Abstract
We describe a novel method of random chimeragenesis based on highly frequent deletion formation in the Escherichia coli ssb-3 strain and a deletion-directed chimera selection system that uses the rpsL+ gene as a reporter. It enables the selection of chimeras without target gene expression and can therefore be applied to cytotoxic targets. When this system was applied to phospholipase D genes from Streptomyces septatus TH-2 and Streptomyces halstedii subsp. scabies K6 (examples of cytotoxic targets), chimeragenesis occurred between short identical sequences at the corresponding position of the parental genes with large variations. Chimeragenesis was >1,000 times more frequent in the ssb-3 background than in the ssb+ background. We called this system repeat-length-independent broad-spectrum shuffling. It enables the convenient chimeragenesis and functional study of chimeric proteins. In fact, we found two amino acid residues related to the thermostability of phospholipase D (Phe426 and Thr433) by comparing thermostability among the chimeric enzymes obtained.
Techniques for obtaining random chimeric proteins opened a new field in protein engineering, particularly, that of the laboratory-based evolution of protein molecules, even without biochemical and three-dimensional structural information. However, the chimeric gene library can be prepared both in vitro and in vivo, and most studies are carried out by the in vitro method called DNA shuffling that is achieved by the random fragmentation of closely related sequences and the reassembly of these fragments into full-length genes via self-priming PCR (4, 24, 25). It enabled the combination of multiple mutation alleles in the target protein. However, DNA shuffling is hard to apply to targets that are difficult to express or screen, such as cytotoxic proteins, because it requires the functional screening of chimeric genes to isolate chimeric proteins with altered specificities.
In the conventional in vivo random chimeragenesis (10, 11, 16), two homologous genes are cloned tandemly on a plasmid and then digested between them. The transformation of the Escherichia coli or Saccharomyces cerevisiae host with such a linearized plasmid induces recombination between two parental genes, resulting in chimera formation and plasmid circularization. It allows plasmid replication and the selection of chimeras in the presence of an appropriate selective marker independent of target gene expression.
In contrast to in vitro DNA shuffling, a chimera obtained from in vivo chimeragenesis is composed of two segments with a single junction. Although this system is not directly comparable to DNA shuffling by PCR, it has potential advantages for cytotoxic targets. However, the variation of chimeras is limited in in vivo chimeragenesis, since this system is based on RecA-dependent homologous recombination. Some E. coli ssb alleles were reported to enhance RecA-independent deletion formation between two homologous genes on a plasmid, thus increasing the variety of chimeras obtained (17). This system requires the expression of a target gene fused to lacZ to detect recombination and is therefore unsuitable for the functional study of chimeric proteins. Moreover, the need to express the target gene makes this system difficult to apply to cytotoxic targets.
Here, we describe a novel in vivo method of generating a random chimera library based on the combination of highly frequent deletion formation in the E. coli ssb-3 strain with an rpsL-based chimera selection system. This system enables the selection of chimeras without the target gene expression and can therefore be applied to the chimeragenesis of cytotoxic targets even in their intact form. We selected Streptomyces phospholipase D (PLD) as the cytotoxic target. PLD is also interesting as an industrial enzyme because of its transphosphatidylation activity, i.e., the replacement of a head group of phospholipids, aside from its hydrolytic activity (26). The former activity is useful for the synthesis of rare phospholipids and also artificial ones (6, 22).
Recently, we isolated several Streptomyces strains that produce PLDs (7) and cloned their PLD genes (8, 9). PLD from Streptomyces septatus TH-2 showed the highest transphosphatidylation activity (7) and thermostability (8) among them. We applied it to random chimeragenesis with another thermostable PLD from Streptomyces halstedii subsp. scabies K6. We also investigated the amino acid residues related to the thermostability of PLD by comparing the chimeras obtained.
MATERIALS AND METHODS
Materials.
Phosphatidyl-p-nitrophenol was prepared from soybean phosphatidic acid and p-nitrophenol according to the procedure of D'Arrigo et al. (5). Plasmids pACYC184 (3) and pET-22b(+) were purchased from Nippon Gene (Japan) and Novagen, respectively. pETKmS2 (15) was kindly provided by T. Yamane (Nagoya University, Nagoya, Japan). Other chemicals were commercial products of the highest grade available and were used without further purification.
Strains.
E. coli MK1018 and MK1019 were rpsL(Smr) mutant derivatives of E. coli EJ2848 (lacI3 ΔlacZ lacY+ ΔfliC) (17) and EJ2885 (lacI3 ΔlacZ lacY+ ΔfliC ssb-3), respectively. E. coli EJ2885 was a tetracycline-sensitive derivative of E. coli EJ2882 (lacI3 ΔlacZ lacY+ ΔfliC ssb-3 zjc::Tn10) (17).
Construction of plasmids.
A 150-bp DNA fragment containing the 3′-terminal region of the PLD gene from strain K6 (PD K6) was amplified by PCR with the primers TCCAGGACTTCGGGTACGTGGTG and ACCATGGGGTACCGGATCCGAGGCCGGGCAGAGG (BamHI, KpnI, and NcoI sites added to the 3′-end of PD K6 are italic) and cloned into pGEM-T Easy (Promega) by TA cloning. The resultant plasmid was linearized by ApaI and BamHI and then ligated with a 2.6-kb ApaI-BamHI fragment containing the 5′ half of lacIq, the T7 lac promoter, the pelB signal sequence, and PD K6 of pETKmS2(K6), yielding pGEM(T7/K6). A 210-bp DNA fragment containing the hexahistidine (His6) tag coding region to the T7 terminator sequence of pET-22b(+) was amplified by PCR with the primers TCGGATCCGAATTCGAGCTCCG and AGATATCTCCGGATATAGTTCCTCCTTTC (the EcoRV site added is italic). After TA cloning, this region was excised by BamHI and SpeI and inserted between the BamHI and SpeI sites of pBSII(TH-2), a pBluescript II KS(+)-derived plasmid harboring the PLD gene from strain TH-2 (PD TH-2), yielding pBSII(TH-2/His).
The 1.3-kb ApaI-PmaCI fragment (containing the portion from the 5′ half of lacIq to the 5′-terminal region of PD K6) and 1.4-kb PmaCI-NcoI fragment (containing PD K6 excluding the 5′-terminal region) from pGEM(T7/K6) were inserted between the ApaI and NcoI sites of pBSII(TH-2/His), yielding pBSII(T7/K6/TH-2/His). The 3.6-kb EcoRV-Aor51HI fragment from pBSII(T7/K6/TH-2/His) containing the portion from the 5′ half of lacIq to the T7 terminator was inserted between the EcoRV and NruI sites of pACYC184 (3), yielding pACT(K6/TH-2). The 6.0-kb NdeI-PshAI fragment from pACT(K6/TH-2) was ligated with the 1.7-kb NdeI-PshAI fragment containing the lacIq and the T7 lac promoters of pET-22b(+), yielding pACTI(K6/TH-2). The 760-bp BamHI-PstI fragment including ccdAB of the mini-F plasmid pTN1117 was blunted by the Klenow fragment and T4 DNA polymerase and ligated with the PshAI digest of pACTI(K6/TH-2), yielding pACTIS1(K6/TH-2) and pACTIS2(K6/TH-2) (ccdAB in the same and opposite directions as lacIq, respectively). In this study, pACTIS2(K6/TH-2) was used since the production of TH-2 PLD in the T7 expression system was higher with pACTIS2 as the vector than with pACTIS1 in a preliminary experiment.
rpsL+ of E. coli was amplified by PCR. After TA cloning, rpsL+ was recloned between the tac promoter and the rrnB transcriptional terminator of pKK223-3 (2), yielding pNC121. The 1-kb DNA fragment containing the gentamicin (Gm) resistance gene from pMS255 (1) was cloned into pMCL210 (18) with SacI and SalI, yielding pNC122. The 1.9-kb SalI fragment from pNC121 was inserted downstream of the Gmr gene of pNC122 at the SalI site, yielding pNC124, which carries rpsL+ in the same direction as the Gmr gene. The 2.8-kb KpnI fragment from pNC124 was inserted into the KpnI site between PD K6 and PD TH-2 in pACTIS2(K6/TH-2), yielding pACTIS2(K6/Gmr-rpsL+/TH-2), which harbors the Gmr and rpsL+ genes in the same direction as the PLD genes.
PD K6, PD TH-2, and the mutants of chimeras H and M were cloned in the expression vector pETKmS2 (15) by the NcoI and BamHI sites (PLD genes were fused to the pelB signal sequence and C-terminal His6 tag sequence). For the convenient construction of the mutants of chimeras H and M, we introduced the silent substitution of nucleotides 1287 to 1292 (TAGCTC) for the XhoI site (CTCGAG). This silent substitution and the site-directed mutations were introduced by PCR with the corresponding primers.
Random chimeragenesis.
All cultivations described below were carried out at 37°C. Chloramphenicol, gentamicin, and streptomycin were used at 50, 20, and 50 μg/ml, respectively. E. coli MK1018 and MK1019 were transformed with pACTIS2(K6/Gmr-rpsL+/TH-2) by electroporation, and transformants were selected on Luria-Bertani (LB) plates containing chloramphenicol and gentamicin. The sensitivities of Cmr/Gmr transformants to streptomycin (Sms)were checked on LB plates containing chloramphenicol, gentamicin, and streptomycin. The expression of rpsL+ from the tac promoter was sufficient for this test even without induction.
Eight Sms transformants from each host were cultivated in LB medium containing chloramphenicol. After appropriate dilutions, cultures were spread on LB plates containing chloramphenicol with or without streptomycin. The occurrence of Smr revertants was expressed as the ratio of colony number on the plate containing chloramphenicol and streptomycin versus chloramphenicol alone. Plasmids isolated from 48 selected Smr revertants (six clones × eight runs) from each host were analyzed by agarose gel electrophoresis. The 1.6-kb DNA fragment containing the PLD gene on isolated plasmids was amplified by PCR with the GC-Rich PCR system (Roche) with primers corresponding to the coding regions of the pelB signal (CGACCGCTGCTGCTGGTCTGC) and the His6 tag (TGGTGGTGGTGCTCGAGTGCGGC). Recombination sites were mapped by the change in the pattern of digestion of these fragments with several restriction endonucleases and determined in detail by DNA sequencing.
Expression and purification of PLDs.
Escherichia coli BL21-Gold(DE3) (Invitrogen) was transformed with PLD expression plasmids. Expression was carried out according to a method described previously (8) in the presence of appropriate antibiotics (chloramphenicol at 50 μg/ml and kanamycin at 50 μg/ml for strains harboring plasmids obtained from the random chimeragenesis and plasmids derived from pETKmS2, respectively) except for the addition of premixed protease inhibitor tablet (Complete Mini, EDTA-free; Roche) at the induction period (one tablet per culture). Cultures were centrifuged for 60 min at 3,500 × g. Resultant supernatants were concentrated to ≈1 ml by ultrafiltration with Amicon Ultra (Millipore) and dialyzed overnight against 4 liters of 20 mM Tris-HCl (pH 8.0) with one buffer change. PLDs were purified from dialysates by nickel affinity chromatography with a MagExtractor His tag (Toyobo) according to the manufacturer's instructions. Purified samples were dialyzed against 3 liters of 10 mM sodium acetate buffer (pH 6.0) and checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Assay for PLD activity and thermostability.
PLD activity was determined on the basis of the hydrolysis of phosphatidyl-p-nitrophenol as described by D'Arrigo et al. (5) with several modifications. The reaction was performed at 37°C with 2 mM phosphatidyl-p-nitrophenol in 0.1 M sodium acetate buffer (pH 5.5). The thermostability of PLD was determined by a method reported previously based on residual hydrolytic activity for phosphatidyl-p-nitrophenol after appropriate heat treatment (8).
Nucleotide sequence accession number.
The accession numbers of the PD genes from K6 and TH-2 in the GenBank/EMBL/DDBJ databases are AB074305 and AB058783, respectively.
RESULTS
Strategy of random chimeragenesis.
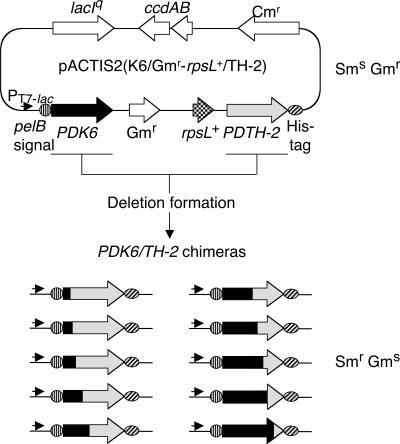
Plasmid pACTIS2(K6/Gmr-rpsL+/TH-2) was constructed for chimeragenesis (Fig. 1). Two homologous genes (PD K6 and PD TH-2) were placed in same direction, and a cassette containing the Gmr and E. coli rpsL+ genes was inserted between them. rpsL+ encodes the ribosomal protein S12 (20), the target of streptomycin. E. coli MK1019 (ssb-3 rpsL[Smr]) was prepared as a host for chimeragenesis. A certain mutation in the genomic rpsL gene led this strain to Smr. The transformation of MK1019 with pACTIS2(K6/Gmr-rpsL+/TH-2) altered the phenotype of cells from Smr to Sms (and also Gms to Gmr), since the Sms ribosome was reconstituted with the wild-type RpsL protein encoded on the plasmid.
FIG. 1.
Strategy of random chimeragenesis. The transformation of E. coli strain MK1019 (ssb-3 rpsL[Smr]) with plasmid pACTIS2(K6/Gmr-rpsL+/TH-2) altered the phenotype of this host from Smr to Sms by the wild-type RpsL protein expressed from the plasmid (rpsL+ and rpsL[Smr] represent genes for Sms wild-type and Smr mutant ribosomal protein S12 of E. coli, respectively). Then, Sms transformants were cultivated overnight at 37°C in medium containing chloramphenicol. When an in-frame deletion between two PLD genes (PD K6 and PD TH-2), i.e., the formation of a chimeric PLD gene (PD K6/TH-2) concomitant with the deletion of the Gmr-rpsL+ cassette occurs, the phenotype of the cells is reversed from Sms to Smr. Therefore, strains carrying chimeric PLD genes are selectable on streptomycin-chloramphenicol plates. The resulting chimera can be expressed in an appropriate E. coli strain for the T7 expression system and purified with the C-terminal His6 tag by nickel affinity chromatography. pelB and lacIq encode a secretion signal peptide of Erwinia carotovora pectate lyase and a repressor protein of the E. coli lac operon, respectively. ccdA and ccdB compose the stabilizing system of the E. coli F plasmid.
When recombination occurs between two homologous genes, the Gmr-rpsL+ cassette is simultaneously deleted from the plasmid, and the cells reverse their phenotype from Sms/Gmr to Smr/Gms. The Gmr gene excludes the accidental deletion of the Gmr-rpsL+ cassette. In strain MK1019, the expression levels of PD K6 in the parental plasmid and chimeric PLD genes in the resulting plasmids are negligible, since these genes are controlled by the T7 lac promoter; however, this strain does not have T7 RNA polymerase. Moreover, leaky expression must be repressed by LacI encoded by the plasmid. Thus, the intact form of chimeric gene is selectable without expression. On the other hand, when the isolated plasmids are transferred into the host of the T7 expression system, chimeric genes can be expressed from the T7 lac promoter. These features are advantageous for applying this system to the functional study of cytotoxic proteins such as PLDs. The ccdAB genes are also suitable for applications to cytotoxic targets, since they stabilize plasmids in E. coli cells (15). The pelB signal peptide and the His6 tag fused to the N and C termini of targets allow secretion of the resulting chimeric proteins and their easy purification, respectively.
Random chimeragenesis of PLD genes.
Strain MK1019 (ssb-3 rpsL[Smr]) was transformed with pACTIS2(K6/Gmr-rpsL+/TH-2). To evaluate the effect of the ssb-3 mutation, strain MK1018 (ssb+ rpsL[Smr]) was also used. All the clones tested had been altered to Sms. To induce chimeragenesis, eight Sms clones from each transformant were cultivated in the absence of gentamicin. Then Smr revertants were selected on the plate containing streptomycin. However, the occurrence of Smr revertants from the ssb-3 strain varied among runs (225 ± 293 per 106 cells), and it was typically >1,000 times more frequent than that from the ssb+ strain (0.14 ± 0.06 per 106 cells). The plasmids from 33 of the 48 Smr revertants (six clones per eight runs) in the ssb-3 background were smaller than the parental plasmid but identical in size to the plasmid harboring a single PLD gene on agarose gel electrophoresis (data not shown). This result suggests that ≈70% of the Smr revertants from the ssb-3 strain carry chimeric PLD genes. The remaining 15 clones carried plasmids of various sizes that were probably formed by deletion in an undesired manner. Similarly, 30 (≈60%) of the 48 Smr revertants in the ssb+ background carried chimeric PLD plasmids; however, the occurrence of chimeragenesis was >1,000 times lower than that in the ssb-3 background.
Distribution of recombination sites.
Recombination occurred between identical sequences at the corresponding position in the entire region of the PLD genes (≈1,530 nucleotides) (Fig. 2) without a significant bias in either background (Table 1). Some identical chimeras appeared within each run, but this behavior could be overcome by replicate studies (eight runs in this study) at the chimera-forming step. In total, in the ssb-3 background, recombination occurred at identical 6- to 29-nucleotide sequences.
FIG.2.
Distribution of recombination sites. Conserved nucleotides in the parental genes are indicated by asterisks between two sequences. Recombination sites observed in ssb-3, ssb+, and both backgrounds are indicated by shading, a frame, and both, respectively. The length of identical sequences in recombination sites is designated above the sequences (in some cases with alphabetical suffixes to distinguish sites). Recombination sites that correspond to the same expression product are indicated by underlining. The recombination sites of chimeras used in this study (chimeras A to I and M to O) are indicated with parentheses. An unintentional point mutation not associated with chimeragenesis had been introduced in PD TH-2 (position 1035, G to A) prior to construction of pACTIS2(K6/Gmr-rpsL+/TH-2). This mutation does not alter the primary structure of the protein.
TABLE 1.
Number of chimeras formed at each recombination site
| Recombination site size (nucleotides)a | No. of isolates
|
|
|---|---|---|
| MK1019 (ssb-3) | MK1018 (ssb+) | |
| 50 | 1 | |
| 32 | 2 | |
| 29 | 3 | 1 |
| 26 | 1 | |
| 21a | 2 | |
| 21b | 3 | |
| 20 | 1 | |
| 18a | 2 | |
| 18b | 2 | |
| 16 | 2 | 3 |
| 15 | 1 | |
| 14a | 1 | |
| 14b | 6 | 1 |
| 14c | 3 | |
| 13a | 1 | 2 |
| 13b | 1 | |
| 12a | 1 | |
| 12b | 4 | 2 |
| 11a | 1 | |
| 11b | 1 | |
| 11c | 1 | |
| 10 | 1 | |
| 9 | 2 | |
| 8a | 1 | |
| 8b | 1 | |
| 8c | 1 | |
| 8d | 1 | |
| 7 | 1 | |
| 6 | 1 | |
| 5 | 1 | |
| Total | 31 | 28 |
The position of each recombination site is designated by the length of identical sequence, with alphabetical suffixes as used in Fig. 2 to distinguish the sites.
The number of chimeras obtained did not apparently correlate with the length of the recombination junctions (Table 1). In the ssb+ background, recombination occurred in slightly longer identical sequences (5 to 50 nucleotides) than in the ssb-3 background (Table 1). However, no hot spots were observed in the recombination sites (Table 1). As suggested by agarose gel electrophoresis of chimeric plasmids, correct chimera formation was confirmed in >90% of plasmids. Rarely, recombination occurred between identical sequences in different but proximal positions of parental genes, resulting in short deletions or insertions (2 of 33 clones in the ssb-3 background and 2 of 30 clones in the ssb+ background; data not shown). Other undesired errors were not observed. Variations of chimeric genes in the ssb-3 and ssb+ backgrounds were 16 and 19, respectively, excluding incorrect chimeras (Table 1). These corresponded to 13 and 17 kinds of chimeric PLD enzyme, respectively, 23 kinds in total (Fig. 2).
Characterization of thermostability of chimeric PLDs.
We expressed the chimeric PLDs obtained with the T7 expression system. Chimeric PLD was translated with an N-terminal pelB signal and a C-terminal His6 tag (Fig. 1) and secreted into the culture medium as a ≈57-kDa protein concomitant with the scission of the pelB signal peptide. Chimeric PLDs were purified by nickel affinity chromatography (data not shown). To verify the applicability of our system to the functional study of chimeric protein, we examined the thermostability of chimeric PLDs. TH-2 PLD and K6 PLD consist of 509 and 511 amino acids, respectively (excluding the N-terminal Met and the C-terminal His6 tag region in the recombinant enzymes; Fig. 2B) with 80% identity in primary structure, and have similar thermostability (8).
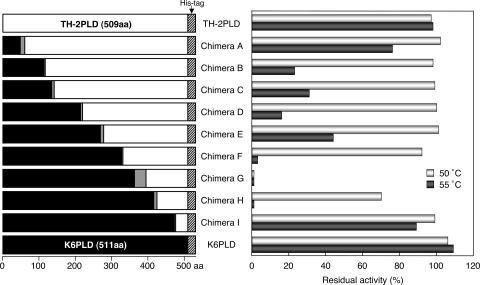
To determine the region related to thermostability, we first analyzed the thermostability of the parental and nine chimeric PLDs (chimeras A to I, Fig. 3) at 50 and 55°C. Interestingly, in spite of the similar thermostability of the parental enzymes, thermostability varied among their chimeras. From TH-2 PLD to chimera G in Fig. 3, thermostability decreased. In contrast, from chimera G to K6 PLD, thermostability increased. Particularly, chimera G was quite unstable even at 50°C.
FIG. 3.
Thermostability of chimeric PLDs. Primary structures of TH-2 PLD, chimeras A to I, and K6 PLD are illustrated schematically on the left. The regions derived from TH-2 PLD, K6 PLD, chimera-forming junctions, and the C-terminal His6 tag region are indicated by white, black, gray, and hatched rectangles, respectively. The map is drawn to scale (aa, amino acids). The thermostability of the PLDs is indicated on the right. Purified PLDs were incubated at 50 and 55°C for 10 min, and then residual activity was determined after appropriate dilution with 0.1% bovine serum albumin. Residual activity is represented as a percentage of that of the untreated enzyme.
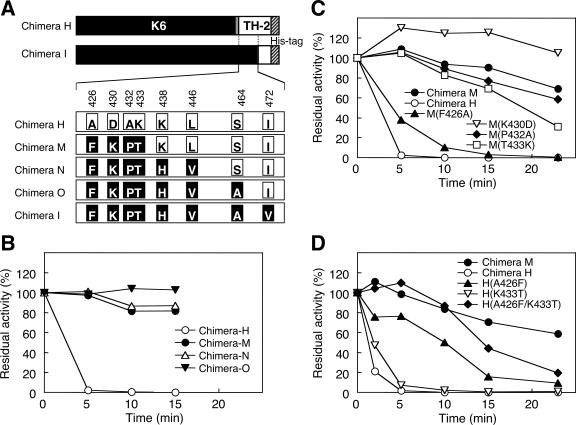
In the present study, we focused on the difference in thermostability between chimeras H and I. They differ by eight residues in their primary structure (Fig. 4A). This suggests that some of these residues are related to the difference in thermostability. To restrict residues, we analyzed the thermostability of three additional chimeras (chimeras M, N, and O, Fig. 4A and B). Their recombination junctions were present between those of chimeras H and I. At 60°C, chimera H was almost inactivated after 5 min, whereas >80% of the enzyme activities of chimeras M, N, and O remained even after 15 min. Chimeras H and M differ by four amino acid residues, Ala426, Asp430, Ala432, and Lys433 (TH-2 PLD type) in chimera H instead of Phe426, Lys430, Pro432, and Thr433 (K6 PLD type) in chimera M (residues are numbered in the TH-2 PLD manner).
FIG. 4.
Identification of amino acid residues related to thermostability. (A) Primary structures of chimeras H and I are illustrated in the same manner as in Fig. 3. Eight residues that differed among chimeras H, M, N, O, and I are depicted with the K6 PLD-type residues indicated by black boxes and TH-2 PLD-type residues by white boxes (residues are numbered in as for TH-2 PLD). The thermostability of (B) chimeras H, M, N, O, and I, (C) chimera M mutants, and (D) chimera H mutants was determined. Purified PLDs were incubated at 60°C for the indicated times, and then residual activity was determined after appropriate dilution with 0.1% bovine serum albumin (B to D). Residual activity is represented as a percentage of that of the untreated enzyme.
Identification of residues related to thermostability.
To determine the contribution of each residue, the thermostability of chimera M mutants that had a substitution of one of each of the residues from the K6 PLD type to the TH-2 PLD type were studied at 60°C (Fig. 4C). After 23 min, ≈70% of the enzyme activity of chimera M remained; however, the activities of chimeras M(F426A) and M(T433K) decreased to ≈0 and ≈30%, respectively. Chimera M(P432A) and chimera M had similar thermostability, and chimera M(K430D) had higher thermostability than chimera M. For the reverse investigation, the thermostability of chimera H mutants H(A426F), H(K433T), and H(A426F/K433T) were studied at 60°C (Fig. 4D). After 10 min, chimera H was completely inactivated, whereas, ≈50% of the PLD activity of chimera H(A426F) was maintained. The K433T mutation slightly stabilized chimera H. Chimera H(A426F/K433T) had the highest thermostability among the chimera H mutants. Therefore, we conclude that Ala426/Phe426 and Lys433/Thr433 are the residues related to the thermal instability and stability of PLDs. The thermal stabilization of TH-2 PLD by the A426F and/or K433T mutation supports this conclusion (data not shown).
DISCUSSION
We called our novel random chimeragenesis system repeat-length-independent broad-spectrum shuffling (RIBS) (Fig. 1), since chimeragenesis occurred at short identical sequences that are repeated between two parental genes with a large variation (Table 1 and Fig. 2). The success of chimeragenesis for Streptomyces PLDs verified the advantage of the application of this system to cytotoxic targets. Moreover, the chimeric PLD genes obtained from this system could be expressed easily and applied directly to functional studies (Fig. 3).
The ssb gene encodes single-stranded DNA-binding protein (SSB) (21). E. coli SSB binds single-stranded DNA formed transiently during DNA metabolic processes, i.e., replication, recombination, and repair (12, 14). It has been proposed that ssb mutations (ssb-1, ssb-3, and ssb-113) lower exonuclease activity and lead to the stabilization of misannealed intermediates and therefore enhance deletion formation between two tandem homologous genes in vivo (17). We consider that both the high homology (82% identity) and high GC content (≈70%) of parental Streptomyces PLD genes stabilized the heteroduplex intermediate and led to the disappearance of hot spots for chimeragenesis even in the ssb+ background. The frequency of chimeragenesis in the ssb+ background (typically >1,000 times lower than that in the ssb-3 background) is insufficient for the preparation of many chimeras. Therefore, it is expected that both the frequency and variation of chimeragenesis in the ssb+ background decrease when it is applied to genes with low homology and/or a low GC content. Thus, the ssb-3 strain must be more useful than the ssb+ strain as a host for RIBS.
Recently, several homology-independent protein recombination methods, e.g., incremental truncation for the creation of hybrid enzyme (ITCHY) (13, 19) and sequence homology-independent protein recombination (SHIPREC) (23), have been reported. In contrast, RIBS needs some sequence homology in target genes; although the recombination is RecA independent, it occurs between short direct repeats present between target genes (Table 1 and Fig. 2). This homology dependency is a major drawback of RIBS in comparison with the above-mentioned methods. However, the homology required in RIBS is lower than that in traditional RecA-dependent random chimeragenesis (10, 11, 16). From another point of view, homologous recombination has an advantage, since it ensures the formation of correct chimeras. In fact, most of chimeric PLD plasmids formed in this study carried correct chimeric genes (Table 1 and Fig. 2). On the other hand, in homology-independent methods, many nonfunctional chimeric genes must be formed, and therefore a system for selecting functional chimeras is needed.
In spite of the similar thermostability of K6 PLD and TH-2 PLD, the thermostability of their chimeras varied (Fig. 3). This finding enabled us to identify potential amino acid residues related to the thermostability from parental proteins having similar thermostabilities. On the basis of the comparisons of the thermostability of the chimeras and their mutants, we identified amino acid residues 426 and 433 as the residues related to thermostability (Fig. 4). Further studies of these residues must answer these questions. Interestingly, in spite of the presence of Ala426 and Lys433, the residues destabilizing PLD, these residues seem to destabilize PLD, but TH-2 PLD and chimera A were quite stable. Furthermore, thermostability decreased from TH-2 PLD to chimera G and increased from chimera G to K6 PLD in Fig. 3. These results indicate the presence of amino acid residues associated with thermostability besides residues 426 and 433.
Finally, RIBS has the advantage of conventional in vivo random chimeragenesis over PCR-based DNA shuffling (convenient chimera selection without expression of the target gene); furthermore, it overcomes the disadvantages of the former (low frequency of chimeragenesis and small variation of chimeras). This method may increase the chance of carrying out molecular evolution for proteins that had not been targets for DNA shuffling and supply a novel strategy for analyzing the function of individual amino acid residues or sequences.
Acknowledgments
K. Mori was supported in this work by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
We gratefully acknowledge N. Miyata for support in the study and T. Negishi for helpful discussion.
REFERENCES
- 1.Becker, A., M. Schmidt, W. Jäger., and A. Pühler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162:37-39. [DOI] [PubMed] [Google Scholar]
- 2.Brosius, J., and A. Holy. 1984. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. USA 81:6929-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crameri, A., S.-A. Raillard, E. Bermudez, and W. P. C. Stemmer. 1998. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391:288-291. [DOI] [PubMed] [Google Scholar]
- 5.D'Arrigo, P., V. Piergianni, D. Scarcelli, and S. Servi. 1995. A spectrophotometric assay for phospholipase D. Anal. Chim. Acta 304:249-254. [Google Scholar]
- 6.D'Arrigo, P., and S. Servi. 1997. Using phospholipases for phospholipid modification. Trends Biotechnol. 15:90-96. [Google Scholar]
- 7.Hagishita, T., M. Nishikawa, and T. Hatanaka. 2000. Isolation of phospholipase D producing microorganism with high transphosphatidylation activity. Biotechnol. Lett. 22:1587-1590. [Google Scholar]
- 8.Hatanaka, T., T. Negishi, M. Kubota-Akizawa, and T. Hagishita. 2002. Study on thermostability of phospholipase D from Streptomyces sp. Biochim. Biophys. Acta 1598:146-154. [DOI] [PubMed] [Google Scholar]
- 9.Hatanaka, T., T. Negishi, M. Kubota-Akizawa, and T. Hagishita. 2002. Purification, characterization, cloning and sequencing of phospholipase D from Streptomyces septatus TH-2. Enzyme Microb. Technol. 31:233-241. [Google Scholar]
- 10.Kim, J.-Y., and P. N. Devreotes. 1994. Random chimeragenesis of G-protein-coupled receptors. Mapping the affinity of the cAMP chemoattractant receptors in Dictyostelium. J. Biol. Chem. 269:28724-28731. [PubMed] [Google Scholar]
- 11.Levin, L. R., and R. R. Reed. 1995. Identification of functional domains of adenylyl cyclase using in vivo chimeras. J. Biol. Chem. 270:7573-7579. [DOI] [PubMed] [Google Scholar]
- 12.Lohman, T. M., and M. E. Ferrari. 1994. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63:527-570. [DOI] [PubMed] [Google Scholar]
- 13.Lutz, S., M. Ostermeier, and S. J. Benkovic. 2001. Rapid generation of incremental truncation libraries for protein engineering using α-phosphothioate nucleotides. Nucleic Acids Res. 29:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer, R. R., and P. S. Laine. 1990. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 54:342-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishima, N., K. Mizumoto, Y. Iwasaki, H. Nakano, and T. Yamane. 1997. Insertion of stabilizing loci in vectors of T7 RNA polymerase-mediated Escherichia coli expression systems: a case study on the plasmids involving foreign phospholipase D gene. Biotechnol. Prog. 13:864-868. [DOI] [PubMed] [Google Scholar]
- 16.Moore, K. R., and R. D. Blakely. 1994. Restriction site-independent formation of chimeras from homologous neurotransmitter-transporter cDNAs. BioTechniques 135:137. [PubMed] [Google Scholar]
- 17.Mukaihara, T., and M. Enomoto. 1997. Deletion formation between the two Salmonella typhimurium flagellin genes encoded on the mini F plasmid: Escherichia coli ssb alleles enhance deletion rates and change hot-spot preference for deletion endpoints. Genetics 145:563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano, Y., Y. Yoshida, Y. Yamashita, and T. Koga. 1995. Construction of a series of pACYC-derived plasmid vectors. Gene 162:157-158. [DOI] [PubMed] [Google Scholar]
- 19.Ostermeier, M., J. H. Shim, and S. J. Benkovic. 1999. A combinatorial approach to hybrid enzymes independent of DNA homology. Nat. Biotechnol. 17:1205-1209. [DOI] [PubMed] [Google Scholar]
- 20.Post, L. E., and M. Nomura. 1980. DNA sequences from the str operon of Escherichia coli. J. Biol. Chem. 255:4660-4666. [PubMed] [Google Scholar]
- 21.Sancar, A., K. R. Williams, J. W. Chase, and W. D. Rupp. 1981. Sequences of the ssb gene and protein. Proc. Natl. Acad. Sci. USA 78:4274-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Servi, S. 1999. Phospholipases as synthetic catalysts. Top. Curr. Chem. 200:127-158. [Google Scholar]
- 23.Sieber, V., C. A. Martinez, and F. H. Arnold. 2001. Libraries of hybrid proteins from distantly related sequences. Nat. Biotechnol. 19:456-460. [DOI] [PubMed] [Google Scholar]
- 24.Stemmer, W. P. C. 1994. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370:389-391. [DOI] [PubMed] [Google Scholar]
- 25.Stemmer, W. P. C. 1994. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91:10747-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, S. F., S. Freer, and A. A. Benson. 1967. Transphosphatidylation by phospholipase D. J. Biol. Chem. 242:477-484. [PubMed] [Google Scholar]