Abstract
To date, anti-CTLA-4 (ipilimumab) or anti-PD-1 (nivolumab) monotherapy has not demonstrated significant clinical benefit in patients with prostate cancer. To identify additional immune inhibitory pathways in the prostate tumor microenvironment, we evaluated untreated and ipilimumab-treated tumors from patients on a pre-surgical clinical trial. PD-L1 and VISTA inhibitory molecules increased on independent subsets of macrophages. Our data suggest that VISTA is another compensatory inhibitory pathway in prostate tumors after ipilimumab therapy.
MAIN TEXT
Immune checkpoint therapies, including anti-CTLA-4 and anti-PD-1, that block T cell inhibitory pathways have led to durable anti-tumor responses and clinical benefit in a significant number of cancer patients 1,2. However, prostate cancer has proven poorly responsive to immune checkpoint monotherapy 3–5. To better understand the immune profile within prostate tumors and potential compensatory immune inhibitory pathways that may arise in the setting of immune checkpoint monotherapy, we conducted a clinical trial (NCT01194271) with ipilimumab plus androgen deprivation therapy (ADT) prior to surgery in patients with localized prostate cancer (Supplementary Fig. 1a–c, Supplementary Tables 1 and 2).
We compared post-treatment blood and baseline samples (Supplementary Fig. 1a), with evaluation of CD4 and CD8 T cells (Supplementary Fig. 2a) as well as T cell subsets expressing inducible costimulator (ICOS), OX40, 4-1BB, PD-1, CTLA-4, and FoxP3 (Supplementary Fig. 2a–b). We observed an increase in CD4 and CD8 T cells, including PD-1+ and ICOS+ subsets after ipilimumab therapy, which is similar to our previous finding with ipilimumab monotherapy in patients with melanoma and bladder cancer 6–8. We also compared post-treatment tumor tissues (Supplementary Fig. 1a) to stage-matched untreated tumors from another cohort of patients (Supplementary Fig. 1b). Flow cytometric studies revealed a significantly higher frequency of CD4, CD8, and ICOS+ T cells in post-treatment tumors (Fig. 1a). Immunohistochemical (IHC) studies also demonstrated significant increases in tumor infiltrating immune cells, including CD4+, CD8+, ICOS+, CD45RO+, granzyme-B (GrB)+, and CD68+ cells (Supplementary Fig. 3). We found a significantly greater immune cell infiltration in prostate tumors after ipilimumab therapy but not ADT, although ADT monotherapy was associated with significantly higher ICOS+ and granzyme-B+ cells, which may reflect an activated T cell subset (Fig. 1b). Taken together, our data suggest that the immunologic changes in post-treatment tumors were mostly due to ipilimumab therapy, as opposed to ADT. However, we cannot discount a possible synergistic effect between ipilimumab and ADT.
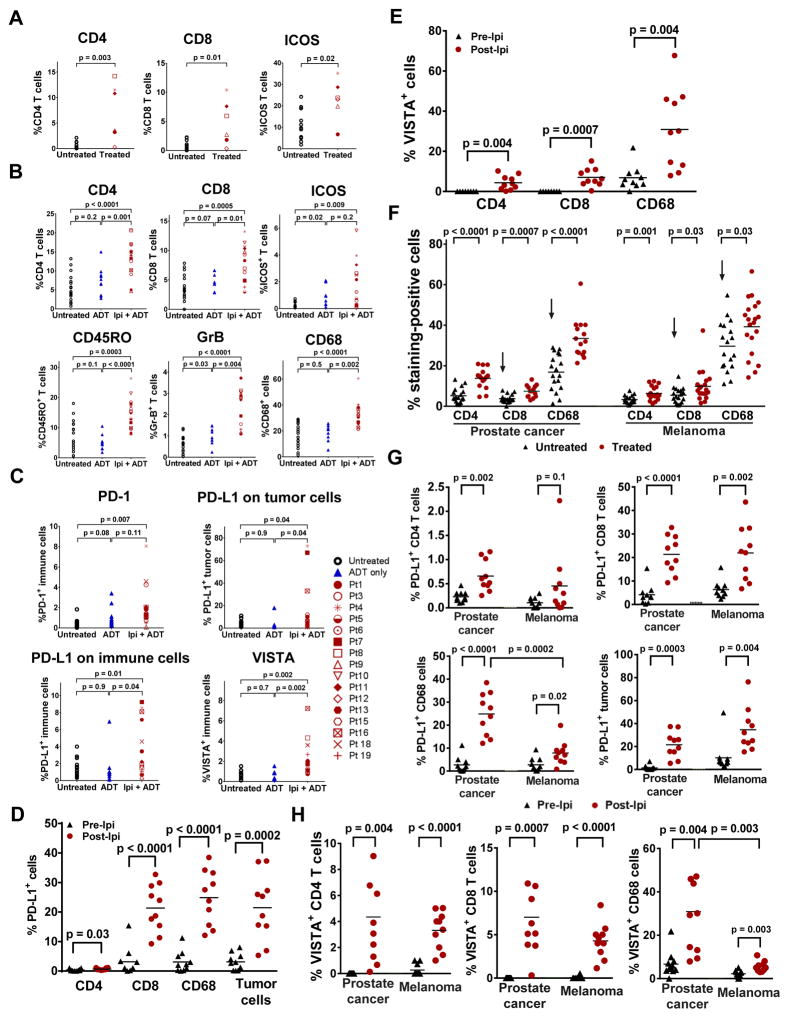
Figure 1. Treatment with ipilimumab increases immune cell infiltration, as well as expression of PD-L1 and VISTA in prostate tumors.
(a) Frequency of CD4, CD8 and ICOS+ T cells in untreated (N=11) and treated (N=6) tumors. (b) IHC analyses of CD4, CD8, ICOS+, CD45RO+, and GrB+ T cells, as well as CD68+ macrophages. (c) IHC of PD-L1, PD-1, and VISTA in tumor cells and tumor-infiltrating immune cells, with b-c comprised of tumors from 3 different cohorts of stage-matched patients: untreated (N=18), ADT-treated (N=10), and ipilimumab + ADT (N=16). Asterisk indicates patients received high dose steroids with surgery delay. (d) Frequency of PD-L1 expression on CD4 T cells, CD8 T cells, CD68+ macrophages, and tumor cells. (e) Frequency of VISTA expression on CD4 T cells, CD8 T cells, and CD68+ macrophages, with d-e comprised of matched pre-treatment (N=10) and post-treatment tumors (N=10). (f) IHC staining of CD4 and CD8 T cells, and CD68+ macrophages in stage-matched untreated (N=18) and ipilimumab + ADT-treated (N=15) prostate tumors as compared to stage-matched untreated (N=18) and ipilimumab-treated (N=20) metastatic melanomas. Arrows indicate significant difference of CD8 T cells and CD68+ macrophages between untreated prostate tumors and untreated melanomas. (g) Frequency of PD-L1 expression on CD4 T cells, CD8 T cells, and CD68+ macrophages. (h) Frequency of VISTA expression on CD4 T cells, CD8 T cells, and CD68+ macrophages, with g-h comprised of matched pre-treatment (N=10) and post-treatment prostate tumors (N=10) as compared to matched pre-treatment (N=10) and post-treatment melanomas (N=10).
We did not observe clinical responses consisting of pathologic complete responses as we did previously for patients with bladder cancer 8. To identify potential mechanisms that may explain this lack of response, we performed an unbiased gene expression study and found that ipilimumab therapy resulted in significant changes in expression of a total of 690 genes (FDR<0.2; p<0.028; log2 fold change >0.5) (Supplementary Table 3), most of which are related to immune responses (Supplementary Fig. 4a). We focused our analyses on a subset of genes that represent inhibitory immune checkpoints and identified increased PD-L1 and VISTA expression in post-treatment tumors (Supplementary Fig. 4b). Both PD-L1 and VISTA were previously reported as inhibitory molecules that can inhibit murine and human T cell responses 9,10. Here we found significantly greater protein expression of PD1, PD-L1 and VISTA in prostate tumors after ipilimumab therapy (Supplementary Fig. 5a, Fig. 1c).
We also evaluated metastatic tumors and blood samples from patients with metastatic prostate cancer from a separate clinical trial (NCT02113657) who received treatment with ipilimumab and found an increase in PD-L1 and VISTA expression in tumor tissues (Supplementary Fig. 5b) as well as on monocytes in blood (Supplementary Fig. 6a), which was similar to data from a mouse model of prostate cancer (Supplementary Fig. 6b). We suggest that PD-L1 and VISTA are likely to be relevant inhibitory immune checkpoints in both localized and metastatic prostate cancer.
We evaluated PD-L1 and VISTA expression in different cell subtypes from matched pre- and post-treatment prostate tumors and observed significant increases in PD-L1 expression on CD4 T cells, CD8 T cells and CD68+ macrophages (Supplementary Fig. 7a). On average, we observed an increase of approximately 3-fold (from 0.2% to 0.7%) in PD-L1 expression on CD4 T cells (Fig. 1d). More strikingly, we observed an average increase of approximately 5-fold (from 4.4% to 21.3%) in PD-L1 expression on CD8 T cells and an increase of approximately 10-fold (from 2.5% to 25%) in PD-L1 expression on CD68+ macrophages (Fig. 1d). We also observed, on average, an increase of approximately 12-fold (from 1.8% to 21.5%) in PD-L1 expression on tumor cells (Fig. 1d). Similarly, ipilimumab therapy resulted in significant increases in VISTA expression on CD4 T cells, CD8 T cells and CD68+ macrophages (Supplementary Fig. 7b). To our knowledge, this represents the first report of VISTA expression on T cells in human tumors 10. Detectable VISTA expression was not observed on CD4 and CD8 T cells in pre-treatment tumor tissues but was detected at approximately 4% on CD4 T cells and 7% on CD8 T cells after ipilimumab therapy (Fig. 1e). VISTA expression on CD68+ macrophages was observed to increase approximately 4-fold (from 7% to 31%) (Fig. 1e).
Next, we compared stage-matched untreated and post-therapy prostate tumors with stage-matched untreated and post-therapy metastatic melanomas. We found that untreated melanomas had significantly higher CD8 T cells and CD68+ macrophages than untreated prostate tumors (p=0.04 and p=0.0005, respectively, Fig. 1f). Both melanoma and prostate post-treatment tumors demonstrated an approximate 2-fold higher frequency of CD4 T cells and an approximate 1.5-fold higher frequency of CD8 T cells and CD68+ macrophages as compared to the respective stage-matched untreated tumors (Fig. 1f). Our data indicate an increase in PD-L1 expression on T cells and tumor cells in post-treatment tumor tissues for both tumor types (Fig. 1g). However, we found that CD68+ macrophages had significantly greater PD-L1 expression in post-treatment prostate tumors, with an approximate 3-fold greater expression of PD-L1 on CD68+ macrophages (Fig. 1g). Since PD-L1 expression is known to be regulated by IFN-γ and ipilimumab therapy is known to increase IFN-γ production by T cells 6,11, we evaluated potential correlations between PD-L1 and IFN-γ responsive genes (IL-15, IFNAR2, CXCL10, IRF1) and found a high degree of correlation between expression of PD-L1 and IFN-γ responsive genes in post-treatment prostate tumors and melanomas (Supplementary Fig. 8).
We also found a higher frequency of VISTA expression on T cells and CD68+ macrophages in post-treatment melanoma and prostate tumors as compared to matched pre-treatment tumors (Fig. 1h). However, we found that CD68+ macrophages had significantly greater VISTA expression in post-treatment prostate tumors, with an approximate 5-fold greater expression of VISTA on CD68+ macrophages in prostate tumors based on percent expression (Fig. 1h). Unlike PD-L1, there was no correlation between VISTA expression and IFN-γ responsive genes (Supplementary Table 4).
We sought to determine whether PD-L1 and VISTA were co-expressed on CD68+ macrophages or represented individual subsets of inhibitory cells in post-treatment prostate tumor tissues (Fig. 2a). We found that PD-L1+CD68+ cells comprised about 29.4% of all CD68+ macrophages, while VISTA+CD68+ cells had a similar frequency of 26.5% and PD-L1+VISTA+ double positive CD68+ cells comprised only 2% of the total CD68+ population in post-treatment tumor tissues (Fig. 2b). CyTOF analysis on fresh tumors from 2 patients who received ipilimumab therapy on a separate protocol (NCT02113657) also demonstrated PD-L1 and VISTA expression on predominantly distinct subsets of CD68+ macrophages (Supplementary Fig. 9).
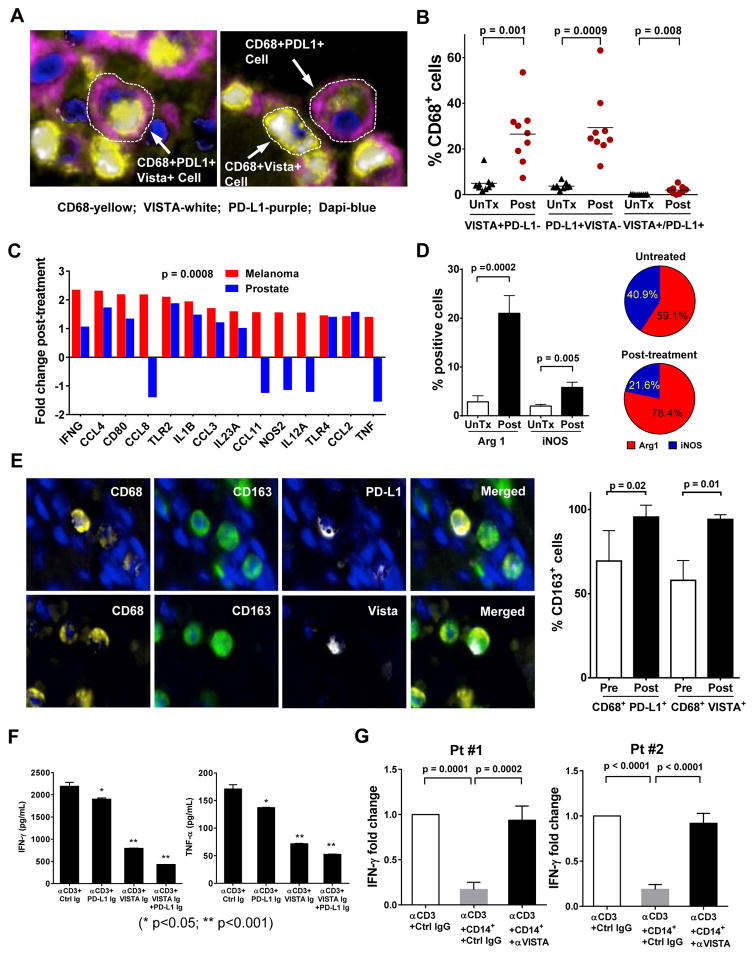
Figure 2. PD-L1+ and VISTA+ macrophages (CD68+) manifest an M2-phenotype and suppress T cell function.
(a) Representative photographs from immunofluorescence (IF) multiplex staining of PD-L1 (purple), VISTA (white), CD68 (yellow), and tumor nuclei (blue) in post-treatment prostate tumors. (b) Frequency of PD-L1 and VISTA expression on CD68+ macrophages from stage-matched untreated (UnTx; N=9) and post-treatment prostate tumors (N=9); (c) Fold induction of M1-like genes in post-treatment prostate tumors (N=6) and post-treatment melanomas (N=20) as compared to respective untreated samples. (d) Quantitative IHC analysis of Arg1+ cells in untreated (N=10) and post-treatment (N=15) prostate tumors (left panel), as well as ratio of Arg1/iNOS in untreated and post-treatment tumors (right panel). (e) Representative photographs from multiplex IF staining of tumor (blue), CD68 (yellow) and CD163 (green) with PD-L1 (white) or VISTA (white) in post-treatment prostate tumors (upper panel), as well as quantitative analysis of CD163 expression by CD68+PD-L1+ or CD68+VISTA+ cells from pre- (N=5) and post-treatment (N=5) prostate tumors (lower panel). (f) IFN-γ (left panel) and TNF-α (right panel) production by patients’ (N=7) peripheral T cells after CD3 stimulation in the presence of plate-bound control Ig (Ctrl Ig), PD-L1-Ig, VISTA-Ig protein or a combination of both. (g) IFN-γ production by patient’s peripheral T cells after CD3 stimulation without co-culture of monocytes or in the presence of either untreated monocytes or monocytes pretreated with anti-VISTA antibody. Experiments were performed in triplicates.
Since macrophages can be classified as M1 or M2 subtypes, which function to promote anti-tumor responses or tumor growth, respectively genes 12, we evaluated post-treatment prostate and melanoma tumors for changes in expression of M1-like genes. We found higher fold induction of M1-like genes in post-treatment melanomas as compared to post-treatment prostate tumors (Fig. 2c). Additional studies also demonstrated a significant increase (7.4-fold) in Arg1 expression (M2-like) in post-treatment prostate tumor tissues (Fig. 2d). The ratio of Arg1 (M2-like) to iNOS (M1-like) changed from 1.4 to 3.6 in pretreatment vs. post-treatment tumors (Figure 2d). Furthermore, we found that both CD68+PD-L1+ and CD68+VISTA+ cells had a significant increase in expression of CD163 (M2-like) in post-treatment samples (Fig. 2e). Overall, our data show an increase in PD-L1+ and VISTA+ macrophages, with higher expression of CD163 and Arg1 suggestive of M2-like phenotype and function, respectively. In addition, in vitro studies with plate-bound protein of PD-L1 or VISTA led to a significant decrease in production of IFN-γ and TNF-α by T cells from prostate cancer patients (Fig. 2f). In contrast, pretreatment of monocytes with anti-VISTA resulted in significant reversal of monocyte suppression of T cell IFN-γ production (Fig. 2g).
The identification of multiple immune checkpoints, such as PD-L1 and VISTA, indicates the importance of understanding the expression of these molecules within the tumor microenvironment in order to design rational combination treatment strategies. Our data strongly suggest that therapies with anti-CTLA-4 can enhance immune cell infiltration into tumors, including prostate cancer. These data highlight the importance of on-treatment biopsies, possibly after 2 doses of therapy, to identify the immunologic changes that occur in the tumor microenvironment. Our data suggest that an increase in immune cell infiltration may be insufficient to generate anti-tumor responses and blockade of other immune checkpoints such as PD1/PD-L1 and/or VISTA may be necessary to provide significant clinical benefit for patients with prostate cancer. To our knowledge, our data represent the first finding of VISTA as a compensatory inhibitory pathway in the setting of ipilimumab therapy in prostate cancer but, future studies will need to elucidate the role of VISTA as a potential resistance mechanism and determine whether VISTA can be targeted to improve anti-tumor responses in patients.
ONLINE METHODS
Clinical trial
Patients with localized, high-risk (T3, PSA>20, or Gleason score 7–10) prostatic adenocarcinoma who were candidates for radical prostatectomy were consented on a M. D. Anderson Cancer Center Institutional Review Board (IRB)-approved protocol (MDACC 2009-0135, NCT01194271) to receive one intramuscular injection of ADT (leuprolide acetate, 22.5 mg intramuscular) on week 0 and ipilimumab at 10 mg/kg/dose on weeks 1 and 4. Patients were then scheduled for surgery at week 8 (Supplementary Fig. 1a). Optional biopsies of tumor tissues were collected prior to treatment and tumor tissues were then collected at the time of surgery for immune monitoring studies. Blood was also collected prior to each therapeutic intervention for immune monitoring studies. A total of 20 patients were evaluated for this trial. Patient #2 and 20 failed to meet eligibility criteria; patients #14 withdrew from consent prior to treatment; the other 17 patients received ADT plus ipilimumab treatment and completed analyses for safety. Of these 17 patients, 16 completed surgery. One patient died of a cardiac complication prior to surgery 13. All 17 patients experienced some type of immune-related adverse event (irAE); however, grade 3–4 irAEs occurred in 8/17 patients and consisted of colitis/diarrhea, hypophysitis, pancreatitis, and transaminitis (Supplementary Table 2), which were treated with corticosteroids and other immune-suppressive agents as previously described 13. Only two patients (#15 and #16) experienced grade 3–4 irAEs prior to surgery, which required high dose corticosteroids with subsequent delay of surgery. We did not observe any surgical or post-operative complications as a result of treatment on the clinical trial. Of the 16 patients, 6 developed biochemically recurrent disease as measured by elevated blood prostate specific antigen (PSA) levels. Among these 6 patients with biochemical recurrence, 3 were found to have detectable metastatic disease on imaging studies. Ten patients were noted to remain without any evidence of disease for at least 3.5 years. One patient was lost to follow-up and 15 of 16 patients remain alive 3.5 years after completion of last surgery as of November 2016. Healthy donor blood and additional tissue samples from untreated patients with localized prostate cancer (Supplementary Fig. 1b) and ADT-treated prostatectomy samples (Supplementary Fig. 1c) were obtained for comparison as per IRB-approved laboratory and clinical protocols MDACC 2005-0027, 2009-0322, and PA13-0291. Tumor tissues from metastatic castration resistant prostate cancer (CRPC) patients pre- and post-ipilimumab therapy were obtained from IRB-approved clinical protocol MDACC 2013-0444 (NCT02113657).
IHC
For IHC, primary antibodies were used to detect CD4 (Novocastra, CD4-368-L-A), CD8 (Thermo Scientific, MS-457-S), CD45RO (Novocastra, PA0146), GrB (Leica Microsystems, PA0291), ICOS (Spring Bioscience, M3980), CD68 (Dako, M0876), PD-L1 (Cell Signaling, 13684S), PD-1 (Epitomicsm, Abcam, ab137132), Arginase-1 (Cell Signaling, 93668), iNOS (Abcam, ab95866) and VISTA (proprietary antibody from Janssen). Slides were scanned and digitalized using the scanscope system (Scanscope XT, Aperio/Leica). Quantitative analyses of IHC staining were conducted using image analysis software (ImageScope-Aperio/Leica). Five random areas (at least 1 mm2 each) were selected and then a customized algorithm specific for each marker was used for quantification (percent of positive cells). Only samples that had greater than 1% tumor cells present in the sample were used for analyses. Two pathologists (IIW and JB) performed independent review of IHC and immunofluorescence data. PD-L1 staining was carried out as described previously 14. Representative pictures for positive and negative controls that were analyzed for VISTA antibody are shown in Supplementary Fig. 10. VISTA antibody development and validation was performed according to Janssen’s patent description at https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2015097536. Parental human leukemia cell line K562 (Supplementary Fig. 10a) was used as a negative control; VISTA-transfected K562 cell line (Supplementary Fig. 10b) and human tonsil tissue (Supplementary Fig. 10c) were used as positive controls for our studies and demonstrated cell surface staining (Supplementary Fig. 10d). The anti-VISTA antibody used for our IHC studies was obtained from Janssen and will be available via a material transfer agreement between interested researchers and Janssen.
Multiplex immunofluorescence assay and multispectral analysis
Multiplex staining was performed per the Opal protocol staining method 15 for markers: CD4 (1:25, CM153BK, Biocare) with subsequent visualization using fluorescein AF-488 (1:50); CD8 (1:200, M7103, Dako) with visualization using AF-594 (1:50); CD68 (1:100, M0876, Dako) with visualization using AF-647; VISTA (1:100, Janssen) with visualization using coumarin (1:50); CD163 (1:100, NCL-L-CD163, Leica) with visualization using AF-488 and PD-L1 (1:100, 13684, Cell Signaling Technology) with visualization using AF-555 (1:50). Nuclei were visualized with DAPI (1:2000). All sections were cover-slipped using Vectashield Hardset 895 mounting media.
For multispectral analysis, each individually stained section (CD4/AF-488, CD8/AF-594, CD68/AX-647, VISTA/coumarin, PD-L1/AF-555, CD163/AF-488, and DAPI) was utilized to establish the spectral library of fluorophores. Slides were scanned using the Vectra slide scanner (PerkinElmer). For each marker, the mean fluorescent intensity per case was determined as a base point from which positive calls could be established. The co-localization algorithm was used to determine percent of PD-L1 and VISTA staining on each cellular subset. Five random areas on each sample were analyzed blindly by a pathologist at 20× magnification.
Cytometry by time of flight (CyTOF)
Fresh metastatic prostate tumor samples were used for the CyTOF analysis. The tumor samples were dissociated with the GentleMACS system (Miltenyi Biotec; Bergisch Gladbach, Germany) and cultured overnight in RPMI complete media. The cells were then collected and stained with 36 analytes including CD68, PD-L1, VISTA, CD70, Foxp3, BTLA, 41BBL, ICOSL, CD80, B7-H4, CTLA-4, CD3, TIM-3, CD27, CD86, PD-1, CD28, KI67, 41BB, TIGIT, CD73, CD4, CD8, OX40, CD326, ICOS, LAG3, Galectin-9, HVEM, B7-H3, CD45, GITR, PD-L2, OX40L, HLA-DR, and CD56. The data was analyzed using ViSNE program 16 and Phenograph algorithm 17 programs.
Flow cytometry analysis
Fresh peripheral blood mononuclear cells (PBMC) preparation, tumor single cell suspensions (> 100,000 total cells at different time points), and multiparameter flow cytometric analysis of T cell subsets including CD4, CD8, ICOS, Foxp3, ICOS (Biolegend, CA), PD-1 (Biolegend, CA), 4-1BB, OX40, CTLA-4, CD14 (eBioscience, CA), CD16 (BD Biosciences, CA), PD-L1 (BD Biosciences, CA), and VISTA (kindly provided by Janssen, CA), were carried out as we previously reported 6.
Microarray and nanostring analyses
Pre- and post-therapy prostate tumor samples were collected for total RNA isolation using the Qiagen RNeasy Mini Kit. Total RNA was used for Affymetrix GeneChip Human Gene 1.0 ST Array analysis. Microarray data were preprocessed using the aroma.affymetrix bioconductor package. Background correction and quantile normalization were performed using Robust Multi-chip Average. Two sample Welch t-test was used to compare the gene expression of the pre-therapy and post-therapy groups. The beta-uniform mixture model was used to fit the p-value distribution for multiple testing adjustments. Pathway analysis was performed using IPA (Ingenuity Pathway Analysis) (Redwood City, CA). The Pearson correlation was calculated between PD-L1 and each IFN-response gene in melanoma and in prostate respectively. The microarray data have been submitted in MIME-compliant format to GEO with GSE number of GSE77910. The nanostring data have been reported previously 18.
In vitro human T cell function analyses
PBMCs from 7 patients with prostate cancer who received ipilimumab on protocol NCT02113657 were used for these studies. Briefly, 96-well plates were coated overnight with 2.5 μg/ml anti-CD3 (BD Bioscience), together with Fc fusion proteins: 10 μg/ml VISTA Ig (R&D Systems), 10 μg/ml PDL-1 Ig (R&D Systems), 10 μg/ml Control Ig (R&D Systems) or a VISTA/PDL-1 Ig combination. CD3 T cells were plated at 2×105 cells per well in complete TCCM media (IMDM-500ml Human AB Serum (5%), Pen/Strep, HEPES, 2-Mercaptoethanol and Gentamicin) and incubated for 48 hours. The supernatants were collected after 48 hours and cytokines (IFN-γ and TNF-α) were measured using the Mesoscale Discovery system (Rockville, MD).
For co-culture studies, blood samples were obtained from 2 patients treated with ipilimumab. A total of 100,000 purified CD3+ T cells were cultured with or without 500,000 CD14+ monocytes in 96-well plates in triplicate wells. Fifty microliters of anti-CD3 antibody (0.5 μg/mL) was added to each well for 2 hours at 37°C prior to adding cells. For blocking cell surface VISTA, CD14+ monocytes were incubated with 5 μg anti-VISTA antibody (kindly provided by Janssen, CA) for 2 hours at 37°C prior to co-culture with T cells. Cells were cultured for 8 hours at 37°C and then treated with Brefeldin A (BD Biosciences, CA) prior to harvesting for flow cytometric analysis of intracellular IFN-γ. Antibodies used were specific to IFN-γ (BD Biosciences, CA) and CD3 (eBiosciences, CA).
Murine TRAMP-C2 prostate tumor model
Male C57BL/6 mice were purchased from the National Cancer Institute (Frederick, MD). All mice were kept in specific pathogen-free conditions in the Animal Resource Center at The University of Texas MD Anderson Cancer Center (MDACC). Animal protocols were approved by the Institutional Animal Care and Use Committee of MDACC. The prostate cancer cell line TRAMP-C2 was maintained as described previously 19. The cell lines was tested for mycoplasma and fingerprinted before use. Tumor cell inoculation and treatment were performed as we previously reported 20. Briefly, mice were inoculated subcutaneously in the right flank with 1×106 TRAMP-C2 cells on day 0. Mice were then treated with α-CTLA-4 or isotype control antibody via intraperitoneal injection on day 6, 9, 12, 18, 21 and 24. The first treatment was 200 μg/mouse and all the subsequent treatments were 100 μg/mouse. Mice were euthanized within 2 days after the last treatment to collect draining lymph nodes for flow cytometric analysis using anti-VISTA (MH5A; Biolegend), anti-PD-L1 (10F.9G2; eBioscience) and anti-CD11b (M1/70; Biolegend) and anti-F4-80 (BM8; Biolegend). The experiment was performed three times with 3–5 mice per cohort.
Statistical analyses
All group results are expressed as mean with standard deviation. Comparisons between unmatched groups were made using the Mann-Whitney-Wilcoxon rank-sum test or t-test. Comparisons between paired groups were made using two-sided Wilcoxon matched-pairs signed rank test. P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank the clinical and research team for this study including: P. Corn, J. Araujo, B. Chapin, S. Matin, A. Zurita-Saavedra, J. Davis, L. Pisters and A. Aparicio for patient treatment and care. We thank M. Campbell for assistance with clinical data collection; L. Xiong, B. Guan, J. Kim, M. Higa, J. Bustillos, D. Ng Tang, and S. Basu for technical support; M. Curran and M. Ai for help with the TRAMP-C2 mouse models; and Dr. C. G. Liu of Sequencing & Non-coding RNA Program for microarray studies. The clinical trial was supported by Bristol-Myers-Squibb (BMS). The research work was also supported by funding from the American Association for Cancer Research (AACR) Stand Up To Cancer-Cancer Research Institute Cancer Immunology Dream Team Translational Research Grant (SU2C-AACR-DT1012; J.P.A., P.S., S.K.S., J.G.); Prostate Cancer Foundation (PCF) Challenge Grant in Immunology (J.P.A., P.S.); PCF 2014 Young Investigator Award (S.K.S.); Conquer Cancer Foundation-American Society of Clinical Oncology (ASCO) 2012 Young Investigator Award (J.G.); Cancer Prevention Research in Texas (CPRIT) RP120108 (P.S.); NIH/NCI R01 CA163793 (P.S.); NIH/NCI K12 CA088084 (J.G.) and NIH/NCI P30CA016672 (M.D. Anderson Cancer Center institutional grant).
Footnotes
Accession Codes. All microarray data are accessible under the accession code GSE77910.
Data Availability Statement. Any supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
P.S. designed the study; L.Z.S., J.B., L.M.V., J.C., I.I.W., M.A.S., J.S., and H.C. performed the experiments; J.G., J.F.W., C.A.P., L.Z.S., J.C., J.B., L.M.V., I.I.W., J.S., H.Z., J.W., E.E. and P.T. analyzed the data; J.G., P.S., J.P.A., S.K.S, J.W. and C.J.L. drafted the manuscript; and all authors reviewed the final draft of the manuscript.
COMPETING FINANCIAL INTERESTS
Drs. Sharma and Allison are founders and advisors for Jounce Therapeutics. Drs. Sharma and Allison are members of the Parker Institute for Cancer Immunotherapy. Dr. Sharma also serves as a consultant for BMS, AstraZeneca, Amgen and Glaxo SmithKline. Dr. Allison is an inventor of intellectual property owned by the University of California, Berkeley, and licensed to BMS and has received royalties from BMS. Dr. Allison is also inventor of intellectual property owned by Memorial-Sloan Kettering Cancer Center and licensed to Merck. Dr. Gao serves as a consultant for Genentech. Dr. Wistuba serves as a consultant for BMS. Dr. Efstathiou serves as a consultant for Janssen, Bayer, Medivation, Astellas and Sanofi Takeda.
Contributor Information
Jianjun Gao, Department of Genitourinary Medical Oncology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
John F Ward, Department of Urology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Curtis A Pettaway, Department of Urology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Lewis Z Shi, Department of Genitourinary Medical Oncology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Sumit K Subudhi, Department of Genitourinary Medical Oncology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Luis M Vence, The Immunotherapy Platform, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Hao Zhao, The Immunotherapy Platform, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Jianfeng Chen, Department of Genitourinary Medical Oncology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Hong Chen, The Immunotherapy Platform, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Eleni Efstathiou, Department of Genitourinary Medical Oncology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Patricia Troncoso, Department of Pathology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
James P Allison, Department of Immunology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA. The Immunotherapy Platform, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Christopher J Logothetis, Department of Genitourinary Medical Oncology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Ignacio I Wistuba, Department of Translational Molecular Pathology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Manuel A Sepulveda, Janssen Oncology Therapeutic Area, Janssen Research and Development, LLC, Pharmaceutical Companies of Johnson & Johnson, Spring House, PA 19477.
Jingjing Sun, The Immunotherapy Platform, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Jennifer Wargo, Department of Surgical Oncology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Jorge Blando, The Immunotherapy Platform, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Padmanee Sharma, The Immunotherapy Platform, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA. Department of Immunology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
References
- 1.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P, Allison JP. The future of immune checkpoint therapy. Science (New York, NY) 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 3.Kwon ED, et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2014;15:700–712. doi: 10.1016/S1470-2045(14)70189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subudhi SK, et al. Clonal expansion of CD8 T cells in the systemic circulation precedes development of ipilimumab-induced toxicities. Proc Natl Acad Sci U S A. 2016;113:11919–11924. doi: 10.1073/pnas.1611421113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liakou CI, et al. CTLA-4 blockade increases IFNgamma-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc Natl Acad Sci U S A. 2008;105:14987–14992. doi: 10.1073/pnas.0806075105. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng Tang D, et al. Increased frequency of ICOS+ CD4 T cells as a pharmacodynamic biomarker for anti-CTLA-4 therapy. Cancer immunology research. 2013;1:229–234. doi: 10.1158/2326-6066.CIR-13-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carthon BC, et al. Preoperative CTLA-4 blockade: tolerability and immune monitoring in the setting of a presurgical clinical trial. Clin Cancer Res. 2010;16:2861–2871. doi: 10.1158/1078-0432.CCR-10-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeman GJ, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lines JL, Sempere LF, Broughton T, Wang L, Noelle R. VISTA is a novel broad-spectrum negative checkpoint regulator for cancer immunotherapy. Cancer immunology research. 2014;2:510–517. doi: 10.1158/2326-6066.CIR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, et al. Anti-CTLA-4 therapy results in higher CD4+ICOShi T cell frequency and IFN-gamma levels in both nonmalignant and malignant prostate tissues. Proc Natl Acad Sci U S A. 2009;106:2729–2734. doi: 10.1073/pnas.0813175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nature immunology. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 13.Gao J, et al. Review of immune-related adverse events in prostate cancer patients treated with ipilimumab: MD Anderson experience. Oncogene. 2015;34:5411–5417. doi: 10.1038/onc.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wimberly H, et al. PD-L1 Expression Correlates with Tumor-Infiltrating Lymphocytes and Response to Neoadjuvant Chemotherapy in Breast Cancer. Cancer immunology research. 2015;3:326–332. doi: 10.1158/2326-6066.CIR-14-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods (San Diego, Calif) 2014;70:46–58. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Amir el AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature biotechnology. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine JH, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen PL, et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer discovery. 2016;6:827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster BA, Gingrich JR, Kwon ED, Madias C, Greenberg NM. Characterization of prostatic epithelial cell lines derived from transgenic adenocarcinoma of the mouse prostate (TRAMP) model. Cancer Res. 1997;57:3325–3330. [PubMed] [Google Scholar]
- 20.Shi LZ, et al. Interdependent IL-7 and IFN-gamma signalling in T-cell controls tumour eradication by combined alpha-CTLA-4+alpha-PD-1 therapy. Nature communications. 2016;7:12335. doi: 10.1038/ncomms12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.