Abstract
Shewanella oneidensis MR-1 is a mesophilic bacterium with a maximum growth temperature of ≈35°C but the ability to grow over a wide range of temperatures, including temperatures near zero. At room temperature (≈22°C) MR-1 grows with a doubling time of about 40 min, but when moved from 22°C to 3°C, MR-1 cells display a very long lag phase of more than 100 h followed by very slow growth, with a doubling time of ≈67 h. In comparison to cells grown at 22°C, the cold-grown cells formed long, motile filaments, showed many spheroplast-like structures, produced an array of proteins not seen at higher temperature, and synthesized a different pattern of cellular lipids. Frequent pilus-like structures were observed during the transition from 3 to 22°C.
Shewanella oneidensis MR-1 was first isolated from sediments of Lake Oneida in New York State (20). The cells are gram-negative straight rods capable of moving by means of a single polar flagellum. S. oneidensis MR-1 is a mesophilic, facultative anaerobe having a respiratory type of metabolism, with an optimal growth temperature of ≈30°C. Lake Oneida is a shallow freshwater system that freezes over completely during the winter (from early to mid-December to mid- or late January, with complete ice-out in March or April), with ice thickness reaching up to 70 cm (19). During May, water temperatures begin to rise, reaching a maximum of 25°C in midsummer (19).
Because of their ability to metabolically reduce metals such as U(VI), Tc(VII), and Cr(VI), organisms such as Shewanella are considered candidates for bioremediation of subsurface metal-contaminated areas. However, subsurface temperatures are often low, and predictions of the utility of Shewanella in such environments require an understanding of the effects of low temperatures on their metabolism and general ecology. Recently, it was shown that S. oneidensis MR-1, while capable of growth at low temperature, exhibited a growth transition at about 10°C, and below these temperatures showed a differential increase in rRNA synthesis compared to DNA synthesis (4). Here we report on other properties of MR-1 grown at low temperature (3°C), including a dramatically different phenotype with changes in morphology, growth rate, ultrastructure, and protein and lipid composition.
MATERIALS AND METHODS
Cells of Shewanella oneidensis strain MR-1 (ATCC 700550) were grown overnight aerobically in batch cultures at 22°C in 150-ml flasks containing 50 ml of Luria-Bertani (LB) broth Miller (Difco) at 130 rpm. For growth measurements, new cultures were started by transferring 1.0 ml of the original culture to a 250-ml Erlenmeyer flask containing 100 ml of LB and incubated at 130 rpm at two temperatures (3 and 22°C). All incubations were done in triplicate. Cells analyzed for proteins and phospholipid fatty acids were harvested by centrifugation and washed with sterile phosphate-buffered saline buffer, wet weight was determined, and the cells were frozen at −70°C until analysis.
The turbidity of the cultures was measured at 600 nm. For measuring cell mass the cell pellets were washed with saline solution and dried at 105°C for 24 h. For measuring the protein content 0.5 ml of washed cell suspension was mixed with 2 ml of 0.5 N HClO4 and 1.5 ml of distilled H2O and incubated for 10 min at 0°C. The precipitates were separated by centrifugation (5 min, 5,000 × g., 20°C), resuspended in 5 ml of 0.5 N HClO4 and incubated at 70°C for 20 min. The acid-insoluble fraction was separated by centrifugation (10 min, 5,000 × g, 20°C) and suspended in 0.2 N NaOH, and the total protein content was determined by the Lowry protein assay (17).
For epifluorescent microscopy observations, cells were stained for 5 min with acridine orange (50 μl of 0.1% [wt/vol] acridine orange in 2 ml of a 1:100 dilution of the cell culture) and filtered on a Nuclepore polycarbonate membrane (0.2 μm pore size) (Osmonics Inc.). For electron microscopy of whole-cell suspensions, cells were fixed for 30 min with the paraformaldehyde-glutaraldehyde Karnovsky fixative in sodium cacodylate buffer (0.2 M, pH 7.4). After fixation the cells were rinsed with cacodylate buffer and washed with phosphate-buffered saline buffer (pH 7.4). One hundred microliters of the fixed cells were placed on Formvar carbon grids (Ted Pella Inc.) and stained for 30 s with 1% uranyl acetate. The samples were observed with a Jeol 100 CX II transmission electron microscope.
For electron microscopy of ultrathin sections, the cells were fixed for 3 h with 2.5% glutaraldehyde in HEPES buffer (0.1 M, pH 7.4) and then washed with HEPES buffer and pelleted by centrifugation (14,000 × g, 2 min, 20°C). The pellets were embedded in 1.5% agar and fixed with 1% OsO4. Traces of fixative were removed by several rinses with HEPES buffer. The samples were dehydrated in a graded ethanol series starting with 5% uranyl acetate in 30% ethanol followed by 50, 70, 90, and 100% ethanol. Progressive infiltration of the samples with LR-White resin (London Resin Company Ltd.) was done for 12 h prior to polymerization at 65°C for 24 h. The ultrathin sections were stained with uranyl acetate for 25 min followed by lead citrate for 30 min and viewed on the transmission electron microscope at 80 keV.
For protein analysis, subsamples of the frozen cells were mixed with 2 volumes of a solution containing 9 M urea, 2% 2-mercaptoethanol, 2% ampholytes (pH 8 to 10), and 4% (vol/vol) Nonidet P40 (Bio-Rad). The soluble, denatured proteins were recovered in the supernatants after centrifugation of the samples (435,000 × g, 10 min) with a Beckman TL100 ultracentrifuge. Protein concentrations were determined with a modification of the Bradford protein assay (23). Subsamples containing 40 μg of protein were separated in the first dimension by isoelectric focusing with polyacrylamide gels containing 50% pH 5 to 7 and 50% pH 3 to 10 carrier ampholytes (1). After 14,000 Vh, the first-dimension gels were equilibrated with sodium dodecyl sulfate, and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described by O'Farrell (21) with a linear gradient of 10 to 17% acrylamide (2).
Proteins were detected by staining with silver nitrate (12). The two-dimensional images were digitized with an Eikonix1412 scanner interfaced with a VAX 4000-90 workstation. The images were processed for spot detection and pattern matching with the Progenesis software (Nonlinear). One two-dimensional image of proteins from cells grown at 22°C was used as a reference pattern for the experiment. All patterns in the experiment (four gels for 3°C and four gels for 22°C) were matched to the reference pattern so that the protein spots were given identification numbers. Statistical analysis of the relative abundance of each matched protein spot across the data set was done with a two-tailed Student t test (12).
The original identifications of S. oneidensis MR-1 proteins were obtained by in-gel tryptic digestion followed by liquid chromatography and tandem mass spectrometry to obtain peptide masses for database searches (5, 27). Protein identifications were obtained by comparing the two-dimensional images from these experiments with two-dimensional images of whole-lysate proteins from S. oneidensis MR-1 available in the Argonne proteome database (http://gelbank.anl.gov) (3) and with published two-dimensional images that included identifications (5, 27).
For lipid analysis, cells were removed from centrifuge tubes while frozen with sequential washes of methanol and phosphate buffer. Dichloromethane was added to these solvents to a ratio of 1:2:0.8 (dichloromethane-methanol-phosphate buffer). This extraction mixture formed the first phase of a modified Bligh and Dyer lipid extraction (6, 11). Phospholipid fatty acids were analyzed following the protocols detailed in Findlay (9), which involved conversion of the phospholipid fatty acids to fatty acid methyl ester by treatment to 0.2 N KOH in methanol and analyzed by gas chromatography (Agilent Technologies, model 6890N). Fatty acids were identified by coelution with known standards and gas chromatographic/mass spectrometric analysis. The fatty acid nomenclature used in this study followed that of Findlay and Dobbs (10).
RESULTS
Overnight cultures of MR-1 grown at 22°C inoculated into fresh medium (LB) and grown at 22°C began growing exponentially after a lag phase of 45 min. In contrast, cells inoculated into fresh medium (LB) and grown at 3°C began exponential growth after a lag phase of ≈135 h. If cells grown at 3°C were used as the inoculum, the lag phase for growth at 3°C was reduced to 48 h. However, cells grown at 3°C inoculated in LB at 22°C showed a lag period of 3 h. The doubling time of MR-1 grown at 22°C was ≈45 min (≈1.4 divisions h−1) while the doubling time at 3°C was ≈67 h (0.015 divisions h−1).
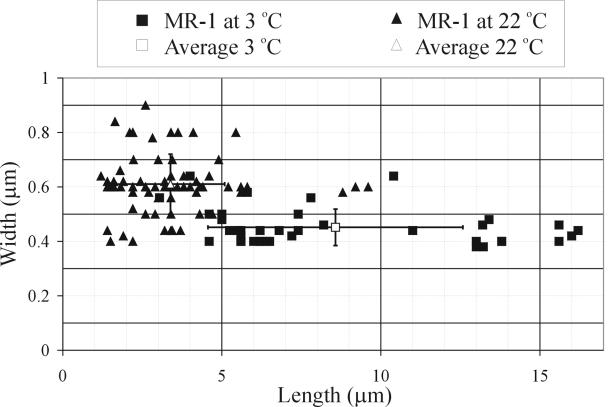
During exponential growth at 3°C and 22°C, the relationships between optical density and both biomass (dry weight) and protein content were linear (not shown). Cells of MR-1 grown at 22°C were rods measuring between 0.4 and 0.9 μm (average, 0.61 ± 0.11 μm) in diameter and 1.2 to 9.6 μm (average, 3.38 ± 1.7 μm) in length (measured from transmission electron microscope images), whereas cells growing at 3°C were filamentous, measuring between 0.38 and 0.64 μm (average, 0.45 ± 0.067 μm) in diameter and between 3.04 and 16 μm (average, 8.57 ± 4 μm) in length (Fig. 1). These averages are significantly different based on a Student t test for unequal variances and α = 0.01. The transmission electron microscope results underestimated the length of the 3°C-grown filaments because many filaments exceeded in length the dimensions of the transmission electron microscope pictures. Under optical microscopy, some filamentous cells were much longer (exceeding 70 μm). These filamentous cells were motile, and some aggregated into groups of up to 10 to 20 entangled filaments.
FIG. 1.
Size variability of S. oneidensis MR-1 grown at 3°C and at 22°C measured from transmission electron microscopy images.
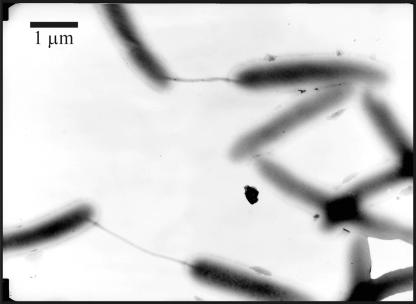
When filamentous cells grown at the low temperature were transferred to the high temperature, their morphology changed. After ≈100 min, cells became shorter (2.9 to 16 μm [average, 5.33 ± 2.45 μm]) and thinner (0.24 to 0.44 μm [average, 0.39 ± 0.06 μm]), and numerous pilus-like structures were observed between cells (Fig. 2).
FIG. 2.
Transmission electron microscopy image showing frequent pilus-like intercellular connections among cells of S. oneidensis MR-1 during the transition from growth at 3°C to 22°C.
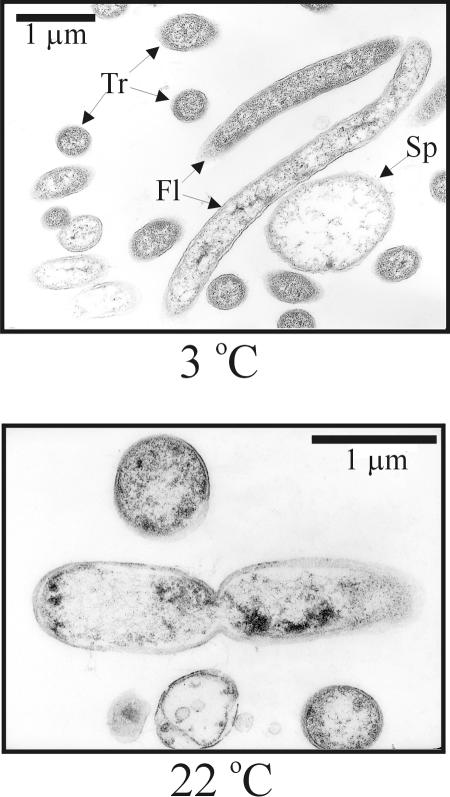
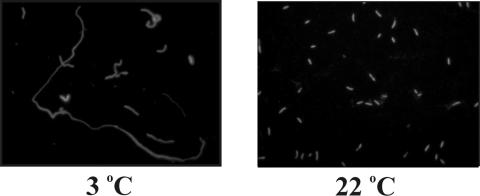
Ultrastructural (thin section) analysis of MR-1 cells grown at 22°C revealed normal dividing cells exhibiting a constriction that separated the longest cells in half (Fig. 3) and electron-dense deposits near the poles of some cells. In contrast, cells grown at 3°C had no median constrictions and formed long filaments. Light microscopy analysis revealed that most cells grown at 3°C were filamentous and fluoresced red after staining with acridine orange (Fig. 4) with only a few cells showing green fluorescence. During early exponential growth, some filaments exhibited terminal and/or lateral spheroids (0.8 to 1.3 μm in diameter) resembling spheroplasts (Fig. 5). Only about 10% of the filaments displayed such structures, and < 50% of the spheroplasts observed were independent of the filaments. These spheroids stained gram-negative, were not refractive under phase contrast microscopy, were not heat resistant, and did not stain with malachite green. The spheroids and their associated filaments always stained orange-red with acridine orange. No green sphere-carrying filaments were observed. When acridine orange-stained cells are examined at 440 to 480 nm, it is generally assumed that double-stranded DNA fluoresces green (520 nm), while RNA or single-stranded DNA fluoresces orange-red (650 nm) (24-26). Thus, orange-red fluorescence in an exponentially growing cell may be regarded as a sign of high metabolic activity (i.e., cells producing more ribosomes), while yellow-green fluorescence may be a sign of dormant cells.
FIG. 3.
Ultrastructure of S. oneidensis MR-1 growing exponentially at two different temperatures (3°C and 22°C) observed by transmission electron microscopy. Most filaments in the image at 3°C are sectioned transversally (Tr). Two filamentous cells are shown (Fl). About 50% of the filaments at 3°C show large vesicles that are similar to spheroplasts (Sp).
FIG. 4.
Cell morphology of S. oneidensis MR-1 grown at two different temperatures (3°C and 22°C), observed by epifluorescent microscopy (×1,000) after staining with acridine orange.
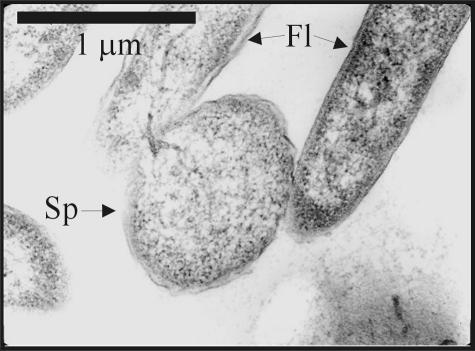
FIG. 5.
Ultrastructure of S. oneidensis MR-1 growing exponentially at 3°C observed by transmission electron microscopy. Two filamentous cells are shown (Fl), one of them with a spheroplast-like structure attached (Sp).
Two-dimensional gel analysis of whole-lysate proteins from cells grown at 3°C and 22°C revealed 17 proteins that were overexpressed and 33 proteins that were significantly underexpressed at 3°C (Table 1). Thirteen of these proteins were identified with the S. oneidensis proteome database, and are listed in Table 2.
TABLE 1.
S. oneidensis MR-1 proteins (four replicates were used) varying significantly (P < 0.001, two-tailed Student t test) in abundance in cells grown at 3 or 22°C
| Spot no.a | Avg integrated density
|
|
|---|---|---|
| 3°C | 22°C | |
| 101 | 37,685 | 72,408 |
| 166 | 21,580 | 58,101 |
| 243 | 60,561 | 21,117 |
| 285 | 33,829 | 62,025 |
| 293 | 78,834 | 33,645 |
| 309 | 0 | 32,895 |
| 310 | 0 | 57,920 |
| 390 | 0 | 34,535 |
| 434 | 56,147 | 27,025 |
| 447 | 76,224 | 29,889 |
| 480 | 0 | 39,392 |
| 481 | 23,179 | 70,452 |
| 524 | 11,831 | 19,872 |
| 543 | 34,234 | 61,477 |
| 548 | 107,889 | 51,836 |
| 579 | 47,128 | 23,780 |
| 583 | 31,161 | 97,196 |
| 584 | 46,592 | 68,581 |
| 585 | 26,032 | 46,312 |
| 609 | 98,501 | 59,919 |
| 624 | 140,439 | 59,529 |
| 647 | 28,946 | 81,954 |
| 694 | 80,455 | 35,591 |
| 720 | 48,961 | 93,055 |
| 753 | 80,315 | 141,705 |
| 808 | 60,477 | 93,153 |
| 869 | 126,461 | 72,214 |
| 874 | 20,189 | 44,896 |
| 893 | 77,472 | 107,077 |
| 896 | 50,433 | 100,508 |
| 909 | 145,848 | 65,118 |
| 920 | 82,232 | 42,842 |
| 926 | 33,842 | 62,129 |
| 927 | 73,102 | 128,701 |
| 943 | 24,643 | 40,599 |
| 988 | 48,029 | 94,485 |
| 1033 | 124,488 | 74,256 |
| 1034 | 31,754 | 227,128 |
| 1054 | 51,567 | 105,946 |
| 1061 | 0 | 166,223 |
| 1070 | 23,301 | 37,900 |
| 1402 | 31,302 | 0 |
| 1570 | 42,617 | 0 |
| 1655 | 0 | 257,104 |
| 1656 | 228,488 | 93,359 |
| 1657 | 0 | 54,707 |
| 1660 | 336,397 | 97,161 |
| 1676 | 0 | 269,291 |
| 1692 | 73,113 | 323,724 |
| 1695 | 36,808 | 253,874 |
Correspond to numbers on gels.
TABLE 2.
Identifications for a subset of proteins from MR-1
| Spot no. | General function | Name of protein | Regulation
|
|
|---|---|---|---|---|
| 3°C | 22°C | |||
| 1676 | Anaerobic respiration | Fumarate reductase | − | + |
| 1655 | Transport | Outer membrane protein OmpW (transport Fe) | − | + |
| 1692 | Small-molecule metabolism | Alcohol dehydrogenase | − | + |
| 753 | Malate dehydrogenase | − | + | |
| 584 | Lipid metabolism | Glutamate-1-semialdehyde-2,1-aminomutase | − | + |
| 896 | Enoyl-CoA hydratase | − | + | |
| 583 | Nucleotide metabolism | Adenylosuccinate synthetase | − | + |
| 874 | Deoxyribosephosphate aldolase | − | + | |
| 1660 | Uracl phosphoribosyltransferase | − | + | |
| 243 | Molybdopterin | Molybdopterin biosynthesis (MoeA) | + | − |
| 1034 | Stress and DNA repair | Universal stress protein | − | + |
| 1054 | DNA-binding protein H-NS family | − | + | |
| 624 | RecA protein | + | − | |
Phospholipids of cells of S. oneidensis MR-1 grown at 3°C and 22°C contained the same complement of fatty acids, although growth temperature was correlated with significant shifts in composition (Table 3). Phospholipid fatty acid profiles of cells grown at 22°C contained three major fatty acids (16:1ω7, i15:0, and 16:0), while profiles of cells grown at 3°C were dominated by i15:0. The changes in composition were also evident as a decrease in fatty acid chain length (20.57 ± 5.70 versus 66.79 ± 2.19% of the fatty acids were 16 carbons or greater in length at 3°C and 22°C, respectively), a decrease in the proportion of unsaturated fatty acids (13.49 ± 5.99 versus 44.57 ± 1.85; 3°C versus 22°C), and an increase in the proportion of branched fatty acids (82.44 ± 5.99 versus 27.21 ± 1.62 for 3°C versus 22°C).
TABLE 3.
Fatty acid composition of Shewanella oneidensis at 3 and 22°C
| Fatty acid | % of total wt
|
|
|---|---|---|
| 3 °C | 22 °C | |
| 12:0 | 0.02 ± 0.01 | 0.04 ± 0.01 |
| i13:0 | 3.60 ± 2.00 | 0.25 ± 0.06 |
| 13:0 | 0.03 ± 0.02 | 0.07 ± 0.02 |
| i14:0 | 1.65 ± 0.62 | 0.48 ± 0.09 |
| 14:1a | 0.19 ± 0.09 | 0.21 ± 0.03 |
| 14:1b | 0.14 ± 0.05 | 0.08 ± 0.01 |
| 14:0 | 0.58 ± 0.26 | 4.03 ± 0.43 |
| br15:1 | 0.47 ± 0.24 | 0.01 ± 0.01 |
| i15:0 | 70.09 ± 3.83 | 24.38 ± 1.37 |
| a15:0 | 2.02 ± 0.47 | 0.83 ± 0.13 |
| 15:1a | 0.20 ± 0.10 | 0.19 ± 0.02 |
| 15:0 | 0.38 ± 0.18 | 2.65 ± 0.18 |
| i16:0 | 0.57 ± 0.21 | 0.29 ± 0.02 |
| 16:1ω9 | 1.48 ± 0.85 | 3.09 ± 0.06 |
| 16:1ω7 | 5.52 ± 2.26 | 29.89 ± 1.37 |
| 16:1ω7t | 0.05 ± 0.05 | 0.21 ± 0.06 |
| 16:1ω5 | 0.43 ± 0.20 | 0.09 ± 0.03 |
| 16:0 | 2.00 ± 0.69 | 20.38 ± 0.36 |
| br17:1 | 0.30 ± 0.10 | 0.04 ± 0.01 |
| 10me16:0 | 0.18 ± 0.05 | 0.04 ± 0.01 |
| i17:0 | 3.51 ± 0.37 | 0.89 ± 0.06 |
| 17:1a | 0.15 ± 0.08 | 0.27 ± 0.02 |
| 17:1ω8 | 1.01 ± 0.33 | 3.24 ± 0.34 |
| cy17:0 | 1.29 ± 0.84 | 0.31 ± 0.02 |
| 17:1ω6 | 0.06 ± 0.06 | 0.02 ± 0.02 |
| 17:0 | 0.19 ± 0.10 | 0.70 ± 0.61 |
| 18:1ω9 | 0.42 ± 0.39 | 2.55 ± 0.19 |
| 18:1ω7 | 2.93 ± 2.95 | 4.11 ± 0.24 |
| 18:1ω7t | 0.00 ± 0.00 | 0.01 ± 0.01 |
| 18:1ω5 | 0.31 ± 0.24 | 0.05 ± 0.01 |
| 18:0 | 0.11 ± 0.06 | 0.46 ± 0.03 |
| cy19:0 | 0.03 ± 0.01 | 0.03 ± 0.02 |
| 20:5ω3 | 0.04 ± 0.03 | 0.09 ± 0.06 |
DISCUSSION
Not only do S. oneidensis MR-1 cells grow at 3°C (μ = 0.015 divisions h−1), they also exhibit a number of unusual features, including filamentous cells, smaller diameter, presence of spheroplasts, some proteins being differently expressed, and extensive changes in cellular lipids. Cells growing at 3°C are considerably smaller in diameter (≈50%) and many are filamentous, reaching lengths ≈10 to 70 times longer than cells grown at 22°C. Upon transfer to the low temperature, a long lag period (>100 h) is seen, followed by exponential but very slow growth. These slow-growing cells are notably different, based on cell morphology, cellular proteins, and cellular lipids.
The meaning of the filamentous phenotype is unclear. One can speculate that since S. oneidensis faces large seasonal temperature changes, this phenotype may represent an adaptation to ease predation by protozoans, as suggested previously (7, 13). Another explanation may be that because temperature influences the kinetics of differently catalyzed reactions differently, large changes in temperature (within limits appropriate for growth) may result in significant changes in the state of biological systems. The filamentous phenotype may simply be the result of low-temperature disequilibrium between growth and septation. At this point our results cannot support either explanation.
Many bacteria alter membrane fatty acid composition in response to changes in growth temperature as a mechanism for maintaining membranes in a fluid state necessary for biological functioning. Previously observed changes in growth at lower temperatures include an increased proportion of unsaturated fatty acids, an increased proportion of branched fatty acids, and a decrease in fatty acid chain length (8). With the shift in growth temperature from 22°C to 3°C, MR-1 showed an increased proportion of branched fatty acids and a decrease in fatty acid chain length but a decreased proportion of unsaturated fatty acids. The 3.3-fold decrease in unsaturation is likely offset by approximately 80% of the phospholipid fatty acids being branched and 15 carbons long or shorter. These changes in phospholipid fatty acid composition, which occur without changes in the fatty acid complement, may provide the homeophasic adaptation necessary for active growth and metabolism at low temperatures.
With the changes in morphology and lipid content, one should expect to see major differences in protein content, and this was seen. Some proteins, such as RecA, MoeA, (molybdopterin biosynthesis), and T-protein glycine cleavage system (amino acid metabolism), were overexpressed at 3°C. A variety of other proteins are downregulated at 3°C, such as proteins used in respiration, transport, synthesis of small metabolites, amino acid synthesis, lipid metabolism, and nucleotide synthesis. These results are in concert with several other reports about cold acclimation proteins in other mesophiles (16, 22). However, given the preliminary nature of the two-dimensional gel analyses, most of the variable proteins are surely not yet elucidated, and the reason for this response can only be speculated upon at this time. Further studies of both genomic and proteomic responses of MR-1 to changes in temperature are now under way.
The changes that occur upon transfer back to high temperature have not been studied in detail. Rapid cell division is seen without an increase in biomass, suggesting that sufficient DNA is present in the long filaments. Equally interesting is the appearance of numerous pilus-like structures upon short-term shift from growth at 3°C to 22°C that are similar to structures found in some species of Clostridium (14, 15, 18). At this point we have insufficient information to understand the meaning of this. Are these sex pili? Is there some reason to enhance genetic exchange under such conditions, or are they somehow involved with exchange of nutrients? Genomic and proteomic (global analyses) now under way may well provide insight into the reason for these responses to low temperature.
Acknowledgments
This research was supported by grants from the U.S. Department of Energy, Office of Biological and Environmental Research Genomics, GTL Program, to K.H.N. and C.S.G. (contract W-31-109-ENG-38). V.S.-E. was supported by a postdoctoral grant from Secretaría de Estado de Educación y Universidades (Spain).
REFERENCES
- 1.Anderson, N. G., and N. L. Anderson. 1978. Analytical techniques for cell fractions. 21. 2-dimensional analysis of serum and tissue proteins-multiple isoelectric-focusing. Anal. Biochem. 85:331-340. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, N. L., and N. G. Anderson. 1978. Analytical techniques for cell fractions. 22. 2-dimensional analysis of serum and tissue proteins-multiple gradient-slab gel-electrophoresis. Anal. Biochem. 85:341-354. [DOI] [PubMed] [Google Scholar]
- 3.Babnigg, G., and C. S. Giometti. 2003. GELBANK: a database of annotated two-dimensional gel electrophoresis patterns of biological systems with completed genomes. Nucleic Acids Res. 32:D582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakermans, C., and K. H. Nealson. 2004. Relationship of critical temperature to macromolecular synthesis and growth yield in Psychrobacter cryopegella. J. Bacteriol. 186:2340-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beliaev, A. S., D. K. Thompson, M. W. Fields, L. Y. Wu, D. P. Lies, K. H. Nealson, and J. Z. Zhou. 2002. Microarray transcription profiling of a Shewanella oneidensis etrA mutant. J. Bacteriol. 184:4612-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Med. Sci. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 7.Caron, D. A., J. C. Goldman, and M. R. Dennett. 1988. Experimental demonstration of the roles of bacteria and bacterivorous protozoa in plankton nutrient cycles. Hydrobiology 159:27-40. [Google Scholar]
- 8.Denich, T. J., L. A. Beaudette, H. Lee, and J. T. Trevors. 2003. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J. Microbiol. Methods 52:149-182. [DOI] [PubMed] [Google Scholar]
- 9.Findlay, R. H. Determination of microbial community structure using phospholipid fatty acid profiles. In G. A. Kolalchuk, F. J. Bruijn, I. M. Head, A. D. Akkermans, and J. D. Elsas (ed.), Molecular microbial ecology manual, 2nd ed., in press. Kluywer Academic Publishers, Dordrecht, The Netherlands.
- 10.Findlay, R. H., and F. C. Dobbs. 1993. Quantitative description of microbial communities using lipid analysis, p. 271-284. In P. F. Kemp et al. (ed.), Current methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 11.Findlay, R. H., G. M. King, and L. Watling. 1989. Efficacy of phospholipid analysis in determining microbial biomass in sediments. Appl. Environ. Microbiol. 55:2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giometti, C. S., M. A. Gemmell, S. L. Tollaksen, and J. Taylor. 1991. Quantitation of human-leukocyte proteins after silver staining-a study with 2-dimensional electrophoresis. Electrophoresis 12:536-543. [DOI] [PubMed] [Google Scholar]
- 13.Guede, H. 1979. Grazing by protozoa as selection factor for activated sludge bacteria. Microb. Ecol. 5:225-237. [DOI] [PubMed] [Google Scholar]
- 14.Kuesel, K., A. Karnholz, T. Trinkwalter, R. Devereux, G. Acker, and H. L. Drake. 2001. Physiological ecology of Clostridium glycolicum RD-1, an aerotolerant acetogen isolated from sea grass roots. Appl. Environ. Microbiol. 67:4734-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhner, C. H., C. Matthies, G. Acker, M. Schmittroth, A. S. Goessner, and H. L. Drake. 2000. Clostridium akagii sp. nov. and Clostridium acidisoli sp nov., acid-tolerant, N2-fixing clostridia isolated from acidic forest soil and litter. Int. J. Syst. Evol. Microbiol. 50:873-881. [DOI] [PubMed] [Google Scholar]
- 16.Liu, S. Q., J. E. Graham, L. Bigelow, P. D. Morse, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 18.Matthies, C., C. H. Kuhner, G. Acker, and H. L. Drake. 2001. Clostridium uliginosum sp. nov., a novel acid-tolerant anaerobic bacterium with connecting filaments. Int. J. Syst. Evol. Microbiol. 51:1119-1125. [DOI] [PubMed] [Google Scholar]
- 19.Mills, E. L., and K. T. Holeck. 2001. Oneida lake: undergoing ecological change. Clearwaters 31:4. [Google Scholar]
- 20.Myers, C. R., and K. H. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319-1321. [DOI] [PubMed] [Google Scholar]
- 21.O'Farrell, P. H. 1975. High resolution two-dimensional polyacrylamide gel electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 22.Panoff, J. M., D. Corroler, B. Thammavongs, and P. Boutibonnes. 1997. Differentiation between cold shock proteins and cold acclimation proteins in a mesophilic gram-positive bacterium, Enterococcus faecalis JH2-2. J. Bacteriol. 179:4451-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramagli, L. S., and L. V. Rodriguez. 1985. Quantitation of microgram amounts of protein in two-dimensional polyacrylamide-gel electrophoresis sample buffer. Electrophoresis 6:559-563. [Google Scholar]
- 24.Romeis, B. (ed.). 1968. Mikroskopische Technik. Oldenbourg Verlag, Munich, Germany.
- 25.Shapiro, H. M. 1988. Practical flow cytometry, 2nd ed. A. R. Liss, New York, N.Y.
- 26.Stockert, J. C., and J. A. Lisanti. 1972. Acridine orange differential fluorescence of fast-and slow-reassociating chromosomal DNA after in-situ DNA denaturation and reassociation. Chromosoma (Berlin) 37:117-130. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, D. K., A. S. Beliaev, C. S. Giometti, S. L. Tollaksen, T. Khare, D. P. Lies, K. H. Nealson, J. Lim, J. Yates, C. C. Brandt, J. M. Tiedje, and J. Z. Zhou. 2002. Transcriptional and proteomic analysis of a ferric uptake regulator (fur) mutant of Shewanella oneidensis: possible involvement of fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68:881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]