Abstract
Cryptosporidium parvum and C. hominis have been the cause of large and serious outbreaks of waterborne cryptosporidiosis. A specific and sensitive recovery-detection method is required for control of this pathogen in drinking water. In the present study, nested PCR-restriction fragment length polymorphism (RFLP), which targets the divergent Cpgp40/15 gene, was developed. This nested PCR detected only the gene derived from C. parvum and C. hominis strains, and RFLP was able to discriminate between the PCR products from C. parvum and C. hominis. To evaluate the sensitivity of nested PCR, C. parvum oocysts inoculated in water samples of two different turbidities were recovered by immunomagnetic separation (IMS) and detected by nested PCR and fluorescent antibody assay (FA). Genetic detection by nested PCR and oocyst number confirmed by FA were compared, and the results suggested that detection by nested PCR depends on the confirmed oocyst number and that nested PCR in combination with IMS has the ability to detect a single oocyst in a water sample. We applied an agitation procedure with river water solids to which oocysts were added to evaluate the recovery and detection by the procedure in environmental samples and found some decrease in the rate of detection by IMS.
Cryptosporidium is an important waterborne pathogen that infects the gastrointestinal or respiratory tract of mammals, including humans. The Cryptosporidium oocyst has enough resistance to drinking water disinfectants such as chlorine and chloramines to survive in many water treatment and distribution systems (6, 7). The 50% infective dose of the pathogen in healthy human volunteers has been reported to be 9 to 1,042 oocysts, showing that a low dose of the parasite oocyst is sufficient to cause infection (2, 14). In particular, infection with Cryptosporidium in immunocompromised person results in chronic and sometimes lethal diarrhea. Contamination of drinking water by infectious oocysts has been the cause of large and serious outbreaks of cryptosporidiosis such as the Milwaukee outbreak in 1993 (9). Consequently, investigation of the presence of Cryptosporidium in source and drinking water is indispensable for the effective control of waterborne cryptosporidiosis outbreaks.
The genus Cryptosporidium is composed of at least 13 species (23). In this genus, waterborne outbreaks are only known to be caused by C. parvum and C. hominis, which were previously classified into C. parvum bovine and human genotypes, respectively (13, 15, 16, 23). Only C. parvum and C. hominis have been confirmed to infect humans, though at least three species of Cryptosporidium, C. meleagridis, C. felis, and C. canis, have been detected and/or isolated from immunocompromised persons or children (12, 21). Thus, C. parvum and C. hominis are the most notable species to be controlled for safe drinking water as well as the water environment. In addition, discrimination between C. parvum and C. hominis is useful for elucidation of the contamination source by the pathogens in the water environment.
In the present study, we aimed to develop nested PCR which specifically detects the gene derived from C. parvum and C. hominis and discriminates the amplified product between these species. The Cpgp40/15 gene, which encodes the sporozoite membrane glycoproteins and shows high diversity between species (18), was suggested to be suitable as the amplification target. The generated product was analyzed by restriction fragment length polymorphism (RFLP) for discrimination between C. parvum and C. hominis. To investigate the sensitivity of nested PCR, the detection method was applied to oocyst inoculation experiments with concentrates of suspended solids recovered from river water, in which PCR inhibitors would be present (4). C. parvum oocysts inoculated into water samples of two different turbidities were recovered by immunomagnetic separation (IMS). The recovered oocysts were detected by both nested PCR and fluorescent antibody assays (FA). By comparison of the results, we discuss the evaluation of nested PCR for detection of C. parvum from an environmental sample.
MATERIALS AND METHODS
Parasites.
Oocysts of the C. parvum Iowa strain and the C. muris RN66 strain and cysts of the Giardia lamblia H3 strain and the G. muris Robert-Thompson strain were obtained from Waterborne, Inc. Oocysts of the C. parvum HNJ-1 strain were kind gifts from Tsuyoshi Hirata and Shigemitsu Morita (Azabu University, Japan). Frozen oocysts of C. hominis isolated from the feces of a patient in the waterborne outbreak of Ogose, Saitama Prefecture, Japan, 1996 (Ogose strain), were kind gifts from Norishige Yamamoto (Saitama Institute of Public Health, Japan). The oocysts and cysts except for the Ogose strain were stored at 4°C, whereas the oocysts of the Ogose strain were stored at −20°C.
DNA extraction.
Chromosomal DNA was extracted and purified from 3 × 106 oocysts of the C. parvum Iowa strain as described by Maniatis et al. (10). The resulting DNA, which was 1.7 × 10−6 g in weight, was resolved in 100 μl of Tris-EDTA buffer. The chromosomal DNA was diluted with 10-fold serial dilutions to determine the sensitivity of nested PCR.
Chromosomal DNA of the C. parvum NHJ-1 strain, C. hominis, C. muris, G. lamblia, and G. muris was extracted as described by Johnson et al. (5) with some modifications. Briefly, the oocysts or cysts of the parasites were pelleted by centrifugation (1,200 × g at room temperature for 10 min) and then suspended in 50 μl of Tris-EDTA buffer. The suspension was frozen at −20°C for 15 min followed by thawing in boiling water for 3 min. These manipulations were performed for six cycles. The resultant was centrifuged at 20,000 × g for 1 min. The supernatant was recovered and then used as a template for the first PCR.
Nested PCR-RFLP analysis.
Design of the primers was referenced to the nucleotide sequences of the Cpgp40/15 genes as determined by Strong et al. (18). The GenBank accession numbers for the sequences cited in the present study are AF022929, AF164487 to AF164505, AF164508, AF164509, AF178690 to AF178697, and AF224462 to AF224464. The primers for the amplification of the Cpgp40/15 gene used in the present study are listed in Table 1. In order to amplify the Cpgp40/15 gene, the first PCR was performed with the primer set gp40/15-51 and gp40/15-31, and nested PCR was carried out with a pair of primers, gp40/15-52 and gp40/15-32. The first and nested PCR was performed in a 50-μl reaction system of 1x buffer, 0.2 mM deoxynucleoside triphosphate mix, 0.5 μM each of the primers, 0.2 units of Ex Taq polymerase (Takara Shuzo, Shiga, Japan), and 1 μl of the template. The reaction of the first and nested amplification for the Cpgp40/15 gene was programmed at initial denaturation at 94°C for 3 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min, and a final extension at 72°C for 7 min. Nested PCR for the small-subunit (SSU) rRNA gene was performed as described by Xiao et al. (22, 24). The amplified product was separated by electrophoresis on 3% agarose gel, stained with ethidium bromide, and visualized under a UV transilluminator.
TABLE 1.
Primers used in this study
| Primera | Nucleotide sequence |
|---|---|
| gp40/15-51 | 5′-TCCGCTGTATTCTCAGCCCCA-3′ |
| gp40/15-31 | 5′-AGCAGAGGAACCAGCATCCTT-3′ |
| gp40/15-52 | 5′-TGTTCCTGTTGAGGGCTCATC-3′ |
| gp40/15-32 | 5′-GGCAAACAAATCGACGGTTGC-3′ |
| SSUrRNA-51 | 5′-AACCTGGTTGATCCTGCCAGTAGTC-3′ |
| SSUrRNA-31 | 5′-TGATCCTTCTGCAGGTTCACCTACG-3′ |
| SSUrRNA-52 | 5′-TTCTAGAGCTAATACATGCG-3′ |
| SSUrRNA-32 | 5′-CCCTAATCCTTCGAAACAGGA-3′ |
Primers for amplification of the SSU rRNA gene were from a report described by Xiao et al. (22).
For RFLP analysis, the target PCR product was excised from the agarose gel and purified with a QIAquick gel extraction kit (Qiagen, Tokyo, Japan). The fragment of the Cpgp40/15 gene was digested with HindIII, MflI (Takara Shuzo), and RsaI (Toyobo, Osaka, Japan), whereas that of the SSU rRNA gene was digested with SspI and VspI (Takara Shuzo) as described by Xiao et al. (22). The digestion was performed at 37°C for 60 min. The reactant was separated by electrophoresis on 3% agarose gels, stained with ethidium bromide, and visualized under a UV transilluminator.
Recovery of inoculated C. parvum oocysts from turbid water by IMS.
River water was collected on a rainy day in January 2002 from Tama River, Tokyo. The river water was filtrated with hydrophilic polytetrafluoroethylene (PTFE) membrane filters (pore size, 5 μm; Advantec, Inc., Tokyo, Japan). The membrane-trapped solids were eluted by ultrasonication into phosphate-buffered saline and centrifuged at 1,500 × g for 10 min. The recovered pellet was suspended into phosphate-buffered saline and stored at 4°C before use.
The concentrates recovered from 0.24 and 1.2 liters of the original river water, which were 0.05 and 0.3 g in wet weight, respectively, were washed three times with distilled water. The concentrates obtained were resuspended into 50 ml of distilled water, which resulted in turbidities of about 200 and 1,000 nephelometric turbidity units (NTU), respectively. These turbid water samples were prepared in two tubes, and each pair of tubes was inoculated with 50, 100, 200, and 600 oocysts of the C. parvum Iowa strain. The mixtures of turbid water and oocysts in 50-ml polypropylene tubes were agitated with a vertical rotator overnight. After overnight agitation, the samples were centrifuged at 1,500 × g for 10 min, and the pellets were used for the oocyst recovery procedure. The inoculated oocysts in the two tubes were separately recovered by IMS from each tube with a commercial kit (Dynabeads GC-combo; Dynal Biotech, Oslo, Norway). The recovered oocysts from one of the two tubes were detected with the Cpgp40/15 gene by nested PCR and that from another was investigated for the number of Cryptosporidium oocysts by FA. For nested PCR, chromosomal DNA from the recovered oocysts was extracted by the freeze-thaw method as described above, and three reaction systems were prepared for each trial. The inoculation trial was performed in duplicate or triplicate.
FA staining of the recovered oocysts.
The purified oocysts were stained on hydrophilic PTFE membrane filters (diameter, 25 mm; pore size, 0.2 μm; Advantec, Inc.) with a vacuum filtration unit (Advantec, Inc.). Staining of the oocysts was performed with a commercial kit according to the instructions (Hydrofluoro Combo Kit; Strategic Diagnostics Inc., Newark, Del.). The stained samples were observed with an epifluorescence differential-interference microscope (BX-50; Olympus, Tokyo, Japan) at a magnification of ×400. The confirmed oocysts, which are 4 to 6 μm in diameter with a specific fluorescence at the edge of the spherical bodies, were counted on each membrane filter.
RESULTS
Specificity of PCR and RFLP analysis.
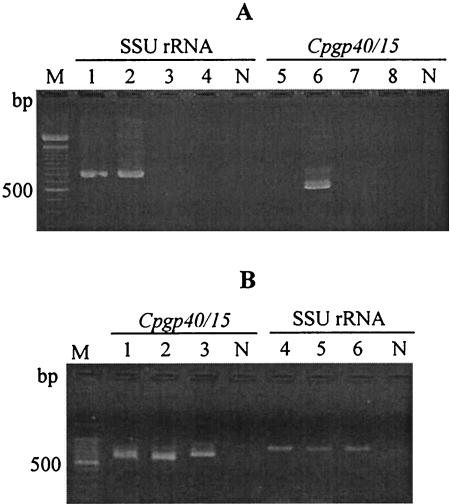
Nested PCR for the Cpgp40/15 gene amplified a product that is the almost same size as we expected (607 bp) from C. parvum Iowa strain (Fig. 1A). No amplified products were obtained from C. muris, G. lamblia, and G. muris templates. Nested PCR for amplification of the SSU rRNA gene, however, generated a product not only from C. parvum but also from C. muris. Amplified products from the two C. parvum strains and the C. hominis Ogose strain were generated by nested PCR targeting the Cpgp40/15 gene and the SSU rRNA gene (Fig. 1B). The DNA band size of the Cpgp40/15 gene generated from the C. parvum strains was about 600 bp, whereas that from the C. hominis strain was about 670 bp.
FIG. 1.
Nested PCR for the Cpgp40/15 gene and the SSU rRNA. (A) Nested PCR with templates of four kinds of parasites. Nested PCRs for the SSU rRNA gene and the Cpgp40/15 gene are shown in lanes 1 to 4 and 5 to 8, respectively. Lanes 1 and 5, C. muris RN66 strain; lanes 2 and 6, C. parvum Iowa strain; lanes 3 and 7, G. lamblia H3 strain; lanes 4 and 8, G. muris Roberts-Thompson strain. (B) Nested PCR with templates of C. parvum and C. hominis strains. Nested PCRs for the Cpgp40/15 and SSU rRNA are shown in lanes 1 to 3 and 4 to 6, respectively. Lanes 1 and 4, C. parvum Iowa strain; lanes 2 and 5, C. parvum HNJ-1 strain; lanes 3 and 6, C. hominis Ogose strain. Lanes M and N show DNA size standards (100-bp DNA ladder; Takara Shuzo) and the PCR negative control, respectively.
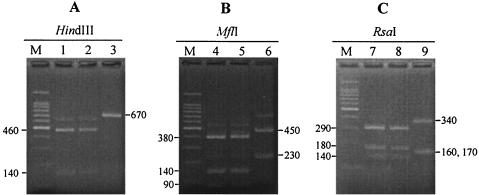
Digestion of the C. parvum products with HindIII resulted in two fragments of about 140 bp and 460 bp, whereas the C. hominis product was not digested with the restriction enzyme (Fig. 2A). Digestion of C. parvum and C. hominis products with MflI resulted in three or two fragments of about 90, 140, and 380 bp and 230 and 450 bp, respectively (Fig. 2B). Digestion of C. parvum and C. hominis products with RsaI resulted in three fragments of about 140, 180, and 290 bp and 160, 170, and 340 bp, respectively (Fig. 2C).
FIG. 2.
RFLP of the Cpgp40/15 PCR product generated from three C. parvum and C. hominis strains. The products digested with HindIII (A), MflI (B), and RsaI (C) are in lanes 1 to 3, 4 to 6, and 7 to 9, respectively. Lanes 1, 4, and 7, C. parvum Iowa strain; lanes 2, 5, and 8, C. parvum HNJ-1 strain; lanes 3, 6, and 9, C. hominis Ogose strain. Lane M shows a 100-bp DNA ladder. The lower band of the Ogose strain digested with RsaI (lane 9) consists of two fragments of 160 and 170 bp.
Sensitivity of PCR.
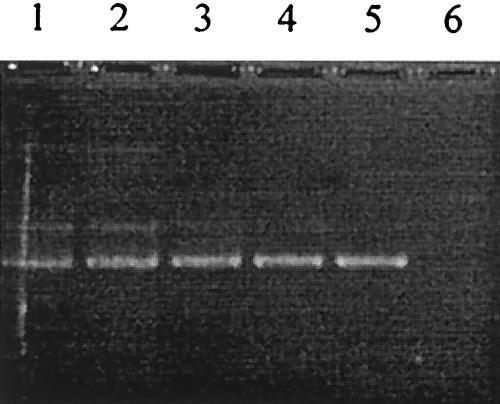
Nested PCR was performed with a template of 10-fold-diluted chromosomal DNA ranging from 5 × 10−9 g to 5 × 10−14 g. The detectable limit of nested PCR was 5 × 10−13 g, as shown in Fig. 3, which was almost equivalent to the weight of chromosomal DNA corresponding a single oocyst.
FIG. 3.
Detectable limit of nested PCR. Nested PCR was performed with 10-fold serially diluted chromosomal DNA template of the C. parvum Iowa strain ranging from 5 × 10−9 g (lane 1) to 5 × 10−14 g (lane 6).
Detection of inoculated oocysts from turbid water by IMS-PCR and IMS-FA.
At first, it was confirmed that neither the C. parvum and C. hominis gene nor the Cryptosporidium oocysts were detected from the concentrate of suspended solids recovered from 1.2 liters of river water (data not shown). In addition, to investigate the oocyst-inoculated trials without suspended solids, distilled water was inoculated with oocysts in a 50-ml tube. The preparation was agitated overnight as described in materials and methods, and oocyst detection was performed by IMS-FA. The resulting recovery rates were more than 85%.
The results obtained from the inoculation trials are shown in Table 2. In the trials in low turbidity water of about 200 NTU, the amplified product was detected from all trials with the 600 oocyst-inoculated water samples and two of three trials with the 100 and 200 oocyst-inoculated samples. In these trials, the PCR product of the 200 and 600 oocyst-inoculated trials was obtained from all three reaction systems, whereas that of the 100 oocyst-inoculated trials was obtained from one or two of the three reaction systems in each trial. In the trials with high turbidity water of about 1,000 NTU, amplified products were detected from one trial inoculated with 200 and 600 oocysts, in which the product was obtained from one and two of the three reaction systems.
TABLE 2.
Recovery and detection of C. parvum by IMS-PCR and IMS-FA from oocyst-inoculated turbid water samples
| Sample type | Trial | Detection by nested PCR and FA at inoculation dose:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 600 oocysts
|
200 oocysts
|
100 oocysts
|
50 oocysts
|
||||||
| Detection | No. (%) | Detection | No. (%) | Detection | No. (%) | Detection | No. (%) | ||
| Low turbidity (200 NTU) | First | +++ | 312 (52%) | +++ | 76 (38%) | ++ | 58 (58%) | − | 17 (34%) |
| Second | +++ | 318 (53%) | − | 125 (63%) | − | 47 (47%) | − | 18 (36%) | |
| Third | NT | +++ | 117 (59%) | + | 67 (67%) | NT | |||
| Avg | 315 (53%) | 106 (53%) | 57 (57%) | 18 (36%) | |||||
| High turbidity (1,000 NTU) | First | ++ | 89 (15%) | + | 38 (19%) | − | 30 (30%) | − | 13 (26%) |
| Second | − | 134 (22%) | − | 39 (20%) | − | 35 (35%) | − | 15 (30%) | |
| Third | NT | − | 62 (31%) | − | 19 (19%) | NT | |||
| Avg | 112 (19%) | 46 (23%) | 28 (28%) | 14 (28%) | |||||
Detection: +++, detection in all three reaction systems; ++, detection in two reaction systems; +, detection in one reaction system; −, no detection. No. (%): number of oocysts confirmed by FA (% recovery). NT, not tested.
In the IMS-FA experiments, the oocyst numbers and recovery rates are also shown in Table 2. The recovered oocyst number depended on the inoculated oocyst numbers and turbidity of each sample. The rates obtained from the trials of the low-turbidity water sample were 34 to 67%, with a mean of 51% ± 12%, whereas those of the high-turbidity water sample were 15 to 35%, with a mean of 25% ± 7%.
Detection of oocysts from inoculated water sample without overnight agitation.
The IMS-PCR and IMS-FA assays were also applied to the oocyst-inoculated turbid water samples without the overnight agitation procedure to evaluate the effect of agitation on the recovery step by IMS followed by nested PCR and FA. One hundred oocysts were inoculated into water samples of two different turbidities at about 200 and 1,000 NTU. The trial was performed in duplicate for each turbid water sample and the results are shown in Table 3. PCR products were detected from all trials of both turbidities, and the product was obtained from all three reaction systems except for a single trial of high turbidity. The oocyst recovery rates confirmed by FA were 87 to 91% for the trials with the low-turbidity water sample and 52 to 71% for those of the high-turbidity water sample.
TABLE 3.
Recovery and detection of C. parvum by IMS-PCR and IMS-FA from oocyst-inoculated turbid water samples without agitationa
| Trial | Detection by nested PCR and FA
|
|||
|---|---|---|---|---|
| Low turbidity (200 NTU)
|
High turbidity (1,000 NTU)
|
|||
| Detection | No. (%) | Detection | No. (%) | |
| First | +++ | 91 (91%) | + | 71 (71%) |
| Second | +++ | 87 (87%) | +++ | 52 (52%) |
| Avg | 89 (89%) | 62 (62%) | ||
The dose was 100 oocysts. See also Table 2, footnote a.
DISCUSSION
For control of pathogenic Cryptosporidium organisms in water environments, various recovery and/or detection methods have been developed and evaluated after waterborne cryptosporidiosis outbreaks have been documented. IMS-FA based on the United States Environmental Protection Agency method 1623 (20) is applied for protozoan analysis in drinking water in the United States and other countries. IMS-FA, however, is not able to discriminate Cryptosporidium species because IMS and FA use the genus-specific antibody for the capture and detection of the oocysts, respectively (19). Thus, we developed the nested PCR-RFLP method for the highly divergent Cpgp40/15 gene to detect the C. parvum and C. hominis genes and discriminate between the two species.
A specific investigation showed that the nested PCR-RFLP developed in the present study detects only the C. parvum and C. hominis genes. The PCR product of C. hominis was digested with RsaI and MflI but not with HindIII, whereas that of C. parvum was digested with all the enzymes. A distinct RFLP patterns were obtained between the PCR products of C. parvum and C. hominis digested with MflI and RsaI. In addition, the combination of the PCR product length and RFLP patterns of MflI and RsaI is likely to enable to classification of C. hominis into four subtypes (Ia to Id), which was suggested by Strong et al. (18). The Ogose strain was found to be classified as genotype Ia.
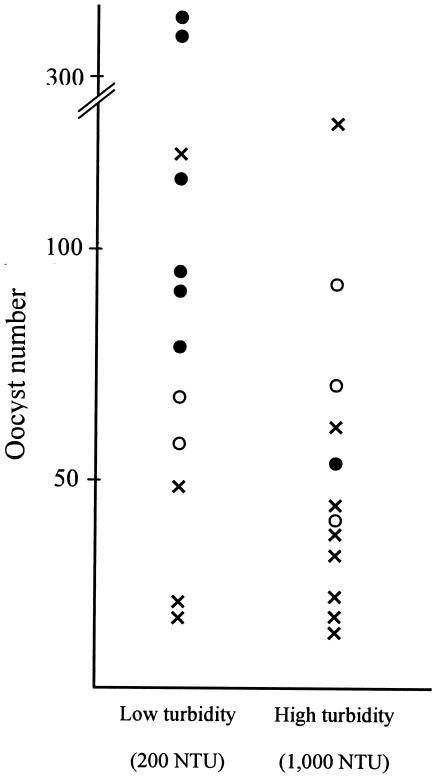
In order to evaluate the application of nested PCR for environmental water samples, all the results obtained from the oocyst inoculation experiments using water samples of two different turbidities are shown in Fig. 4. The comparison between the genetic detection by nested PCR and oocyst numbers confirmed by FA suggests that the detection of C. parvum by nested PCR depends on the recovered oocyst number and that the detectable limit of nested PCR is about 50 oocysts in one trial. In the present study, however, one μl in 50 μl of DNA solution extracted from all the recovered oocysts was used as the template of the first PCR, whereas all the recovered sample was investigated for oocyst number by FA. Thus, nested PCR combined with IMS is suggested to have the ability to detect a single oocyst in an environmental sample. The detectable limit of nested PCR with purified C. parvum chromosomal DNA was 5 × 10−13 g and was found to correspond to a single oocyst. Thus, the detection limit of nested PCR with oocysts recovered from a water sample by IMS was suggested to be almost equivalent to that with the purified DNA sample.
FIG. 4.
Relationship between genetic detection by nested PCR and oocyst number confirmed by FA from application experiments with oocyst-inoculated turbid water samples. Circles show successful detection by nested PCR. Solid circles show successful detection in all three reaction systems, and open circles show detection in at least one of the reaction systems. Crosses show failed detection by nested PCR.
The oocyst recovery results obtained by IMS-FA were found to depend on the turbidity of the water sample and inoculated oocyst number as described by various reports (1, 3, 8, 11). The recovery rates, however, were lower than the rates reported in other studies (1, 3, 17). Filtration procedures, which have been shown to result in a decrease of the recovery rate by Feng et al. (3), were not performed in the present study. We evaluated the IMS sample preparation procedure by allowing the oocysts to contact suspended solids concentrated from river water. To evaluate the influence of overnight agitation on the recovery rate, the results were compared between experiments with and without overnight agitation. This comparison showed that higher recovery rates were obtained from the trials without overnight agitation. Overnight agitation in small-scale tubes with a vertical rotator was suggested to allow frequent contacts between the oocysts and the suspended solids and to result in the lower recovery rate. In addition, it is noteworthy that the efficient association of the oocysts with the suspended solids was observed in distilled water as the suspending medium, which may bias the association compared to what might be observed in the presence of soluble organic materials and salts. The association mechanism remains unclear. As a possibility, however, the association may be mediated by polymerized organic substances such as extracellular polysaccharides produced by bacteria or algae in the solids. The viscosity of the organic substances may play a role in the association. Concentrates from the river water used in the present study were suggested to contain the substance because the river water was collected downstream of effluent sewage, where numerous bacteria and algae are present. The association is a critical problem for the oocyst recovery procedure by IMS because the antibody cannot recognize the antigen on the surface of the oocyst wall. The oocysts are suggested to frequently contact and attach to the suspended solids in environmental water, including river water and effluents from sewage treatment facilities. The reproduction of the environmental conditions in which the oocysts must be present is suggested to be necessary for exact evaluation of the application of the oocyst recovery and detection method to environmental water.
In conclusion, we showed that the nested PCR developed in the present study was able to specifically detect a selected gene of C. parvum and C. hominis and that the resulting product has the ability to discriminate between the two Cryptosporidium species by RFLP analysis. Our data also suggest that gene amplification in combination with IMS has the ability to detect the gene from a single oocyst in an environmental water sample. However, the application experiments showed that association of the oocysts with the suspended solids, which must also occur in environmental water, is a cause of the decrease in the oocyst recovery by IMS. We propose that the agitation procedure, which allows the oocysts to associate with suspended solids, should be performed in the application experiments for evaluation of the recovery and detection methods of the pathogens from environmental water samples.
Acknowledgments
We thank Tsuyoshi Hirata, Shigemitsu Morita, and Norishige Yamamoto for providing C. parvum and C. hominis isolates. We also thank Clarence J. Peters for helpful advice and comments on the manuscript.
REFERENCES
- 1.Bukhari, Z., R. M. McCuin, C. R. Fricker, and J. L. Clancy. 1998. Immunomagnetic separation of Cryptosporidium parvum from source water samples of various turbidities. Appl. Environ. Microbiol. 64:4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DuPont, H. L., C. L. Chappell, C. R. Sterling, P. C. Okhuysen, J. B. Rose, and W. Jakubowski. 1995. The infectivity of Cryptosporidium parvum in healthy volunteers. N. Engl. J. Med. 332:855-859. [DOI] [PubMed] [Google Scholar]
- 3.Feng, Y. Y., S. L. Ong, J. Y. Hu, L. F. Song, X. L. Tan, and W. J. Ng. 2003. Effect of particles on the recovery of Cryptosporidium oocysts from source water samples of various turbidities. Appl. Environ. Microbiol. 69:1898-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ijzerman, M. M., D. R. Dahling, and G. S. Fout. 1997. A method to remove environmental inhibitors prior to the detection of waterborne enteric viruses by reverse transcription-polymerase chain reaction. J. Virol. Methods 63:145-153. [DOI] [PubMed] [Google Scholar]
- 5.Johnson, D. W., N. L. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl. Environ. Microbiol. 61:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korich, D. G., J. R. Mead, M. S. Madore, N. A. Sinclair, and C. R. Sterling. 1990. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 56:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lisle, J. T., and J. B. Rose. 1995. Cryptosporidium contamination of water in the USA and UK: a mini review. Aqua 44:103-117. [Google Scholar]
- 8.Lowery, C. J., J. E. Moore, B. C. Millar, D. P. Burke, K. A. J. McCorry, E. Crothers, and J. S. G. Dooley. 2000. Detection and speciation of Cryptosporidium spp. in environmental water samples by immunomagnetic separation, PCR and endonuclease restriction. J. Med. Microbiol. 49:779-785. [DOI] [PubMed] [Google Scholar]
- 9.MacKenzie, W. R., N. J. Hoxie, M. E. Proctor, M. S. Gradus, K. A. Blair, D. E. Peterson, J. J. Kazmierczak, D. G. Addiss, K. R. Fox, J. B. Rose, and J. P. Davis. 1994. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 331:161-167. [DOI] [PubMed] [Google Scholar]
- 10.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.McCuin, R. M., Z. Bukhari, J. Sobrinho, and J. L. Clancy. 2001. Recovery of Cryptosporidium oocysts and Giardia cysts from source water concentrates using immunomagnetic separation. J. Microbiol. Methods 45:69-76. [DOI] [PubMed] [Google Scholar]
- 12.Morgan, U., R. Weber, L. Xiao, I. Sulaiman, R. C. A. Thompson, W. Ndiritu, A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan-Ryan, U. M., A. Fall, L. A. Ward, N. Hijjawi, I. Sulaiman, R. Fayer, R. C. Thompson, M. Olson, A. Lai, and L. Xiao. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433-440. [DOI] [PubMed] [Google Scholar]
- 14.Okhuysen, P. C., C. L. Chappell, J. H. Crabb, C. R. Sterling, and H. L. DuPont. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 180:1275-1281. [DOI] [PubMed] [Google Scholar]
- 15.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. L. Ong, W. R. MacKenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira S. J., N. E. Ramirez, L, Xiao, and L. A. Ward. 2002. Pathogenesis of human and bovine Cryptosporidium parvum in gnotobiotic pigs. J. Infect. Dis. 186:715-718. [DOI] [PubMed] [Google Scholar]
- 17.Stanfield, G., E. Carrington, F. Albinet, B. Compagnon, N. Dumoutier, B. Hambsch, A. Lorthioy, G. Medema, H. Pezoldt, M. R. Roubin, A. Lohman, and T. Whitmore. 2000. An optimized and standardized test to determine the presence of the protozoa Cryptosporidium and Giardia in water. Water Sci. Technol. 41:103-110. [Google Scholar]
- 18.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturbaum, G. D., P. T. Klonicki, M. M. Marshall, B. H. Jost, B. L. Clay, and C. R. Sterling. 2002. Immunomagnetic separation (IMS)-fluorescent antibody detection and IMS-PCR detection of seeded Cryptosporidium parvum oocysts in natural waters and their limitations. Appl. Environ. Microbiol. 68:2991-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.U.S. Environmental Protection Agency. 1999. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. Publication EPA-821-R-99-006. U.S. Environmental Protection Agency, Washington, D.C.
- 21.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 22.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao, L., R. Fayer, U. Ryan, and S. J. Upton. 2004. Cryptosporidium Taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]