disease condition are unknown.
Our investigation into the G208C NFU1 mutation has elucidated key facts regarding the actual structural and functional impairments caused by the mutant NFU1 in MMDS1. While several questions remain to be answered, such as the exact link between NFU1 and lipoic acid synthase or succinate dehydrogenase [21, 42], we postulate that the mutation alters the structure of the NFU1 protein such that the protein exists in a primarily dimeric state, which prevents it from being able to be reconstituted effectively with an iron-sulfur cluster in vivo. The kinetics of [2Fe-2S] cluster transfer have been perturbed due to the mutation, but the connection between the mutant NFU1 and how it interacts with other potential target proteins remains under investigation.
Materials and Methods
Materials
PD10 desalting columns were purchased from GE Healthcare. Ferric chloride, sodium sulfide, DTT (Dithiothreitol), TCEP (Tris (2-carboxy-ethyl) phosphine) and L-cysteine were purchased from Fisher.
Mutagenesis
Stratagene QuikChange Mutagenesis was employed to introduce the p.Gly208Cys point mutation in full length human NFU1. PCR reactions contained 50 ng of native NFU1 DNA in the pET28b+plasmid, 2 units of Phusion DNA polymerase (New England Biolabs), 10x Phusion buffer (New England Biolabs), 125 ng of each primer (Integrated DNA Technologies), 0.2 mM dNTPs, and 3% DMSO. The forward primer sequence was 5’-TACAGCTGAAACTCCAGTGTTCTTGTACCAGCTGC-3’ and the reverse primer sequence was 5’-GCAGCTGGTACAAGAACACTGGAGTTTCAGCTGTA-3’ with the substituted nucleotide in bold. The thermocycle was identical to that described in the QuikChange manual: the sample was melted by heating to 95 C for 1 min, followed by 16 cycles of 95 C for 30 sec, 55 C for 1 min and 72 C for 6.5 min (Stratagene). Following amplification, samples were incubated with 7.5 units of DpnI at 37 C for 4 hours. Subsequently, CaCl2-competent BL21 (DE3) cells were transformed via heat shock with the mutant constructs. Mutagenesis results were confirmed by nucleotide sequencing from GENEWIZ.
Protein Expression and Purification
Full length human G208C NFU1 in a pET28(b+) vector in E. coli strain BL21(DE3) host cells was grown overnight at 37 °C in 10 mL of Luria–Bertani (LB) broth media containing kanamycin (50 μM) [29]. The overnight cultures were diluted 1:1000 in LB media containing 50 μM kanamycin until the OD600 reached 0.6. At this point, protein expression was induced with 0.5 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG), and cultures were incubated overnight at 37 °C. Cell pellets were collected by centrifugation at 4,330g for 15 min at 4 °C, and resuspended in 30 mL of 50 mM HEPES, 100 mM NaCl, and pH 7.5. Resuspended pellets were incubated with 30 mg lysozyme and 0.6 mg DNase I for 30 min at 4 °C, and then lysed by use of a dismembranator. Cell lysate was centrifuged at 28,982g for 50 min at 4 °C, and the supernatant was applied to a TALON column. Protein was eluted with a buffer containing 50 mM HEPES, 100 mM NaCl, 150 mM imidazole, pH 7.5 and concentrated by Amicon ultrafiltration over a 10 kDa membrane.
Purification of Hs IscU and Thermatoga maritima (Tm) Nifs was performed as previously reported [67–69]. The expression vector for human ferredoxin-1 (Hs Fdx1) was kindly provided by J. Markley and protein was expressed and purified according to literature procedures [70]. Purification for human ferredoxin-2 (Hs Fdx2) was performed as previously reported [71]. Briefly, Hs Fdx1 was purified by use of DE-52 anion exchange column chromatography followed by FPLC purification with a size exclusion Superose-12 column (HR 16/50, Pharmacia) run at 0.2 mL/min with 50 mM HEPES, 100 mM NaCl, pH 7.5 at 40 C. All colored fractions were collected and combined. Purification for human ferredoxin-2 (Hs Fdx2) was performed as previously reported [71], by use of a TALON column. Protein was eluted with a buffer containing 50 mM HEPES, 100 mM NaCl, 150 mM imidazole, pH 7.5 and concentrated by amicon ultrafiltration over a 10 kDa membrane. The ferredoxins purified as holo proteins and were then subsequently converted to apo forms by treatment with 100 mM EDTA, 5mM DTT and 8M urea in a buffered solution, pH 7.5. A construct of human Grx2 (comprising residues 56–161), with a tobacco etch virus cleavable N-terminal His6 tag in expression vector pNic-Bsa4, was kindly provided by Drs. Kavanagh, Muller-Knapp and Oppermann and protein was expressed and purified as previously reported [72]. Schizosaccharomyces pombe Isa1 protein was expressed and purified as previously reported [73]. Yeast Grx3 (Δ1–35) in pET28b(+) E. coli BL21 (DE3) was purified as noted [44]. Human HSPA9 with a N-terminal 6X-His tag was purified as previously described [74], as was human Hsc20 [75].
In all cases, protein purity was assayed by use of a 12 % SDS-PAGE gel that was visualized with Coomassie Blue staining. Imidazole was removed by dialysis at 4 °C against a buffer containing 50 mM HEPES, 100 mM NaCl, pH 7.5, and protein concentration was determined by use of the Bradford assay and confirmed by ε280.
Differential Scanning Calorimetry (DSC)
DSC samples (0.1 mM – 0.3 mM) were dialyzed against 50 mM HEPES, 100 mM NaCl, pH 7.5 at 4 C with Spectra/Por dialysis membrane (MWCO 10 000; Spectrum Laboratories, Inc.). The resulting dialysis buffers were used as reference cell buffers for precision and repeatability. Reference buffers and protein samples were thoroughly degassed using a MicroCal Thermovac2 (Malvern Instruments, Inc.) prior to analysis on a MicroCal VP-DSC (Malvern Instruments, Inc.). The data were obtained using a differential mode at a rate of 1.0 C per minute from 10 C to 100 C and analyzed using Origin software (Origin Labs) and fit to a non-two state model [4].
Variable Temperature Circular Dichroism characterization
Circular dichroism (CD) samples (10 μM) were dialyzed in phosphate buffer (40 mM phosphate, pH 7.4) with Spectra/Por dialysis membrane (MWCO 10 000; Spectrum Laboratories, Inc.). The resulting dialysis buffers were used as reference cell solvents for precision and repeatability. Prior to analyses, all sample and reference solutions were rigorously degassed with a Microcal Thermovac2 device (Malvern Instruments, Inc.). All CD data acquisitions were obtained on a JASCO J-815 CD spectrometer (JASCO) equipped with quartz cells with a 0.1 cm path length. Variable temperature studies were performed at a rate of 0.4 ° C min−1 from 20 to 95 °C. All data were processed with Origin 7 (Origin Laboratories). VTCD data were fit to equation 3 [4], where R is the ideal gas constant in calories per mole, Tm is the melting temperature in Kelvin, ΔHV is the van’t Hoff enthalpy, and Cp is the heat capacity. F and U represent the mean residue ellipticities (θmr) of the folded and unfolded protein, respectively.
| (3) |
Secondary Structure Prediction from Circular Dichroism
Protein samples at 10 μM in 40 mM phosphate, pH 7.4 were placed in a 1 mm quartz cuvette and the signal monitored from 300–165 nm. The buffer baseline signal was subtracted from the data sets before conversion to Δε. The resulting CD spectra were processed using the analysis program CDSSTR [2] using reference set 7 [76] found on the online server Dichroweb [8, 9].
Reconstitution of apo proteins
NifS-mediated in vitro reconstitution of G208C NFU1 was completed as previously described [1, 77] . Briefly, ferric chloride and L-cysteine were added to an anaerobic mixture of approximately 200 μM purified NFU1, 2 μM T. maritima (Tm) NifS, and 5 mM DTT to final concentrations of 1.6 mM FeCl3 and 3.2 mM L-cysteine. Alternatively, sodium sulfide and ferric chloride were added to an anaerobic mixture of approximately 200 μM purified NFU1, 2.1 mM urea, and 5 mM DTT to final concentrations of 1 mM FeCl3 and 1 mM Na2S [44]. The final solution was incubated for 1 h with stirring at room temperature, before separation of excess iron and sulfide through a PD-10 column that was equilibrated with an argon-purged solution of 50 mM HEPES, 100 mM NaCl, pH 7.5. Reconstituted protein was eluted with 3.5 mL of the equilibration buffer. The protein concentration was determined via the Bradford assay and the reconstitution of protein was confirmed by absorbance at 330 nm and 420 nm on a Cary WinUV spectrophotometer.
For IscU and Isa1 [78], 200 μM apo protein was incubated with 50 mM DTT, argon purged for 30 min, and then made up to 1 mM in Fe2+ and 1 mM in S2-. After incubation for 1 h, the reaction mixture was concentrated and passed through a PD10 desalting column to remove excess of Fe2+ and S2−, and the UV-vis spectrum was obtained for both apo and holo IscU and Isa1.
Reconstitution Analysis by Circular Dichroism
CD scans of apo and holo proteins were recorded on a JASCO J-815 CD spectrometer in a quartz 1 cm anaerobic cuvette. CD scans from 300 nm to 600 nm were collected to analyze signature cluster-bound protein peaks at a scan rate of 200 nm/min at 25 °C. Data were processed using JASCO Spectramanager II Analysis software and were represented in Origin 7.0.
Oligomerization State Determination by analytical ultracentrifugation (AUC)
Apo G208C NFU1 at 50 μM protein (OD280 = 1.0), in the presence or absence of 1 mM TCEP, was loaded into the ultracentrifugation chambers and sealed, using 50 mM HEPES, 100 mM NaCl, pH 7.5 as a reference, with the addition of 1 mM TCEP where needed. Reconstituted holo G208C NFU1 at 269 μM (OD420 = 1.5) was loaded in the same manner with the same reference buffer. Samples were centrifuged at 45,000 rpm for 6 hours to reach complete sedimentation. The sedimentation profiles were fit using SEDFIT to the Lamm equation [6, 7].
[Fe2S2](GS)4] Synthesis
The cluster used was synthesized as previously reported [43]. Briefly, ferric chloride (20 mM) and sodium sulfide (20 mM) were added to 10 mL 40 mM glutathione solution, pH 8.6. A volume (40 mL) of ethanol was added to the mixture and mixed by vortexing. The precipitate was collected by centrifugation at 13,000 rpm for 10 min, washed twice with ethanol and dried under vacuum.
Iron Quantitation [78, 79]
A solution of [Fe2S2](GS)4 (0.05 mM, 200 μL) in H2O or holo protein was acidified by concentrated HCl (60 μL) and heated to 100 oC for 15 min. The resulting suspension was centrifuged at 14,000 rpm for 2 min and the supernatant (100 μL) was diluted with Tris-HCl (0.5 M, 1.3 mL, pH 8.5). Solutions of sodium ascorbate (0.1 mL, 5%) and bathophenanthroline-disulfonate (0.4 mL, 0.1%) were sequentially added to the neutralized reaction solution with mixing between each addition. The solution was incubated at 25 °C for 1 h and iron was quantitated by measuring the absorbance at 535 nm on a UV-Vis spectrophotometer and calculated from a calibration curve made with 0.01–0.2 mM FeCl3 standard solutions (Fig. S1).
Iron-sulfur Cluster Uptake Monitored by CD
The ability of NFU1 to take up an iron-sulfur cluster from the [2Fe-2S](GS)4 complex was examined by circular dichroism (CD). CD scans were recorded on a JASCO J-815 CD spectrometer in a 1 cm anaerobic quartz cuvette from 600–300 nm at a scan rate of 200 nm/min at 25°C, with a 2 min interval between each accumulation. NFU1 (50 μM) in 50 mM HEPES, 100 mM NaCl pH 7.5, was thoroughly degassed in the presence of 5 mM DTT and transferred to the anaerobic cuvette. Solid [2Fe-2S](GS)4 was resuspended in degassed 50 mM HEPES, 100 mM NaCl pH 7.5 and added to the argon-purged anaerobic cuvette via a gas-tight syringe to a final concentration of 400 μM to initiate the reaction. Data were processed using JASCO Spectramanager II Analysis software and analyzed in Origin 7.0. The deconvolution function from Spectramanager II analysis software was used for analysis of bands in the spectra that contained overlapping Lorentzian curves having the same full width at half maximum value that accurately distinguishes the peak positions for each band.
Kinetics of Fe-S cluster extraction from holo NFU1 by glutathione
Glutathione has been previously shown to extract the iron-sulfur cluster from various holo proteins to form the [2Fe-2S](GS)4 complex by monitoring the change in the charge transfer bands at 330 nm and 420 nm by UV-Vis spectrophotometry [43, 44] . As was done for native holo NFU1 [1], degassed, reconstituted holo G208C NFU1 in 50 mM HEPES, 100 mM NaCl, pH 7.5 was incubated with a 4- to 10-fold excess of GSH in an anaerobic cuvette and the absorbance at 420 nm on a Cary Win UV spectrophotometer was monitored every 2 min over the course of 1 h. The change in absorbance at 420 nm was plotted against time and fit to an exponential decay to obtain the kobs. A control reaction for holo G208C NFU1 in the absence of excess GSH was carried out under the same conditions to account for inherent cluster instability. Cluster extraction by GSH to form the [2Fe-2S](GS)4 complex was confirmed using ESI mass spectrometry on a Bruker Micro-TOF (ESI) spectrometer and data was analyzed by use of DataAnalysis software (Bruker) [43, 46].
Kinetic cluster transfer experiments
Kinetic cluster transfer experiments were designed based on the cluster transfer experiments by Johnson and coworkers [49, 59], and refined by our own group [1, 44]. Reactions were performed on a JASCO J-815 CD spectrophotometer in a 1 cm anaerobic quartz cuvette from 600–300 nm at a scan rate of 200 nm/min at 25°C, with a 2 min interval between each accumulation. Reactions that reached completion within the first 10 min were analyzed over a 10 nm wavelength scale based on the peak of interest with 10 second intervals between accumulations. Spectra were processed using JASCO Spectramanager II Analysis software and were represented in Origin 7.0.
Reactions in 50 mM HEPES, 100 mM NaCl, pH 7.5 were prepared by degassing a mixture of 40 μM apo protein in 5 mM DTT, and transferred to an anaerobic cuvette via a gas tight syringe. For reactions with the co-chaperone system, 22 μM HSPA9 and 22 μM Hsc20 were included. MgCl2 (40 mM) and ATP (2 mM) were degassed separately and added immediately before addition of holo IscU [50, 56, 59–61]. Degassed holo protein at 40 μM was added to the cuvette to initiate the reaction. The concentration of [2Fe-2S] in the reaction for each holo protein was determined via standard iron quantitation methods. Kinetics of cluster transfer was analyzed by converting the change in CD signal to the percentage of cluster transferred fit using DynaFit [3] to determine the second-order rate constants for the various reactions by best-fit simulation to second-order kinetics.
Supplementary Material
Highlights.
A point mutation (G208C) on human NFU1 results in a disease phenotype: MMDS1
The G208C mutation introduces a minor structural change that promotes dimerization
The dimerization impairs iron-sulfur cluster transfer capabilities
Mutant NFU1 cannot accept cluster, preventing function and downstream delivery
Acknowledgments
We thank Dr. Marina Bakhtina for her assistance with analytical ultracentrifugation experiments and Dr. Tom Magliery for providing a thermocycler. This work was supported by a grant from the National Institutes of Health [AI072443]. Christine Wachnowsky was supported by an NIH Chemistry/Biology Interface training grant (T32 GM095450), as well an Ohio State University Presidential Fellowship. Nathaniel Wesley was supported by scholarships from the Ohio State Undergraduate Research Office and the Honors College of Arts and Sciences.
Abbreviations
- MMDS1
Multiple Mitochondrial Dysfunctions Syndrome 1
- GSH
Glutathione
- DSC
Differential Scanning Calorimetry
- VTCD
Variable Temperature Circular Dichroism
- ESI-MS
Electrospray Ionization Mass Spectrometry
- CD
Circular Dichroism
- AUC
Analytical Ultracentrifugation
- IscU
Iron-sulfur cluster scaffold protein
- Isa1
Iron-sulfur cluster assembly protein
- Fdx
Ferredoxin
- Grx
Glutaredoxin
- LIAS
Lipoate synthase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wachnowsky C, Fidai I, Cowan JA. Iron-sulfur cluster exchange reactions mediated by the human Nfu protein. J Biol Inorg Chem. 2016;21:825–36. doi: 10.1007/s00775-016-1381-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sreerama N, Woody RW. Estimation of protein secondary structure from CD spectra: Comparison of CONTIN, SELCON and CDSSTR methods with an expanded reference set. Anal Chem. 2000;287:252–60. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 3.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237:260–73. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Ding S, Cowan JA. Thermodynamic and structural analysis of human NFU conformational chemistry. Biochemistry. 2013;52:4904–13. doi: 10.1021/bi400320s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai K, Liu G, Frederick Ronnie O, Xiao R, Montelione Gaetano T, Markley John L. Structural/functional properties of human NFU1, an intermediate [4Fe-4S] carrier in human mitochondrial iron-sulfur cluster biogenesis. Structure. 2016 doi: 10.1016/j.str.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamm O. Die Differentialgleichung der Ultrazentrifugierung. Ark Mat Astr Fys. 1929:1–4. [Google Scholar]
- 8.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W73. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 10.Fidai I, Wachnowsky C, Cowan JA. Mapping cellular Fe–S cluster uptake and exchange reactions – divergent pathways for iron – sulfur cluster delivery to human ferredoxins. Metallomics. 2016;8:1283–93. doi: 10.1039/c6mt00193a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasserre JP, Dautant A, Aiyar RS, Kucharczyk R, Glatigny A, Tribouillard-Tanvier D, et al. Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis Model Mech. 2015;8:509–26. doi: 10.1242/dmm.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G. Mitochondrial Control of Cellular Life, Stress, and Death. Circul Res. 2012;111:1198–207. doi: 10.1161/CIRCRESAHA.112.268946. [DOI] [PubMed] [Google Scholar]
- 13.Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, et al. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta. 2012;1823:1491–508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Wachnowsky C, Fidai I, Cowan JA. Cytosolic iron-sulfur cluster transfer: a proposed kinetic pathway for the reconstitution of glutaredoxin 3. FEBS Lett. 2016;590:4531–40. doi: 10.1002/1873-3468.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tort F, Ferrer-Cortes X, Ribes A. Differential diagnosis of lipoic acid synthesis defects. J Inherit Metab Dis. 2016;39:781–93. doi: 10.1007/s10545-016-9975-4. [DOI] [PubMed] [Google Scholar]
- 16.Stehling O, Wilbrecht C, Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. doi: 10.1016/j.biochi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Cameron JM, Janer A, Levandovskiy V, Mackay N, Rouault TA, Tong WH, et al. Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am J Hum Genet. 2011;89:486–95. doi: 10.1016/j.ajhg.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Invernizzi F, Ardissone A, Lamantea E, Garavaglia B, Zeviani M, Farina L, et al. Cavitating leukoencephalopathy with multiple mitochondrial dysfunction syndrome and NFU1 mutations. Front Genet. 2014:5. doi: 10.3389/fgene.2014.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nizon M, Boutron A, Boddaert N, Slama A, Delpech H, Sardet C, et al. Leukoencephalopathy with cysts and hyperglycinemia may result from NFU1 deficiency. Mitochondrion. 2014;15:59–64. doi: 10.1016/j.mito.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Ahting U, Mayr JA, Vanlander AV, Hardy SA, Santra S, Makowski C, et al. Clinical, biochemical, and genetic spectrum of seven patients with NFU1 deficiency. Front Genet. 2015:06. doi: 10.3389/fgene.2015.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarro-Sastre A, Tort F, Stehling O, Uzarska Marta A, Arranz José A, del Toro M, et al. A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. Am J Hum Genet. 2011;89:656–67. doi: 10.1016/j.ajhg.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ajit Bolar N, Vanlander AV, Wilbrecht C, Van der Aa N, Smet J, De Paepe B, et al. Mutation of the iron-sulfur cluster assembly gene IBA57 causes severe myopathy and encephalopathy. Hum Mol Genet. 2013;22:2590–602. doi: 10.1093/hmg/ddt107. [DOI] [PubMed] [Google Scholar]
- 23.Debray F-G, Stümpfig C, Vanlander AV, Dideberg V, Josse C, Caberg J-H, et al. Mutation of the iron-sulfur cluster assembly gene IBA57 causes fatal infantile leukodystrophy. J Inherit Metab Dis. 2015;38:1147–53. doi: 10.1007/s10545-015-9857-1. [DOI] [PubMed] [Google Scholar]
- 24.Al-Hassnan ZN, Al-Dosary M, Alfadhel M, Faqeih EA, Alsagob M, Kenana R, et al. ISCA2 mutation causes infantile neurodegenerative mitochondrial disorder. J Med Genet. 2015;52:186–94. doi: 10.1136/jmedgenet-2014-102592. [DOI] [PubMed] [Google Scholar]
- 25.Baker PR, Friederich MW, Swanson MA, Shaikh T, Bhattacharya K, Scharer GH, et al. Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5. Brain. 2013;137:366–79. doi: 10.1093/brain/awt328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lossos A, Stümpfig C, Stevanin G, Gaussen M, Zimmerman B-E, Mundwiller E, et al. Fe/S protein assembly gene IBA57 mutation causes hereditary spastic paraplegia. Neurology. 2015:84. doi: 10.1212/WNL.0000000000001270. [DOI] [PubMed] [Google Scholar]
- 27.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–81. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 28.Lill R, Muehlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Cowan JA. Iron sulfur cluster biosynthesis. Human NFU mediates sulfide delivery to ISU in the final step of [2Fe-2S] cluster assembly. Chem Commun. 2007:3192–4. doi: 10.1039/b704928e. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Cowan JA. Iron sulfur cluster biosynthesis: characterization of a molten globule domain in human NFU. Biochemistry. 2009;48:7512–8. doi: 10.1021/bi9002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong WH, Jameson GNL, Huynh BH, Rouault TA. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc Natl Acad Sci USA. 2003;100:9762–7. doi: 10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angelini S, Gerez C, Choudens SOd, Sanakis Y, Fontecave M, Barras F, et al. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J Biol Chem. 2008;283:14084–91. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- 33.Gao H, Subramanian S, Couturier J, Naik SG, Kim S-K, Leustek T, et al. Arabidopsis thaliana Nfu2 accommodates [2Fe–2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins. Biochemistry. 2013;52:6633–45. doi: 10.1021/bi4007622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishio K, Nakai M. Transfer of iron-sulfur cluster from NifU to apoferredoxin. J Biol Chem. 2000;275:22615–8. doi: 10.1074/jbc.C000279200. [DOI] [PubMed] [Google Scholar]
- 35.Py B, Gerez C, Angelini S, Planel R, Vinella D, Loiseau L, et al. Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Mol Microbiol. 2012;86:155–71. doi: 10.1111/j.1365-2958.2012.08181.x. [DOI] [PubMed] [Google Scholar]
- 36.Yabe T. The Arabidopsis chloroplastic NifU-like protein CNFU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell. 2004;16:993–1007. doi: 10.1105/tpc.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maio N, Rouault TA. Mammalian Fe–S proteins: definition of a consensus motif recognized by the co-chaperone HSC20. Metallomics. 2016;8:1032–46. doi: 10.1039/c6mt00167j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganesh S. The Lafora disease gene product laforin interacts with HIRIP5, a phylogenetically conserved protein containing a NifU-like domain. Hum Mol Gen. 2003;12:2359–68. doi: 10.1093/hmg/ddg253. [DOI] [PubMed] [Google Scholar]
- 39.Melber A, Na U, Vashisht A, Weiler BD, Lill R, Wohlschlegel JA, et al. Role of Nfu1 and Bol3 in Iiron-sulfur cluster transfer to mitochondrial clients. eLife. 2016;5:e15991. doi: 10.7554/eLife.15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Touraine B, Boutin J-P, Marion-Poll A, Briat J-F, Peltier G, Lobréaux S. Nfu2: a scaffold protein required for [4Fe-4S] and ferredoxin iron-sulphur cluster assembly in Arabidopsis chloroplasts. Plant J. 2004;40:101–11. doi: 10.1111/j.1365-313X.2004.02189.x. [DOI] [PubMed] [Google Scholar]
- 41.Bandyopadhyay S, Naik SG, O'Carroll IP, Huynh BH, Dean DR, Johnson MK, et al. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J Biol Chem. 2008;283:14092–9. doi: 10.1074/jbc.M709161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrer-Cortès X, Font A, Bujan N, Navarro-Sastre A, Matalonga L, Arranz JA, et al. Protein expression profiles in patients carrying NFU1 mutations. Contribution to the pathophysiology of the disease. J Inherit Metab Dis. 2012;36:841–7. doi: 10.1007/s10545-012-9565-z. [DOI] [PubMed] [Google Scholar]
- 43.Qi W, Li J, Chain CY, Pasquevich GA, Pasquevich AF, Cowan JA. Glutathione complexed Fe-S centers. J Am Chem Soc. 2012;134:10745–8. doi: 10.1021/ja302186j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fidai I, Wachnowsky C, Cowan JA. Glutathione-complexed [2Fe-2S] clusters function in Fe-S cluster storage and trafficking. J Biol Inorg Chem. 2016;21:887–901. doi: 10.1007/s00775-016-1387-2. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Cowan JA. Glutathione-coordinated [2Fe–2S] cluster: a viable physiological substrate for mitochondrial ABCB7 transport. Chem Commun (Camb) 2015;51:2253–5. doi: 10.1039/c4cc09175b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi W, Li J, Chain CY, Pasquevich GA, Pasquevich AF, Cowan JA. Glutathione-complexed iron–sulfur clusters. Reaction intermediates and evidence for a template effect promoting assembly and stability. Chem Commun. 2013;49:6313. doi: 10.1039/c3cc43620a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JH, Bothe JR, Alderson TR, Markley JL. Tangled web of interactions among proteins involved in iron–sulfur cluster assembly as unraveled by NMR, SAXS, chemical crosslinking, and functional studies. Biochim Biophys Acta. 2015;1853:1416–28. doi: 10.1016/j.bbamcr.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephens PJ, Thomson AJ, Dunn JBR, Keiderling TA, Rawlings J, Rao KK, et al. Circular dichroism and magnetic circular dichroism of iron-sulfur proteins. Biochemistry. 1978;22:4770–8. doi: 10.1021/bi00615a026. [DOI] [PubMed] [Google Scholar]
- 49.Mapolelo DT, Zhang B, Randeniya S, Albetel A-N, Li H, Couturier J, et al. Monothiol glutaredoxins and A-type proteins: partners in Fe–S cluster trafficking. Dalton Trans. 2013;42:3107. doi: 10.1039/c2dt32263c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandramouli K, Johnson MK. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry. 2006;45:11087–95. doi: 10.1021/bi061237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broom HR, Vassall KA, Rumfeldt JAO, Doyle CM, Tong MS, Bonner JM, et al. Combined isothermal titration and differential scanning calorimetry define three-state thermodynamics of fALS-associated mutant apo SOD1 dimers and an increased population of folded monomer. Biochemistry. 2016;55:519–33. doi: 10.1021/acs.biochem.5b01187. [DOI] [PubMed] [Google Scholar]
- 52.Doyle CM, Rumfeldt JA, Broom HR, Broom A, Stathopulos PB, Vassall KA, et al. Energetics of oligomeric protein folding and association. Arch Biochem Biophys. 2013;531:44–64. doi: 10.1016/j.abb.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 53.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and protein. Biophys J. 1998;75:422–7. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maio N, Rouault TA. Iron–sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim Biophys Acta. 2015;1853:1493–512. doi: 10.1016/j.bbamcr.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu S-P, Mansy SS, Cowan JA. Iron-sulfur cluster biosynthesis. Molecular chaperone DnaK promotes IscU-bound [2Fe-2S] cluster stability and inhibits cluster transfer activity. Biochemistry. 2005;44:4284–93. doi: 10.1021/bi0483007. [DOI] [PubMed] [Google Scholar]
- 57.Shan Y, Cortopassi G. Mitochondrial Hspa9/Mortalin regulates erythroid differentiation via iron-sulfur cluster assembly. Mitochondrion. 2016;26:94–103. doi: 10.1016/j.mito.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 58.Maio N, Singh A, Uhrigshardt H, Saxena N, Tong W-H, Rouault Tracey A. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014;19:445–57. doi: 10.1016/j.cmet.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shakamuri P, Zhang B, Johnson MK. Monothiol glutaredoxins function in storing and transporting [2Fe–2S] clusters assembled on IscU scaffold proteins. J Am Chem Soc. 2012;134:15213–6. doi: 10.1021/ja306061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones. Biochemistry. 2008;47:12795–801. doi: 10.1021/bi801565j. [DOI] [PubMed] [Google Scholar]
- 61.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Facilitated transfer of IscU–[2Fe2S] clusters by chaperone-mediated ligand exchange. Biochemistry. 2011;50:9641–50. doi: 10.1021/bi201123z. [DOI] [PubMed] [Google Scholar]
- 62.Uzarska MA, Nasta V, Weiler BD, Spantgar F, Ciofi-Baffoni S, Saviello MR, et al. Mitochondrial Bol1 and Bol3 function as assembly factors for specific iron-sulfur proteins. eLife. 2016;5:e15991. doi: 10.7554/eLife.16673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muhlenhoff U, Richhardt N, Gerber J, Lill R. Characterization of iron-sulfur protein assembly in isolated mitochondria. A requirement for ATP, NADH, and reduced iron. J Biol Chem. 2002;277:29810–6. doi: 10.1074/jbc.M204675200. [DOI] [PubMed] [Google Scholar]
- 64.Lundberg M, Johansson C, Chandra J, Enoksson M, Jacobsson G, Ljung J, et al. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J Biol Chem. 2001;276:26269–75. doi: 10.1074/jbc.M011605200. [DOI] [PubMed] [Google Scholar]
- 65.Chung WH, Kim KD, Roe JH. Localization and function of three monothiol glutaredoxins in Schizosaccharomyces pombe. Biochem Biophys Res Commun. 2005;330:604–10. doi: 10.1016/j.bbrc.2005.02.183. [DOI] [PubMed] [Google Scholar]
- 66.Seyda A, Newbold RF, Hudson TJ, Verner A, MacKay N, Winter S, et al. A Novel Syndrome Affecting Multiple Mitochondrial Functions, Located by Microcell-Mediated Transfer to Chromosome 2p14–2p13. Am J Hum Genet. 2001;68:386–96. doi: 10.1086/318196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foster MW, Mansy SS, Hwang J, Penner-Hahn JE, Surerus KK, Cowan JA. A mutant human IscU protein contains a stable [2Fe-2S]2+ center of possible functional significance. J Am Chem Soc. 2000;122:6805–6. [Google Scholar]
- 68.Mansy SS, Xiong Y, Hemann C, Hille R, Sundaralingam M, Cowan JA. Crystal structure and stability studies of C77S HiPIP: a serine ligated [4Fe-4S] cluster. Biochemistry. 2002;41:1195–201. doi: 10.1021/bi011811y. [DOI] [PubMed] [Google Scholar]
- 69.Nuth M, Yoon T, Cowan JA. Iron-sulfur cluster biosynthesis: characterization of iron nucleation sites for assembly of the [2Fe-2S]2+ cluster core in IscU proteins. J Am Chem Soc. 2002;124:8774–5. doi: 10.1021/ja0264596. [DOI] [PubMed] [Google Scholar]
- 70.Xia B, Cheng H, Bandarian V, Reed GH, Markley JL. Human ferredoxin: overproduction in Escherichia coli, reconstitution in vitro, and spectroscopic studies of iron-sulfur cluster ligand cysteine-to-serine mutants. Biochemistry. 1996;35:9488–95. doi: 10.1021/bi960467f. [DOI] [PubMed] [Google Scholar]
- 71.Qi W, Li J, Cowan JA. Human ferredoxin-2 displays a unique conformational change. Dalton Trans. 2013;42:3088–91. doi: 10.1039/c2dt32018e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qi W, Cowan JA. Mechanism of glutaredoxin-ISU [2Fe-2S] cluster exchange. Chem Commun. 2011;47:4989–91. doi: 10.1039/c0cc05079b. [DOI] [PubMed] [Google Scholar]
- 73.Wu G, Mansy SS, Hemann C, Hille R, Surerus KK, Cowan JA. Iron-sulfur cluster biosynthesis: characterization of Schizosaccharomyces pombe Isa1. J Biol Inorg Chem. 2002;7:526–32. doi: 10.1007/s00775-001-0330-2. [DOI] [PubMed] [Google Scholar]
- 74.Luo W-I, Dizin E, Yoon T, Cowan JA. Kinetic and structural characterization of human mortalin. Protein Expr Purif. 2010;72:75–81. doi: 10.1016/j.pep.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoon T. Functional and structural studies of human frataxin: An iron chaperone protein for mitochondrial iron-sulfur cluster and heme biosynthesis. Columbus: The Ohio State University; 2005. [Google Scholar]
- 76.Janes RW. Modern Techniques in Circular Dichroism and Synchrotron Radiation Circular Dichroism Spectroscopy. IOS Press; 2008. Reference Datasets Circular Dichroism and Synchrotron Radiation Circular Dichroism Spectroscopy of Proteins. [Google Scholar]
- 77.Krebs C, Agar JN, Smith AD, Frazzon J, Dean DR, Huynh BH, et al. IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry. 2001;40:14069–80. doi: 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- 78.Wu S-p, Wu G, Surerus KK, Cowan JA. Iron-sulfur cluster biosynthesis. Kinetic analysis of [2Fe-2S] cluster transfer from holo ISU to apo Fd: role of redox chemistry and a conserved aspartate. Biochemistry. 2002;41:8876–85. doi: 10.1021/bi0256781. [DOI] [PubMed] [Google Scholar]
- 79.Moulis J-M, Meyer J. Characterization of the selenium-substituted 2[4Fe-4Se] ferredoxin from Clostridium pasteurianum. Biochemistry. 1982;21:4762–71. doi: 10.1021/bi00262a037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.