The first step for the cure of multiple myeloma (MM) is to achieve a complete response (CR);1, 2 however, regardless of CR improvement, most patients experience disease progression or relapse in part due to the persistence of low levels of clonal plasma cells after treatment (minimal residual disease (MRD)).3 In addition, many patients who achieve MRD-negative status also relapse, indicating that the sensitivity and specificity of traditional techniques for MRD assessment can be improved.4 At present, multiparameter flow cytometry (MFC), allele-specific oligonucleotide PCR and high-throughput sequencing (NGS) are the three high-sensitivity techniques available for MRD quantification in MM.3, 5, 6 A growing body of evidence strongly suggests that detection of subclinical levels of MRD with this high-sensitivity methods provides powerful independent prognostic information.7, 8 MFC is the most widely available technique with excellent sensitivity and applicability; however, there is variability in sensitivity, panels and performance between laboratories, and requires high level of expertise.9 On the other hand, in previous work, we have shown that NGS of immunoglobulin (IG) rearranged genes is an effective technology to identify and quantify pathological clonal cells in MM with a sensitivity of at least 0.001%.7, 10 However, this NGS technology is a proprietary multiplex PCR that is performed at centralized laboratories increasing the turn-around time for the results. Furthermore, the NGS method for MRD quantification in MM needs additional clinical validation to prove its utility for patient risk stratification, as well as to evaluate the efficacy of different treatment schemes.
In the present study, we describe and analytically validate a simplified in-house deep-sequencing method to identify and quantify MRD in MM patients from 1 μg of DNA. The method uses the standardized primers developed by the Biomed-2 concerted action to amplify all IGH or IGK sequences in a patient sample.11 Libraries were prepared by ligation of specific adaptor oligos and sequenced either on an Ion S5 (ThermoFisher Scientifc, Palo Alto, CA, USA) or on a MiSeq sequencer (Illumina, San Diego, CA, USA). The sequencing data were analyzed with a set of specific mathematical and bioinformatics tools to identify and quantitate the clone-specific sequence (clonotype) present on each sample (code patent pending). A clonotype was identified when at least 400 identical sequencing reads were obtained, or was present at a frequency of >1%.
We analyzed MRD by deep sequencing in bone marrow (BM) samples from 73 MM patients of which DNA was available for testing (Bioproject PRJNA360043). Patients were enrolled in the phase 2 trial for newly diagnosed elderly MM patients PETHEMA/GEM2010MAS65 study (www.clinicaltrials.gov as #NCT01237249); patients were randomized to receive 9 cycles of bortezomib/melphalan/prednisone (VMP) followed by 9 cycles of lenalidomide/dexamethasone (Rd; sequential arm, n=38) vs 18 alternating cycles of VMP/Rd (alternating arm, n=35).12 Nighty-five percent (n=69) of the patients experienced a CR according to the International Myeloma Working Group (IMWG).13 Median follow-up of the series was 3 years.
Statistical analyses were performed with the SPSS program version 21.0 (IBM, Armonk, NY, USA). Linear regression was used to compare different dilutions of the DNA samples. The Spearman correlation coefficient was used to compare MFC data with NGS data. Overall survival (OS) and progression-free survival (PFS) were estimated by Kaplan–Meier survival analysis and statistical differences assessed via log-rank and Wilcoxon analyses.
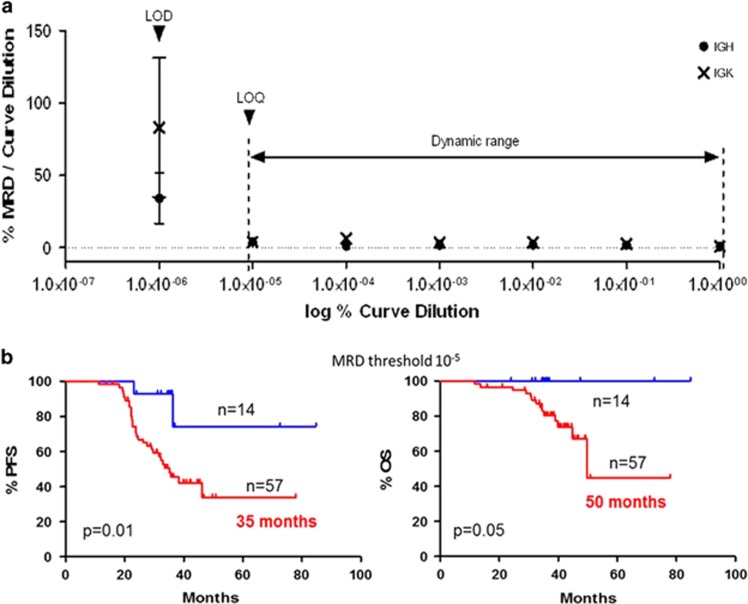
With our approach of deep sequencing, a clonotype was detected in the 97% (71 out of 73) of MM patients, indicating a specificity similar to the clinical specificity reported in the original validation studies of the Biomed-2 method (98%),11 slightly higher to the previously described NGS assay for IG quantification (91%),7 and comparable to 8c-MFC.8 Clonotypes were not detected in normal tonsil and BM samples, indicating the high specificity of the assay. This method also presents a very good analytical sensitivity of at least 10−5, as determined from a 10-fold dilution curve of commercial control monoclonal and polyclonal DNAs (Figure 1a). With the same primer combinations, the analytical sensitivity of Biomed-2 amplification to detect a clonal population was 10−2 for IGH and IGK,11 consequently, applying deep-sequencing technologies to the Biomed-2 design highly improves the sensitivity of clonal identification. In addition, the method also shows high reproducibility between runs and different NGS platforms (99.2%, 91 samples tested in duplicate in different sequencing runs), and is very precise for samples with MRD-negative values (median CV 8.1%, range 3.9–39).
Figure 1.
Performance characteristics of the NGS assay. (a) Plot of the dynamic range, limit of quantification (LOQ) and limit of detection (LOD) of the method from a 10-fold dilution curve. The vertical axis represents the ratio of MRD values to curve dilution and the horizontal axis represents the logarithm value for curve dilution. R2 = 0.98 and 0.96 for IGH and IGK genes, respectively, P<0.0001. (b) Progression-free survival (PFS) and overall survival (OS) plots according to MRD levels. MRD-negative values (blue) and MRD-positive values (Red). The threshold for negative values was 10−5.
As demonstrated in several trials, a prolonged therapy is an effective approach to improve survival in elderly patients. This was the basis for the 18 cycles explored in the GEM10mas65 clinical trial (VMP+Ld combination), which yielded excellent clinical results.12, 14 When we analyzed the molecular response in these elderly MM patients, we found that the proportion of patients achieving an MRD-negative status is significantly higher after 18 cycles of treatment (27% (n=19) vs 11.5% (n=8) after 18 and 9 cycles, respectively, P=0.04), confirming that a prolonged treatment improves the rate of molecular responses. Similar to the data obtained with other MRD methods,7, 8, 15 the achievement of molecular responses measured by our NGS method is able to predict 3-year survival in patients enrolled in the GEM10 clinical trial. Median PFS was 35 months vs not reached for patients with MRD-positive and -negative values, respectively (hazard ratio (HR)=2.76, 95% confidence interval (CI) 1.21–6.25, P=0.01; Figure 1b). Median OS was also prolonged in MRD-negative patients compared with MRD-positive patients, with 3-year OS rates of 100% and 45%, respectively (median OS of 50 months vs not reached respectively, HR=3.66, 95% CI 0.98–13.67, P=0.05). Hence, achieving a molecular response as determined by this new deep-sequencing method results in improved PFS and OS.
The high efficacy of the treatment based on VMP and Rd in a sequential or alternating scheme (CR rates of 42% and 40%, and PFS of 74% and 80%, respectively) was demonstrated previously.8, 12 In our present study, when we applied the NGS method to assess MRD negativity, more patients in the sequential treatment than in the alternating arm achieved a molecular response (36% (n=13) vs 20% (n=7), respectively). Nonetheless, we observed no significant difference in OS between both treatment arms. Taken together, the data show that patients could not only benefit from a prolonged treatment of 18 cycles but also suggest that 18 cycles of treatment in a sequential scheme can be associated with higher number of molecular responses and prolonged PFS. However, these results should be interpreted carefully because the low number of patients analyzed in the study, and further testing is needed in order to draw stronger conclusions.
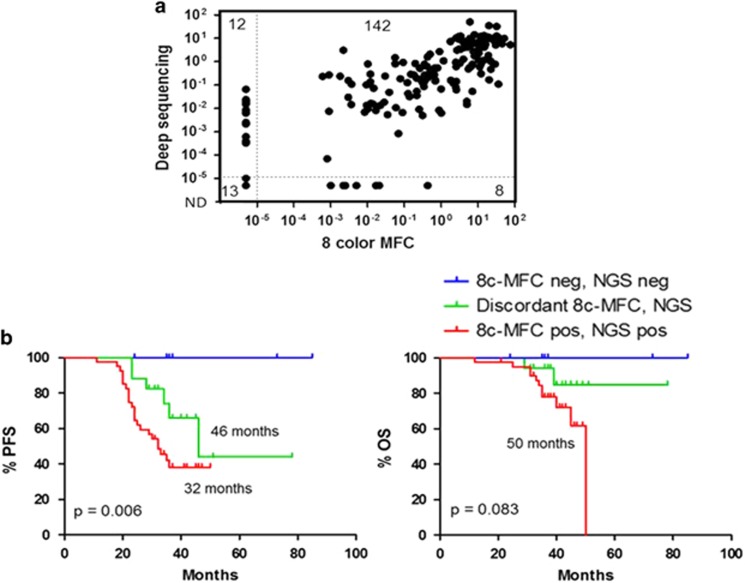
Sixty-six of these patients were also analyzed for MRD by MFC using an eight-color monoclonal antibody combination.8 The levels of MRD obtained with our method have a high degree of correlation with those assessed by 8c-MFC (total 66 patients, n=175 samples, Spearman coefficient R=0.7917, P<0.0001; n=109 post-treatment samples, R=0.6388, P<0.0001), with a global 89% concordance between MFC and NGS data (Figure 2a). Accordingly, there were no significant differences in terms of PFS and OS between the data obtained by our in-house method and MFC. Nevertheless, patients with discordant results between these two technologies, show an intermediate median PFS (46 months) as compared to double-positive (32 months) or double-negative (not reached) MRD values (P=0.0063; Figure 2b). At the time of the analysis, the OS for patients with discordant results was similar to that of NGS MRD-negative patients (not reached) versus a median of 50-month survival for MRD-positive patients (P=0.0835).
Figure 2.
Comparison between deep-sequencing (NGS) and multiparameter flow cytometry (8c-MFC) data. (a) Scatter plot showing correlation of MRD values between deep sequencing and eight-color MFC. Numbers on upper left and lower right indicates samples with discordant results. Spearman correlation coefficient R=0.7917, P<0.0001. (b) Progression-free survival (PFS) and overall survival (OS) plots of patients grouped according to concordance of MRD levels between NGS and 8c-MFC. Data corresponds to patients with MRD-negative values (MRD<10−5) by both methods (blue); MRD-positive values (MRD>10−5) by both methods (red); and MRD with discrepant data between NGS and 8c-FCM (green).
Due to the need of new response criteria that allows the identification of deeper responses than the now defined as clinical CR, the International Myeloma Working Group (IMGW) has defined new response categories of MRD negativity.13 One of them is sequencing MRD negative, reflecting the importance of the sensitivity of deep-sequencing methodology applied to the detection of very low numbers of tumor cells. In the new criteria, the IMGW recommends both deep-sequencing or next-generation flow to assess MRD in the BM, depending on the availability of the techniques at each center. As shown in the high degree of correlation of this study, this new method to measure MRD in MM by deep sequencing could be used to define MRD negativity by sequencing as defined in the new criteria of the IMWG.
In summary, our data confirm the clinical application of quantifying MRD levels by our in-house deep-sequencing method in MM patients. Our method shows a high analytical reproducibility and can be implemented in any laboratory with NGS capability, can be applied to the majority of MM patients with a short turn-round time, has a sensitivity of 10−5 and can be fully automated (from DNA extraction to data analysis), and thus easily standardized minimizing lab-to-lab variation.
Acknowledgments
This study was supported by the Cooperative Research Thematic Network grant RD12/0036/0061 of the Red de Cancer (Cancer Network of Excellence RTICC); the Subdirección General de Investigación Sanitaria (Instituto de Salud Carlos III, Spain) grants PI15/02062 and PI15/01484; the CRIS against Cancer foundation grant 2014/0120; Joan Rodés grant (JR 14/00016); and Undergraduate Fellowship IFI14/00008.
Footnotes
This study has been presented in part at the Annual Meeting of the American Society of Hematology in 2015 (Orlando, FL, USA) and at the 20th Congress of European Hematology Association (EHA) in 2015 (Vienna, Austria).
The authors declare no conflict of interest.
References
- Martinez-Lopez J, Blade J, Mateos M-V, Grande C, Alegre A, García-Laraña J et al. Long-term prognostic significance of response in multiple myeloma after stem cell transplantation. Blood 2011; 118: 529–534. [DOI] [PubMed] [Google Scholar]
- Gay F, Larocca A, Wijermans P, Cavallo F, Rossi D, Schaafsma R et al. Complete response correlates with long-term progression-free and overall survival in elderly myeloma treated with novel agents: analysis of 1175 patients. Blood 2011; 117: 3025–3031. [DOI] [PubMed] [Google Scholar]
- Paiva B, Gutiérrez NC, Rosiñol L, Vídriales M-B, Montalbán M-Á, Martínez-López J et al. High-risk cytogenetics and persistent minimal residual disease by multiparameter flow cytometry predict unsustained complete response after autologous stem cell transplantation in multiple myeloma. Blood 2012; 119: 687–691. [DOI] [PubMed] [Google Scholar]
- Martínez-López J, Paiva B, López-Anglada L, Mateos M-V, Cedena T, Vidríales M-B et al. Critical analysis of the stringent complete response in multiple myeloma: contribution of sFLC and bone marrow clonality. Blood 2015; 126: 858–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailankody S, Korde N, Lesokhin AM, Lendvai N, Hassoun H, Stetler-Stevenson M et al. Minimal residual disease in multiple myeloma: bringing the bench to the bedside. Nat Rev Clin Oncol 2015; 12: 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez J, Fernández-Redondo E, García-Sánz R, Montalbán MA, Martínez-Sánchez P, Pavia B et al. Clinical applicability and prognostic significance of molecular response assessed by fluorescent-PCR of immunoglobulin genes in multiple myeloma. Results from a GEM/PETHEMA study. Br J Haematol 2013; 163: 581–589. [DOI] [PubMed] [Google Scholar]
- Martinez-Lopez J, Lahuerta JJ, Pepin F, González M, Barrio S, Ayala R et al. Prognostic value of deep sequencing method for minimal residual disease detection in multiple myeloma. Blood 2014; 123: 3073–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva B, Cedena M-T, Puig N, Arana P, Vidriales M-B, Cordon L et al. Minimal residual disease monitoring and immune profiling using second generation flow cytometry in elderly multiple myeloma. Blood 2016; 127: 3165–3174. [DOI] [PubMed] [Google Scholar]
- Roschewski M, Stetler-Stevenson M, Yuan C, Mailankody S, Korde N, Landgren O. Minimal residual disease: what are the minimum requirements? J Clin Oncol 2014; 32: 475–476. [DOI] [PubMed] [Google Scholar]
- Faham M, Zheng J, Moorhead M, Carlton VEH, Stow P, Coustan-Smith E et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood 2012; 120: 5173–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JJM, Langerak AW, Brüggemann M, Evans PAS, Hummel M, Lavender FL et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003; 17: 2257–2317. [DOI] [PubMed] [Google Scholar]
- Mateos M-V, Martínez-López J, Hernández M-T, Ocio E-M, Rosiñol L, Martínez R et al. Sequential vs alternating administration of VMP and Rd in elderly patients with newly diagnosed MM. Blood 2016; 127: 420–425. [DOI] [PubMed] [Google Scholar]
- Kumar S, Paiva B, Anderson KC, Durie B, Landgren O, Moreau P et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol 2016; 17: e328–e346. [DOI] [PubMed] [Google Scholar]
- Palumbo A, Mina R, Cerrato C, Cavallo F. Role of onsolidation/maintenance therapy in multiple myeloma. Clin Lymphoma Myeloma Leuk 2013; 13 (Suppl 2): S349–S354. [DOI] [PubMed] [Google Scholar]
- Ladetto M, Pagliano G, Ferrero S, Cavallo F, Drandi D, Santo L et al. Major tumor shrinking and persistent molecular remissions after consolidation with bortezomib, thalidomide, and dexamethasone in patients with autografted myeloma. J Clin Oncol 2010; 28: 2077–2084. [DOI] [PubMed] [Google Scholar]