Abstract
Introduction
While contamination of mobile phones in the hospital has been found to be common in several studies, little information about bacterial abundance on phones used in the community is available. Our aim was to quantitatively determine the bacterial contamination of secondary school students’ mobile phones.
Methods
Altogether 27 mobile phones were studied. The contact plate method and microbial identification using MALDI-TOF mass spectrometer were used for culture studies. Quantitative PCR reaction for detection of universal 16S rRNA, Enterococcus faecalis 16S rRNA and Escherichia coli allantoin permease were performed, and the presence of tetracycline (tetA, tetB, tetM), erythromycin (ermB) and sulphonamide (sul1) resistance genes was assessed.
Results
We found a high median bacterial count on secondary school students’ mobile phones (10.5 CFU/cm2) and a median of 17,032 bacterial 16S rRNA gene copies per phone. Potentially pathogenic microbes (Staphylococcus aureus, Acinetobacter spp., Pseudomonas spp., Bacillus cereus and Neisseria flavescens) were found among dominant microbes more often on phones with higher percentage of E. faecalis in total bacterial 16S rRNA. No differences in contamination level or dominating bacterial species between phone owner’s gender and between phone types (touch screen/keypad) were found. No antibiotic resistance genes were detected on mobile phone surfaces.
Conclusion
Quantitative study methods revealed high level bacterial contamination of secondary school students’ mobile phones.
Keywords: Mobile phones, contact plate, quantitative, potentially pathogenic microbes, children
Introduction
Mobile phones are the most frequently owned and used electronic devices worldwide. Disinfection or even cleaning of phones is problematic as the excess moisture could damage them. Mobile phones contaminated with nosocomial pathogens in hospital settings have been acknowledged as potential vectors for transferring nosocomial germs inside the hospital and also to the community.1 Still the contamination of schoolchildren’s mobile phones with bacteria and the consequent possible occurrence of antibiotic resistance has not been studied in sufficient detail yet.
The aim of our study was to evaluate the microbial contamination and the presence of resistance genes on the surfaces of mobile phones owned by secondary school students in Estonia.
Methods
The samples were collected from 27 mobile phones of secondary school students aged 16 to 18 years. One side of the phone (the back) was investigated using the blood agar contact plate (diameter 60 mm, with grid) method for cultivable bacteria2 and a 30 square cm area of the other side (the front) was sampled using sterile swabs for molecular studies. Additionally, mobile phone characteristics (touch screen/keypad) and the gender of the owner was registered. For quantitative cultures the contact plates were pressed onto the back side of the mobile phone, covered, transported to the laboratory and incubated at 37 °C for 48 hours in aerobic atmosphere. Bacterial colonies were counted and average colony count in 1 cm2 was calculated; three different colony types represented in highest numbers (dominant colony types) were identified using MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). The cotton swab sticks were rubbed onto the front side of the phone and transported to the laboratory. The swabs were soaked in phosphate buffer and DNA was extracted from the buffer using commercial kit QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). DNA concentration was measured using NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Quantitative PCR reaction for detection of universal 16S rRNA,3 Enterococcus faecalis 16S rRNA4 and Escherichia coli allantoin permease5 was carried out. Briefly, qPCR mix in final volume of 10 µL containing 1 µL DNA, 0,0004 mM primers,3-5 5 µL Maxima™ SYBR Green qPCR Master Mix (2X) (MBI Fermentas, Vilnius, Lithuania) and 3.6 µL sterile distilled water was prepared and qPCR reactions were completed using Rotor Gene Q thermocycler (Qiagen, Hilden, Germany).
Additionally, the presence of genes encoding tetracycline (tetA, tetB, tetM), erythromycin (ermB) and sulphonamide (sul1) resistance was assessed according to methods used by Peak et al.,6 Knapp et al.,7 Heuer et al.,8 and Borjesson et al.9
Statistical analysis was performed using SigmaStat (Jandel Scientific, San Rafael, CA, USA) and Excel (Microsoft Corp., Redmond, OR, USA). Fisher exact test, t-test, Mann-Whitney U test and Chi squared test were used to compare group differences. P values of < 0.05 were considered statistically significant.
Results and discussion
The culture method revealed bacterial contamination in all samples. This was different from previous studies where not all phones were found to be contaminated,10,11 although in these studies methods other than contact plate were used. On the other hand a study by Lee et al.12 using the agar touch method revealed bacteria in all phones. In our study, a high median microbial colony count [median 10.5 (IQR 3-16.8) colony forming units (CFU)/cm2] was detected (Figure 1). There are insufficient comparative data as only few studies have determined phone contamination quantitatively. Still this value is approximately ten times higher than that found in a similar study among university students in Germany where the average bacterial load on phones was 1.37 CFU/cm2.13
Figure 1. High level bacterial contamination of secondary school students’ mobile phone revealed by contact plate (ø 60 mm) method.
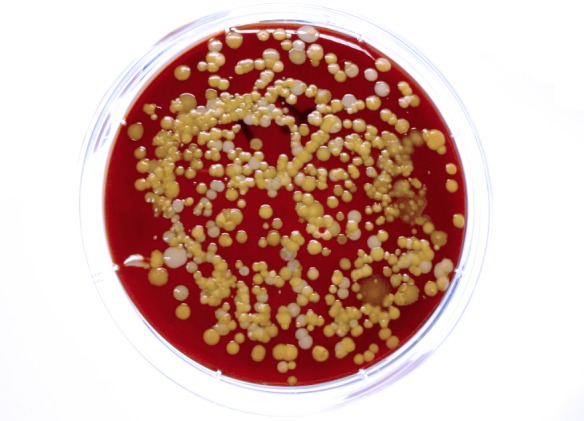
Since the overall hygiene behavior in the two European countries is similar, differences in contamination of mobile phones may be explained by possible different hygiene habits between teenagers and young adults. Microbes on owner’s hands play an important role in the contamination of mobile phone surfaces.14 Still the count of bacteria on mobile phones was lower than that previously found in skin touch samples where the median colony count was 480 per cm2.15
Altogether more than 20 different dominant microbial species were detected. Gram-negative bacteria were found in 41% (n=11) of cultured phone surfaces. The most frequent contaminants of mobile phones were Micrococcus luteus (n=15; 63%), Acinetobacter lwoffii (n=9; 33%), Staphylococcus epidermidis (n= 8; 30%) and Staphylococcus hominis (n= 5; 19%). Other germs like S. aureus, Pseudomonas luteola, and Neisseria flavescens were found each in one and Bacillus cereus on two mobile phones among dominant microbial species.
A study in teaching hospitals of South Korea revealed the touch screen phones but not the button phones to be a significant risk factor for contamination by potentially pathogenic bacteria.12 In our study smart phones were contaminated with a median of 15.8 (IQR 5.8-20.0) CFU/cm2 and buttoned phones with a median of 7.4 (IQR 4.6-16.0) CFU/cm2. We did not find any differences in contamination between phone types among our studied 27 phones (the mean ranks of buttoned phones and smart phones were 3.587 and 8.413, respectively; U=46.5, Z=-0.84, p=0.401).
Previous investigations have found significant differences in microbiota on phones belonging to adult males and females.10,14 However Lee et al.12 found gender not to be a risk factor for phone contamination. In our study the phones owned by girls were contaminated with a median of 9 (IQR 3-14.3) CFU/cm2 and the phones owned by boys with a median of 15.6 (IQR 5.8-20.4) CFU/cm2. No differences in microbial count on mobile phones between phone owner genders were found (the mean ranks of girls’ and boys’ phones were 5.56 and 7.44, respectively; U=48, Z=-1.61, p=0.108). This may be caused by the relatively low number of studied phones in our study or by the method used, in which only back sides of the phones were cultured.
We detected a median of 17032 (IQR 6628-22086) bacterial 16S rRNA gene copies per mobile phone. Although universal primers targeting the 16S rRNA gene cannot measure the exact number of bacterial cells, it has been used to estimate total bacterial abundance.16 Although Escherichia coli and Enterococcus faecalis were not among the dominant bacterial species, molecular studies showed the presence of these fecal indicator bacteria on secondary school students’ mobile phones. Fecal indicator bacteria E. coli and Enterococcus spp. are acknowledged for their putative relationship with risk of infection and are used as surrogates for pathogen contamination in general.17 Their absence among the dominant bacterial species in our study indicates no considerable fecal contamination of mobile phones. As most of the bacterial species detected in our study are present in the human skin microbiome, they most likely originate from student’s hands.18 Molecular studies revealed E. coli and E. faecalis in median values of 3.1% (IQR 1.5-8.8) and 1.0% (IQR 0.4-2.3), respectively, from total bacterial 16S rRNA of all studied phone surfaces. There was good correlation between counts of E. coli and E. faecalis (R=0.674, p<0.001). Fecal enterococci but not coliforms in fomites have been associated with the risk of respiratory infection in child care facilities.19 Similar associations have not been studied for mobile phones. We found more frequently the presence of potentially pathogenic S. aureus, Acinetobacter, Pseudomonas, Bacillus cereus or Neisseria flavescens in case of more than one percent of E. faecalis DNA compared to less than one percent in total bacterial 16S rRNA of studied 27 mobile phones (10/15 vs. 3/12, respectively; p=0.031). The non-pathogenic skin microbe M. luteus frequently found in large amounts on circulating currencies20,21 was also found more frequently on phones with more than one compared to less than one percent (11/15 vs. 4/12, respectively; p=0.038) of E. faecalis DNA (Table 1). Although in our study an evaluation of the health condition of schoolchildren was not addressed, E. faecalis appeared to be a good environmental contamination indicator. Kumar et al.22 have also found E. faecalis on mobile phones from hospital inpatients and have associated it with possible contamination with intestinal microbes. In our study a similar association between finding E. coli DNA on phone surfaces and contamination with aforementioned potentially pathogenic microbes was not found. The limitation of the current study was the lack of the opportunity to evaluate bacterial contamination using culture and molecular methods on the same side of the phone at the same time.
Table 1. Dominating bacterial species revealed by culture method and MALDI-TOF identification on mobile phones according to the percentage of Enterococcus faecalis in total bacterial 16S rRNA (Group 1 - >1%; Group 2 - <1%) found on the particular mobile phone surface.
| Dominating bacteria | No and % of phones contaminated with bacteria | p | |||
|---|---|---|---|---|---|
| Group 1 (15 phones) | Group 2 (12 phones) | ||||
| n | % | n | % | ||
| Coagulase-negative staphylococci | 12 | 80 | 8 | 67 | 0.432 |
| Micrococcus luteus | 11 | 73 | 4 | 33 | 0.038 |
| Acinetobacter lwoffii | 6 | 40 | 3 | 25 | 0.411 |
| Kocuria spp. | 3 | 20 | 2 | 17 | 0.825 |
| Bacillus spp. | 1 | 7 | 3 | 25 | 0.183 |
| Corynebacterium spp. | 1 | 7 | 3 | 25 | 0.183 |
| Paenibacillus lactis | 1 | 7 | 1 | 8 | 0.869 |
| Bacillus cereus | 1 | 7 | 1 | 8 | 0.869 |
| Staphylococcus aureus | 1 | 7 | 0 | 0 | 0.362 |
| Pseudomonas luteola | 1 | 7 | 0 | 0 | 0.362 |
| Neisseria flavescens | 1 | 7 | 0 | 0 | 0.362 |
| Rothia dentocariosa | 0 | 0 | 1 | 8 | 0.255 |
Species identification was performed through culturing methods and MALDI TOF; comparison is based on percentage (>1%/<1%) of E. faecalis DNA in total 16S rRNA
Due to extensive use of antibiotics in medicine and agriculture, antibiotic resistant bacteria are widely spread in the environment and there is a threat of resistance gene transfer to microbial pathogens via horizontal gene transfer.23 Sulphonamide, macrolide, and tetracycline resistance genes have often been used as markers of environmental contamination, although not concerning the contamination of mobile phones so far.24 In our study, no tetracycline, erythromycin and sulphonamide resistance encoding genes were found on mobile phone surfaces. Therefore in communities of children, mobile phones, despite being contaminated with commensal, opportunistic or potentially pathogenic bacteria, are probably not important vectors for resistance genes.
Conclusions
Our study showed high level contamination of secondary school students’ mobile phones with potentially pathogenic bacteria to be common, and we hypothesize that this may play a role in the spread of infectious agents in the community. However, based on our results, the mobile phones of secondary school students do not appear to be considerable vectors for the spread of antibacterial resistance.
Acknowledgments
We would like to thank Jaak Truu and Riinu Kiiker for their help in molecular studies.
Footnotes
Authors’ contribution statement: SK, RaM, ReM, TS contributed to study design, data analysis and interpretation, literature review; RaM, TS, TR contributed to data collection and laboratory analysis. All authors read and approved the final version of the manuscript.
Conflicts of interest: All authors – none to declare.
Funding: This study was supported by the Estonian Research Council (Grant No. IUT34-19) and the Estonian Ministry of Education and Research (Grant No. KOGU-HUMB).
References
- 1.Ustun C, Cihangiroglu M. Health care workers’ mobile phones: a potential cause of microbial cross-contamination between hospitals and community. J Occup Environ Hyg. 2012;9:538–42. doi: 10.1080/15459624.2012.697419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibfelt T, Foged C, Andersen LP. Validation of dipslides as a tool for environmental sampling in a real-life hospital setting. Eur J Clin Microbiol Infect Dis. 2014;33:809–13. doi: 10.1007/s10096-013-2018-2. [DOI] [PubMed] [Google Scholar]
- 3.Gloor GB, Hummelen R, Macklaim JM, et al. Microbiome profiling by illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One. 2010;5:e15406. doi: 10.1371/journal.pone.0015406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosch S, Fite A, Macfarlane GT, McMurdo ME. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol. 2004;70:3575–81. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chetta M, Bafunno V, Grillo R, et al. SYBR green real time-polymerase chain reaction as a rapid and alternative assay for the efficient identification of all existing Escherichia coli biotypes approved directly in wastewater samples. Biotechnol Prog. 2012;28:1106–13. doi: 10.1002/btpr.1573. [DOI] [PubMed] [Google Scholar]
- 6.Peak N, Knapp CW, Yang RK, et al. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ Microbiol. 2007;9:143–51. doi: 10.1111/j.1462-2920.2006.01123.x. [DOI] [PubMed] [Google Scholar]
- 7.Knapp CW, Zhang W, Sturm BS, Graham DW. Differential fate of erythromycin and beta-lactam resistance genes from swine lagoon waste under different aquatic conditions. Environ Pollut. 2010;158:1506–12. doi: 10.1016/j.envpol.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 8.Heuer H, Smalla K. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ Microbiol. 2007;9:657–66. doi: 10.1111/j.1462-2920.2006.01185.x. [DOI] [PubMed] [Google Scholar]
- 9.Borjesson S, Dienues O, Jarnheimer PA, Olsen B, Matussek A, Lindgren PE. Quantification of genes encoding resistance to aminoglycosides, beta-lactams and tetracyclines in wastewater environments by real-time PCR. Int J Environ Health Res. 2009;19:219–30. doi: 10.1080/09603120802449593. [DOI] [PubMed] [Google Scholar]
- 10.Bhoonderowa A, Gookool S, Biranjia-Hurdoyal SD. The importance of mobile phones in the possible transmission of bacterial infections in the community. J Community Health. 2014;39:965–7. doi: 10.1007/s10900-014-9838-6. [DOI] [PubMed] [Google Scholar]
- 11.Ulger F, Esen S, Dilek A, Yanik K, Gunaydin M, Leblebicioglu H. Are we aware how contaminated our mobile phones with nosocomial pathogens? Ann Clin Microbiol Antimicrob. 2009;8:7. doi: 10.1186/1476-0711-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee YJ, Yoo CG, Lee CT, et al. Contamination rates between smart cell phones and non-smart cell phones of healthcare workers. J Hosp Med. 2013;8:144–7. doi: 10.1002/jhm.2011. [DOI] [PubMed] [Google Scholar]
- 13.Egert M, Späth K, Weik K, et al. Bacteria on smartphone touchscreens in a German university setting and evaluation of two popular cleaning methods using commercially available cleaning products. Folia Microbiol (Praha) 2015;60:159–64. doi: 10.1007/s12223-014-0350-2. [DOI] [PubMed] [Google Scholar]
- 14.Meadow JF, Altrichter AE, Green JL. Mobile phones carry the personal microbiome of their owners. PeerJ. 2014;2:e447. doi: 10.7717/peerj.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macias JH, Alvarez MF, Arreguin V, Muñoz JM, Macias AE, Alvarez JA. Chlorhexidine avoids skin bacteria recolonization more than triclosan. Am J Infect Control. 2016;44:1530–4. doi: 10.1016/j.ajic.2016.04.235. [DOI] [PubMed] [Google Scholar]
- 16.Hanna EM, Hamp TJ, McKillop IH, et al. Comparison of culture and molecular techniques for microbial community characterization in infected necrotizing pancreatitis. J Surg Res. 2014;191:362–9. doi: 10.1016/j.jss.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Griffith JF, Weisberg SB, Arnold BF, Cao Y, Schiff KC, Colford JM., Jr Epidemiologic evaluation of multiple alternate microbial water quality monitoring indicators at three California beaches. Water Res. 2016;94:371–81. doi: 10.1016/j.watres.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 18.Patra V, Byrne SN, Wolf P. The skin microbiome: is it affected by UV-induced immune suppression? Front Microbiol. 2016;7:1235. doi: 10.3389/fmicb.2016.01235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julian TR, Pickering AJ, Leckie JO, Boehm AB. Enterococcus spp on fomites and hands indicate increased risk of respiratory illness in child care centers. Am J Infect Control. 2013;41:728–33. doi: 10.1016/j.ajic.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Maritz JM, Sullivan SA, Prill RJ, Aksoy E, Scheid P, Carlton JM. Filthy lucre: A metagenomic pilot study of microbes found on circulating currency in New York City. PLoS One. 2017;12:e0175527. doi: 10.1371/journal.pone.0175527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mändar K, Sõber T, Kõljalg S, Rööp T, Mändar R, Sepp E. Microbiological contamination of the euro currency in Estonia. Infect Dis (Lond) 2016;48:772–4. doi: 10.1080/23744235.2016.1201725. [DOI] [PubMed] [Google Scholar]
- 22.Vinod Kumar B, Hobani YH, Abdulhaq A, et al. Prevalence of antibacterial resistant bacterial contaminants from mobile phones of hospital inpatients. Libyan J Med. 2014;9:25451. doi: 10.3402/ljm.v9.25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao W, Zhang XX, Zhao F, et al. High levels of antibiotic resistance genes and their correlations with bacterial community and mobile genetic elements in pharmaceutical wastewater treatment bioreactors. PLoS One. 2016;11:e0156854. doi: 10.1371/journal.pone.0156854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Y, Wilson CA, Novak JT, et al. Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class I integrons. Environ Sci Technol. 2011;45:7855–61. doi: 10.1021/es200827t. [DOI] [PubMed] [Google Scholar]


