Abstract
Viral inactivation and adhesion-aggregation in water are often studied as separate phenomena. When the focus is placed on viral adhesion-aggregation, inactivation is neglected because the phenomena under investigation occur over a short period measured in days. When viral inactivation is studied, adhesion-aggregation phenomena are considered to be negligible because viral survival is traced over several days or months. In the present work, we took a global approach, examining the relative contributions of each of these processes in a complex system composed of groundwater, Poliovirus 1, and a hydrophobic container (polypropylene) maintained in a dark environment at 20°C. We demonstrated that infectious viral load fell off 2.8 log10 during the first 20 days. During this time, adhesion was far from negligible because it accounted for most of the decline, 1.5 log10. Adhesion was undoubtedly favored by the presence of divalent ions in the groundwater. After 20 days, aggregation may also have been the cause of 0.66 to 0.92 log10 of viral loss. Finally, viral inactivation was quantitatively the lowest phenomena because it only explained 0.38 to 0.64 log10 of the viral loss. This study thus clearly demonstrated that estimates of viral survival in a given system must always take into account adhesion-aggregation phenomena which may be responsible for the majority of viral loss in the aqueous phase. Adhesion and aggregation are reversible processes which may lead to an underestimation of viral load in certain studies.
More than 150 serotypes of enteric viruses (gastrointestinal viruses, hepatitis A and E viruses, enteroviruses) can be transmitted to humans from environmental sources. In water, the decrease of viral particles basically depends on two processes: inactivation and adhesion-aggregation. Both of these processes lead to a real (inactivation-adhesion) or apparent (aggregation) reduction in the concentration of infectious virus in the aqueous phase.
Viral inactivation results from an alteration of the viral capsid or genome. The result is a reduction in the number of infectious units which requires cell culture for quantification (16, 30, 39). The capacity of a virus to survive depends on its serotype and on the physical (e.g., temperature), chemical (e.g., pH and salinity), and biological (e.g., microorganisms) properties of the environment (9, 13, 14, 18, 20, 32, 46). For example, temperature is recognized as the principal factor affecting viral survival (20, 47). Studies devoted to viral survival are classically conducted in vitro using an appropriate viral model (e.g., Poliovirus 1, Adenovirus 40 and 41, hepatitis A virus), a natural (e.g., drinking water, seawater, river water) or artificial (e.g., buffer) aqueous environment, and a container (e.g., polypropylene, glass). Viral inactivation is then defined by following the time course of infectious virus in the aqueous phase. The time required for 90% of the infectious viral units to disappear (T90) is determined when the survival curve is log linear. More generally, viral survival is expressed as a rate of loss in function of time. Using this approach, adhesion-aggregation phenomena are either considered negligible or ignored.
Adhesion-aggregation depends (i) on the viral serotype and, more particularly, the viral surface charge (8, 12, 17, 21, 27, 43), (ii) on chemical properties of the medium such as pH, ionic strength, and the nature of the ions present (3, 11, 23, 26, 40, 43, 48), and finally, in the event of adhesion, (iii) on the nature of the adsorbing surface which in turn is described in terms of surface charge and hydrophobicity (17, 44). Viral adhesion-aggregation processes are reversible (3, 10, 25, 27, 43), and certain authors have described increased survival for adsorbed or aggregated viruses (2, 10, 17, 37). The relative significance of each of these adhesion-aggregation processes can be estimated from cell culture by quantifying the decline in infectious viral particles present in the aqueous phase (11, 29, 40). This is generally conducted in vitro over a short time period and under conditions in which viral inactivation can be neglected.
There have been few studies that have considered viral inactivation and viral adhesion-aggregation simultaneously, making it difficult to obtain a reliable estimate of the relative contribution of each process in a given system. When their effects are studied separately, all three phenomena, adhesion, aggregation, and inactivation, result in a decrease of infectious viral units in water. Consequently, in the presence of adhesion-aggregation phenomena, measuring viral decrease could produce an overestimation of inactivation processes (i.e., an underestimation of viral concentration). The same type of overestimation could result from measuring adhesion-aggregation in the presence of inactivation phenomena.
From a purely technical point of view, it would also be interesting to examine viral behavior in water held in a hydrophobic container because (i) marketed waters are generally stored in hydrophobic containers (e.g., terephthalate polyethylene) (19) and (ii) polypropylene containers have often been used for definitions of viral survival in different waters (1, 9, 16, 41, 45, 47).
The purpose of this work was thus to define the relative importance of inactivation, adhesion, and aggregation phenomena in a reference system composed of groundwater (GW), Poliovirus 1, and a polypropylene container. The overall behavior of viral particles in the reference system at 20°C was first compared with that in a system where the GW was replaced by phosphate buffer. Adhesion-aggregation phenomena in GW were then demonstrated using two approaches. The first consisted of an attempt to reproduce in phosphate buffer the kinetics observed in GW by adjusting parameters (pH, divalent ions) recognized as having an effect on the adhesion-aggregation processes. The second consisted of an attempt to reverse these processes in GW. Finally the kinetics of viral adhesion to a polypropylene container were examined.
MATERIALS AND METHODS
Virus and cells.
Poliovirus type 1 (Lsc2ab strain) was propagated in buffalo green monkey kidney cells growing in minimum essential medium (catalog no. M5650; Sigma) containing 5% fetal calf serum and 1% l-glutamine (catalog no. G7513; Sigma). For this cultivable strain, viral advanced cytopathic effects were observed after 24 to 48 h of incubation at 37°C. Stocks of viral inoculum was prepared by freezing and thawing them three times. The viral suspension was then centrifuged (10,000 × g, 60 min, 4°C). The supernatant stored at −70°C constituted the viral stock. It was quantified by the most probable number of cytopathic units (MPNCU) to 2 × 108 MPNCU/ml.
Quantification of infectious virus by cell culture.
Each sample was treated with an antibiotic and antimycotic solution (catalog no. A5955; Sigma) for 2 h at 37°C. Quantification of infectious Poliovirus 1 was performed in 96-well microplates with buffalo green monkey kidney cells. A total of 50 μl of three successive logarithmic dilutions of each sample was added to 200 μl of 7.5 × 104 cells/ml in minimum essential medium containing 2% fetal calf serum. Each dilution was seeded in 40 wells. The plates were incubated for 6 days at 37°C in a 5% CO2-95% air atmosphere and then examined for cytopathic effect. The number of wells with a cytopathic effect were counted, and the MPNCU was calculated (28).
Viral genome quantification. (i) Viral RNA extraction.
Viral RNA was extracted using a QIAmp viral RNA kit (catalog no. 52904; QIAGEN). In aqueous phase, viral RNA was extracted from the total volume (1 ml). This required some adaptation of the manufacturer's instructions. Lysis buffer (AVL) and ethanol (96 to 100%) volumes were proportionally increased. On polypropylene support, RNA was extracted by adding 1 ml of lysis buffer into the vial after the aqueous solution had ben removed. After shaking (vortexing) and contact time of 15 min, the suspension was transferred in 1 ml of 95% ethanol. For both aqueous phase and support, the suspension was filtered on the QIAmp spin column in a vacuum to adsorb RNA. After washing steps, RNA was eluted in 60 μl of elution buffer.
(ii) Primers and probes.
The primers and probe (Table 1) were previously described (16).
TABLE 1.
Primers and probe designed with Primer Express software version 1.0 and used in the Poliovirus 1 fluorogenic RT-PCR assay
| Primer or probe | Sequence (5′ to 3′) | Localization on Poliovirus 1 genome |
|---|---|---|
| Ent-r | 5′-CAAGATTGGTTCCTGCTTGATCTT-3′ | 5648-5625 |
| Ent-f | 5′-AGCATTGTGATCGATGGCAA-3′ | 5573-5592 |
| Ent-p | 5′-f-AGATCTTGGATGCCAAAGCGCTCG-t-3′a | 5601-5624 |
f, 6-carboxyfluorescein reporter dye; t, 6-carboxytetramethylrhodamine quencher dye.
(iii) cDNA synthesis.
cDNA was synthesized from the extracted RNA by use of the reverse primer (Ent-r) at a final concentration of 0.5 μM in a mix of 20 μl containing 4 μl of 5× reverse transcription (RT) buffer (250 mM Tris-HCl [pH 8.4], 50 mM MgCl2, 350 mM KCl, 15 mM dithiothreitol, 2.5 mM spermidine), 40 U of RNase inhibitor (catalog no. N2111; Promega), 0.25 μM of each deoxynucleoside triphosphate (catalog no. N8080260; Perkin Elmer), 10 U of reverse transcriptase (catalog no. M5108; Promega), 6 μl of free DNase, RNase water (catalog no. W4502; Sigma), and 5 μl of extracted product heated 3 min at 95°C. RT was performed at 42°C for 60 min. RNA-DNA hybrids were denatured, and reverse transcriptase was inactivated by heating to 95°C for 5 min. The resulting cDNA was then quantified by real-time PCR.
(iv) Real-time PCR.
PCR assay was done using 5 μl of cDNA in a final volume of 50 μl containing 25 μl of Taqman Universal Master Mix (catalog no. 4304437; PE Biosystems), 14.5 μl of nuclease-free water, forward (Ent-f) and reverse (Ent-r) primers at a final concentration of 0.5 μM, and a Taqman probe (Ent-p) at a final concentration of 0.3 μM. Amplification and detection were performed with an ABI Prism 7700 sequence detection system (Perkin Elmer Inc.). The amplification ramp included two hold programs, (i) 2 min at 50°C to activate the uracil N′-glycosylase followed by (ii) 10 min at 95°C to release the activity of the hot start DNA polymerase, followed by the repetition of 50 cycles of 15 s at 95°C and 60 s at 60°C. Real-time fluorescence measurements were obtained and directly analyzed using ABI prism 7700 sequence detection system software. The threshold cycle (CT) value for each sample generated was calculated by determining the point at which fluorescence exceeded a threshold limit.
Media.
Phosphate-buffered saline (PBS) (catalog no. 75511; Biomérieux) containing 7.650 g of NaCl/liter, 0.724 g of disodium phosphate/liter, and 0.210 g of monopotassium phosphate/liter was adjusted to different pH levels (5.1, 6.6, 7.2, 7.8, and 8.3) and sterilized by autoclaving. In some cases sterile MgCl2 was added at a final concentration of 0.033, 0.005, 0.0033, or 0.00033 M. In this case pH was readjusted after MgCl2 addition.
The concentrations of ions and compounds of the GW used in this study and determined by International Organization for Standardization procedures were as follows: Ca2+, 97 mg/liter; Mg2+, 21 mg/liter; Na+, 8.68 mg/liter; K+, 5.24 mg/liter; Fe2+, 0.390 mg/liter; Mn2+, 0.040 mg/liter; SO42−, 128 mg/liter; HCO3−, 256 mg/liter; Cl−, 4.6 mg/liter; PO43−, <0.01 mg/liter; SiO2, 8.1 mg/liter; NO3−, <0.1 mg/liter; NO2−, <0.01 mg/liter, and NH4+, 0.01 mg/liter. The conductivity of this water was 645 μS/cm, and the pH was measured in the laboratory at 7.8. This water comes from a deep well in the Bundsandstein sandstone situated in northeast France. By analogy with the mineral water classification scheme (31), this water can be considered to be of low-mineral-water content.
The same GW with the autochthonous microbial flora was previously used in a study examining virus survival (16). The autochthonous flora was not characterized, but the influence of this flora was defined by comparing Poliovirus 1 behavior in GW filtered using a 0.22-μm-pore-size filter (Millipore MillexR-GS) to that seen with unfiltered GW.
STE.
In a previous study (16), the persistence of Poliovirus 1 in comparison to that of Norovirus coming from human stools was estimated. To study the effect of this STE on virus behavior, the same extract used in a previous study (16) and kept frozen at −70°C was added to the PBS or GW. This STE was prepared as follows. The stool sample was diluted to 10% suspension in PBS, mixed with an equal volume of 1,1.2-trichloro-1,2.2-trifluoroethane, and clarified by centrifugation for 20 min at 500 × g. The supernatant was diluted 1/100 in GW or in PBS.
Experimental protocol.
A total of 100 ml of PBS or GW was seeded with Poliovirus 1 to obtain a final concentration of 105 MPNCU/ml. The 100-ml samples were homogenized 15 min at 20°C and distributed in 1-ml polypropylene vials (catalog no. 33 606 01; Polylabo), which were placed in the dark at 20°C. At sampling time, vials were removed from incubation and stored at −70°C until analysis by cell culture and real-time RT-PCR were performed. In cases in which the viral decrease as a function of time could be represented with a linear model, the slopes were statistically compared using Student's t test and a threshold of 5%.
To study reversibility of adhesion-aggregation processes, several vials were taken after 30 days of incubation and submitted to different treatments. Several surfactants were tested: sodium dodecyl sulfate (SDS) (catalog no. 27926238; Prolabo), an anionic detergent, was used at a final concentration of 10 mM, and Tween 80 (catalog no. 28830291; Prolabo) and Triton X-100 (catalog no. T-8787; Sigma), nonionic detergents, were used at a final concentration of 0.1%. In addition, fetal calf serum (catalog no. 010062; Eurobio) was used at a final concentration of 10% and urea (catalog no. 018228; Eurobio) was used at a final concentration of 10 mM. The desired pH was obtained by adding 1 M HCl and 1 M NaOH. After each treatment, tubes were briefly shaken with a vortex and kept at 20°C for 3 h before storage at −70°C until analysis by cell culture. Some of the treatments applied (SDS and Triton X-100) are toxic for cells used for quantifying infectious Poliovirus 1 at up to a 10-fold dilution (a 100-fold dilution has no toxic effect).
RESULTS
Global behavior of viral particles.
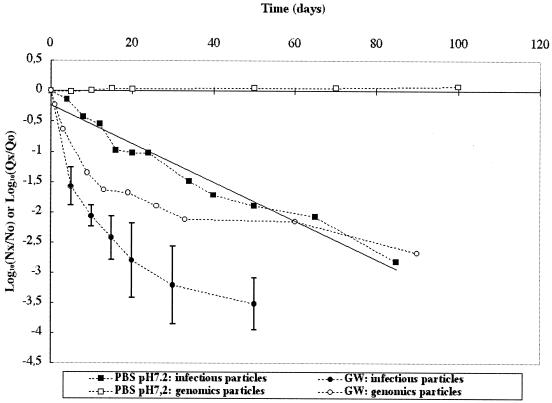
Preliminary results show that Poliovirus 1 behavior data were similar for 0.22-μm-pore-size-filtered GW and unfiltered GW, suggesting that autochthonous floras had no significant effect under our experimental conditions (data not shown). Therefore, only unfiltered GW is considered in the following results. Behavior of Poliovirus 1 was studied in an aqueous phase composed of GW or PBS at pH 7.2 and 20°C. In these two media, concentrations of infectious particles (cell culture) and viral RNA (real time RT-PCR) as a function of time were established (Fig. 1). Significant differences were observed between the results obtained with the two media. In PBS, infectious virus displayed a regular decline over an 85-day period, describing a classical linear regression equation (r2 = 0.96) with a T90 of 31.3 days. In GW, in contrast, virus loss monitored for 50 days in 4 replicates was not linear. At 20 days, the loss of infectious virus was approximately 0.9 log10 in PBS and 2.8 log10 in GW. In PBS, viral genome remained stable over the analyzed period (100 days), whereas in GW a nonlinear decrease was observed over a 90-day period. After 20 days, there was no loss of viral genome in PBS whereas a 1.7 log10 loss in GW was observed.
FIG. 1.
Persistence of infectious Poliovirus 1 and its genome at 20°C in GW and PBS at pH 7.2 (PBS). Nx/N0 is the ratio of the quantity of infectious viral particles at time x over the quantity of infectious viral particles at time 0. Qx/Q0 is the ratio of the quantity of viral genomes at time x over the quantity of viral genomes at time 0.
Significant differences were also observed between the behaviors of infectious particles and viral genomes in a same medium, with greater loss of infectious virus than of viral genome. Loss of infectious virus in aqueous phase as measured by cell culture can result from inactivation, adhesion to the container, and aggregation, whereas for viral genomes, loss measured by RT-PCR can only result from adhesion to the container or degradation because the viral capsid (and therefore viral aggregates) is completely degraded by RNA extraction performed before the detection step.
Occurrence of adhesion-aggregation processes.
Two strategies were developed to determine whether adhesion-aggregation processes were responsible for a significant part of the viral particle loss in GW.
The first strategy consisted of an attempt to reproduce in PBS the kinetic pattern observed in GW. This was done by adjusting parameters (pH, divalent ions) recognized as affecting adhesion-aggregation processes. In addition, the effect of STE was studied.
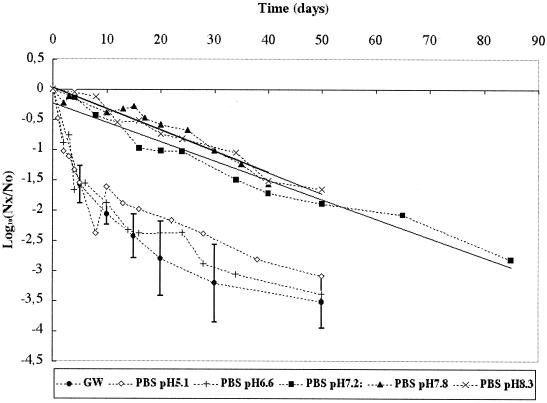
Loss of infectious Poliovirus 1 was monitored in PBS at pH 5.1, 6.6 (the isoelectric point of Poliovirus 1), 7.2, 7.8 (pH of GW), and 8.3. The results presented in Fig. 2 show that the patterns of viral loss were not significantly different (P > 0.05) when pH was held at 7.2, 7.8, and 8.3 (r2 values were 0.96, 0.93, and 0.97 and T90 values were 31.3, 28.6, and 28.5 days, respectively). The patterns observed at pH 6.6 and 5.1 were significantly different from the patterns observed at the other pH values and similar to the pattern observed in GW. Therefore, holding the pH at the isoelectric point of the virus, a factor known to favor adhesion-aggregation processes, or at a level below the isoelectric point induced viral particle behavior in PBS similar to that observed in GW. Nevertheless, the pattern of viral loss observed in GW cannot be explained by pH because the pH of GW is 7.8.
FIG. 2.
Persistence of infectious Poliovirus 1 at 20°C in GW and PBS at different pHs (5.1, 6.6, 7.2, 7.8, and 8.3). Nx/No is the ratio of the quantity of infectious viral particles at time x over the quantity of infectious viral particles at time 0.
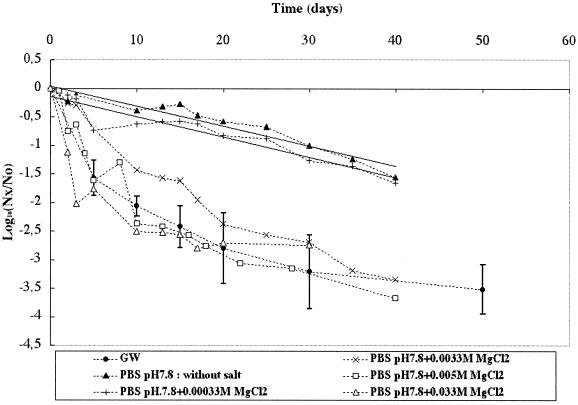
Loss of infectious Poliovirus 1 was then monitored in PBS at pH 7.8 (the pH of the GW) in the presence of a divalent salt (MgCl2) at increasing concentrations of 0.00033, 0.0033 (which corresponds to the sum of all divalent cations [Ca2+ plus Mg2+ plus Fe2+ plus Mn2+] present in GW), 0.005, and 0.033 M and without MgCl2. As illustrated in Fig. 3, the loss of infectious particles was significantly affected by the concentration of MgCl2. The linear loss observed at the lower concentration or without divalent salt (r2 values were 0.90 and 0.93 and T90 values were 28 and 28.6 days, respectively) became nonlinear for the higher MgCl2 concentrations. The pattern was similar to that observed in GW for an MgCl2 concentration near the concentration (0.0033 M) of divalent cations found in GW.
FIG. 3.
Persistence of infectious Poliovirus 1 at 20°C in GW and in PBS at pH 7.8 in the presence of increasing concentrations of MgCl2 (0.00033 to 0.033 M). Nx/No is the ratio of the quantity of infectious viral particles at time x over the quantity of infectious viral particles at time 0.
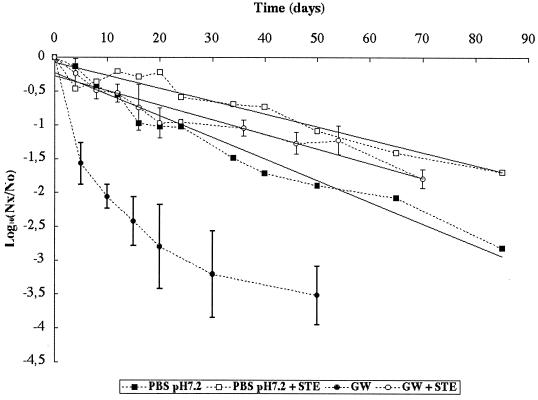
Loss of infectious Poliovirus 1 was also monitored in PBS at pH 7.2 and in GW in the presence or absence of the STE. The results are given in Fig. 4. The presence of STE significantly modified the behavior of infectious particles both in PBS and in GW. In PBS at 85 days, loss of infectious virus with and without STE (r2 = 0.92 and 0.96, respectively) fit a linear-regression model, but the T90 was significantly (P < 0.05) higher with (52.6 days) than without (31.3 days) STE. In GW, viral loss exhibited a nonlinear pattern without STE and a linear pattern at 70 days with STE (r2 = 0.91) for the two replicates. With STE, the T90 (46.1 days) with GW was not significantly (P > 0.05) different from that observed with PBS (52.6 days).
FIG. 4.
Persistence of infectious Poliovirus 1 at 20°C in GW and in PBS in the presence or absence of STE. Nx/No is the ratio of the quantity of infectious viral particles at time x over the quantity of infectious viral particles at time 0.
The second strategy consisted of an attempt to reverse the adhesion-aggregation processes. After 30 days of incubation in GW, during which time the concentration of infectious Poliovirus 1 fell off approximately 3 log10 (Fig. 1), samples were drawn and submitted to different treatments before viral concentration by cell culture was measured. The results are summarized in Table 2. Reducing the pH to 4 or 7.2 (initial pH in GW = 7.8) or adding urea had no significant effect on infectious viral concentrations. All other treatments had a significant effect. Adding SDS, Triton X-100, or Tween 80 increased the viral concentration 1 log10 unit or more; adding fetal calf serum increased the concentration about 0.8 log10 units; finally, raising the pH to 9 also increased viral concentration about 0.3 log10 units. These results demonstrate the partial reversibility of infectious viral loss in GW stored in hydrophobic containers.
TABLE 2.
Viral concentration after 30 days of incubation in GW at 20°C with different treatments before analysis by cell culturea
| No treatment |
Poliovirus 1 concentration (MPNCU/ml) after treatment with:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Tween 80 | Triton X-100 | SDS | Fetal calf serum | Urea | pH
|
|||
| 4 | 7.2 | 9 | ||||||
| 41 (± 9) | 434 (± 295) | 647 (± 161) | 1,220 (± 269) | 247 (± 37) | 65 (± 14) | 46 (± 24.3) | 40 (± 6) | 86 (± 26) |
Experiments are realized in triplicate (n = 3).
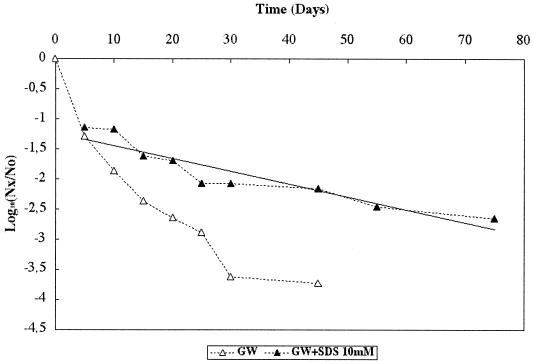
SDS at a final concentration of 10 mM provided the best reversal of the viral loss phenomena. Thus, when the samples (drawn between day 5 and day 75) used to study viral behavior in GW (Fig. 5) were treated with SDS before viral concentrations were determined by cell culture, the observed viral concentration increased for all the samples. The linear regression equation fitting this decrease curve gave r2 = 0.87 and T90 = 46.7 days, which was not significantly (P > 0.05) different from the T90 values observed for GW and PBS in the presence of STE (46.1 and 52.6 days, respectively). Loss of infectious virus was not, however, totally reversed with SDS, since loss at day 5 was 1.1 log10, which could be attributed either to viral inactivation or to nonreversible adhesion-aggregation.
FIG. 5.
Decrease of infectious Poliovirus 1 levels at 20°C in GW with (GW+SDS) or without an SDS treatment before analysis by cell culture. Nx/No is the ratio of the quantity of infectious viral particles at time x over the quantity of infectious viral particles at time 0.
Kinetics of viral adhesion to the polypropylene container.
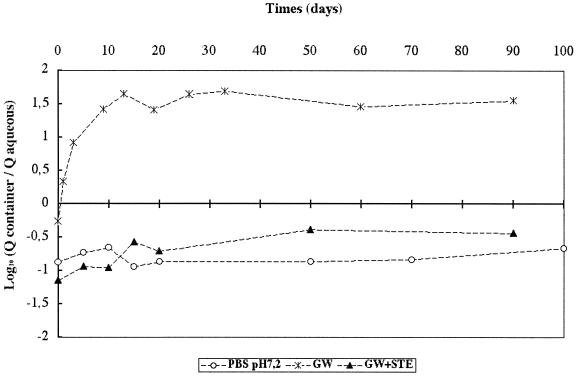
To investigate viral adhesion to the polypropylene container, the time course of genome present in the entire aqueous phase (1 ml) was compared with the total quantity of viral genome present on the walls of the polypropylene tube. The relative proportions of genome present in the aqueous phase and adhering to the polypropylene container were monitored in GW and in PBS at pH 7.2 and also in GW with STE. The results are presented in Fig. 6. In GW, the genome quantity initially distributed in water was two times greater than that distributed onto the container. Then, between day 0 and 20, the majority of the genome moved from the aqueous phase onto the walls of the container, with more than 30 times more genome adhering to the container at day 20 than was found in the aqueous phase. At this time point, the quantity of genome in the aqueous phase was negligible compared with the quantity adhering to the container. There was an equilibrium phase from day 20 to day 90, during which levels of genome loss were equivalent in the aqueous phase and on the container. The two other media (PBS at pH 7.2 and GW with STE) exhibited similar adhesion patterns which were very different from that observed in GW alone. For PBS and GW with STE, 8 to 13 times more genome was found in the aqueous phase than on the container walls at day 0, a distribution pattern that remained relatively stable over the 100 days of the test. This analysis demonstrated that the majority of the viral particles in a Poliovirus 1-GW-polypropylene container system adhere to the walls of the container (1.5 log10) and that in the same system but with PBS at pH 7.2 or GW with STE, very few viral particles adhere on the container.
FIG. 6.
Relative proportions of viral genome in the aqueous phase (Q-aqueous) and adhering to the container (Q-container) at 20°C in GW, PBS at pH 7.2, and GW with STE (GW+STE).
DISCUSSION
Our study of Poliovirus 1 behavior in GW held in a polypropylene container demonstrated that both adhesion-aggregation and inactivation phenomena have a significant effect on loss of infectious Poliovirus 1. Quantifying one process without taking into account the effect of the other process necessarily produces a measurement bias.
Adhesion of viral particles to the hydrophobic walls of the container was clearly the major phenomenon operating in the system studied. Quantitatively significant (1.5 log10) viral transfer from the aqueous phase onto the hydrophobic walls occurred during the first 20 days of incubation. The process was undoubtedly favored by the presence of divalent ions in our GW, but in other GW such a process can be favored by a pH below or equivalent to the viral isoelectric point. Major adhesion of viruses to the walls of different container materials has been already reported (7, 29, 40, 42, 45). Taylor et al. (40) observed in a synthetic water-Poliovirus 2-polypropylene support system that when the pH fell below 7, 90% of the viral particles adhered to the supporting walls. Ward and Winston (45) reported the same phenomenon with Poliovirus 1 and GW in a polypropylene container. In contrast, Biziagos et al. (5), who also worked with Poliovirus 1 and GW, were unable to demonstrate adherence. The explanation might lie in the difference in divalent cation concentrations (16.4 mg/liter versus 119 mg/liter in the GW used for the present study). The type of salts involved and their concentration in the medium are known to affect adsorption (3, 17, 26, 36).
Earlier work on the behavior of different viruses (Poliovirus 1, Norovirus, and Feline Calicivirus f9) has been conducted with the same GW used for the present study (16). These authors, who monitored viral concentration in water in the presence of diluted STE, did not examine adhesion-aggregation processes, interpreting their survival curves solely as the result of the effects of viral inactivation or genome degradation. In the light of our present findings, this was justified because our results show that the presence of STE (which was exactly the same as used by Gassilloud et al.) (16) inhibits adhesion to the container. Inhibition of viral adhesion to the hydrophobic container could have two explanations. Organic matter present in the STE might saturate sites of adsorption on the polypropylene walls, thus preventing viral adhesion. The saturation would result from a competition between the organic matter and the virus for adhesion sites (7, 17, 24, 33). This mechanism was used to elute viruses adsorbed on membranes (27) or in protocols for concentrating viruses from water, as a lot of elution buffer contains beef extract (6, 34). Another explanation could be that suspended matter in the STE could also be preferential adsorption sites for the virus (4, 7, 38). Here, the viruses remain in the aqueous phase but are adsorbed on suspended matter contained in the STE. In fact, under some conditions, such as those we used, it is clear that adding the STE reduced viral loss as a function of time, not because of a protective effect but just because it inhibits adhesion processes.
In our experimental conditions, viral inactivation also played a significant role in viral loss. There are two ways to estimate its effect.
The first is to consider the PBS medium at pH 7.2 as a reference system in which adhesion-aggregation phenomena are negligible. This is present practice when the viral survival curve is log linear. In our case, the coefficient was 0.96 and the T90 was 31.3 days. Thus, if adhesion-aggregation were considered negligible, the observed improvement in viral persistence after adding STE (T90 = 52.6 days) would correspond to another protective effect. In this case, adhesion-aggregation processes also provide a protective effect, as the T90 obtained after SDS treatment (46.7 days) is higher than that obtained with PBS (31.3 days). A protective effect due to viral adsorption or aggregation (2, 10, 17, 21, 37) or to the presence of organic matter (4, 9, 22) has been previously suggested. But this protective effect can be discussed, since other researchers have been unable to demonstrate a significant improvement in the survival of adsorbed viruses exposed to inactivating agents such as heat (15, 35). With PBS used as a reference for inactivation at 20°C, it can be considered that if the inactivation process was similar in GW it was responsible for 0.64 log10 viral loss (calculated using T90 in PBS) during the first 20 days.
Our study also led to a second hypothesis, which considers that adhesion-aggregation phenomena are less significant in PBS than in GW but are still active and sufficient to lower the T90. This would explain why adhesion-aggregation processes in GW inhibited by STE (T90 = 46.1 days) or reversed by SDS (T90 = 46.7 days) yielded a T90 equivalent to that of PBS+STE (T90 = 52.6 days; nonsignificant adhesion-aggregation) but not to that of PBS alone (T90 = 31.3 days; significant adhesion-aggregation). Consequently, the systems allowing evaluation of the proportional effect of viral inactivation were the systems containing STE or treated by SDS (T90 = 46.1 or 52.6 days). The inhibition of adhesion-aggregation processes could explain the contradictory results with respect to the possible protective effect of organic matter or suspended solids.
Whatever the hypothesis under consideration, it can be assumed that viral inactivation after 20 days represents a decrease between 0.38 log10 (medium containing STE as reference) and 0.64 log10 (PBS pH 7.2 as reference).
The greater part of the 2.8 log10 loss in infectious Poliovirus 1 observed at 20 days in GW held at 20°C can be considered to result from adhesion phenomena (1.5 log10). Since viral inactivation accounted for 0.38 to 0.64 log10 loss, aggregation can be considered to be responsible for 0.66 or 0.92 log10 loss.
In conclusion, this study demonstrates clearly that estimating viral survival in a given system must always take into consideration adhesion-aggregation phenomena. Future experiments examining viral inactivation should be designed to minimize or correct for adsorptive and aggregative losses. These processes may be the major mechanism of viral loss in the aqueous phase, and since they are reversible, may be a source of underestimation of viral survival in certain studies.
From a practical point of view, the reality of massive adhesion of viruses to the hydrophobic container in a GW implies taking into account adsorbed viruses when virological analysis of GW bottled in a hydrophobic material (terephthalate polyethylene) is conducted. Protocols should be defined for practical applications.
Acknowledgments
This work was funded by la chambre syndicale des eaux minerales.
REFERENCES
- 1.Alvarez, M. E., M. Aguilar, A. Fountain, N. Gonzalez, O. Rascon, and D. Saenz. 2000. Inactivation of MS-2 and poliovirus in groundwater. Can. J. Microbiol. 46:159-165. [DOI] [PubMed] [Google Scholar]
- 2.Babich, H. B., and G. Stotzky. 1980. Reduction in inactivation rates of bacteriophages by clay minerals in lake water. Water Res. 14:185-187. [Google Scholar]
- 3.Bales, R. C., S. R. Hinkle, T. W. Kroeger, K. Stocking, and C. P. Gerba. 1991. Bacteriophage adsorption during transport through porous media: chemical perturbations and reversibility. Environ. Sci. Technol. 25:2088-2095. [Google Scholar]
- 4.Bixby, R. L., and D. J. O'Brien. 1979. Influence of fulvic acid on bacteriophage adsorption and complexation in soil. Appl. Environ. Microbiol. 38:840-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biziagos, E., J. Passagot, J.-M. Crance, and R. Deloince. 1988. Long-term survival of hepatitis A virus and poliovirus type 1 in mineral water. Appl. Environ. Microbiol. 54:2705-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrego, J. J., R. Cornax, D. R. Preston, S. R. Farrah, B. McElhaney, and G. Bitton. 1991. Development and application of new positively charged filters for recovery of bacteriophages from water. Appl. Environ. Microbiol. 57:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay, D., S. Chattopadhyay, W. G. Lyon, and J. T. Wilson. 2002. Effect of surfactants on the survival and sorption of viruses. Environ. Sci. Technol. 36:4017-4024. [DOI] [PubMed] [Google Scholar]
- 8.Dowd, S. E., S. D. Pillai, S. Wang, and M. Y. Corapcioglu. 1998. Delineating the specific influence of virus isoelectric point and size on virus adsorption and transport through sandy soils. Appl. Environ. Microbiol. 64:405-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enriquez, C. E., C. J. Hurst, and C. P. Gerba. 1995. Survival of the enteric adenoviruses 40 and 41 in tap, sea, and waste water. Water Res. 29:2548-2553. [Google Scholar]
- 10.Floyd, R., and D. G. Sharp. 1977. Aggregation of poliovirus and reovirus by dilution in water. Appl. Environ. Microbiol. 33:159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Floyd, R., and D. G. Sharp. 1978. Viral aggregation: effects of salts on the aggregation of poliovirus and reovirus at low pH. Appl. Environ. Microbiol. 35:1084-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Floyd, R., and D. G. Sharp. 1979. Viral aggregation: buffer effects in the aggregation of poliovirus and reovirus at low and high pH. Appl. Environ. Microbiol. 38:395-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujioka, R. S., P. C. Loh, and L. S. Lau. 1980. Survival of human enteroviruses in the Hawaiian ocean environment: evidence for virus-inactivating microorganisms. Appl. Environ. Microbiol. 39:1105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujioka, R. S., and B. S. Yoneyama. 2002. Sunlight inactivation of human enteric viruses and fecal bacteria. Water Sci. Technol. 46:291-295. [PubMed] [Google Scholar]
- 15.Gantzer, C., A. Maul, Y. Levi, and L. Schwartzbrod. 1998. Fate of the genome and infectious units of coxsackie B3 virus in phosphate buffered saline. Water Res. 32:1329-1333. [Google Scholar]
- 16.Gassilloud, B., L. Schwartzbrod, and C. Gantzer. 2003. Presence of viral genomes in mineral water: a sufficient condition to assume infectious risk? Appl. Environ. Microbiol. 69:3965-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerba, C. P. 1984. Applied and theoretical aspects of virus adsorption to surfaces. Adv. Appl. Microbiol. 30:133-168. [DOI] [PubMed] [Google Scholar]
- 18.Hurst, C. J., W. H. Benton, and K. A. McClellan. 1989. Thermal and water source effects upon the stability of enteroviruses in surface freshwater. Can. J. Microbiol. 35:474-480. [DOI] [PubMed] [Google Scholar]
- 19.Jones, C. R., M. R. Adams, P. A. Zhdan, and A. H. L. Chamberlain. 1999. The role of surface physicochemical properties in determining the distribution of the autochthonous microflora in mineral water bottles. J. Appl. Microbiol. 86:917-927. [DOI] [PubMed] [Google Scholar]
- 20.Kutz, S. M., and C. P. Gerba. 1988. Comparison of virus survival in freshwater sources. Water Sci. Technol. 20:467-471. [Google Scholar]
- 21.LaBelle, R. L., and C. P. Gerba. 1979. Influence of pH, salinity, and organic matter on the adsorption of enteric viruses to estuarine sediments. Appl. Environ. Microbiol. 38:93-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaBelle, R. L., and C. P. Gerba. 1982. Investigations into the protective effect of estuarine sediment on virus survival. Water Res. 16:469-478. [Google Scholar]
- 23.Lance, J. C., and C. P. Gerba. 1984. Effect of ionic composition of suspending solution on virus adsorption by a soil column. Appl. Environ. Microbiol. 47:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipson, S. M., and G. Stotzky. 1984. Effect of proteins on reovirus adsorption to clay minerals. Appl. Environ. Microbiol. 48:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loveland, J. P., J. N. Ryan, G. L. Amy, and R. W. Harvey. 1996. The reversibility of virus attachment to mineral surfaces. Colloids Surf. 107:205-221. [Google Scholar]
- 26.Lukasik, J., T. M. Scott, D. Andryshak, and S. R. Farrah. 2000. Influence of salts on virus adsorption to microporous filters. Appl. Environ. Microbiol. 66:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lytle, D. C., and L. B. Routson. 1995. Minimized virus binding for tests of barrier materials. Appl. Environ. Microbiol. 61:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maul, A. 1991. Aspects statistiques des methodes de quantification en virologie, p. 143-171. In L. Schwartzbrod (ed.), Virologie des milieux hydriques. Lavoisier, Paris, France.
- 29.Moore, R. S., D. H. Taylor, L. S. Sturman, M. M. Reddy, and G. W. Fuhs. 1981. Poliovirus adsorption by 34 minerals and soils. Appl. Environ. Microbiol. 42:963-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuanualsuwan, S., and D. O. Cliver. 2002. Pretreatment to avoid positive RT-PCR results with inactivated viruses. J. Virol. Methods 104:217-225. [DOI] [PubMed] [Google Scholar]
- 31.Official Journal of the European Communities. 1980. Council directive 80/777/EEC of 15 July 1980 on the approximation of the laws of the member states relating to the exploitation and marketing of natural mineral waters. Annex III. Indications and criteria laid down in article 9. Off. J. Eur. Commun. 229:1-10.
- 32.Pancorbo, O. C., B. G. Evanshen, W. F. Campbell, S. Lambert, S. K. Curtis, and T. W. Woolley. 1987. Infectivity and antigenicity reduction rates of human rotavirus strain Wa in fresh waters. Appl. Environ. Microbiol. 53:1803-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powelson, D. K., J. R. Simpson, and C. P. Gerba. 1991. Effects of organic matter on virus transport in unsaturated flow. Appl. Environ. Microbiol. 57:2192-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preston, D. R., T. V. Vasudevan, G. Bitton, S. R. Farrah, and J.-L. Morel. 1988. Novel approach for modifying micoporous filters for virus concentration from water. Appl. Environ. Microbiol. 54:1325-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quignon, F., and L. Schwartzbrod. 1995. Influence of salts and montmorillonite upon heat inactivation of poliovirus in sterile water. Water Sci. Technol. 31:177-180. [Google Scholar]
- 36.Redman, J. A., S. B. Grant, T. M. Olson, J. M. Adkins, J. L. Jackson, M. S. Castillo, and W. A. Yanko. 1999. Physicochemical mechanisms responsible for the filtration and mobilization of a filamentous bacteriophage in quartz sand. Water Res. 33:43-53. [Google Scholar]
- 37.Sharp, D. G., R. Floyd, and J. D. Johnson. 1975. Nature of the surviving plaque-forming unit of reovirus in water containing bromine. Appl. Microbiol. 29:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobsey, M. D., and A. R. Hickey. 1985. Effects of humic and fulvic acids on poliovirus concentration from water by microporous filtration. Appl. Environ. Microbiol. 49:259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobsey, M. D., D. A. Battigelli, G. A. Shin, and S. Newland. 1998. RT-PCR amplification detects inactivated viruses in water and wastewater. Water Sci. Technol. 38:91-94. [Google Scholar]
- 40.Taylor, D. H., R. S. Moore, and L. S. Sturman. 1981. Influence of pH and electrolyte composition on adsorption of poliovirus by soils and minerals. Appl. Environ. Microbiol. 42:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, S. S., M. Flury, M. V. Yates, and W. A. Jury. 1998. Role of the air-water-solid interface in bacteriophage sorption experiments. Appl. Environ. Microbiol. 64:304-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, S. S., and M. V. Yates. 1999. Bacteriophage inactivation at the air-water-solid interface in dynamic batch systems. Appl. Environ. Microbiol. 65:1186-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Totsuka, A., K. Ohtaki, and I. Tagaya. 1978. Aggregation of enterovirus small plaque variants and polioviruses under low ionic strength conditions. J. Gen. Virol. 38:519-533. [DOI] [PubMed] [Google Scholar]
- 44.Voorthuizen, E. M., N. J. Ashbolt, and A. I. Schäfer. 2001. Role of hydrophobic and electrostatic interactions for initial enteric virus retention by MF membranes. J. Membr. Sci. 194:69-79. [Google Scholar]
- 45.Ward, R. L., and P. E. Winston. 1985. Development of methods to measure virus inactivation in fresh waters. Appl. Environ. Microbiol. 50:1144-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward, R. L., D. R. Knowlton, and P. E. Winston. 1986. Mechanism of inactivation of enteric viruses in fresh water. Appl. Environ. Microbiol. 52:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yates, M. V., C. P. Gerba, and L. M. Kelley. 1985. Virus persistence in groundwater. Appl. Environ. Microbiol. 49:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhuang, J., and Y. Jin. 2003. Virus retention and transport through Al-oxide coated sand columns: effects of ionic strength and composition. J. Contam. Hydrol. 60:193-209. [DOI] [PubMed] [Google Scholar]