To the editor
Janus kinase 3 (JAK3) is a cytosolic tyrosine kinase that is primarily expressed in hematopoietic cells.1 Null mutations in JAK3 typically manifest as T−B+NK− severe combined immunodeficiency (SCID), while hypomorphic mutations can present with a broader range of immune dysregulation.1,2 Here we report two siblings with a homozygous synonymous exonic mutation in JAK3 that creates a dominant splice site and dramatically decreases expression of WT protein. This mutation resulted in T−B+NKlow SCID in one patient and in a combined immunodeficiency (CID) with granulomatous skin disease in her sibling.
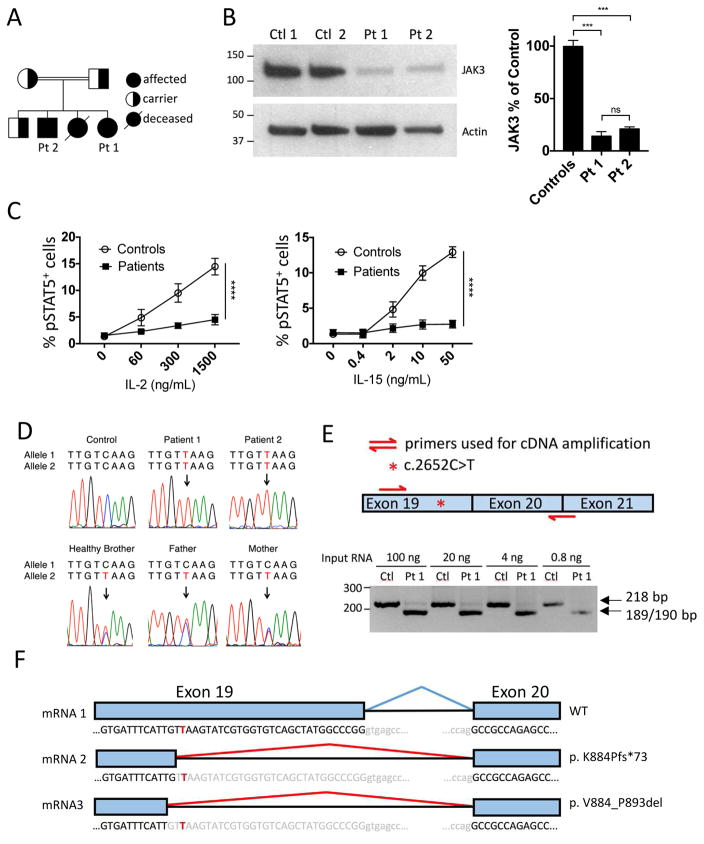
Four children resulted from the consanguineous union of two parents from Saudi Arabia (Fig 1A). Given a family history of SCID in an older sibling, the family’s fourth child (Patient 1) was screened shortly after birth by lymphocyte flow cytometry and diagnosed with T−B+NKlow SCID. Her 6-year-old brother (Patient 2) had a persistent violaceous, granulomatous dermatitis on his arms since 2 years of age (see Fig E1 in this article’s Online Repository at www.jacionline.org) but no history of frequent or unusual infections. Skin biopsy revealed a prominent dermal nodular infiltrate and poorly formed granulomas extending to the deep dermis (see Fig E2 in this article’s Online Repository at www.jacionline.org). Special stains for organisms performed with Periodic acid–Schiff, Gram, Grocott’s methenamine silver, and Fite were negative (data not shown). As granulomatous skin disease has been associated with CID2,3 an immunologic evaluation of both siblings was performed.
Fig 1. Characterization of the JAK3 mutation.
(A) Pedigree. (B) Immunoblot of lysates from BLCLs derived from patients and controls (left). JAK3 expression relative to controls (right). ***p<0.001, ns=not significant (one-way ANOVA). (C) Percentage of pSTAT5+ BLCLs after stimulation for 10 minutes with IL-2 or IL-15. ****p<0.0001 (two-way ANOVA). (D) Sanger sequencing of c.2652C>T variant. (E) RT-PCR amplification of JAK3 mRNA encompassing the patient mutation site. (F) Diagram of mRNA splice variants induced by mutation. For B and C n=2 for controls, n=2 for patients; data pooled from 2 independent experiments. Columns/lines and bars represent means and SEM.
Immunological workup of Patient 1 revealed nearly absent T cells (56 cells/μl), normal B cells (1600 cells/μl), low NK cells (54 cells/μl) and negligible T cell proliferation to PHA, ConA and anti-CD3 (Table 1). All detectable T cells were of maternal origin (data not shown). Patient 2 had low numbers of T cells (566 cells/μl), B cells (120 cells/μl) and NK cells (17 cells/μl), consistent with a CID. He also had reduced percentages of naïve T cells, and inversion of the CD4:CD8 ratio (Table 1). Proliferation to mitogens and antigens was below the lower limit of normal (Table 1). There was no evidence of maternal engraftment (data not shown). Patient 2 had normal TCR Vβ usage that did not suggest a restricted T cell repertoire (see Fig E3 in this article’s Online Repository at www.jacionline.org), and normal immunoglobulins (Table 1), but poor responses to tetanus and pneumococcal vaccines (data not shown).
Table 1.
Immune profile of patients
| Patient 1 | Patient 2 | |||
|---|---|---|---|---|
|
| ||||
| Age 3 months | Normal Values for Agea | Age 6 years | Normal Values for Agea | |
|
| ||||
| Lymphocyte subsets (cells per μl) | ||||
|
| ||||
| CD3+ | 56 | 2500–5600 | 566 | 1000–2600 |
| CD3+CD4+ | 7 | 1600–4000 | 154 | 530–1500 |
| CD4+CD45RA+CCR7− (TEMRA) | 16.90% | 0.1–1.9% | 3.90% | 0.4%–2.6% |
| CD4+CD45RA+CCR7+ (naïve) | 11.90% | 76.7–91.4% | 5.60% | 57.1%–84.9% |
| CD4+CD45RA−CCR7− (effector memory) | 54.20% | 1.1–5.3% | 38.50% | 3.3%–15.2% |
| CD4+CD45RA−CCR7+ (central memory) | 16.90% | 6.7–15.6% | 52% | 11.3%–26.7% |
| CD4+CD31+CD45RA+ (recent thymic emigrants) | 34.30% | 59.7–82.4% | 6.60% | 45.3–63.6% |
| CD3+CD8+ | 37 | 500–1700 | 344 | 330–1100 |
| CD8+CD45RA+CCR7− (TEMRA) | 0.90% | 1.5–22.7% | 15.40% | 9.1–49.1% |
| CD8+CD45RA+CCR7+ (naïve) | 0.30% | 62.1–94.0% | 2.30% | 28.4–80.6% |
| CD8+CD45RA−CCR7− (effector memory) | 96.30% | 1.3–19.5% | 71.30% | 6.2–29.3% |
| CD8+CD45RA−CCR7+ (central memory) | 2.50% | 0.9–5.6% | 11.00% | 1.0–4.5% |
| CD19+ | 1600 | 300–2000 | 120 | 270–860 |
| IgD+CD27− (naïve) | n.d. | 94.10% | 47.30–77.00% | |
| IgD+CD27+ (unswitched memory) | n.d. | 3.00% | 5.20–20.40% | |
| IgD−CD27+ (switched memory) | n.d. | 1.00% | 10.90–30.40% | |
| CD24hiCD38hi (transitional) | n.d. | 19.90% | 7.2–23.8% | |
| CD24lowCD38hi (plasmablasts) | n.d. | 0.00% | 0.4–5.2% | |
| CD19+CD21lowCD38low | n.d. | 5.60% | 2.5–9.4% | |
| CD24hiCD38low (marginal zone-like) | n.d. | 9.80% | 12.6–36.0% | |
| CD16+/CD56+ | 54 | 160–1100 | 17 | 70–480 |
|
| ||||
| Immunoglobulins | ||||
|
| ||||
| IgG (mg/dL) | 809* | 280–750 | 775 | 639–1344 |
| IgA (mg/dL) | <7 | 6–50 | 104 | 70–312 |
| IgM (mg/dL) | 39 | 15–70 | 171 | 34–210 |
| IgE (kU/L) | <1 | 0–30 | 5 | 0–200 |
|
| ||||
| T cell proliferation (counts per minute) | ||||
|
| ||||
| Phytohemagglutinin | 750 | 96,090–358,179 | 26,802 | 96,090–358,179 |
| Concanavalin A | 657 | 65,699–239,344 | 24,681 | 65,699–239,344 |
| Anti-CD3 | 1966 | 62,927–217,761 | 34,629 | 62,927–217,761 |
| Tetanus | n.d. | 1239 | 8544 –102,895 | |
| Candida | n.d. | 17348 | 6231–197,940 | |
| Background | 1273 | 204–2104 | 322 | 204–2104 |
normal values from age-matched controls in the Boston Children’s Hospital clinical immunology laboratory
on IVIG
n.d. = not done
To determine the underlying genetic defect, genomic DNA from Patient 1 was evaluated in a commercial laboratory using massive parallel sequencing of 46 genes associated with SCID/CID that included JAK3. While no pathogenic variants were reported, the possibility of a non-coding mutation in JAK3 that affects protein expression was pursued, as mutations in this gene typically present with T−B+NK− SCID. Immunoblotting of lysates from Epstein Barr virus-transformed B-lymphoblastoid cell lines (BLCLs) revealed a dramatic decrease in JAK3 expression in both patients (Fig 1B). STAT5 phosphorylation in response to IL-2 and IL-15 stimulation of BLCLs was severely diminished in the patients, confirming a functional defect in JAK3 signaling (Fig 1C).
A comprehensive list of all exonic variants found by the commercial laboratory was requested. It included a single homozygous, synonymous variant in exon 19 of JAK3 (c.2652C>T; pV884V) that had been presumed benign as it did not alter the predicted amino acid sequence. However, in silico analysis using the independent splice site prediction tools MaxEntScan4 and Human Splicing Finder5 indicated that the variant likely created a donor splice site (see Table E1 in this article’s Online Repository at www.jacionline.org). The variant was confirmed by Sanger sequencing to be homozygous in both patients, and heterozygous in the parents and healthy sibling (Fig 1D).
To evaluate whether the mutation impacted splicing of JAK3 mRNA, a 218 bp cDNA fragment surrounding the mutation site was amplified by RT-PCR (Fig 1E, top panel). mRNA purified from control PBMCs produced a single band of the expected molecular weight (Fig 1E, bottom panel). In contrast, two bands were amplified from PBMC mRNA isolated from Patient 1: a barely detectable full-length band, and a much more abundant, lower molecular weight band (Fig 1E, bottom panel). Sanger sequencing revealed that the full-length band contained WT cDNA, while the lower band contained two cDNA species with deletions of either 29 or 30 bp at the 3′ end of exon 19 (Fig 1F). The 29 bp deletion resulted in a frameshift with a premature stop codon (p.K884Pfs*73). Immunoblotting of patient BLCL lysates using a mAb specific for the N-terminus of JAK3 revealed no truncated protein at the predicted molecular weight of approximately 95 kDa, indicating that the mutant protein was not expressed or was rapidly degraded (Fig 1B). The 30 bp deletion removes 10 terminal amino acids of exon 19 (p.V884_P893del), thereby encoding a mutant protein ~1.2 kDa smaller than WT JAK3. Due to the closeness of their predicted molecular weight (115 kDa versus 114 kDa), the WT and p.V884_P893del mutant protein would be expected to migrate as a single band of ~115 kDa on gel electrophoresis. The dramatic decrease in the intensity of this band strongly suggests that the p.V884_P893del mutation decreases protein stability.
It is notable that Patient 1 and Patient 2 had identical homozygous mutations in JAK3 with similar reductions in protein expression but different phenotypes (see Table E2 in this article’s Online Repository at www.jacionline.org for additional clinical details on all siblings). The relatively normal TCR Vβ repertoire of Patient 2, residual proliferation to mitogens, and lack of significant infections indicates that his T cells retain some function and are not merely expanded from a limited number of clones. Such phenotypic variability has been documented in a variety of other PID genes6. Modifier genes, epigenetic factors, or environmental exposures may all play a role in determining the clinical phenotype.6
Synonymous codon variants are generally thought to have little significance7 and are often filtered out during analysis of the vast amount of data generated though next generation sequencing.6 Nevertheless, synonymous mutations in genes such as IL10RA and MRE11A have previously been reported to cause disease through disruption of known splice sites.8,9 Our case illustrates the importance of evaluating synonymous mutations even when they are distant from known splice sites, as they may create new donor or acceptor splice sites.
The significance of a synonymous variant can be determined through a series of steps. First, splicing prediction tools can determine the probability that a variant is pathogenic. Next, cDNA amplification and sequencing can definitively demonstrate that splicing is affected. Finally, functional analysis is necessary to prove that that a change in splicing is responsible for the patient phenotype. As whole exome sequencing and targeted gene panels become an increasingly important part of the clinical immunologist’s toolbox, the need to evaluate the biologic relevance of synonymous variants will continue to increase.
Supplementary Material
Acknowledgments
Supported by: National Institute of Health grants T32 AI007512 (C.P.), K12 HD052896-10 (C.P.), and the Perkins Fund (R.S.G.).
Abbreviations
- BLCL
B-lymphoblastoid cell line
- CID
combined immunodeficiency
- JAK3
Janus kinase 3
- SCID
severe combined immunodeficiency
- HSCT
hematopoietic stem cell transplantation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Casanova JL, Holland SM, Notarangelo LD. Inborn Errors of Human JAKs and STATs. Immunity. 2012;36:515–28. doi: 10.1016/j.immuni.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scarselli A, Di Cesare S, Di Matteo G, De Matteis A, Ariganello P, Romiti ML, et al. Combined immunodeficiency due to JAK3 mutation in a child presenting with skin granuloma. J Allergy Clin Immunol. 2016;137:648–51. doi: 10.1016/j.jaci.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Harp J, Coggshall K, Ruben BS, Ramirez-Valle F, He SY, Berger TG. Cutaneous granulomas in the setting of primary immunodeficiency: A report of four cases and review of the literature. Int J Dermatol. 2015;54:617–25. doi: 10.1111/ijd.12765. [DOI] [PubMed] [Google Scholar]
- 4.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 5.Desmet FO, Hamroun D, Lalande M, Collod-Bëroud G, Claustres M, Béroud C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:1–14. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platt C, Geha RS, Chou J. Gene hunting in the genomic era: Approaches to diagnostic dilemmas in patients with primary immunodeficiencies. J Allergy Clin Immunol. 2014;134:262–8. doi: 10.1016/j.jaci.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt RC, Simhadri VL, Iandoli M, Sauna ZE, Kimchi-Sarfaty C. Exposing synonymous mutations. Trends Genet. 2014;30:308–21. doi: 10.1016/j.tig.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto Y, Miyamoto T, Sakamoto H, Izumi H, Nakazawa Y, Ogi T, et al. Two unrelated patients with MRE11A mutations and Nijmegen breakage syndrome-like severe microcephaly. DNA Repair. 2011;10:314–21. doi: 10.1016/j.dnarep.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Oh SH, Baek J, Liany H, Foo JN, Kim KM, Yang SC-O, et al. A Synonymous Variant in IL10RA Affects RNA Splicing in Pediatric Patients with Refractory Inflammatory Bowel Disease. J Crohns Colitis. 2016 doi: 10.1093/ecco-jcc/jjw102. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.