Abstract
The ATPase p97 is a ubiquitin targeted segregase that uses the energy of ATP binding and hydrolysis to extract ubiquitylated substrates from biological membranes, from other proteins, or from protein complexes to carry out myriad tasks in eukaryotes. Increased p97 activity has been linked to a poor prognosis in cancer patients, making p97 an anti-neoplastic target. In the present study, we show that dehydrocurvularin (DHC) and its chlorinated variants are covalent inhibitors of p97, interfering with its ATPase acitivity. Interestingly, cellular studies revealed both DHC and its monochloro analogue interfere with both the proteasome and p97, whereas its dichloro analogue showed p97 specificity.
Graphical abstract
The AAA+ (ATPase associated with diverse cellular activities) chaperone p97 is a homohexameric protein that utilizes the energy derived from ATP binding and hydrolysis to structurally remodel target substrates, often by segregating a ubiquitylated protein from another biomolecule such as another protein or a membrane.1 For this reason, p97 has been dubbed a segregase.2 In the functional state, p97 is comprised of six subunits arranged in a ring. Each subunit contains three domains: an N-domain that binds to a collection of cofactors to assist with p97’s biological functions; a D1 domain that is necessary and sufficient to form the functional hexamer; and a D2 domain that is quite dynamic and has been proposed to generate the force needed to carry out p97’s machine function.3
p97 is an essential chaperone involved in diverse biological processes that include ubiquitin proteasome system (UPS) mediated degradation, endoplasmic reticulum associated degradation (ERAD), cell-cycle progression, transcription factor regulation, and autophagy.4–6 These diverse p97 actions implicate it in a variety of pathological states including protein misfolding disorders and cancer.4 In addition, clinical studies have shown elevated p97 levels to correlate with a poor clinical outcome. Consequently, there is much interest in developing strategies aimed at targeting p97.7–8 In fact, a compound targeting p97 from Cleave Biosciences has recently entered clinical trials.9
In an ongoing effort to discover molecules that modulate p97 function for potential therapeutic leads or as chemical biological agents, we evaluated a small collection of fungal andplant derived extracts (1760) and purified natural products (88). Some of the extracts and purified products were known to have biological activity, but this was not a prerequisite of screening, as we were using a biochemically targeted procedure. To do so, we have adapted a simple colorimetric ATPase assay. This assay measures ATP hydrolysis by quantifying liberated inorganic phosphate after forming a phosphomolybdate complex, which reacts with malachite green.10 We then applied this assay in both 96- and 384-well plate format. Initial screening was carried out at 10 μg/mL for extracts and 20 μM for purified compounds in a 100 μL reaction containing 50 nM p97 and 100 μM ATP. These concentrations for enzyme and substrate were chosen because they gave a Z-factor > 0.8 in both 96-well and 384-well format using DMSO and EDTA as a negative and positive control, respectively.11 These controls were also used in our screening assays. One of the natural products that showed p97 inhibitory activity was 10(11)-dehydrocurvularin (DHC) (2).12 This prompted us to evaluate its analogues 1 and 3–4 (Fig. 1, S1, and S2).13–16
Fig. 1.
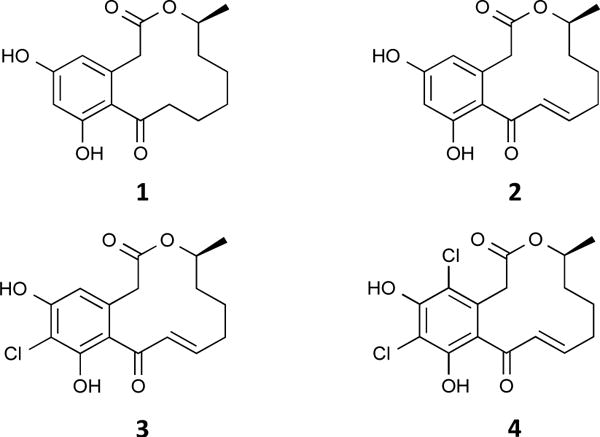
Structures of curvularin (1) and its analogues 10(11)-dehydrocurvularin (2), 6-chloro-10(11)-dehydrocurvularin (3), and 4,6-dichloro-10(11)-dehydrocurvularin (4).
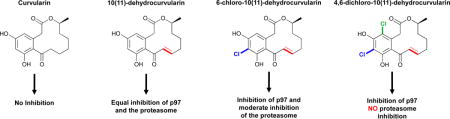
Curvularins are macrocyclic lactones that are produced by a variety of fungal species, such as those from the genera Penicillium.12 These compounds have been shown to display various biological activities including inhibition of cell division, inhibition of expression of human inducible nitric oxide synthase, and antifungal activities17–19; however, the underlying mechanisms by which they produce their biological effects have yet to be elucidated. In the present study, we discovered that unsaturated curvularin analogues 2–4 (Fig. 1) exhibit inhibition of p97 ATPase activity by covalent modification of the cysteine in the D2-ATP binding pocket, while curvularin (1) displayed no activity against p97. Excitingly, we found that DHC (2) inhibited both p97 and the 26S proteasome in cellular assays, but its 4,6-dichloro analogue (4) exhibited specific inhibitory activity for p97 in cellular assays.
After initial singlicate screening, to confirm that 2 was a valid hit candidate, the compound was screened in triplicate, followed by a 9-point dose-response [0.137 μM, 0.411 μM, 1.23 μM, 3.70 μM, 11.1 μM, 33.3 μM, 66.7 μM, 100 μM, and 200 μM]. The results confirmed 2 was a genuine p97 inhibitor with an IC50 value of 15.3 ± 9.9 μM (Fig. 2 and Table S1). To gain insight into the mechanism of DHC interaction with p97, two naturally-occurring DHC analogues, 3 and 4 were tested as well as the parent compound, 1. Compound 1 showed no inhibition of p97 at concentrations as high as 200 μM whereas 3 and 4 showed IC50 values about equal to 2 (24.3 and 13.9 μM, respectively – See Fig. 2 and table S1). These data suggested that the unsaturated ketone was critical to the function of the hit compounds. Next, because 2, 3, and 4 were identified as hits from an ATPase screen, the concentration of ATP was increased and the IC50 measured again to determine if the compounds were competitive-like (Fig. 2 and Table S1). As shown the IC50 values were independent of ATP concentration, arguing these compounds are not competitive-like, but see below.
Fig. 2.
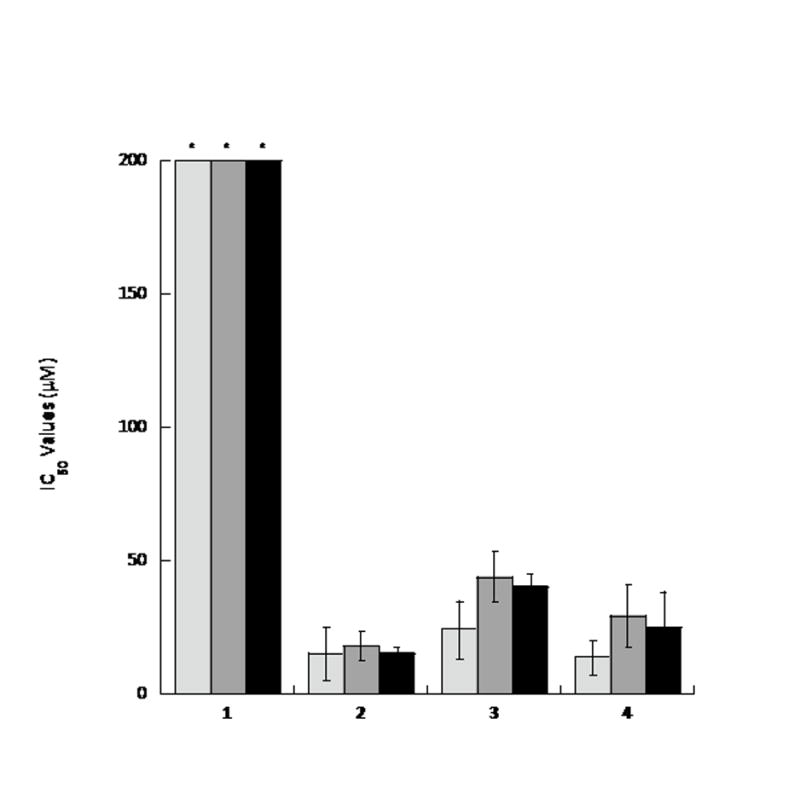
Dehydrocurvularin and its chlorinated analogues are p97 inhibitors. The IC50 values for curvularin (1) and its 10(11)-unsaturated analogues (2–4) were determined at varying ATP concentrations: 100 μM (light gray), 500 μM (gray), and 1000 μM (black). In each case, experiments were repeated in triplicate and error bars express standard deviation from the mean. An asterisk (*) indicates IC50 values greater than 200 μM.
Given the presence of a Michael acceptor moiety (compare compound 1 to compounds 2, 3, and 4) in 10(11)-dehyrocurvularins, it was reasoned that the interaction between these compounds and p97 might be covalent. In order to test this, a cysteineless p97 variant was constructed (p97-Cys0). This construct was generated by converting each of the structural cysteines to valine and the two ATP-binding pocket cysteines to alanine.20 The IC50 values were then measured against p97-Cys0 for each of the compounds. Compounds 1–4 showed no inhibition at concentrations up to 200 μM (Fig. 3b and Table S1). Since compounds 2–4 were shown to inhibit the ATPase activity of p97, a potential interaction between these compounds and one of the two cysteine residues of the ATP-binding pockets, C209 in the D1 domain pocket and C522 in the D2 domain pocket (Fig 3A) was considered. To test this, single cysteine mutants p97-C209A and p97-C522A were generated. The C209A mutant showed inhibition of ATPase activity with similar IC50 values as wild-type p97 (Fig. 3C and Table S1); conversely, the C522A mutant showed no inhibition (Fig. 3D and Table S1), indicating that 2–4 bind in the D2 ATPase pocket and require Cys522 for activity.
Fig. 3.
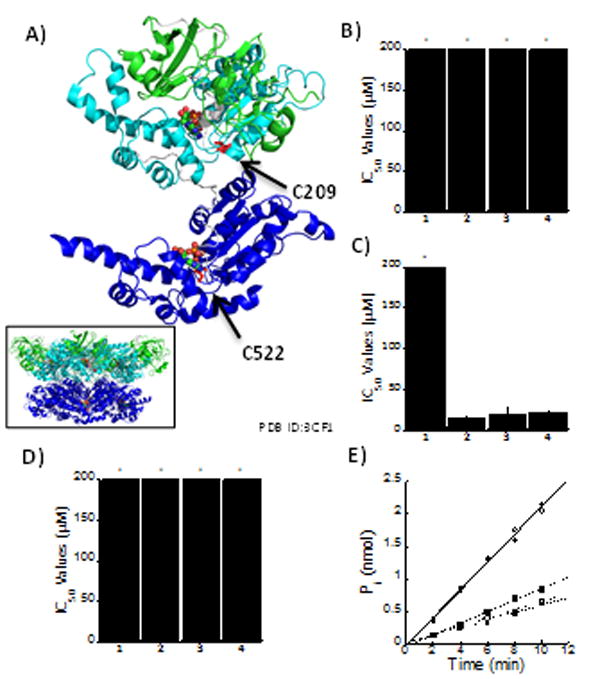
Curvularin derivatives bind in the D2 ATPase pocket. (A) Schematic representation of p97 monomer (Inset: p97 hexamer) in complex with ADP/ADP (colored by atom). The N domain is indicated by green, D1 domain by cyan, D2 domain by blue, and the labelled cysteine residues in red. IC50 values for each of the curvularins against ATPase activity of: (B) cysteine-less variant of p97, p97-Cys0; (C) p97-C209A; and (D) p97-C522A. (E) Covalent modification was tested by incubation of curvularins with p97 followed by extensive dialysis and subsequent measurement of ATPase activity. DMSO is represented by black circles; 1 by white circles; 2 by black squares; 3 by white squares; and 4 by black triangles.
Given the stringent cysteine requirement and the required Michael acceptor in the active curvularins, the reversibility of their binding was examined by incubating wild-type p97 in the presence of all four compounds followed by extensive dialysis to remove any unbound molecule. This ensured that non-reversibly attached compounds would not interfere with downstream biochemical analysis. Finally, the ATPase activity of p97 was measured using the dialysed p97 and our malachite green assay. As expected, incubation with 1 did not inhibit p97 activity; however, inhibition of p97 persisted in the presence of 10(11)-dehydrocurvularins 2–4, even after extensive dialysis (Fig. 3E). These data suggest that these inhibitors are covalent modifiers of p97 at position 522.
Given the performance of 2–4 in the biochemical assays, we set out to study their respective cellular activities and to examine cellular specificities. To determine if the curvularins were acting on the UPS pathway, we examined the level of total ubiquitylated proteins, which have been shown to increase with p97 inhibition as well as with inhibition of the proteasome.10 HEK293 cells were incubated with three different doses of each curvularin for 24 h before total protein extracts were prepared and analysed by immunoblotting. The doses selected were based on the predetermined LD50 values (Table S2). As predicted, total ubiquitin increased in the presence of curvularins compared to DMSO control (Fig. 4A), suggesting that some aspect of the UPS was being inhibited.
Fig. 4.
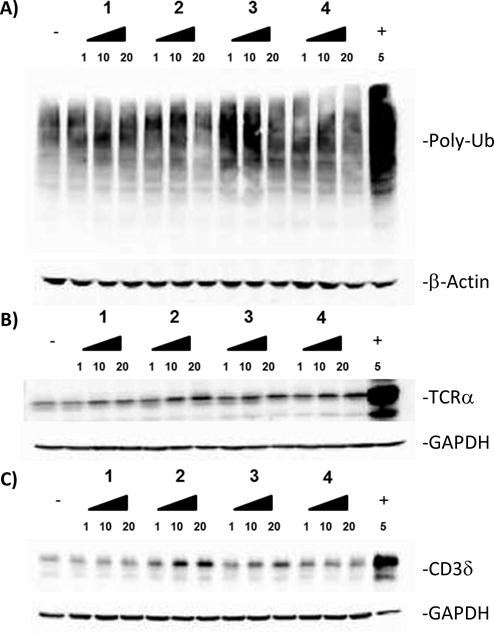
Compound 4 is selective for p97. (A–C) HEK293 cells were treated with indicated concentrations of curvularins for 24 h. (A) Total ubiquitin, (B) TCRα-GFP (green fluorescent protein) and (C) hemagglutinin (HA)-CD3δ were analyzed by immunoblot. The negative (−) and positive (+) controls used were DMSO and MG132 (a proteasome inhibitor), respectively. Loading controls are β-actin and GAPDH.
Because an increase in total ubiquitin does not necessarily arise from p97 inhibition, we sought to distinguish between p97 and proteasome inhibition. To do this, we turned to two different endoplasmic reticulum (ER) transmembrane subunits of the T-cell receptor, which are both subject to endoplasmic reticulum associated degradation (ERAD): TCRα and CD3δ.3, 21–22 TCRα has been shown to be a substrate of p97 and impairment of p97 activity prevents its degradation.3 Interestingly, a deglycosylated form of TCRα is observable only in the presence of proteasome inhibitors such as MG132.19 The second reporter, CD3δ, has been shown to be degraded by the proteasome, but independent of p97.21–22
Using these two reporters, we were able to differentiate between p97-dependent and p97-independent ERAD pathways. TCRα-green fluorescent protein (GFP) and hemagglutinin (HA)-CD3δ were expressed in HEK293 cells and analysed by immunoblotting. Similar to the negative control, 1 showed no significant increase of either TCRα (Fig. 4B) or CD3δ (Fig. 4C). In contrast, both 2 and 3 showed an increase in both TCRα (Fig. 4B) and CD3δ (Fig. 4C) levels, indicating that these compounds inhibit both the proteasome and p97. However, the increase in CD3δ was less pronounced for 3 than for 2. Excitingly, 4 showed a dose-dependent increase in TCRα (Fig. 4b) but surprisingly, showed no increase in the levels of CD3δ (Fig. 4C), suggesting that 4,6-dichloro-10(11)-dehydrocurvularin 4 is selective for p97.
Conclusions
In the current study we established that analogues of the natural product, 10(11)-dehydrocurvularin (DHC), inhibit the ATPase activity of p97 by binding in the D2 ATP binding pocket specifically and likely form a covalent adduct with Cys522 (Fig. 5A). These docking studies also indicated curvularin (1) but not the dehydrated adducts (2–4) sterically clash with the binding pocket (Fig. 5B and C), explaining the complete lack of activity for 1. Moreover, the calculated inhibition constants from these docking experiments indicate 4 binds with much higher affinity than 1 (Table S3). Further cellular investigations of the four curvularin analogues showed that compounds 2–4 each increased the amount of total ubiquitin in cells and increased the amount of TCRα indicating inhibition of some aspect of the UPS and likely p97. To differentiate between p97 and the proteasome, a second, p97 independent reporter was used, CD3δ. Excitingly, 2 inhibited both the proteasome and p97, 3 inhibited p97 and showed less activity against the proteasome, and 4 was p97 selective. Although not predicted at the outset, it seems likely this difference in selectivity is due to the electron withdrawing nature of the chlorines on the aromatic rings of 3 and 4. These will act to decrease the reactivity of the Michael acceptor, making the D2-ATP pocket specific binding critical for facile reactivity.
Fig. 5.
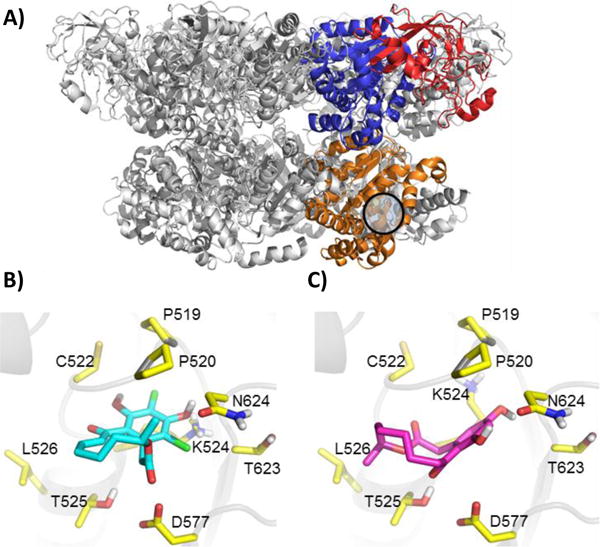
Compound 4 docks in the D2 ATPase pocket. (A) The p97 hexamer with one subunit highlighting domain architecture. The N-domain is indicated by red, the D1 domain by blue, and the D2 domain by orange. PDBID: 3CF3 (B) Compound 1 docked into p97 hexamer. (C) Compound 4 docked into the p97 hexamer.
Supplementary Material
Acknowledgments
This work was supported by the U.S. National Institute of Environmental Health Sciences Grant R01 ES023758, National Institute of Environmental Health Sciences Training Grant T32 ES007091, and the U.S. National Cancer Institute Grant R01 CA090265.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Erzberger JP, Berger JM. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 2.Meyer H, Bug M, Bremer S. Nat Cell Biol. 2012;14(2):117–23. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 3.DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Mol Cell. 2006;22(4):451–62. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 4.Chapman E, Fry AN, Kang M. Mol Biosyst. 2011;7(3):700–10. doi: 10.1039/c0mb00176g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer H, Weihl CC. J Cell Sci. 2014;127(Pt 18):3877–83. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bug M, Meyer H. J Struct Biol. 2012;179(2):78–82. doi: 10.1016/j.jsb.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Kang MJ, Wu T, Wijeratne EM, Lau EC, Mason DJ, Mesa C, Tillotson J, Zhang DD, Gunatilaka AAL, La Clair JJ, Chapman E. Chembiochem. 2014;15(14):2125–31. doi: 10.1002/cbic.201402258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman E, Maksim N, de la Cruz F, La Clair JJ. Molecules. 2015;20(2):3027–49. doi: 10.3390/molecules20023027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M. Cancer Cell. 2015;28(5):653–65. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geladopoulos TP, Sotiroudis TG, Evangelopoulos AE. Anal Biochem. 1991;192(1):112–6. doi: 10.1016/0003-2697(91)90194-x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang JH, Chung TD, Oldenburg KR. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 12.Zhan J, Gunatilaka AAL. J Nat Prod. 2005;68(8):1271–3. doi: 10.1021/np0580309. [DOI] [PubMed] [Google Scholar]
- 13.Curvularin (1) was prepared by the catalytic hydrogenation of 2; 10(11)-Dehydrocurvularin (2), 6-chloro-10(11)-dehydrocurvularin (3), and 4,6-dichloro-10(11)-dehydrocurvularin (4)were obtained from the fungal strain, Alternaria sp. nov. AST0039 (B.P. Bashyal et al., manuscript in preparation). See SI for spectral characterization of 4,6-dichloro-10(11)-dehydrocurvularin (4).
- 14.Compound 2:; Zhan J, Wijeratne EMK, Se;oga CJ, Zhang J, Pierson EE, Pierson LS, III, Vanetten HD, Gunatilaka AAL. J Antibiot. 2004;57(5):341–4. doi: 10.7164/antibiotics.57.341. [DOI] [PubMed] [Google Scholar]
- 15.Santagata S, Xu Y, Wijeratne EMK, Kontnik R, Rooney C, Perley CC, Kwon H, Clardy J, Kesari S, Whitesell L, Lindquist S, Gunatilaka AAL. ACS Chem Biol. 2012;7(2):340–9. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Compound 1 and compound 3:; Ghisalberti EL, Rowland CY. J Nat Prod. 1993;56(12):2175–77. doi: 10.1021/np50100a020. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi A, Hino T, Yata S, Itoh TJ, Sato H, Kawazu K. Agric Biol Chem. 1988;52:3119–23. [Google Scholar]
- 18.Yao Y, Hausding M, Erkel G, Anke T, Förstermann U, Kleinert H. Mol Pharmacol. 2003;63(2):383–91. doi: 10.1124/mol.63.2.383. [DOI] [PubMed] [Google Scholar]
- 19.Xie LW, Ouyang YC, Zou K, Wang GH, Chen MJ, Sun HM, Dai SK, Li X. Appl Biochem Biotechnol. 2009;159(1):284–93. doi: 10.1007/s12010-009-8591-2. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L, Baumann U, Reymond JL. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong Y, Fang S. J Biol Chem. 2012;287(33):28057–66. doi: 10.1074/jbc.M112.381798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wójcik C, Rowicka M, Kudlicki A, Nowis D, McConnell E, Kujawa M, DeMartino GN. Mol Biol Cell. 2006;17(11):4606–18. doi: 10.1091/mbc.E06-05-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


