Abstract
A gene coding for a thermostable esterase was isolated by functional screening of Escherichia coli cells that had been transformed with fosmid environmental DNA libraries constructed with metagenomes from thermal environmental samples. The gene conferring esterase activity on E. coli grown on tributyrin agar was composed of 936 bp, corresponding to 311 amino acid residues with a molecular mass of 34 kDa. The enzyme showed significant amino acid similarity (64%) to the enzyme from a hyperthermophilic archaeon, Pyrobaculum calidifontis. An amino acid sequence comparison with other esterases and lipases revealed that the enzyme should be classified as a new member of the hormone-sensitive lipase family. The recombinant esterase that was overexpressed and purified from E. coli was active above 30°C up to 95°C and had a high thermal stability. It displayed a high degree of activity in a pH range of 5.5 to 7.5, with an optimal pH of approximately 6.0. The best substrate for the enzyme among the p-nitrophenyl esters (C4 to C16) examined was p-nitrophenyl caproate (C6), and no lipolytic activity was observed with esters containing an acyl chain length of longer than 10 carbon atoms, indicating that the enzyme is an esterase and not a lipase.
Many lipolytic enzymes, including lipases (EC 3.1.1.3), esterases or carboxylesterases (EC 3.1.1.1), and various types of phospholipases, have been found in a wide range of organisms from bacteria to humans (22). True lipases can be defined as carboxylesterases that catalyze the hydrolysis and synthesis of relatively long-chain acylglycerols with acyl chain lengths of >10 carbon atoms, with trioleoylglycerol as the standard substrate. In contrast, esterases catalyze the hydrolysis of glycerolesters with acyl chain lengths of <10 carbon atoms, with tributylglycerols (tributyrin) as the standard substrate (42), even though lipases are also capable of hydrolyzing esterase substrates. Lipases from prokaryotes share a group of conserved amino acids, including a serine in a highly conserved Gly-X-Ser-X-Gly pentapeptide and an aspartate or glutamate residue that is hydrogen bonded to a histidine to form a catalytic triad. Based on comparisons of amino acid sequences and biological properties, prokaryote-derived lipases have been classified into eight different families (2).
In an effort to isolate novel genes from enormous and largely unexploited gene pools in uncultured microorganisms and/or those that are difficult to culture, the metagenomic library approach has recently been used successfully (14, 25, 29, 36, 38, 41, 43). This type of approach does not require the cultivation of diverse microorganisms from environmental samples, which is often difficult or impossible and can result in an enrichment of dominant strains under a specific selective condition. Thus, more global microbial genetic information can be provided from total microorganisms than from culturable subpopulations or enrichment cultures (10). Two different approaches have been previously used to isolate novel lipase genes. Henne et al. isolated novel genes conferring lipolytic activity in Escherichia coli transformed with metagenomic libraries constructed with temperate environmental soil samples (17). As an alternative approach, Bell et al. used a PCR method for the direct isolation of novel lipase genes from metagenomes to avoid potential difficulties in achieving the expression of a lipase in a heterologous host (5).
Thermophiles are a valuable source of thermostable enzymes with properties that are often associated with stability in solvents and detergents, giving these enzymes considerable potential for many biotechnological and industrial applications (7, 13, 27). One of these enzymes is a thermophilic and thermostable lipolytic enzyme that has been applied to the synthesis of biopolymers and biodiesel and used for the production of pharmaceuticals, agrochemicals, cosmetics, and flavors (13, 23). To date, fewer than one dozen thermostable lipases/esterases have been isolated from thermophiles and hyperthermophiles (13). In particular, lipases and esterases, which are functional at temperatures over 80°C, have been isolated mainly from hyperthermophilic archaea. Currently, three esterases with thermophilic archaeon origins and known amino acid sequences have been biochemically characterized, and all of them belong to family IV. These include esterases from Archaeoglobus fulgidus (31), Sulfolobus solfataricus (33), and Pyrobaculum calidifontis (18). Besides these enzymes, esterases from Sulfolobus acidocaldarius (39, 40), Pyrococcus furiosus (21), and Sulfolobus shibatae (19) have been purified and characterized, but their cognate genes have not been reported.
Recent direct genome shotgun sequencing and molecular phylogenetic studies using metagenomes have indicated substantial microbial diversity even in high-temperature environments (3, 4, 20, 34). Therefore, metagenomic libraries of thermal environments should be useful for screening novel thermostable enzymes, including lipases, but no such libraries have been reported yet. For this work, we have isolated a gene encoding a new esterase with sequence similarity to the hormone-sensitive lipase (HSL) family (family IV) by a functional screening of metagenomic libraries derived from thermal environmental samples from Indonesia. The cloned esterase gene was overexpressed in E. coli. The recombinant esterase was purified to homogeneity, and its biochemical properties were partially characterized.
MATERIALS AND METHODS
DNA extraction from environmental samples.
For the construction of metagenomic libraries, environmental samples (mixtures of mud and sediment-rich water) were collected from hot springs and mud holes in solfataric fields in the Tangkuban Perahu (85 to 92°C, pH 3 to 4), Cisolok (95°C, pH 8), Sileri (80 to 95°C, pH 4 to 6), and Likupang (65 to 80°C, pH 4.5 to 8) regions of Indonesia. After draining of the excess water, the metagenomes from these thermal environmental samples were extracted as described previously (45), with slight modifications. Environmental samples (100 g of wet weight) were mixed well with an equal volume of DNA extraction buffer (100 mM Tris-HCl [pH 8.0], 100 mM EDTA, 100 mM sodium phosphate [pH 8.0], 1.5 M NaCl, 1% hexadecyl methylammonium bromide), to which sodium dodecyl sulfate (SDS) and N-lauroyl sarcosine were each added to a final concentration of 1.5%. After the mixtures were incubated in a 65°C water bath for 1 h with gentle inversion every 15 to 20 min, an equal volume of chloroform-isoamyl alcohol (24:1) was added and gently mixed, followed by further incubation for 1 h at 50°C. The aqueous phase was recovered by centrifugation at 12,000 × g for 20 min. DNAs were precipitated with a 0.6 volume of isopropanol and recovered by centrifugation at 12,000 × g for 20 min at room temperature. Recovered DNAs were extracted once more with phenol-chloroform (1:1). DNAs precipitated with isopropanol were air dried and resuspended in an appropriate volume of distilled water.
Construction of metagenomic libraries.
The environmental genomic DNAs were resolved in 1% agarose (pulse field-certified agarose; Bio-Rad) containing 1% polyvinylpyrrolidone (Sigma) by pulsed-field gel electrophoresis with a CHEF-DRIII system (Bio-Rad) in order to fractionate the isolated DNAs by size. Pulsed-field gel electrophoresis gels were run at 4.5 V/cm with an angle of 120° and with ramping from an initial switch time of 10 s to a final switch time of 100 s at 14°C in 0.5× Tris-borate-EDTA buffer for 18 h. DNAs with approximate lengths of 40 to 50 kbp were recovered by electroelution. A metagenomic library for each sample was constructed by use of a CopyControl fosmid library production kit (Epicentre) according to the manufacturer's instructions. Both ends of the size-fractionated DNA were repaired to create blunt 5′-phosphorylated ends and were ligated into the pCC1FOS fosmid vector (Epicentre). Ligated DNA mixtures were then packaged by use of the supplied lambda packaging extracts and were transformed into an EPI300-T1R phage T1-resistant E. coli host.
Screening of esterase gene.
E. coli BL21(DE3) (Novagen) cells were transformed with each library. An activity-based esterase/lipase screening was performed by plating the transformed cells onto Luria-Bertani (LB) agar plates containing 12.5 μg of chloramphenicol/ml and 1% tributyrin in the presence of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After incubation at 37°C until colonies were observed, the plates were further incubated at 50°C. Transformants with clear halos around colonies were chosen as candidate clones, and their plasmids were isolated and analyzed.
DNA sequencing and sequence analysis.
DNA sequencing was performed by use of an ABI PRISM kit and a model 310 capillary DNA sequencer (Perkin-Elmer Applied Biosystems). Lipase and esterase sequences for comparative study were retrieved from protein and nucleotide databases on the NCBI Entrez server at http://www.ncbi.nlm.nih.gov/Entrez/. Sequence similarity searches were performed with the BLAST 2.0 program (1). Amino acid sequence alignments of EstE1 with homologous proteins and phylogenetic analyses were performed with the Align X program, a component of the Vector NTI suite (InforMax, North Bethesda, Md.) using the blosum62mt2 scoring matrix (16). Phylogenetic trees were constructed by the neighbor-joining method (37) by the use of Molecular Evolutionary Genetics Analysis 2.1 software (MEGA, version 2.1) (26). Bootstrapping (12) was used to estimate the reliability of phylogenetic reconstructions (1,000 replicates).
Expression and purification of recombinant esterase.
The esterase gene was amplified by a PCR with the primers estE1-f (5′-CCCAAGCTTATGCCGCTCGACCCACAAATC-3′) and estE1-r (5′-TTTTCCTTTTGCGGCCGCAGAGGGCTGGAGGCCAGAACG-3′), containing restriction enzyme sites (underlined) for HindIIII and NotI, with the selected fosmid (Mge4) isolated from an esterase-producing transformant as a template, and with Vent DNA polymerase (NEB). The PCR product was digested with HindIII and NotI, ligated into the HindIII- and NotI-linearized expression vector pET-22b(+) (Novagen), and introduced into E. coli BL21(DE3) (Novagen). Transformants were cultivated in LB medium containing 100 μg of ampicillin/ml at 37°C, and protein expression was induced at 25°C by the addition of 1 mM IPTG when the optical density at 660 nm reached 0.8. After induction for 12 h, the cells were harvested, washed twice with phosphate-buffered saline (pH 7.4), resuspended in 10 ml of binding buffer A (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, 10 mM imidazole, 10 mM β-mercaptoethanol, 10% glycerol, and 1% Nonidet P-40), and frozen at −80°C. After thawing, the cells were sonicated on ice and crude cell extracts were centrifuged at 20,000 × g for 30 min to obtain cleared lysates. The supernatant was incubated at 80°C for 10 min and then centrifuged for 30 min at 4°C to discard denatured proteins. The supernatant was then incubated for 2 h on ice with Ni-nitrilotriacetic acid (NTA)-agarose resin (QIAGEN) that had been preequilibrated with binding buffer. After being washed with 20 column volumes of binding buffer, bound esterase proteins were step eluted with binding buffer containing 50 to 450 mM imidazole. Esterase-containing fractions were collected, dialyzed against gel filtration column buffer (50 mM Tris-HCl [pH 7.8], 150 mM NaCl, 1 mM dithiothreitol, 0.5 mM EDTA), and then run at a flow rate of 0.5 ml/min on a Sephacryl S-200HR column (Amersham Pharmacia Biotech) on an ÄKTA Prime instrument (Amersham Pharmacia Biotech). Fractions were collected, dialyzed against binding buffer A, and then further purified and concentrated by passage through a Ni-NTA column (Amersham Pharmacia Biotech). Adsorbed proteins were then eluted with a linear gradient of 50 to 450 mM imidazole in binding buffer A and dialyzed against 50 mM potassium phosphate buffer (pH 7.0) containing 10% glycerol. The protein concentration was determined by use of a Bio-Rad protein assay kit, with bovine serum albumin as a standard.
Western blotting analysis.
After SDS-polyacrylamide gel electrophoresis, the proteins were transferred onto nitrocellulose membranes (Amersham Pharmacia Biotech). The membranes were blocked with 2% nonfat milk in phosphate-buffered saline, incubated with an anti-penta-His antibody (QIAGEN) for 2 h at room temperature, washed, and incubated with the appropriate secondary antibody coupled to horseradish peroxidase (Amersham Pharmacia Biotech). Immunoblots were developed with an ECL detection kit (Amersham Pharmacia Biotech).
Enzyme assays.
Esterase activities against p-nitrophenyl esters were determined by measuring the amount of p-nitrophenol released by esterase-catalyzed hydrolysis (28). The production of p-nitrophenol was continuously monitored at 405 nm by use of a DU-650 spectrophotometer with a thermal controller (Beckman). Unless otherwise described, esterase activity was measured by a standard assay at 70°C, with 1 mM p-nitrophenyl caproate as a substrate in 50 mM morpholineethanesulfonic acid (MES; pH 6.0) containing 1% acetonitrile. After preincubation for 3 min, the reaction was started by the addition of the purified enzyme to a final concentration of 1 nM. Blank reactions were performed with every measurement under different conditions to subtract the appropriate values for nonenzymatic hydrolysis of substrates from the results. The extinction coefficients of p-nitrophenol were also determined under each reaction condition prior to the measurements. The activity was determined by measuring the initial rate of hydrolysis of p-nitrophenyl ester. One unit of enzyme activity was defined as the amount of activity required to release 1 μmol of p-nitrophenol/min from p-nitrophenyl ester.
Modification of enzymes.
The inhibitory effects of modifying reagents for Ser and His were examined by using phenylmethylsulfonyl fluoride (PMSF; Sigma) and diethyl pyrocarbonate (DEPC; Sigma), respectively. The enzyme (0.6 μM), in 50 mM MES (pH 6.0), was incubated with various concentrations of PMSF or DEPC at 37°C for 10 min. The modification reactions were stopped by cooling samples in ice water, and the residual activities were measured by the standard assay described above.
Substrate specificity.
Substrate specificities for p-nitrophenyl esters were determined by using p-nitrophenyl butyrate (0.2 to 3 mM) (Sigma), p-nitrophenyl valerate (0.2 to 3 mM) (Sigma), p-nitrophenyl caproate (0.4 to 3 mM) (Fluka), and p-nitrophenyl caprate (0.2 to 3 mM) (Sigma) as substrates in 50 mM MES (pH 6.0) with 1% acetonitrile at 70°C. The p-nitrophenyl ester substrates with C4 to C10 acyl chains were dissolved in acetonitrile at a final concentration of 100 mM. p-nitrophenyl palmitate (Sigma) was dissolved in a mixture of acetonitrile and 2-propanol (1:4) in order to solubilize the substrate, and reactions were performed in a final concentration of 1% acetonitrile and 4% 2-propanol. Initial reaction velocities measured at various concentrations of substrates were fitted to the Lineweaver-Burk transformation of the Michaelis-Menten equation. Kinetic analyses by curve fitting were performed with the SigmaPlot program (SPSS Science, Chicago, Ill.).
Effect of pH on enzyme activity.
For determination of the optimum pH of the enzyme, esterase activities were measured for a pH range of 3.0 to 9.5 under standard assay conditions. The buffers used were 50 mM sodium acetate (pH 3.0 to 5.5), 50 mM MES (pH 5.5 to 7.0), 50 mM HEPES (pH 7.0 to 7.5), and 50 mM glycine (pH 7.5 to 9.5). The production of p-nitrophenoxide and p-nitrophenol from p-nitrophenyl caproate was monitored at 348 nm (the pH-independent isosbestic wavelength of p-nitrophenoxide and p-nitrophenol).
Effect of temperature on enzyme activity and thermostability.
The optimal temperature for enzyme activity was determined for a temperature range of 30 to 95°C under standard assay conditions. Thermostability was analyzed by measuring the residual activity after incubating the enzyme (6.0 μM in 20 mM potassium phosphate buffer, pH 7.0) at 80, 85, 90, or 95°C for various times in Eppendorf tubes with mineral oil on top to prevent evaporation. For calculation of the half-life of the enzyme, the experimental data were fitted to a single exponential decay curve (y = ae−bx) by a nonlinear regression procedure based on the Marquardt-Levenberg algorithm in SigmaPlot software.
Nucleotide sequence accession number.
The estE1 nucleotide sequence reported here is available in the GenBank database under accession number AY726780.
RESULTS
Screening for thermostable esterases from metagenomic libraries.
We constructed a total of four independent fosmid metagenomic libraries from thermal environmental samples (ca. 300 to 500 g of wet weight) from the Cislock, Likupang, Tangkuban Perahu, and Sileri areas of Indonesia. Environmental DNAs in the size range of 30 to 50 kbp were routinely obtained, with yields of 1 to 4 μg per 100 g of sample. Together, these libraries consisted of approximately 5,000 independent recombinant fosmids with inserts in the range of 20 to 40 kbp (data not shown).
In order to screen for esterase/lipase genes, we individually introduced these metagenomic libraries into E. coli BL21(DE3) cells, which were then plated onto LB medium containing tributyrin and IPTG to induce gene expression from the T7 RNA polymerase promoter located at either the 5′ or the 3′ end of the insert. IPTG was added in order to induce the transcription of genes that lacked a promoter or did not contain a promoter that was functional in a heterologous host. After the growth of E. coli cells transformed with each library at 37°C, followed by a further incubation at 50°C, a total of four esterase/lipase-positive transformants which formed clear halos on tributyrin plates were isolated from a library constructed with an environmental sample from the Sileri hot spring area. This metagenomic library consisted of approximately 2,000 fosmids with various sizes of metagenomic DNA inserts of over 20 kbp.
The four fosmids, named Mge1, -2, -3, and -4, contained metagenomes of 33, 32, 38, and 30 kbp, respectively. All E. coli cells that were transformed with these fosmids, except for those transformed with Mge1, were dependent on IPTG for halo formation on plates with the tributyrin substrate, indicating that transcription in these fosmids relied on the T7 RNA polymerase promoter in the fosmid. However, all four fosmids failed to confer on E. coli transformants a lipolytic activity on the agar plate containing triolein (C18), indicating that there was no lipase activity.
EstE1 is a new member of the hormone-sensitive lipase family.
We used Mge4, which had the smallest insert size, for subcloning of the minimal esterase gene in an E. coli expression vector. We initially attempted, without success, to clone the gene by subcloning of either EcoRI- or BamHI-digested DNA fragments into a T7 RNA polymerase-driven E. coli expression vector and screening for tributyrin-digesting transformants. Therefore, we sequenced both the 5′ and 3′ ends of each EcoRI-digested DNA fragment of Mge4 that was cloned in the expression vector. BLAST searches for the deduced amino acid sequence revealed two DNA fragments encoding the N- and C-terminal portions of EstE1 that were most similar to Pyrobaculum calidifontis esterase (18). The esterase gene, named estE1, contained an open reading frame of 936 bp that was capable of encoding a protein with a predicted molecular mass of 34 kDa and an isoelectric point of 5.82. PCRs with estE1 gene-specific primers and with Mge1-3 as a substrate, followed by restriction enzyme digestion, indicated that all of these fosmids had the same esterase gene (data not shown).
The deduced amino acid sequence of EstE1 was used to perform a BLAST search of the National Center for Biotechnology Information and SwissProt databases. This search revealed a relatively high similarity (over 50%) between EstE1 and other thermostable esterases/lipases from archaea, including an esterase (BAC06606) from Pyrobaculum calidifontis (64%), the lipase LipP-2 (NP_343862) from S. solfataricus (63%), a carboxylesterase (NP_070544) from Archaeoglobus fulgidus DSM 4304 (57%), an esterase (NP_375919) from Sulfolobus tokodaii (57%), the lipase LipP-1 (NP_343839) from S. solfataricus (57%), a carboxylesterase (BAB59879) from Thermoplasma volcanium (53%), and an esterase (NP_111246) from Thermoplasma volcanium (53%). A high similarity was also observed for esterases/lipases from thermophilic bacteria, including the carboxylesterase Est2 (1EVQ_A) from Alicyclobacillus acidocaldarius (51%) and an esterase/lipase (ZP_00057664) from Thermobifida fusca (52%). EstE1 was also relatively similar to human HSL (NP_005348) (28% identity and 43% similarity). Multiple alignments of the entire 313 amino acids of EstE1 with the most closely related sequences of lipases/esterases (>40% similarity) in the National Center for Biotechnology Information database are presented in Fig. 1A.
FIG. 1.
Amino acid sequence blocks conserved in the deduced amino acid sequences of EstE1 and homologous lipases and esterases. (A) Multiple amino acid sequence alignments of EstE1 and its homologs. The accession numbers of the aligned sequences are for the following organisms: 1EVQ_A, carboxylesterase Est2 from Alicyclobacillus acidocaldarius; ZP_00095860, esterase/lipase from Novosphingobium aromaticivorans; AAC38151, lipase from Pseudomonas sp. strain B11-1; ZP_00170489, hypothetical protein from Ralstonia eutropha JMP134; ZP_00215452, hypothetical protein from Burkholderia cepacia R18194; AAC12774, brefeldin A esterase from Bacillus subtilis; CAA37862, triacylglycerol lipase from Moraxella sp.; AAC73578, putative lipase (EC 3.1.1.) from E. coli K-12; NP_005348, hormone-sensitive lipase from Homo sapiens (human); BAB59879, carboxylesterase from Thermoplasma volcanium; BAC06606, esterase from Pyrobaculum calidifontis; NP_343862, lipase (LipP-2) from S. solfataricus; NP_070544, carboxylesterase (EstA) from Archaeoglobus fulgidus DSM 4304; NP_343839, lipase (LipP-1) from S. solfataricus; NP_375919, 303-amino-acid hypothetical esterase from S. tokodaii; NP_243114, lipase (esterase) from Bacillus halodurans C-125; NP_947767, putative lipase/esterase from Rhodopseudomonas palustris CGA009; ZP_00057664, esterase/lipase from Thermobifida fusca; ZP_00215124, hypothetical protein from Burkholderia cepacia R18194; NP_960379, hypothetical protein from Mycobacterium avium subsp. paratuberculosis strain k10; ZP_00214276, hypothetical protein from Burkholderia cepacia R18194; AAC41424, lipase-like enzyme from Wautersia eutropha; ZP_00028572, esterase/lipase from Burkholderia fungorum; ZP_00216007, hypothetical protein from Burkholderia cepacia R18194; ZP_00223864, hypothetical protein from Burkholderia cepacia R1808; AAO17429, unknown protein from Pseudomonas aeruginosa; BAA82510, esterase HDE from Oleomonas sagaranensis. (B) Amino acid sequence alignment of EstE1 with putative esterases identified from Sagasso Sea environmental genomes. The accession numbers are indicated to the left of the amino acid sequences. Identical residues have a gray background. Symbols: •, amino acids forming a catalytic triad; ○, amino acids involved in oxyanion hole formation.
EstE1 contains the lipase-conserved catalytic triad residues Asp251 and His281 and the catalytic nucleophile Ser154 in the consensus pentapeptide GDSAG. The HSL family conserved HGGG motif (amino acids 80 to 83), which is involved in hydrogen bonding interactions for stabilization of the oxyanion hole and plays a role in catalysis (44), was found upstream of the active-site conserved motif (Fig. 1A). These amino acid sequence comparisons indicated that EstE1 should be classified as a new member of the hormone-sensitive lipase (HSL) family. The phylogenetic tree shown in Fig. 2 indicates that EstE1 is most closely related to the esterase of Pyrobaculum calidifontis and clusters with family IV esterases from several species of Sulfolobus and from Archaeoglobus fulgidus.
FIG. 2.
Phylogenetic tree of HSL family homologues to EstE1. The phylogenetic tree was constructed by the neighbor-joining method with MEGA, version 2.1, software. The accession numbers of the aligned sequences shown in parentheses are the same as those described in the legend to Fig. 1A. The numbers associated with the branches refer to bootstrap values (confidence limits) resulting from 1,000 replicate resamplings. Only bootstrap values higher than 50% are shown. The scale represents the number of amino acid substitutions per site.
A sequence alignment of EstE1 with open reading frames from metagenomes which were recently identified by shotgun sequencing of seawater environmental DNA samples from the Sargasso Sea near Bermuda (41) revealed 20 homologous HSL-like esterase/lipase genes with approximately 50% similarity in their amino acid sequences. The presence of HSL family conserved motifs in these genes with hitherto unknown functions indicates that these EstE1 homologs are all novel HSL family esterases or lipases (Fig. 1B).
Overexpression and purification of recombinant EstE1.
In order to investigate the biochemical properties of EstE1, we expressed the EstE1 protein with a six-histidine tag at its C terminus in E. coli and purified the protein to homogeneity. Bacteria transformed with the expression vector and induced with IPTG abundantly expressed the histidine-tagged protein, as observed by the appearance of an extra protein band migrating at 34 kDa in a Coomassie-stained gel (Fig. 3A, compare lane 2 with lane 1). A heat treatment of the bacterial lysates harboring recombinant EstE1 at 80°C resulted in an enrichment of EstE1 (lane 3), which was then further purified by Ni-NTA-agarose chromatography. Additional bands representing degraded and/or nonspecifically bound proteins that appeared in the eluates from Ni-NTA-agarose chromatography (lane 4) were removed by subsequent gel filtration chromatography (GFC). Substantial purification of the protein was achieved by GFC, which yielded the recombinant EstE1 protein purified to homogeneity (lane 5). The fractions from GFC were combined, purified, and concentrated by a second round of Ni-NTA chromatography, resulting in homogenous EstE1, as shown by silver staining (Fig. 3C). This purification protocol routinely yielded >4 mg of homogeneous EstE1 from 1 liter of bacterial culture. In order to verify the purification protocol for recombinant EstE1, we performed a Western blot analysis of the various purification steps by using an anti-penta-His antibody. A protein band of approximately 34 kDa was identified as EstE1 (Fig. 3B).
FIG. 3.
Expression and purification of recombinant EstE1. (A) Proteins recovered during the various purification steps were separated by SDS-12% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. Lane M, molecular weight standards; lane 1, total lysate of E. coli BL21(DE3) transformed with an empty vector; lane 2, proteins as described above, but with the EstE1 expression vector; lane 3, proteins after thermal denaturation; lane 4, purified fraction from Ni-NTA column; lane 5, purified fraction after GFC. (B) Western blot analysis of purification steps probed with an anti-penta-His antibody. The samples are the same as those described for panel A. (C) Silver staining of the purified esterase (1 μg) from Ni-NTA affinity chromatography of the pooled fractions from GFC. Protein size markers are indicated in kilodaltons on the left, and recombinant EstE1 proteins are indicated by the arrowheads on the right.
Since the esterase catalytic triad Asp, His, and Ser residues were found in EstE1 (Fig. 1A), we attempted to confirm the function of Ser and His residues in the enzyme's activity by chemical modification with various concentrations (0.01 to 10 mM) of either PMSF or DEPC. Modification of the enzyme with either 1 mM PMSF or 1 mM DEPC reduced the esterase activity by 88 and 63%, respectively. Less than 5% of the residual activity was observed with each inhibitor at a concentration of 10 mM (data not shown), indicating that these amino acids are probably essential for the enzyme's function. However, we could not completely rule out the possibility that a modification of amino acids other than those in the lipase catalytic triad may affect the protein structure and/or activity of the enzyme.
Substrate specificity of EstE1.
To examine substrate specificity and to find the best substrate for an enzyme activity assay, we tested various p-nitrophenyl esters with acyl chains of different lengths (butyrate, C4; valerate, C5; caproate, C6; caprate, C10; and palmitate, C16). Under our standard assay conditions of pH 6.0 and 70°C, both the Km and kcat values of purified EstE1 decreased with increases in the acyl chain length up to C6 (Table 1). The catalytic efficiency represented by the value of kcat/Km slightly increased with an increase in the acyl chain length. p-Nitrophenyl caproate produced the highest value. No detectable activity was observed at pH 6.0 with p-nitrophenyl caprate or palmitate. The kcat value of EstE1 for the best substrate among the p-nitrophenyl esters tested for this work, which was C6, was approximately 1,600 s−1, with a 1.54- and a 1.77-fold increase compared to the C4 and C5 substrates, respectively. These values are within the range reported for other thermostable esterases, including the esterases from Pyrobaculum calidifontis (2,620 s−1) (18), Alicyclobacillus acidocaldarius (3,420 s−1) (32), and Archaeglobus fulgidus (1,014 s−1) (31). The substrate specificity results shown above and its inability to form halos on triolein agar clearly demonstrated that EstE1 is an esterase and not a lipase.
TABLE 1.
Kinetic parameters for recombinant EstE1
| Substrate (p-nitrophenyl ester) | Km (mM)a | kcat (S−1)a | kcat/Km (s−1mM−1)a |
|---|---|---|---|
| Butyrate (C4) | 2.3 | 2,950 | 1,280 |
| Valerate (C5) | 0.6 | 1,300 | 2,170 |
| Caproate (C6) | 0.7 | 1,600 | 2,290 |
| Caprate (C10) | ND | ND | ND |
| Palmitate (C16) | ND | ND | ND |
ND, not detectable.
Effect of pH on enzyme activity and pH stability of EstE1.
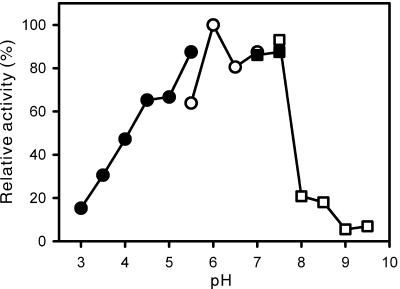
To determine the optimal pH for the esterase, we measured the activity of the purified EstE1 protein at various pH values (pH 3.0 to 9.5), using p-nitrophenyl caproate as a substrate, at 70°C. The amount of released p-nitrophenol for each reaction was measured at 348 nm, the isosbestic point of p-nitrophenol and p-nitrophenoxide (18). EstE1 exhibited >80% of its maximal activity in the pH range of 5.5 to 7.5, with the highest activity at a pH of approximately 6.0 (Fig. 4).
FIG. 4.
pH activity profile of EstE1. Est1 activity was measured at different pH values by a standard assay. The buffers used were sodium acetate (•) (pH 3.0 to 5.5), MES (○) (pH 5.5 to 7.0), HEPES (▪) (pH 7.0 to 7.5), and glycine (□) (pH 7.5 to 9.5).
Effect of temperature on enzyme activity and thermal stability of EstE1.
The effect of temperature on esterase activity was determined by using p-nitrophenyl caproate as a substrate at pH 6.0 in the temperature range of 30 to 95°C. The esterase activity increased with increases in temperature up to 95°C in a reproducible manner. At lower temperatures, the enzyme still showed activity, exhibiting 21 and 30% of the maximal activity at 30 and 40°C, respectively. An Arrhenius plot analysis revealed that the activity was maintained in the temperature range of 30 to 95°C as a linear relationship with a constant activation energy of 20.1 kJ/mol (Fig. 5A). To examine the thermal stability of this esterase, we incubated the enzyme (6.0 μM) for 30 to 120 min and measured its residual activity under standard assay conditions. EstE1 displayed a high thermal stability at 80°C in the absence of any stabilizer. It lost only approximately 20% of its activity, even after incubation for 120 min at 85°C (Fig. 5B). The stability of the enzyme decreased at 90 and 95°C, with half-lives of approximately 20 and 2 min, respectively, but the esterase activity did not decrease at these temperatures (Fig. 5A). These results indicate that even though the apparent optimum activity for the enzyme occurs at 95°C, the purified enzyme in the absence of substrate is not stable at temperatures over 90°C.
FIG. 5.
Temperature dependence and thermostability of EstE1. (A) The effect of temperature on the esterase activity of EstE1 was determined at different temperatures by a standard assay. The logarithms of specific activities were plotted against 1,000/T according to an Arrhenius plot. The correlation coefficient was 0.993. (B) The enzyme (6.0 μM), in 20 mM potassium phosphate buffer (pH 7.0), was incubated at 80°C (•), 85°C (○), 90°C (▴), and 95°C (▵) for the indicated times. The residual activity was measured by a standard assay. The activity of a nonincubated sample was defined as 100%.
DISCUSSION
We isolated a gene (estE1) encoding a thermostable esterase from a fosmid metagenomic DNA library constructed from a thermal environmental sample from Indonesia. Both the nucleotide and amino acid sequences of the EstE1 enzyme were novel. The thermophilic and thermostable properties of EstE1 and its remarkable amino acid sequence similarity to other esterases from thermophilic archaea (Fig. 1A) indicate that EstE1 is likely derived from a hyperthermophilic archaeon. Furthermore, partial sequencing of the Mge4 fosmid revealed that it contains open reading frames that are similar to those of enzymes of hyperthermophilic archaea, including S. solfataricus inorganic pyrophosphatase (63% identity) and a Pyrobaculum aerophilum DNA endonuclease rad2 homolog (70% identity) and acylamino acid-releasing enzyme (48% identity). Therefore, both phylogenetic analysis of EstE1 and comparative sequence analyses of other genes in the Mge4 fosmid strongly supported the idea that the origin of the EstE1 enzyme is a hyperthermophilic archaeon.
Family IV lipases show significant similarity to human HSL, which is involved in lipid metabolism by controlling the release of fatty acids from stored triacylglycerols in adipose tissue. Human HSL contains a catalytic domain and a regulatory module, which is unique to this enzyme, at the N terminus (15, 35). The catalytic domain displays similarity to prokaryote-derived family IV esterases/lipases, indicating that mammalian HSLs probably evolved from prokaryotic family IV enzymes. A homology search of the EstE1 sequence revealed that family IV prokaryote-derived esterases/lipases include the enzymes from Moraxella sp. (11), E. coli (24), Pseudomonas sp. strain B11-1 (6), Alicyclobacillus acidocaldarius (30), Archaeoglobus fulgidus (31), and Pyrobaculum calidifontis (18) (Fig. 1A). EstE1 was strikingly similar to the estPc-encoded carboxylesterase of Pyrobaculum calidifontis (18) (52 and 64% amino acid identity and similarity, respectively). These two proteins are more closely related to each other than to any other members of the HSL family and also show a functional relationship in that both enzymes exhibit only esterase activity with no lipase activity.
EstE1 exhibited thermophilic and thermostable properties. It was active above 30°C up to 95°C (Fig. 5A), while its activity decreased above 95°C. The enzyme was almost 100% stable at 80°C, even after 120 min of incubation (Fig. 5B). However, EstE1 had half-lives of approximately 20 and 2 min at 90 and 95°C, respectively. Thus, the optimal temperature (95°C) may be an artifact of the initial reaction rate measurement of the enzyme activity prior to complete thermal inactivation. Alternatively, this discrepancy may be due to an enhanced stability of the enzyme in the presence of substrate. The activation energy of EstE1 was unchanged between 30 and 95°C and was estimated to be 20.1 kJ/mol from an Arrhenius plot (Fig. 5A). This value is comparable to those for esterases from Pyrobaculum calidifontis (26.4 kJ/mol) (18), Archaeoglobus fulgidus (26 kJ/mol) (31), and Alicyclobacillus acidocaldarius (32 kJ/mol) (30) and is approximately one-half of the value for a cold-adapted lipase from Pseudomonas sp. strain B11-1 measured with p-nitrophenyl butyrate (47 kJ/mol) (6). Thus, EstE1 and the esterases of Pyrobaculum calidifontis and Archaeoglobus fulgidus seem to have comparable catalytic activities considering that the activities were all measured with the same substrate, p-nitrophenyl caproate. In addition, the esterase of Archaeoglobus fulgidus and EstE1 display similar thermostabilities, with approximately 20% of the residual activity present after incubation for 1 h at 90°C (Fig. 5B) (31). They are both, however, less stable than the esterases of Pyrobaculum calidifontis (18) and Pyrococcus furiosus (21), both of which show no activity change after incubation for 2 h, even at 100°C. Even though EstE1 and the esterases of Pyrobaculum calidifontis and Archaeoglobus fulgidus displayed optimal activities above 80°C, these enzymes still exhibited detectable activities at 30°C that were approximately 22, 16, and 25%, respectively, of their activities at their optimal temperatures (Fig. 5A) (18, 31). This is one of the unique properties of extremely thermostable esterases in the HSL family that is not observed for several other thermostable enzymes, including the esterase from Pyrococcus furiosus (21) and the less thermostable HSL family esterase from Alicyclobacillus acidocaldarius (30, 31). This is likely due to their ability to maintain a very stable conformation even at low temperatures. The high degree of thermostability of EstE1 and its functionality at lower temperatures are potentially applicable to biotechnological procedures.
The metagenomes of natural microbial communities contain an immense pool of genes, most of which are not represented by pure and enrichment cultures established under certain selective conditions. Our finding of a new thermostable and thermophilic HSL family esterase emphasizes the importance of thermal environmental metagenomic libraries as a source of isolation of novel genes by functional screening in a heterologous E. coli host. More work could be directed toward direct sequencing of thermal environmental metagenomes to isolate novel thermostable esterases/lipases, as indicated by our finding of 20 novel EstE1 homologs with approximately 50% similarity (Fig. 1B) from metagenomes of the Sargasso Sea near Bermuda that have recently been sequenced (41). A structure determination for EstE1 via X-ray crystallography is under way. Since family IV contains esterases and lipases that originated from psychrophiles, mesophiles, and thermophiles (22), additional three-dimensional information and structural comparisons with the known three-dimensional structures of thermostable esterases of Alicyclobacillus acidocaldarius (8) and Archaeoglobus fulgidus (9) will be useful for finding the determinants of thermal stability of enzymes in this family.
Acknowledgments
We thank Maggy T. Suhartono at Inter University Center for Biotechnology, Bogor Agricultural University, Bogor, Indonesia, and Yu-Ryung Pyun at the Bioproduct Research Center, Yonsei University, for thermal environmental samples.
This work was supported by grant IMT 2000-00016108 from the Korea Ministry of Commerce, Industry, and Energy and by grant 2003-2-0985 from the Korea Science and Engineering Foundation.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arpigny, J. L., and K. E. Jäeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343:177-183. [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, G. C., S. Gaffar, D. A. Cowan, and A. R. Suharto. 2001. Bacterial community analysis of Indonesian hot springs. FEMS Microbiol. Lett. 200:103-109. [DOI] [PubMed] [Google Scholar]
- 4.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, P. J., A. Sunna, M. D. Gibbs, N. C. Curach, H. Nevalainen, and P. L. Bergquist. 2002. Prospecting for novel lipase genes using PCR. Microbiology 148:2283-2291. [DOI] [PubMed] [Google Scholar]
- 6.Choo, D. W., T. Kurihara, T. Suzuki, K. Soda, and N. Esaki. 1998. A cold-adapted lipase of an Alaskan psychrotroph, Pseudomonas sp. strain B11-1: gene cloning and enzyme purification and characterization. Appl. Environ. Microbiol. 64:486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coolbear, T., R. M. Daniel, and H. W. Morgan. 1992. The enzymes from extreme thermophiles: bacterial sources, thermostabilities and industrial relevance. Adv. Biochem. Eng. Biotechnol. 45:57-98. [DOI] [PubMed] [Google Scholar]
- 8.De Simone, G., S. Galdiero, G. Manco, D. Lang, M. Rossi, and C. Pedone. 2000. A snapshot of a transition state analogue of a novel thermophilic esterase belonging to the subfamily of mammalian hormone-sensitive lipase. J. Mol. Biol. 303:761-771. [DOI] [PubMed] [Google Scholar]
- 9.De Simone, G., G. Manco, S. Galdiero, A. Lombardi, M. Rossi, and V. Pavone. 1999. Crystallization and preliminary X-ray diffraction studies of the carboxylesterase EST2 from Alicyclobacillus acidocaldarius. Acta Crystallogr. D 55:1348-1349. [DOI] [PubMed] [Google Scholar]
- 10.Entcheva, P., W. Liebl, A. Johann, T. Hartsch, and W. R. Streit. 2001. Direct cloning from enrichment cultures, a reliable strategy for isolation of complete operons and genes from microbial consortia. Appl. Environ. Microbiol. 67:89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feller, G., M. Thiry, J. L. Arpigny, and C. Gerday. 1991. Cloning and expression in Escherichia coli of three lipase-encoding genes from the psychrotrophic Antarctic strain Moraxella TA144. Gene 102:111-115. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 13.Haki, G. D., and S. K. Rakshit. 2003. Developments in industrially important thermostable enzymes: a review. Bioresour. Technol. 89:17-34. [DOI] [PubMed] [Google Scholar]
- 14.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-R249. [DOI] [PubMed] [Google Scholar]
- 15.Hemila, H., T. T. Koivula, and I. Palva. 1994. Hormone-sensitive lipase is closely related to several bacterial proteins, and distantly related to acetylcholinesterase and lipoprotein lipase: identification of a superfamily of esterases and lipases. Biochim. Biophys. Acta 1210:249-253. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henne, A., R. A. Schmitz, M. Bomeke, G. Gottschalk, and R. Daniel. 2000. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl. Environ. Microbiol. 66:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hotta, Y., S. Ezaki, H. Atomi, and T. Imanaka. 2002. Extremely stable and versatile carboxylesterase from a hyperthermophilic archaeon. Appl. Environ. Microbiol. 68:3925-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huddleston, S., C. A. Yallop, and B. M. Charalambous. 1995. The identification and partial characterisation of a novel inducible extracellular thermostable esterase from the archaeon Sulfolobus shibatae. Biochem. Biophys. Res. Commun. 216:495-500. [DOI] [PubMed] [Google Scholar]
- 20.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division-level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda, M., and D. S. Clark. 1998. Molecular cloning of extremely thermostable esterase gene from hyperthermophilic archaeon Pyrococcus furiosus in Escherichia coli. Biotechnol. Bioeng. 57:624-629. [DOI] [PubMed] [Google Scholar]
- 22.Jäeger, K. E., B. W. Dijkstra, and M. T. Reetz. 1999. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 53:315-351. [DOI] [PubMed] [Google Scholar]
- 23.Jäeger, K. E., and T. Eggert. 2002. Lipases for biotechnology. Curr. Opin. Biotechnol. 13:390-397. [DOI] [PubMed] [Google Scholar]
- 24.Kanaya, S., T. Koyanagi, and E. Kanaya. 1998. An esterase from Escherichia coli with a sequence similarity to hormone-sensitive lipase. Biochem. J. 332:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knietsch, A., T. Waschkowitz, S. Bowien, A. Henne, and R. Daniel. 2003. Metagenomes of complex microbial consortia derived from different soils as sources for novel genes conferring formation of carbonyls from short-chain polyols on Escherichia coli. J. Mol. Microbiol. Biotechnol. 5:46-56. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 27.Lasa, I., and J. Berenguer. 1993. Thermophilic enzymes and their biotechnological potential. Microbiologia 9:77-89. [PubMed] [Google Scholar]
- 28.Lee, Y. P., G. H. Chung, and J. S. Rhee. 1993. Purification and characterization of Pseudomonas fluorescens SIK W1 lipase expressed in Escherichia coli. Biochim. Biophys. Acta 1169:156-164. [DOI] [PubMed] [Google Scholar]
- 29.Lorenz, P., K. Liebeton, F. Niehaus, and J. Eck. 2002. Screening for novel enzymes for biocatalytic processes: accessing the metagenome as a resource of novel functional sequence space. Curr. Opin. Biotechnol. 13:572-577. [DOI] [PubMed] [Google Scholar]
- 30.Manco, G., E. Adinolfi, F. M. Pisani, G. Ottolina, G. Carrea, and M. Rossi. 1998. Overexpression and properties of a new thermophilic and thermostable esterase from Bacillus acidocaldarius with sequence similarity to hormone-sensitive lipase subfamily. Biochem. J. 332:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manco, G., E. Giosue, S. D'Auria, P. Herman, G. Carrea, and M. Rossi. 2000. Cloning, overexpression, and properties of a new thermophilic and thermostable esterase with sequence similarity to hormone-sensitive lipase subfamily from the archaeon Archaeoglobus fulgidus. Arch. Biochem. Biophys. 373:182-192. [DOI] [PubMed] [Google Scholar]
- 32.Manco, G., L. Mandrich, and M. Rossi. 2001. Residues at the active site of the esterase 2 from Alicyclobacillus acidocaldarius involved in substrate specificity and catalytic activity at high temperature. J. Biol. Chem. 276:37482-37490. [DOI] [PubMed] [Google Scholar]
- 33.Morana, A., N. Di Prizito, V. Aurilia, M. Rossi, and R. Cannio. 2002. A carboxylesterase from the hyperthermophilic archaeon Sulfolobus solfataricus: cloning of the gene, characterization of the protein. Gene 283:107-115. [DOI] [PubMed] [Google Scholar]
- 34.Norris, T. B., J. M. Wraith, R. W. Castenholz, and T. R. McDermott. 2002. Soil microbial community structure across a thermal gradient following a geothermal heating event. Appl. Environ. Microbiol. 68:6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osterlund, T., B. Danielsson, E. Degerman, J. A. Contreras, G. Edgren, R. C. Davis, M. C. Schotz, and C. Holm. 1996. Domain-structure analysis of recombinant rat hormone-sensitive lipase. Biochem. J. 319:411-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 38.Schloss, P. D., and J. Handelsman. 2003. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14:303-310. [DOI] [PubMed] [Google Scholar]
- 39.Sobek, H., and H. Gorisch. 1989. Further kinetic and molecular characterization of an extremely heat-stable carboxylesterase from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochem. J. 261:993-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobek, H., and H. Gorisch. 1988. Purification and characterization of a heat-stable esterase from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochem. J. 250:453-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 42.Verger, R. 1997. Interfacial activation of lipases: facts and artifacts. Trends Biotechnol. 15:32-38. [Google Scholar]
- 43.Voget, S., C. Leggewie, A. Uesbeck, C. Raasch, K. E. Jäeger, and W. R. Streit. 2003. Prospecting for novel biocatalysts in a soil metagenome. Appl. Environ. Microbiol. 69:6235-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei, Y., J. A. Contreras, P. Sheffield, T. Osterlund, U. Derewenda, R. E. Kneusel, U. Matern, C. Holm, and Z. S. Derewenda. 1999. Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase. Nat. Struct. Biol. 6:340-345. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]