Abstract
We have combined the culture-independent methods of high-throughput sequencing of chaperonin-60 PCR product libraries and quantitative PCR to profile and quantify the small-intestinal microflora of pigs fed diets based on corn, wheat, or barley. A total of 2,751 chaperonin-60 PCR product clones produced from samples of ileum digesta were examined. The majority (81%) of these clones contained sequences independently recovered from all three libraries; 372 different nucleotide sequences were identified, but only 14% of the 372 different sequences were recovered from all three libraries. Taxonomic assignments of the library sequences were made by comparison to a reference database of chaperonin-60 sequences combined with phylogenetic analysis. The taxa identified are consistent with previous reports of pig ileum microflora. Frequencies of each sequence in each library were calculated to identify taxa that varied in frequency between the corn, barley, and wheat libraries. The chaperonin-60 sequence inventory was used as a basis for designing PCR primer sets for taxon-specific quantitative PCR. Results of quantitative PCR analysis of ileum digesta confirmed the relative abundances of targeted taxa identified with the library sequencing approach. The results of this study indicate that chaperonin-60 clone libraries can be valid profiles of complex microbial communities and can be used as the basis for producing quantitative PCR assays to measure the abundance of taxa of interest during experimentally induced or natural changes in a community.
The development of culture-independent, molecular methods for microbial population profiling has led to a proliferation of studies of diverse microbial ecosystems, and the sizes of these studies, particularly sequence-based studies, have been gradually increasing in scale. Sequence data generated from complex microbial communities are almost exclusively derived from the 16S rRNA gene, due in part to the wealth of reference data available for these genes (24). Despite limitations inherent in the method (32, 32, 42), amplification, cloning, and sequencing of 16S rRNA sequences from microbial consortia has been applied successfully to the study of a wide range of populations, including the gastrointestinal tracts of humans (36) and other animals (16, 22, 35).
We have previously demonstrated that the chaperonin-60 gene is a useful alternative to the 16S rRNA gene as a target for microbial population studies (19). The chaperonin-60 gene (also known as groEL or hsp60) encodes the 60-kDa type I chaperonin found in all eubacteria and eukaryotes. Degenerate PCR primers have been designed that amplify an approximately 550-bp region of the gene corresponding to nucleotides 274 to 828 of the Escherichia coli chaperonin-60 sequence. This primer pair amplifies the chaperonin-60 sequence from eubacteria and eukaryotes and has been successfully applied to the problem of microbial species identification (13-15, 20) and in the creation of PCR product libraries for the characterization of complex microbial communities (19). One advantage of chaperonin-60 as a target is that between closely related taxa, the chaperonin-60 gene contains more phylogenetically informative sequence information than 16S rRNA genes of the same taxa, permitting discrimination of species (14, 21, 25) or subspecies and serotypes (4). A database of chaperonin-60 sequences (cpnDB) is available at http://cpndb.cbr.nrc.ca (18).
The need to identify alternatives to the antimicrobials currently used for growth promotion and prophylactic control of intestinal pathogens in pigs (12, 26) has led to increased interest in understanding the role of the normal intestinal flora in exclusion of epizootic pathogens and promotion of animal health. To this end, the pig gastrointestinal tract microbiota has been the focus of previous 16S rRNA sequence-based studies (22, 31) undertaken with the restricted objective of cataloguing member species of the intestinal tract. The next step in molecular profiling of complex microbial communities is to work toward characterization of population dynamics or community responses to perturbations, which was previously impossible with labor-intensive culture-based methods.
In the current study, we constructed and sequenced chaperonin-60-based PCR product libraries and used phylogenetic analysis to profile the microbial communities of pigs fed diets in which corn, wheat, or barley was the major cereal ingredient. These cereal grains vary markedly in chemical composition (8), significantly modify intestinal microbial colonization (28, 7), impact susceptibility to enteric infection (30), and may affect the efficacy of nonantibiotic feed additives such as direct-fed microbials (2), oligosaccharide prebiotics (9), and organic acids (6). Although the composition of microbial communities along the length of the intestine was of interest, we elected to sequence a large number of clones from a single location in order to maximize the representation of bacteria present in the community and to enable legitimate comparison of library composition among diets. The ileum was targeted as the major site of nutrient absorption in the pig, where microbial communities are less complex than in more distal regions and biologically relevant effects of diet might be more easily identified. For this comparative study, we also wanted to validate the extent to which microbial profiles derived from the library studies, particularly the quantitative value of the frequency of recovery of sequences within a taxon, are true representations of population. We used a quantitative PCR approach to develop molecular tools for the monitoring of taxonomic groups of interest within the pig ileum populations to validate representation of the targeted sequences in the cloned libraries.
MATERIALS AND METHODS
Pigs and diet protocols.
Forty-five weanling pigs (Pig Improvement Canada, Acme AB; Prairie Swine Centre Inc., Saskatoon, Canada) of mixed gender were assigned to one of three treatment groups, which were fed diets containing either yellow dent corn, Laura wheat, or Brier barley as the primary source of energy. Experimental diets were formulated as reported previously (7) to contain similar amounts (3.20 to 3.28 Mcal kg−1) of digestible energy, 3.15 g of digestible lysine per Mcal of digestible energy, and no antibiotics. Diets were fed starting at 35 days of age (2 weeks postweaning) for a period of 3 weeks.
Collection of ileal digesta.
Pigs were euthanized by CO2 asphyxiation and exsanguination, and their intestinal tracts were removed. The small intestine was dissected from the mesentery, and the mid-ileum was identified as 75% of the distance between the duodenum and the ileo-cecal junction. Digesta (approximately 2 to 3 g) from this location were collected and stored on ice until frozen at −80°C prior to analysis.
Genomic DNA isolation.
For each sample, 200 mg of digesta was placed in a bead-beating tube (FastDNA; Bio 101, Carlsbad, Calif.) with Lysis Matrix A (Bio 101) along with 30 μl of 10% (wt/vol) sodium dodecyl sulfate, 3 μl of proteinase K (20 mg/ml), and Tris-EDTA buffer (10 mM Tris-Cl, pH 8.0, 1 mM EDTA) to a total volume of 600 μl. Sample tubes were mixed by vortexing and incubated at 37°C for 2 h; 600 μl of phenol-chloroform-isoamyl alcohol (25:24:1, vol:vol:vol) was added to each tube, followed by processing in a FastPrep Instrument 120 (Bio 101) bead-beating apparatus 10 times for 20-s intervals with periodic cooling of sample tubes on ice. After bead beating, 0.1 volume of 3 M sodium acetate, pH 5.0, was added to each tube, lysates were mixed by inversion and cooled on ice for 10 min. Lysates were clarified by centrifugation at maximum speed in a microcentrifuge for 10 min. Clarified lysates were transferred to fresh tubes, and nucleic acids were precipitated by the addition of 1 volume of cold isopropanol. Nucleic acids were pelleted by centrifugation at maximum speed in a microcentrifuge for 10 min, after which the supernatant was removed and pellets were washed once with 70% (vol:vol) ethanol. Washed pellets were air dried for approximately 20 min and resuspended in 200 μl of Tris-EDTA buffer.
PCR and creation of PCR product libraries.
PCRs were performed for each of the three genomic DNA templates as described previously (19) with the following exceptions: four reactions were performed with each template at annealing temperatures of 42, 46.5, 50.4, or 56°C. PCRs at the four different annealing temperatures were done in parallel on a Stratagene Gradient-40 Robocycler. PCR products produced at each of the four annealing temperatures were pooled by volume, agarose gel purified, and ligated into T-A cloning vector pCR2.1-TOPO (Invitrogen). Ligation mixtures were used to transform E. coli strain JM109. The three resulting libraries, corn, barley, and wheat, were plated on Luria-Bertani (LB) with ampicillin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal); 1,248 white colonies were picked randomly for each library. Colonies were picked into 96-well plates containing 100 μl of LB with 100 μg of ampicillin per ml and incubated overnight at 37°C. After overnight incubation, 100 μl of 30% (vol:vol) glycerol were added to each well and the cultures were stored at −80°C.
Plasmid DNA isolation, sequencing, and analysis.
Template preparation and sequencing reactions were performed robotically as described previously (19). Each template was sequenced with T7 and M13-RP primers. Sequencing reactions were resolved on an ABI 3700 capillary sequencing device. All sequence data assembly, analysis, and storage were done with software available through the Canadian Bioinformatics Resource (http://www.cbr.nrc.ca). Raw sequence data were assembled into contigs with PreGap4 (version 1.1) and Gap4 (version 4.6) in the Staden software package (release 2000.0: J. Bonfield, K. Beal, M. Betts, M. Jordan, and R. Staden, 2000). Contig sequences were compared to cpnDB (18), a database of approximately 3,000 chaperonin-60 sequences, with FASTA and BLASTp. Sequence data, template information, and similarity results were placed in an mySQL database for storage and further analysis. Sequence manipulations such as format changes and amino acid translations were done with the EMBOSS software suite (25). Sequence alignments were done with Clustalw (39) and viewed with GeneDoc (27).
Phylogenetic analysis was done with programs in the PHYLIP software package (J. Felsenstein, 1993, PHYLIP [Phylogeny Inference Package] version 3.5c, distributed by the author, Department of Genetics, University of Washington, Seattle). Specifically, alignments were sampled for bootstrap analysis with Seqboot, distances were calculated with the PAM option of Protdist (for peptide sequences) or the maximum-likelihood option of Dnadist. Dendrograms were constructed from distance data by neighbor joining with neighbor. Consensus trees were calculated with Consense and branch lengths were superimposed on consensus trees with Fitch. Completed trees were viewed with TreeView (28).
Quantitative PCR assays.
Quantitative PCR assays were developed to quantify clusters of bacteria possessing closely related (≥95% identity among members) chaperonin-60 sequences. These were identified by phylogenetic analysis and designated B1 (70% sequence identity with Bacillales family), C1 (80% sequence identity with Clostridium spp.), S1 (95 to 100% identity with Streptococcus alactolyticus ATCC 43077), and L10 (95 to 100% sequence identity with Lactobacillus amylovorus ATCC 33198) as further described in the Results section. Signature Oligo software (LifeIntel Inc., Port Moody, Canada) was used to identify chaperonin-60 nucleotide sequence “signatures” unique to each target cluster compared with neighboring taxa.
Compatible PCR primers were designed to anneal at the location of signature sequences with Oligo 6 primer analysis software (Molecular Biology Insights Inc., Cascade, Colo.). Primer specificity was checked by comparison with corn, barley, and wheat library sequence data and typed strain reference data with BLAST (1) configured for short, nearly exact matches. Specificity was confirmed by PCR amplification of plasmids containing target cluster chaperonin-60 sequences but not closely related sequences.
The forward (f) and reverse (r) primer sequences for each selected cluster were B1f, 5′-TGCAGGAGCAAATCCAATGAT-3′; B1r, 5′-GCATGGCTTCGGCAATTAAA-3′; C1f,,5′-GCTGTTGATGTAGCAGTTGA-3′; C1r, 5′-ATAACCCCTTCGTTTCCTAC-3′; S1f, 5′-TTGACGTGGTTGAAGG-3′; S1r, 5′-GTTTTCAAGACTTCTTCAAGCAA-3′; L10f, 5′-AAGCTGCCGTTGATGAATTAC-3′; and L10r, 5′-AGCGTCAGCGATTAAGTCACC-3′. A TaqMan fluorescent probe was designed for L10 (5′-TTTAGATGCTGATGAAACAGCAGCTACGTT-3′) that was labeled at the 5′ end with 6-carboxyfluoecein and at the 3′ end with Black Hole Quencher-1 (Integrated DNA Technologies, Inc., Coralville, Iowa).
For the C1, B1, and S1 assays, quantitative PCR was performed with the iCycler iQ real-time PCR detection system (Bio-Rad Laboratories, Hercules, Calif.) and a final reaction mix containing iQ SYBRGreen Supermix (Bio-Rad), 500 nM each primer, plasmid calibration standards or genomic DNA isolated from ileal contents, and adjusted to a final volume of 25 μl. Amplification conditions were 95°C for 2 min, followed by 40 cycles of 60 s at 95°C and 60 s at 62°C (B1), 58°C (C1), or 68°C (S1). In the case of the L10 diagnostic, the reaction contained Platinum Quantitative PCR SuperMix-UDG (Invitrogen, Carlsbad, Calif.), 500 nM each primer, 200 nM probe, and genomic calibration standard or ileal contents genomic DNA in a total volume of 25 μl. PCR conditions were 50°C for 3 min and 95°C for 2 min, followed by 40 cycles of 30 s at 95°C and 30 s at 61°C.
Calibration standards for the C1, B1, and S1 assays were developed with a 10-fold dilution series of plasmids containing chaperonin-60 sequence representatives of each target cluster. Plasmid copy number was calculated from plasmid molecular weight, and plasmid concentration was measured with Picogreen (Molecular Probes) with a Fluoroskan Ascent FL fluorometer (Thermo Labsystems). For the L10 assay, calibration standards were developed with a 10-fold dilution series of Lactobacillus amylovorus (ATCC 33198) genomic DNA; the number of target genomes was calculated with an estimated genomic weight of 1.2 × 109 g/mol (based on a genome size of 1,850 kbp) (11). The number of target bacteria genomes per gram of ileal contents was calculated by plotting threshold cycle value versus calibration standard copy number and interpolating the threshold cycle value obtained for genomic DNA isolated from ileal contents with the iCycler iQ optical system software (version 3.0a). Corrections were made for sample dilution during extraction of DNA and PCR. Calibration standards and unknown samples were assayed in triplicate.
Nucleotide sequence accession number.
The sequences of the cloned chaperonin-60 PCR products and relevant reference strain chaperonin-60 sequences have been deposited in GenBank and assigned accession numbers AY562569 to AY562944 inclusive. Sequences can also be retrieved from cpnDB (http://cpndb.cbr.nrc.ca) (18).
RESULTS
Chaperonin-60 gene sequences amplified from ileum contents.
High-quality sequence data covering the entire amplified region of the chaperonin-60 gene were obtained for 2,751 of the clones randomly picked from corn, barley, and wheat libraries prepared from pig ileum contents. Only these completely full-length sequences with no ambiguous positions were analyzed further. As summarized in Table 1, pairwise comparisons of the sequences determined for each library indicated that there were 178 different nucleotide sequences in the corn library (corresponding to 65 unique peptide sequences), 189 different nucleotide sequences in the barley library (corresponding to 58 unique peptide sequences), and 152 different sequences in the wheat library (corresponding to 54 unique peptide sequences). Pooling of data from the three libraries resulted in the identification of 372 different nucleotide sequences (101 unique peptide sequences). Each of the sequences recovered was classified according to the libraries in which it was found; 74% of the sequences were unique to one of the libraries, while only 14% were recovered independently from all three libraries. However, the 14% of sequences recovered independently from all three libraries were recovered in high frequency, accounting for 81% of the total number of clones sequenced.
TABLE 1.
Number of sequences generated for each library
| Library | No. of sequences | No. of unique nucleotide sequences | No. of unique peptide sequences |
|---|---|---|---|
| Corn diet (C) | 909 | 178 | 65 |
| Barley diet (B) | 930 | 189 | 58 |
| Wheat diet (W) | 915 | 152 | 54 |
| C + B + W | 2,751 | 372 | 101 |
Comparison of sequence profiles of corn, barley, and wheat libraries.
Following assembly and editing of the sequences, each was compared to cpnDB, a database of approximately 3,000 chaperonin-60 sequences, with FASTA (for nucleotide sequences) or BLASTp (for peptide sequences). The nearest database neighbor for each clone was recorded. A graphic summary of the contents of each library based on the genus or taxonomic group of the nearest database neighbor for each clone is shown in Fig. 1.
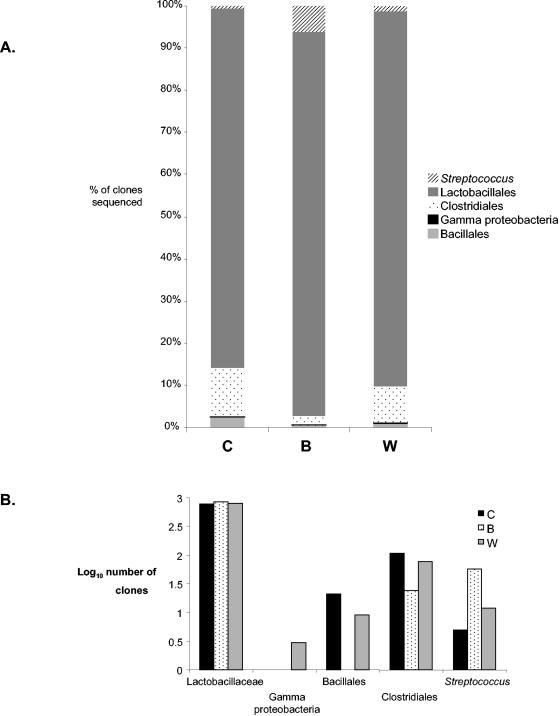
FIG. 1.
Taxonomic composition of corn (C), barley (B), and wheat (W) clone libraries. A. Percentage of clones recovered from corn, barley, and wheat libraries classified by taxonomic group, determined by comparison to the reference database of chaperonin-60 sequences. Total library sizes were 909, 930, and 915 clones for corn, barley, and wheat, respectively. B. Comparison of log10 numbers of clones in each taxonomic category recovered from the corn, barley, and wheat libraries.
In general, all three libraries were dominated by Lactobacillales-like sequences, which constituted 84% of the corn library, 92% of the barley library, and 90% of the wheat library (Fig. 1A). Most of these sequences were 95 to 100% identical to that of Lactobacillus amylovorus ATCC 33198. In fact, sequences 90 to 100% identical to that of L. amylovorous account for 1,399 of the 2,751 clones sequenced. Clostridiales-like sequences were more abundant in the corn library (12% of clones sequenced) compared to the barley library (2% of clones) and the wheat library (8%). Another noteworthy difference between the libraries was the relative prevalence of Streptococcus-like sequences in the barley library (6% of clones) compared to the corn and wheat libraries (1% of each; Fig. 1B). These Streptococcus-like clones included sequences identical to Streptococcus orisratti, S. thermophilus, and S. alactolyticus. Sequences with similarity to the Bacillales family were recovered from the corn and wheat libraries only (17 and 9 clones, respectively). These sequences had relatively weak similarity to Bacillus spp. and Geobacillus spp. (approximately 80% peptide similarity). Only four gamma-proteobacteria-like sequences were recovered from the libraries, all with 80 to 85% nucleotide identity to Actinobacillus spp.
Table 2 summarizes the taxonomic assignments of sequences recovered at least 10 times from the library sequence data set. Although the five most abundant sequences were very similar to each other with pairwise similarities of 99% (maximum of five nucleotide differences between any pair), each one was amplified independently from all three genomic DNA templates. A multiple sequence alignment of these five sequences indicates that of the six variable positions, five are synonymous substitutions, resulting in identical amino acid translations. Of the 33 sequences shown in Table 2, 30 were recovered from all three libraries, one (represented by clone b02_b02) was recovered from barley and wheat only, one (represented by c02_e05) was recovered from corn and wheat only, and one (represented by c13_h07) was recovered from corn and barley only.
TABLE 2.
Names, nearest reference sequences, and frequency of recovery for clones recovered at least 10 times
| Clone name | Nearest cpn60 neighbor | GenBank accession no. | % DNA identity | Frequency (no. of clones)
|
|||
|---|---|---|---|---|---|---|---|
| Corn | Barley | Wheat | Total | ||||
| c10_c06 | Lactobacillus amylovorus ATCC 33198 | AY562571 | 99.8 | 269 | 102 | 208 | 579 |
| c02_a03 | Lactobacillus amylovorus ATCC 33198 | 99.6 | 17 | 97 | 52 | 166 | |
| c01_h12 | Lactobacillus amylovorus ATCC 33198 | 99.3 | 15 | 92 | 37 | 144 | |
| c13_g09 | Lactobacillus amylovorus ATCC 33198 | 99.3 | 26 | 41 | 76 | 143 | |
| c13_h04 | Lactobacillus amylovorus ATCC 33198 | 99.5 | 48 | 37 | 54 | 139 | |
| c02_e07 | Lactobacillus amylovorus ATCC 33198 | 99.1 | 7 | 58 | 26 | 91 | |
| c02_b12 | Lactobacillus johnsonii NCC 533 | NC_005362 | 99.1 | 46 | 22 | 23 | 91 |
| c01_g02 | Clostridium disporicum ATCC 43838 | AY562569 | 84.5 | 43 | 10 | 33 | 86 |
| c03_c06 | Lactobacillus reuteri (Biologics) | AY562572 | 97.8 | 33 | 19 | 21 | 73 |
| c01_f10 | Lactobacillus vaginalis ATCC 49540 | AY123651 | 88.9 | 8 | 21 | 33 | 62 |
| c09_f09 | Lactobacillus amylovorus ATCC 33198 | 99.8 | 5 | 42 | 14 | 61 | |
| c01_c10 | Lactobacillus amylovorus ATCC 33198 | 99.6 | 19 | 19 | 21 | 59 | |
| c13_h06 | Lactobacillus reuteri (Biologics) | 97.6 | 29 | 11 | 19 | 59 | |
| c13_h12 | Lactobacillus amylovorus ATCC 33198 | 99.6 | 17 | 9 | 30 | 56 | |
| c05_h11 | Streptococcus alactolyticus ATCC 4307 | AY123648 | 100.0 | 1 | 25 | 4 | 30 |
| c02_c09 | Clostridium disporicum ATCC 43838 | 84.4 | 18 | 2 | 7 | 27 | |
| c02_b09 | Lactobacillus amylovorus ATCC 33198 | 99.5 | 13 | 4 | 7 | 24 | |
| c13_g07 | Lactobacillus amylovorus ATCC 33198 | 99.6 | 9 | 4 | 9 | 22 | |
| c02_d10 | Lactobacillus johnsonii NCC 533 | 98.9 | 13 | 6 | 3 | 22 | |
| c13_d02 | Lactobacillus amylovorus ATCC 33198 | 99.3 | 4 | 12 | 6 | 22 | |
| c02_d04 | Lactobacillus johnsonii NCC 533 | 99.3 | 11 | 4 | 6 | 21 | |
| c06_a02 | Lactobacillus crispatus CECT 4840T | AY562570 | 99.6 | 3 | 14 | 2 | 19 |
| c09_f02 | Lactobacillus amylovorus ATCC 33198 | 99.6 | 2 | 14 | 2 | 18 | |
| c13_h07 | Lactobacillus vaginalis ATCC 49540 | 88.7 | 6 | 12 | 0 | 18 | |
| c11_g09 | Lactobacillus amylovorus ATCC 33198 | 99.6 | 9 | 3 | 5 | 17 | |
| c02_e05 | Lactobacillus reuteri (Biologics) | 96.6 | 11 | 0 | 5 | 16 | |
| c12_h03 | Lactobacillus reuteri (Biologics) | 97.5 | 7 | 2 | 4 | 13 | |
| c03_b02 | Lactobacillus crispatus CECT 4840T | 99.6 | 1 | 9 | 3 | 13 | |
| b02_b02 | Lactobacillus amylovorus ATCC 33198 | 99.3 | 0 | 8 | 4 | 12 | |
| c12_f06 | Lactobacillus johnsonii NCC 533 | 98.9 | 2 | 5 | 5 | 12 | |
| c01_d01 | Lactobacillus amylovorus ATCC 33198 | 99.8 | 3 | 3 | 4 | 10 | |
| c02_d09 | Lactobacillus johnsonii NCC 533 | 98.7 | 4 | 2 | 4 | 10 | |
| c10_d08 | Lactobacillus amylovorus ATCC 33198 | 99.6 | 5 | 2 | 3 | 10 | |
Phylogenetic analysis of the sequences recovered from each of the libraries indicated that some of the most significant differences between the libraries were in the form of sequence diversity within the major taxonomic groups, in addition to the relative abundances of each group. This is apparent in the results of a phylogenetic analysis of the unique peptide sequence data set for each library (not shown). Approximately five times more Clostridiales-like clones were recovered from the corn library than the barley library. However, rather than simply recovering more clones containing the same sequences, we recovered three times as many different peptide sequences from the corn library compared to the barley library (15 different sequences from corn, five from barley). A similar relationship was observed between the Streptococcus-like sequences recovered from the barley library (10 sequences, 57 clones) and the wheat library (five sequences, 12 clones).
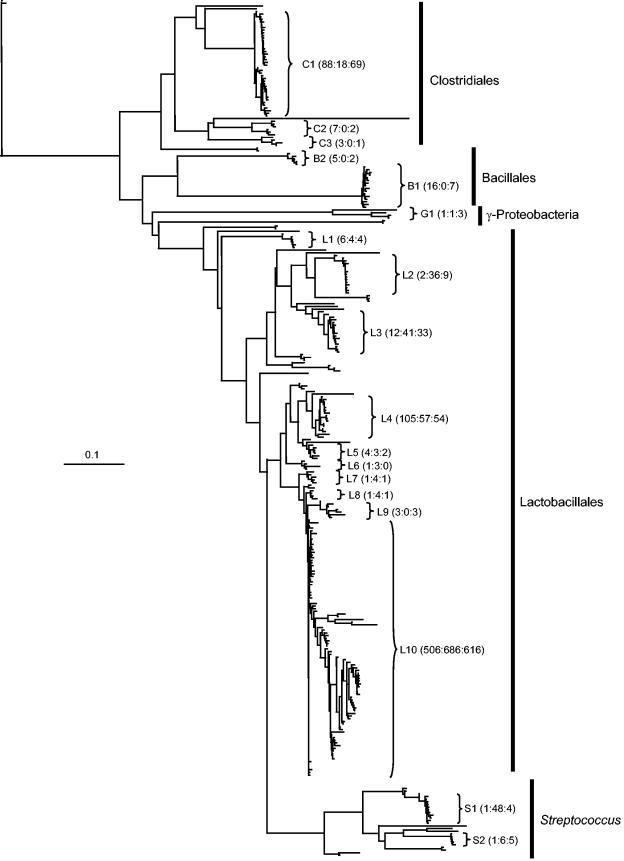
Pooling all sequence data from the three libraries resulted in the identification of 372 different nucleotide sequences. To further compare the contents of the corn, barley, and wheat libraries, a multiple sequence alignment and distance matrix were calculated for representatives of each of the 372 different nucleotide sequences. The resulting DNA distance matrix was neighbor joined to produce the tree in Fig. 2. Sequence clusters were defined as indicated in the figure, based on distance. The cluster name indicates the taxonomic assignment of the cluster: C, Clostridiales; B, Bacillales; G, gamma-proteobacteria; S, Streptococcus; and L, Lactobacillales. For each cluster, the library frequencies of each individual sequence in the cluster were combined to produce a corn:barley:wheat ratio for the cluster. For example, sequences belonging to the Lactobacillales-like cluster L2 were recovered twice from the corn library, 36 times from the barley library, and nine times from the wheat library. Clusters C1 (88:18:69), B1 (16:0:7), S1 (1:48:4), and L10 (506:686:616) were chosen as target groups for quantitative PCR with the goal of validating the clone ratios.
FIG.2.
Phylogenetic analysis of 372 unique nucleotide sequences recovered from the corn, barley, and wheat libraries. The tree was calculated with a maximum-likelihood distance formula and neighbor joining. Clusters of similar sequences within the tree are named according to the identity of the nearest database neighbors of the group (C = Clostridiales, B = Bacillales, G = gamma-proteobacteria, L = Lactobacillaceae, S = Streptococcus). The scale bar indicates 0.1 substitution per site. Numbers following the group names indicate the number of clones falling into that sequence group recovered from the corn:barley:wheat libraries. For example, C1 group sequences were recovered 88 times from the corn library, 18 times from the barley library, and 69 times from the wheat library.
Validation of clone frequency data by quantitative PCR.
The number of C1, B1, S1, and L10 bacterial genomes detected in pooled genomic DNA used to generate the corn, barley, and wheat libraries as determined by quantitative PCR is presented in Table 3. A significant (P < 0.005) positive correlation (r = 0.745) was observed between log number of genomes and the frequency of recovery of corresponding library clones. Although no clones clustering with B1 bacteria were recovered from the barley library, 2.3 × 105 B1 genomes/g of contents were detected by quantitative PCR. Similarly, recovery of fewer than five clones clustering with S1 bacteria in the corn and wheat libraries was associated with the detection of 2.1 × 105 and 7.4 × 105 S1 genomes/g of contents, respectively, by quantitative PCR. The recovery of 686 clones clustering with L10 bacteria from the barley library corresponded with the detection of 6.5 × 109 L10 genomes/g of contents with the quantitative PCR diagnostic. The number of L10 genomes/g of digesta detected in each of the three pools was very high, consistent with the frequency of clone recovery in the libraries. However, the numbers did not correspond perfectly to the clone frequency data; the corn pool, which had the fewest clones (506), contained more L10 genomes/g than did the barley pool, which had the highest number of clones (686). The wheat pool had the highest number of L10 genomes/g and an intermediate number of clones (616). Among the three pools, the numbers of L10 genomes detected were within 0.5 log of one another.
TABLE 3.
PCR quantification of target taxa B1, C1, S1, and L10 in pooled ileal contents of pigs fed corn (C), barley (B), or wheat (W) and frequencies of recovery of corresponding clones
| Target | Ileal samplea | Mean threshold cycleb ± SE | No. of genomesb (log/g of contents) | Frequency of corresponding clonesc |
|---|---|---|---|---|
| B1 | C | 21.3 ± 0.1 | 8.5 ± 0.0 | 16 |
| B | 31.0 ± 0.1 | 5.4 ± 0.0 | 0 | |
| W | 27.7 ± 0.1 | 6.4 ± 0.0 | 7 | |
| C1 | C | 28.8 ± 0.4 | 9.1 ± 0.2 | 88 |
| B | 30.5 ± 0.2 | 8.2 ± 0.1 | 18 | |
| W | 28.3 ± 0.6 | 8.8 ± 0.3 | 69 | |
| S1 | C | 30.9 ± 0.3 | 5.3 ± 0.1 | 1 |
| B | 28.8 ± 0.5 | 6.0 ± 0.2 | 48 | |
| W | 29.3 ± 0.3 | 5.9 ± 0.1 | 4 | |
| L10 | C | 22.6 ± 0.1 | 10.0 ± 0.0 | 506 |
| B | 27.2 ± 0.1 | 9.8 ± 0.0 | 686 | |
| W | 21.9 ± 0.2 | 10.2 ± 0.0 | 616 |
Genomic DNA extracted from ileal contents of 15 pigs fed each diet was pooled.
Mean of triplicate determinations.
Total number of clones recovered within each library clustering within the target bacteria.
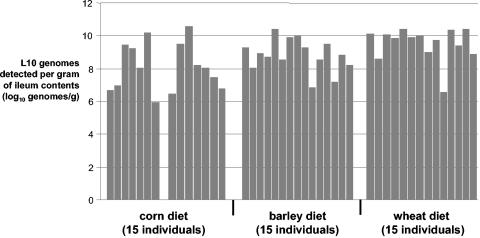
Figure 3 illustrates the number of L10 genomes detected with DNA extracted from ileal contents of individual pigs prior to pooling for library construction. The results suggest marked animal-to-animal variation; for example, in the corn library, the number of L10 genomes/g of ileal contents ranged from undetectable to 4 × 1010. The barley and wheat individuals generally contained higher numbers of L10 genomes, and while there was less variation than was observed in the corn library, genome numbers varied over four orders of magnitude. In the barley and wheat libraries, the mean number of L10 genomes for the 15 individual animals was within a log of the result obtained for the pool (Table 3); in the corn library, the mean of the 15 individuals was 2.42 logs lower than the result obtained for the pool. The latter observation is probably related to the much greater individual variation in L10 counts in samples pooled to generate the corn library.
FIG. 3.
Determination of the prevalence of L10 genomes in each individual animal fed diets based on corn (n = 15), wheat (n = 15), or barley (n = 15). A. Number of genomes (log10) detected in each individual per gram of ileal digesta. The means of the 15 observations within each group were: corn-fed individuals, 7.58; barley-fed individuals, 8.83; and wheat-fed individuals, 9.53.
DISCUSSION
Knowledge regarding microbial ecology of the porcine ileum or the gastrointestinal tract in general is extremely limited. Some targeted studies designed to assay a few taxa by culture-based methods have been conducted (3, 5, 41), but these tend to be narrow in scope and offer general taxonomic information and qualitative descriptions of flora. An extensive 16S rDNA sequence-based study of the swine gastrointestinal tract has been published which describes the 16S rDNA “phylotype” inventory of the gastrointestinal tract of Danish pigs (22). Given the important physiological role of the small intestine and its associated microflora in swine health and performance, these populations are deserving of more attention.
The chaperonin-60 gene is an attractive target both for cataloging microbial diversity with clone libraries and for targeting specific taxa with quantitative PCR methods. The 549- to 567-bp region amplified with chaperonin-60 universal PCR primers is easily sequenced with a single sequencing reaction and is phylogenetically informative, often providing sufficient discriminating information to distinguish subspecies or serotypes of a species (4). Another important feature of the chaperonin-60 universal target region is that it is uniformly variable, lacking the alternating conserved and variable regions that characterize the 16S rDNA sequence, generating significant secondary structure and facilitating PCR artifacts such as chimeras which are a confounding factor in 16S rRNA-based studies (32, 42).
Sequence variation introduced by Taq polymerase infidelity is an unavoidable pitfall of PCR-based methods. However, most of the abundant sequences in the libraries were independently amplified from each of the three template pools (Table 2). Among the clusters of nearly identical sequences, variable positions are significantly skewed towards the third codon position (19; this study). Also, no in-frame stop codons were identified in any of the partial chaperonin-60 sequences. These three observations suggest that while Taq polymerase infidelity no doubt introduces some artificial diversity, it is not a major distorting factor and also that a protein-encoding target sequence offers advantages over structural RNA-encoding targets with respect to monitoring and assessing PCR artifacts.
The overall structure of the ileum microbial population described in this report corresponds to previous characterizations of ileal microflora. The most predominant taxa in this community are the low-G+C gram-positive organisms, particularly the Lactobacillales family, which includes Lactobacillus and Pediococcus spp. among others. Smaller numbers of other low G+C gram-positive bacteria, such as the Clostridiales and Bacillales, and yet smaller numbers of gamma-proteobacteria were also identified. The pattern of relative abundances of these groups illustrated in Fig. 1B corresponds to the colony enumeration results for the samples published previously (7), as well as to the observations made by Leser et al. (22). These results also support observations that the ileal microflora is distinct in composition from populations associated with the cecum, colon, or feces (19, 22, 31, 34), where microbial communities are more diverse and contain higher numbers of gram-negative bacteria, such as the Bacteroides class. The relative lack of microbial diversity in the ileum compared to the more distal regions of the gut is also illustrated by the fact that we observed a relatively small number of sequences (14% of total unique sequences) occurring at high frequency (accounting for 81% of clones examined).
One of the major potential pitfalls of PCR-based studies of microbial communities is biased amplification of different sequences during PCR. The results of quantitative PCR assays of four different targets identified in the ileum populations indicate that the relative abundances of these targets in the three targeted populations are accurately represented by comparison of the frequency of chaperonin-60 clone recovery among the libraries (Table 3). The quantitative PCR study results also give an indication of the sensitivity of the library method. Targets that were detected at levels of 1 to 10 clones in the libraries, such as the S1 group in the corn and wheat libraries, were determined to be present in the digesta samples at 2.1 × 105 and 7.4 × 105 S1 genomes/g of contents, respectively. Consistent with this, the B1 target was not found in the barley library, but 2.3 × 105 B1 genomes/g of ileum contents were detected by quantitative PCR. These results suggest that the sequencing of approximately 1,000 clones from each chaperonin-60 library is sufficient to detect organisms present at >105 CFU/g of sample. This sensitivity level is likely also the explanation for why organisms such as E. coli that are reported to be found in the small intestine at levels at or below this threshold were not detected.
One of the most intriguing results of this study is the observation of variability in the abundance of L10 group organisms in individual pigs. In the genomic DNA pools used to create the corn, barley, and wheat libraries, L10 sequences were by far the most abundant (Table 2 and Fig. 2). However, enormous variability (from undetectable to 4 × 1010 genomes/g of ileum contents) was detected in the corn library (Fig. 3). The L10 quantitative PCR primers were designed to specifically target this group and fail to amplify any related but different targets. Thus, it is possible that minor strain variations in the dominant flora of individual animals account for the differences in L10 detection. It is also possible that variation in sampling location, efficiency of extraction of DNA, and/or variation in copurification of PCR inhibitors could account for some animal-to-animal variation. Others have suggested that the major bacterial groups remain constant among individual pigs fed the same diet (23), but those observations were made based on qualitative profiling methods which, at 106 to 1010 genomes/g, would likely have detected L10 in all pigs in the current study with the exception of the one pig fed the corn diet.
Variation in gut microflora associated with lifestyle and diet in humans and other animals has been described (3, 10, 17, 29), but there is a lack of information regarding differences between individuals. In human subjects persistent individual variation in microbial profile, including individual variation in the dominant Lactobacillus sp., has been noted (37, 38). We were surprised, however, to observe significant individual variation in abundance of the dominant L10 Lactobacillus taxon in pigs of the same age and same genotype, housed in the same room, provided identical diets, and managed according to the same protocol. In agreement, however, considerable variation in denaturing gradient gel electrophoresis banding pattern among individual broiler chickens subject to similar constants of genotype, diet, and housing has been reported (40). It is intriguing to suggest that these differences contribute to animal variation in health or nutrient digestion (e.g., fat metabolism) (12, 37). The development of a broad range of molecular tools for profiling microbial populations is a major step in defining individual variation in microflora composition. These differences are deserving of focused research since they are potentially a major confounding factor in the development of methods and products for the manipulation of these populations for improved health, performance, and pathogen resistance.
We anticipated that diet composition would affect the ileum microflora of pigs and hypothesized that these differences could be detected and quantified with a combination of sequence-based and quantitative PCR-based methods. Differences in the relative abundances of taxa and, perhaps more importantly, the diversity within taxa were indeed detected, particularly within Streptococcus spp. and the Clostridiales. The small sample size and the restriction of sample collection to one time point in this study limit our ability to draw specific conclusions regarding the biological significance of the variation in microbial populations associated with diet composition. However, the sequence libraries and quantitative PCR tools which we continue to develop present the opportunity to conduct more focused in vivo studies aimed at relating microflora shift to specific environmental variables, animal performance, and health outcomes.
Acknowledgments
This work was funded by a Saskatchewan Department of Agriculture, Food and Rural Revitalization strategic research program grant and by the National Research Council of Canada.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Apgar, G. A., E. T. Kornegay, M. D. Lindemann, and C. M. Wood. 1993. The effect of feeding various levels of Bifidobacterium globosum A on the performance, gastrointestinal measurements, and immunity of weanling pigs and on the performance and carcass measurements of growing-finishing pigs. J. Anim.. Sci. 71:2173-2179. [DOI] [PubMed] [Google Scholar]
- 3.Benno, Y., K. Endo, K. Suzuki, T. Mitsuoka, and S. Namioka. 1985. Use of nonprotein nitrogen in pigs: effects of dietary urea on the intestinal microflora. Am. J. Vet. Res. 46:959-962. [PubMed] [Google Scholar]
- 4.Brousseau, R., J. E. Hill, G. Prefontaine, S. H. Goh, J. Harel, and S. M. Hemmingsen. 2001. Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences. Appl. Environ. Microbiol. 67:4828-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butine, T. J., and J. A. Leedle. 1989. Enumeration of selected anaerobic bacterial groups in cecal and colonic contents of growing-finishing pigs. Appl. Environ. Microbiol. 55:1112-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canibe, N., S. H. Steien, M. Overland, and B. B. Jensen. 2001. Effect of K-diformate in starter diets on acidity, microbiota, and the amount of organic acids in the digestive tract of piglets, and on gastric alterations. J. Anim. Sci. 79:2123-2133. [DOI] [PubMed] [Google Scholar]
- 7.Drew, M. D., A. G. Van Kessel, A. E. Estrada, E. D. Ekpe, and R. T. Zijlstra. 2002. Effect of dietary cereal on intestinal bacterial populations in weaned pigs. Can. J. Anim.. Sci. 82:607-609. [Google Scholar]
- 8.Englyst, H. 1989. Classification and measurement of plant polysaccharides. Anim. Feed Sci. Technol. 23:27-42. [Google Scholar]
- 9.Estrada, A., M. D. Drew, and A. G. Van Kessel. 2001. Effect of dietary supplementation of fructooligosacchadies and Bifidobacterium longum to early weaned pigs on performance and fecal bacterial populations. Can. J. Anim.. Sci. 81:141-148. [Google Scholar]
- 10.Finegold, S. M., H. R. Attebery, and V. L. Sutter. 1974. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am. J. Clin. Nutr. 27:1456-1469. [DOI] [PubMed] [Google Scholar]
- 11.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 12.Gaskins, H. R., C. T. Collier, and D. B. Anderson. 2002. Antibiotics as growth promotants: mode of action. Anim. Biotechnol. 13:29-42. [DOI] [PubMed] [Google Scholar]
- 13.Goh, S. H., D. Driedger, S. Gillett, D. E. Low, S. M. Hemmingsen, M. Amos, D. Chan, M. Lovgren, B. M. Willey, C. Shaw, and J. A. Smith. 1998. Streptococcus iniae, a human and animal pathogen: specific identification by the chaperonin-60 gene identification method. J. Clin. Microbiol. 36:2164-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goh, S. H., R. R. Facklam, M. Chang, J. E. Hill, G. J. Tyrrell, E. C. Burns, D. Chan, C. He, T. Rahim, C. Shaw, and S. M. Hemmingsen. 2000. Identification of Enterococcus species and phenotypically similar Lactococcus and Vagococcus species by reverse checkerboard hybridization to chaperonin 60 gene sequences. J. Clin. Microbiol. 38:3953-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goh, S. H., Z. Santucci, W. E. Kloos, M. Faltyn, C. G. George, D. Driedger, and S. M. Hemmingsen. 1997. Identification of Staphylococcus species and subspecies using the chaperonin-60 gene identification method and reverse checkerboard hybridization. J. Clin. Microbiol. 35:3116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, P. M. Sabour, R. Wheatcroft, and S. Chen. 2002. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 208:1-7. [DOI] [PubMed] [Google Scholar]
- 17.Hara, H., N. Orita, S. Hatano, H. Ichikawa, Y. Hara, N. Matsumoto, Y. Kimura, A. Terada, and T. Mitsuoka. 1995. Effect of tea polyphenols on fecal flora and fecal metabolic products of pigs. J. Vet. Med. Sci. 57:45-49. [DOI] [PubMed] [Google Scholar]
- 18.Hill, J. E., S. L. Penny, K. G. Crowell, S. H. Goh, and S. M. Hemmingsen. 2004. cpnDB: a chaperonin sequence database. Genome Res. 14:1669-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill, J. E., R. P. Seipp, M. Betts, L. Hawkins, A. G. Van Kessel, W. L. Crosby, and S. M. Hemmingsen. 2002. Extensive profiling of a complex microbial community by high-throughput sequencing. Appl. Environ. Microbiol. 68:3055-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jian, W., L. Zhu, and X. Dong. 2001. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 51:1633-1638. [DOI] [PubMed] [Google Scholar]
- 21.Kwok, A. Y., J. T. Wilson, M. Coulthart, L. K. Ng, L. Mutharia, and A. W. Chow. 2002. Phylogenetic study and identification of human pathogenic Vibrio species based on partial hsp60 gene sequences. Can. J. Microbiol. 48:903-910. [DOI] [PubMed] [Google Scholar]
- 22.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leser, T. D., R. H. Lindecrona, T. K. Jensen, B. B. Jensen, and K. Moller. 2000. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with Brachyspira hyodysenteriae. Appl. Environ. Microbiol. 66:3290-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marston, E. L., J. W. Sumner, and R. L. Regnery. 1999. Evaluation of intraspecies genetic variation within the 60-kilodalton heat shock protein gene (groEL) of Bartonella species. Int. J. Syst. Bacteriol. 49 Pt. 3:1015-1023. [DOI] [PubMed] [Google Scholar]
- 26.McEwen, S. A., and P. J. Fedorka-Cray. 2002. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl. 3) :S93-S106. [DOI] [PubMed] [Google Scholar]
- 27.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMB News 4:14. [Google Scholar]
- 28.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 29.Peltonen, R., E. Eerola, K. Suomi, H. Aho, M. Kuusisto, and P. Toivanen. 1993. Effect of dietary fish powder on intestinal flora and development of arthritis in the pig. Br. J. Rheumatol. 32:1049-1054. [DOI] [PubMed] [Google Scholar]
- 30.Pluske, J. R., D. W. Pethick, D. E. Hopwood, and D. J. Hampson. 2002. Nutritional influences on some major enteric bacterial diseases of pigs. Nutr. Res. Rev. 15:333-371. [DOI] [PubMed] [Google Scholar]
- 31.Pryde, S. E., A. J. Richardson, C. S. Stewart, and H. J. Flint. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 65:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu, X., L. Wu, H. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 34.Salanitro, J. P., I. G. Blake, and P. A. Muirhead. 1977. Isolation and identification of fecal bacteria from adult swine. Appl. Environ. Microbiol. 33:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzman, N. H., H. de Jong, Y. Paterson, H. J. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 148:3651-3660. [DOI] [PubMed] [Google Scholar]
- 36.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tannock, G. W. 2004. A special fondness for lactobacilli. Appl. Environ. Microbiol. 70:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tannock, G. W., K. Munro, H. J. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Wielen, P. W., D. A. Keuzenkamp, L. J. Lipman, K. F. van, and S. Biesterveld. 2002. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 44:286-293. [DOI] [PubMed] [Google Scholar]
- 41.van Winsen, R. L., B. A. Urlings, L. J. Lipman, J. M. Snijders, D. Keuzenkamp, J. H. Verheijden, and F. van Knapen. 2001. Effect of fermented feed on the microbial population of the gastrointestinal tracts of pigs. Appl. Environ. Microbiol. 67:3071-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, G. C., and Y. Wang. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63:4645-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]