Abstract
Clinical efficacy of antisense oligonucleotides (AONs) for the treatment of neuromuscular disorders depends on efficient cellular uptake and proper intracellular routing to the target. Selection of AONs with highest in vitro efficiencies is usually based on chemical or physical methods for forced cellular delivery. Since these methods largely bypass existing natural mechanisms for membrane passage and intracellular trafficking, spontaneous uptake and distribution of AONs in cells are still poorly understood. Here, we report on the unassisted uptake of naked AONs, so-called gymnosis, in muscle cells in culture. We found that gymnosis works similarly well for proliferating myoblasts as for terminally differentiated myotubes. Cell biological analyses combined with microscopy imaging showed that a phosphorothioate backbone promotes efficient gymnosis, that uptake is clathrin mediated and mainly results in endosomal-lysosomal accumulation. Nuclear localization occurred at a low level, but the gymnotically delivered AONs effectively modulated the expression of their nuclear RNA targets. Chloroquine treatment after gymnotic delivery helped increase nuclear AON levels. In sum, we demonstrate that gymnosis is feasible in proliferating and non-proliferating muscle cells and we confirm the relevance of AON chemistry for uptake and intracellular trafficking with this method, which provides a useful means for bio-activity screening of AONs in vitro.
Keywords: : antisense, delivery, gene silencing, RNA, splicing
Introduction
Antisense oligonucleotide (AON) therapeutics involve a broad range of approaches to adjust levels of RNA targets via effects on posttranscriptional processes [1]. AONs are in (pre)clinical development for many diseases, including cancer, inflammatory conditions, cardiovascular disease, and neurodegenerative and neuromuscular disorders. The clinical efficacy of AONs largely depends on biodistribution to and uptake by target cells and routing to the desired intracellular location (nucleus or cytoplasm). Although this is true for any disease for which AON therapy is an option, it is especially relevant for degenerative muscle disorders. In this class of diseases, muscle tissue throughout the entire body can be affected, requiring a therapeutic effect in as much as 40% of total body mass. Examples of multisystemic neuromuscular disorders for which AON therapy is currently under development are spinal muscular atrophy [2], Pompe disease [3], iron-sulfur cluster deficiency myopathy [4], Duchenne muscular dystrophy (DMD; [5]), and myotonic dystrophy type 1 (DM1; [6,7]).
Here, we focus on DMD and DM1, both of which are muscular dystrophies that are characterized by typically early and highly variable onset, respectively, with progressive general muscle dysfunction [8]. DMD, the most common lethal chromosome X-linked disease in human beings, is caused by mutations in the dystrophin (DMD) gene, creating a premature stop codon that abrogates the synthesis of functional dystrophin protein [9]. As dystrophin is a structural protein that is responsible for connecting the cytoskeleton of muscle fibers to the extracellular basal lamina, its absence affects membrane stability of myofibers, eventually leading to cell death and replacement by connective tissue [10]. DM1, the most common muscular dystrophy in adults [11], has a different etiology. Expansion of an unstable (CTG)n repeat in the 3′ UTR of the DMPK gene is responsible for a highly variable multisystemic phenotype [11,12]. When the number of (CTG)n triplets in the repeat exceeds a certain threshold, expanded DMPK (CUG)n transcripts are produced that accumulate in the cell nucleus and aberrantly bind a variety of ribonucleoprotein factors. Sequestration of these factors causes RNA processing abnormalities of other mRNAs and miRNAs, resulting in a broad spectrum of toxic gain-of-function effects [13].
Therapeutic correction at the RNA level by AON-mediated exon skipping of mutant dystrophin pre-mRNA is a promising disease-modifying treatment option for DMD [5]. Similarly, for DM1, proof-of-concept studies have demonstrated that gene silencing via AON-mediated breakdown and prevention of nuclear accumulation of expanded DMPK (CUG)n mRNA may become a useful therapeutic modality in the future [6,7,14–18]. Notably, although the therapeutic strategies for DMD and DM1 are mechanistically distinct, they share an important aspect: Exon skipping of mutant DMD pre-mRNA as well as breakdown of expanded DMPK (CUG)n RNA must take place in the nucleus of target cells.
To be able to improve AON delivery to the nucleus in diseases such as DM1 and DMD, it is important to study cellular uptake and intracellular distribution aspects on treatment, including the impact that AON chemical modifications have in these processes. Preclinical selection of AON candidates is primarily based on comparative analysis of their bioactivity in cell cultures, typically using transfection or electroporation. Although these methods are useful to study bioactivity, they are less reliable predictors of cellular uptake and trafficking in vivo, since cellular barriers are artificially bypassed.
The uptake of AONs by cells in vitro without the use of transfection reagent or other delivery strategies was coined gymnosis by Stein et al. in 2010 [19]. Actually, 10 years earlier, Kole and coworkers had already started to investigate the free uptake of AONs of different chemistries directly from the culture medium [20,21]. This method was based on the prolonged incubation of cells with AONs that were dissolved in the culture medium, which progressively resulted in cellular internalization by endocytosis, via mechanisms that are currently under debate [22,23]. It is generally accepted that a small fraction of AONs are able to escape from endocytic vesicles and reach their RNA target in the nucleus or cytosol, but this behavior is not well characterized either [24].
To date, the unassisted uptake of AONs has been applied successfully to studies in proliferating cell types [25]. However, less is known about the applicability to non-proliferating cells, such as those that have undergone terminal differentiation. In addition, limited research has been dedicated to addressing how different chemical modifications of AONs influence free uptake and subsequent intracellular trafficking. Phosphorothioate moieties have been suggested to promote gymnosis, presumably through AON binding to heparin-binding proteins on the cell surface and with a role in the subsequent formation of intracellular vesicles [26]. Whether that holds for other types of AON chemistries and to what extent chemical modifications allow or influence endosomal escape, particularly in muscle cells, has remained unclear thus far.
In this study, we used a myogenic cell model derived from DM1 mice to test the effects of AON chemistry and sequence on gymnosis and subsequent nuclear efficacy. Two classes of AONs were compared: (1) analogs of a CAG7 AON for degradation of expanded DMPK (CUG)n transcripts for DM1 [7,15] and (2) variants of DMD23, an AON capable of inducing exon 23 skipping in mouse Dmd transcripts [27] (Table 1). We followed uptake and intracellular distribution of these AONs during gymnosis by means of fluorescence microscopy during myoblast proliferation and differentiation. In parallel, we measured AON activity toward nuclear DM1 and DMD targets. Facilitated uptake using polyethyleneimine (PEI) was usually included as a positive control to bypass cellular uptake and trafficking pathways. Despite the relatively high subcellular AON accumulation, we found that already low levels of AONs in the nucleus may be effective. Thus, gymnosis may be considered a supportive method for the comparative analysis of therapeutic AON candidates in (differentiated) muscle cells in vitro, which takes into account natural uptake and intracellular trafficking.
Table 1.
Antisense Oligonucleotides Used in This Study
| AON | Chemistry | Sequence (5′ > 3′) | Target |
|---|---|---|---|
| CAG7-OMePS (PS58 [15]) | 2′-OMe PS | CAG CAG CAG CAG CAG CAG CAG | Expanded DMPK transcripts |
| CAG7-OMe | 2′-OMe PO | ||
| CAG7-PS [7] | DNA PS | ||
| P4-CAG7-OMePS | 2′-OMe PS | P4-CAG CAG CAG CAG CAG CAG CAG | |
| Cy3-CAG7-OMePS | 2′-OMe PS | Cy3-CAG CAG CAG CAG CAG CAG CAG | |
| Cy3-CAG7-OMe | 2′-OMe PO | ||
| Cy3-CAG7-PS | DNA PS | ||
| FAM-CAG7-OMePS | 2′-OMe PS | FAM-CAG CAG CAG CAG CAG CAG CAG | |
| CAG7-OMePS-Cy3 | 2′-OMe PS | CAG CAG CAG CAG CAG CAG CAG-Cy3 | |
| P4-CAG7-OMePS-Cy3 | 2′-OMe PS | P4-CAG CAG CAG CAG CAG CAG CAG-Cy3 | |
| DMD23-OMePS (M23D(+02–18) [28]) | 2′-OMe PS | GGC CAA ACC UCG GCU UAC CU | Mouse Dmd pre-mRNA (exon 23) |
| DMD23-OMe | 2′-OMe PO | ||
| DMD23-PS | DNA PS | ||
| P4-DMD23-OMePS | 2′-OMe PS | P4-GGC CAA ACC UCG GCU UAC CU | |
| Cy3-DMD23-OMePS | 2′-OMe PS | Cy3-GGC CAA ACC UCG GCU UAC CU | |
| Cy3-DMD23-OMe | 2′-OMe PO | ||
| Cy3-DMD23-PS | DNA PS | ||
| P4-DMD23-OMePS-Cy3 | 2′-OMe PS | P4-GGC CAA ACC UCG GCU UAC CU-Cy3 |
2′-OMe, 2′-O-methyl; AON, antisense oligonucleotide; P4 = LGAQSNF [29]; PS, phosphorothioate; PO, phosphate.
Materials and Methods
Antisense oligonucleotides
AONs used in this study are listed in Table 1 and were synthesized by BioMarin Nederland B.V. The nucleotide sequence of CAG7 AONs is 5′-CAGCAGCAGCAG CAGCAGCAG-3′ and targets the (CUG)n repeat in expanded DMPK transcripts (PS58 [15]). The sequence of DMD23 AONs is 5′-GGCCAAACCUCGGCUUACCU-3′ and targets an exon splicing enhancer in exon 23 of mouse Dmd pre-mRNA (M23D(+02–18) [28]). Three types of chemical modification were used: 2′-O-methyl phosphorothioate (-OMePS), 2′-O-methyl phosphate (-OMe), and DNA phosphorothioate (-PS). Some AONs were conjugated at their 5′ or 3′ end with a Cy3 or FAM fluorophore or with P4, a muscle targeting peptide of sequence LGAQSNF [29]. Cy3-CAG3-ENA is a 5′-CAGCAGCAG-3′ AON with an ethylene-bridged nucleic acid (ENA) phosphate backbone, 5′ conjugated to Cy3.
Cell culture
DM500 myoblasts were derived from a DM1 mouse model as previously described [15]. DM500 myoblasts express human DMPK (CUG)500 transcripts, the disease-causing agent in DM1 [30]. DM500 myoblasts with enhanced green fluorescent protein (EGFP)-stained nuclei were used for most experiments and obtained after stable transduction with a pLZRS retroviral vector expressing an EGFP-hMBNL1 fusion protein (plasmid gift from Dr. D. Brook, Nottingham). Myoblasts were cultured on 0.1% (w/v) gelatin-coated dishes in Dulbecco's modified Eagle's medium (DMEM; GibcoBRL, Gaithersburg, MD) that was supplemented with 20% (v/v) fetal calf serum (FCS), 50 μg/mL gentamycin, 10 U/mL interferon (IFN)-γ (BD Biosciences, San Jose, CA), and 2% (v/v) chicken embryo extract (Sera Laboratories International, Bolney, United Kingdom) at 33°C and 5% CO2.
When myogenic differentiation was applied, myoblasts were seeded on Matrigel® (BD Biosciences, Breda, the Netherlands) and grown until 100% confluency. Subsequently, differentiation medium (DMEM supplemented with 5% (v/v) horse serum and 50 μg/mL gentamycin) was added and the cells were incubated at 37°C for 7–10 days. In some cases, after 3 days of differentiation, 4 μg/mL cytosine β-d-arabinofuranoside (Ara-C) (Sigma, Saint Louis, MO) was added for 2 days to remove undifferentiated myoblasts.
Immortalized human DM1 myoblasts (DM11 cl5, derived from primary myoblasts obtained from a DM1 patient expressing a DMPK allele bearing a (CTG)2600 repeat [31]) were grown and differentiated as previously described [32]. C2C12 mouse myoblasts were cultured on 0.1% (w/v) gelatin-coated dishes in DMEM that was supplemented with 10% (v/v) FCS at 37°C and 7.5% CO2. Differentiation to myotubes was performed under the same conditions as for human DM1 myoblasts.
Gymnosis and transfection
For gymnosis, AONs were supplemented directly in the cell culture medium. A range of concentrations (0.2–5 μM) and incubation times (4 h to 13 days) was tested, as specified for each experiment. During proliferation, cells were seeded at 30%–40% confluency and incubated with AONs in the growth medium. Every time that the medium was refreshed (every 2–3 days) or cells were passaged (before reaching confluency), fresh growth medium containing the same AON concentration was used. In experiments where myogenic differentiation was applied, cells were grown until 100% confluency and incubated with differentiation medium containing AON. Medium was refreshed every 2–3 days, containing the same AON concentration.
Polyethyleneimine (PEI)-mediated transfection of AONs was done as previously described [15] at a final AON concentration of 500 nM. Myoblasts and myotubes were imaged, and RNA was isolated 48 h after transfection.
Use of endocytosis modulators
To block clathrin-dependent endocytosis, cells were supplied with new medium containing 80 μM dynasore (Sigma-Aldrich, Saint Louis, MO) at 1 h before AON treatment. Incubation was then continued in the same medium, but containing the AON, during the entire gymnosis process (24 h). In these experiments, DMEM supplemented with 50 μg/mL gentamycin and 10 U/mL IFN-γ, without serum, was used. Longer incubation periods were not possible due to significant cell death and toxicity.
To induce the release of endosomal and lysosomal contents, cells were treated with 75 μM chloroquine (CHQ) for 4 h at the end of the gymnosis incubation period. No signs of toxicity were observed at this CHQ concentration.
Confocal microscopy and immunochemistry
Myoblasts were grown in μ-Slide 8-well plates (ibidi®, Martinsried, Germany) or in GWSt-3522 WillCo-dish® wells (WillCo Wells B.V., Amsterdam, the Netherlands). After treatment with Cy3-labeled AONs (Table 1), analysis was performed by using a Zeiss LSM510-Meta or a Leica SP5 confocal laser scanning microscope. For live cell imaging, cells were put in a culture chamber that was maintained at a stable temperature of 37°C with a 5% CO2 supply. DM500 myotubes were incubated before analysis with 5 μM carboxyfluorescein succinimidyl ester (CSFE; Invitrogen) for 5 min and with 10 μg/mL Hoechst (Life Technologies) for 30 min to visualize the cytoplasm and nucleus, respectively. Images to evaluate AON distribution were acquired in live cells or immediately after fixation, at different time points after the start of gymnosis. These approaches for image acquisition were adopted because pilot experiments revealed that standard paraformaldehyde protocols did not fix AONs permanently when the cells were kept in phosphate-buffered saline (PBS) after fixation, giving experimental variation and a false impression of their intracellular distribution (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/nat). For fixation, the cells were washed twice with PBS and then incubated for 15 min at 37°C in 0.1 M phosphate buffer (pH 7.5) with 4% (w/v) paraformaldehyde.
For visualization purposes, certain images displayed in the figures were adjusted to the maximum exposure level within the range of a negative background signal. This results in overexposed areas but allows visualization of AONs in other parts of the cell where the concentration is lower, such as the nucleus. For fluorescence quantification (Figs. 1B and Fig. 6), confocal settings were calibrated below Cy3-channel saturation by using the well displaying the highest fluorescence signal (Supplementary Fig. S8). Identical parameters were maintained to quantify all other wells in the same microscopy session, including all treatments (n = 3). Several images of each treatment (25–30 cells) were processed by using ImageJ (Wayne Rasband, National Institute of Health). Masks of either nuclear or cytoplasmic regions (determined by the EGFP signal) were used to select cellular areas of interest from where Cy3 intensities were measured and corrected for background (Fig. 6).
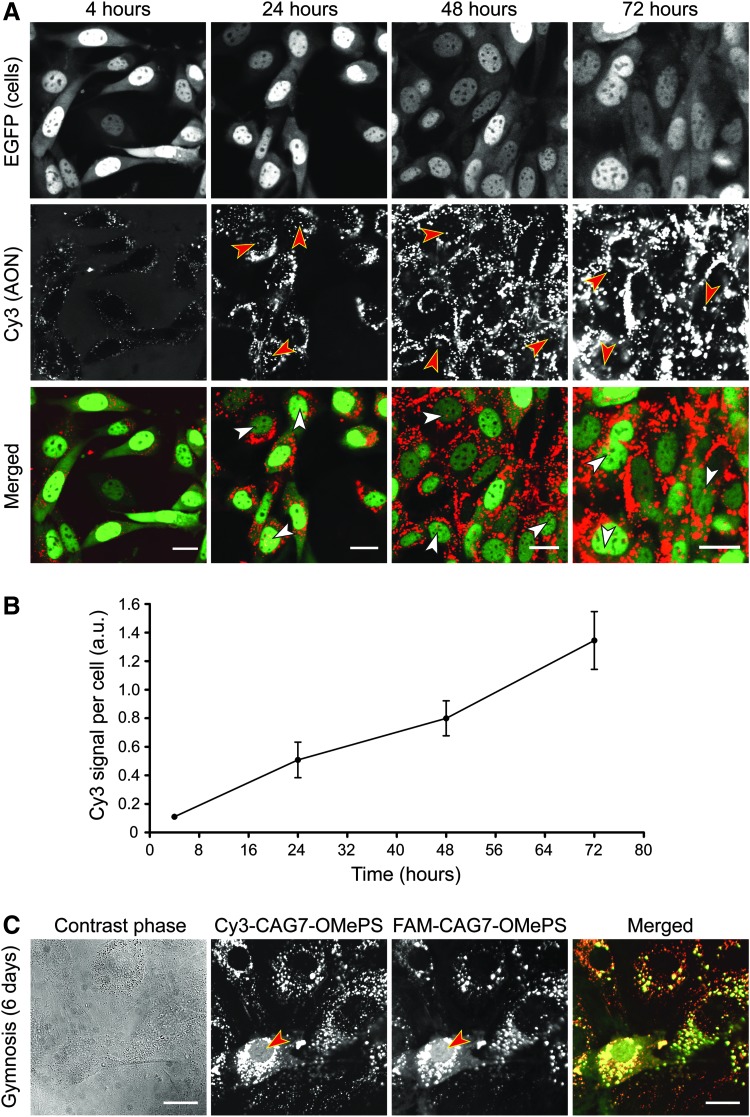
FIG. 1.
Live imaging of gymnosis in proliferating DM500 myoblasts. DM500 EGFP myoblasts were cultured for 3 days in the presence of Cy3-CAG7-OMePS (200 nM). (A) Confocal images showing intracellular localization of the AON (red). Nuclei are green due to the expression of a nuclear EGFP-fusion marker protein. Cy3-CAG7-OMePS quickly accumulated in cytoplasmic vesicle-like structures. The AON accumulated in a fraction of the cells in spots in the nucleus (arrowheads). Scale bars indicate 20 μm. (B) Quantification of red fluorescence per cell (n = 3; 30 cells per experiment) using images under saturation levels. (C) DM500 myoblasts were grown in the presence of a mixture of Cy3- and FAM-conjugated CAG7-OMePS (200 nM each). An identical vesicular, cytoplasmic staining was observed for both AONs after 2 days. After 4 days, a clear diffuse staining in the cytosol and nucleus was detected in a minority (<5%) of the cell population (arrowheads). Scale bars indicate 20 μm. For visualization purposes, images displayed in (A, C) were adjusted to the maximum exposure level within the range of a negative background signal. AON, antisense oligonucleotide; EGFP, enhanced green fluorescent protein.
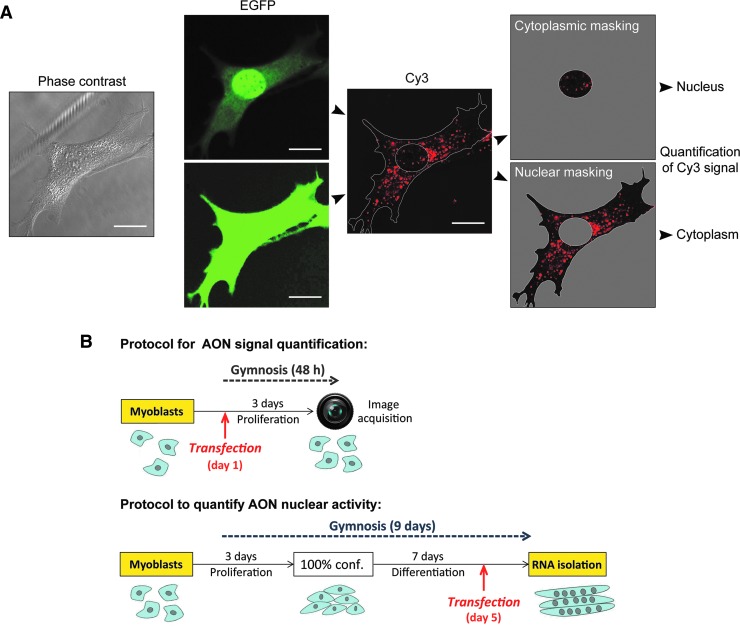
Fig. 6.
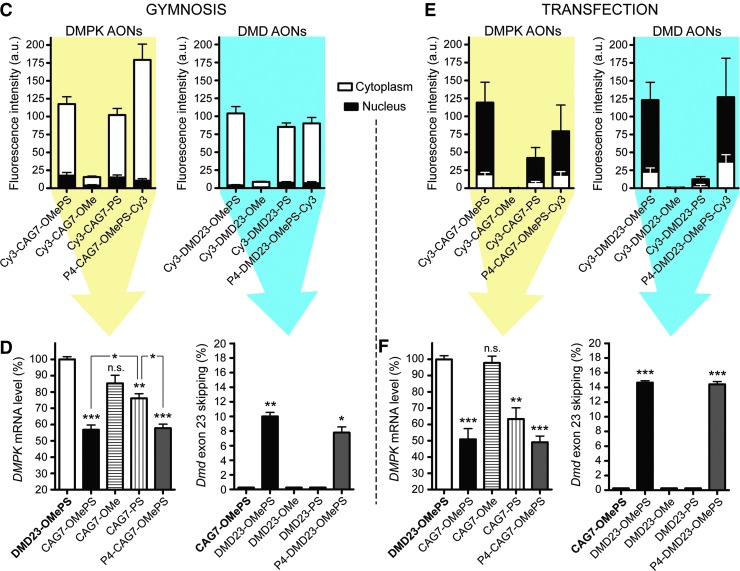
Chemistry effects on AON uptake and activity during gymnosis. (A) Masking procedure used to quantify Cy3-fluorescence in the nucleus and cytoplasm of myoblasts. Scale bars indicate 20 μm. (B) Protocols used for cell culture and gymnosis and subsequent quantification of AON uptake and nuclear activity. (C) Stacked bar charts illustrating mean fluorescence intensity in the cytoplasm (white) and nucleus (black) of DM500 myoblasts after 2 days of gymnosis (500 nM) of Cy3-conjugated CAG7 and DMD23 AONs (n = 3; ∼30 cells per experiment). (D) Effects on expanded DMPK mRNA expression and Dmd exon 23 skipping measured after 7 days of differentiation. Control AONs are depicted in bold. (E) Stacked bar charts illustrating mean fluorescence intensity 2 days after PEI-assisted delivery in the cytoplasm and nucleus of DM500 myoblasts (n = 3; ∼30 cells per experiment). (F) Effects on expanded DMPK mRNA expression and Dmd exon 23 skipping after PEI-assisted delivery in myotubes. Control AONs are depicted in bold. Raw images are shown in Supplementary Figure S8. DMPK qPCR data were compared by using a one-way ANOVA followed by a Bonferroni's Multiple-Comparison Test. Signal of Dmd skipped products was analyzed by t-test against a zero theoretical mean. Comparisons were considered significant when: *p < 0.05, **p < 0.01, and ***p < 0.001. qPCR, quantitative polymerase chain reaction.
For Lamp1 immunochemistry, cells were fixed and blocked with 4% (v/v) animal serum (corresponding to the origin of the secondary antibody) and 0.33% (v/v) Triton-X-100 in PBS for 60 min at room temperature. Rabbit Anti-Lamp1 primary antibody (Sigma, Saint Louis, MO) was diluted in 10 mg/mL bovine serum albumin and 0.33% (v/v) Triton-X-100 and applied overnight at 4°C. Samples were blocked with goat serum and incubated with Goat Anti-Rabbit alexa-647 secondary antibody (Abcam, Cambridge, United Kingdom) for 1 h at room temperature.
RNA isolation and reverse transcriptase polymerase chain reaction (RT-PCR) analysis
RNA was isolated from cultured cells by using the Aurum™ Total RNA Mini Kit (Bio-Rad, Hercules, CA), according to the manufacturer's protocol. Primer sets for polymerase chain reaction (PCR) and quantitative PCR (qPCR) were designed by using Primer-BLAST [33] in the NCBI database and validated in silico by using OligoAnalyzer 3.1 [34] to prevent the formation of hairpins and dimers during amplification (Supplementary Table S1). Resulting products were visualized on agarose gels and sequenced to verify identity.
For reverse transcription, typically 0.5 μg of total RNA was subjected to cDNA synthesis by using the SuperScript® first-strand synthesis system with random hexamer primers in a total volume of 20 μL.
Skipping of exon 23 in Dmd transcripts was analyzed by using conventional PCR with primers annealing in exon 22 (forward primer) and exon 24 (reverse primer; Supplementary Table S1). Per reaction, the mix contained 10.4 μL MilliQ, 4 μL 5 × Q5 Reaction Buffer, 0.2 μL Q5® High Fidelity DNA Polymerase (2 U/μL) (BioLabs®, Inc., Ipswich, MA), 0.4 μL dNTPs (Invitrogen, Eugene, OR), 2 μL forward primer (5 μM), 2 μL reverse primer (5 μM) (Biolegio BV, Nijmegen, the Netherlands), and 1 μL cDNA template. Thermal cycling conditions consisted of 30 s at 98°C followed by 34 amplification cycles of 6 s at 98°C, 20 s at 64°C (annealing), and 10 s at 72°C (elongation). The final elongation step was carried out for 2 min. Fragments were analyzed on 2% agarose gels. Images of ethidium-stained products were acquired under UV light below pixel saturation. Quantification of signals was done by using ImageJ (Wayne Rasband, National Institute of Health), measuring pixel intensity and correcting for background.
For the detection of degradation of expanded DMPK transcripts, real-time qPCR was used, following a protocol previously described [7]. Samples were analyzed by using the CFX96™ Real-time System (Bio-Rad, Hercules, CA). A melting curve was obtained for each sample to confirm single-product amplification. Relative mRNA levels were calculated by using the ΔΔCt method [35]. Gapdh and Actb expression levels were used together for normalization.
qPCR was also used to measure mRNA levels of differentiation markers Aqp1, Myh1, Mylfp and Cav3. In this case, samples were analyzed by the ViiA™ 7 Real-Time PCR System (Life Technologies, Bleiswijk, the Netherlands), by means of commercial TaqMan® assays (Applied Biosystems, Foster City, CA) (Supplementary Table S1), following the manufacturer's instructions. Data were normalized by using Gapdh and Hprt as reference genes. Levels of the group showing the highest expression were set to 100% by using the ΔΔCt method.
Statistical analysis
We performed unpaired Student's t-tests to test whether DMPK levels after CAG7-OMePS treatment (either alone or in the presence of dynasore or chloroquine) differed significantly from the levels that remained after treatment with a control AON. To determine whether the average signals of Dmd skipped products were significantly different from the background, we performed a t-test analysis against a zero theoretical mean. The effect of Ara-C treatment in the expression of differentiation markers was analyzed by unpaired t-tests in each gene group. To compare the effect of AONs of different chemistries on DMPK transcript levels, we used a one-way ANOVA followed by a Bonferroni's Multiple-Comparison Test. To test whether Cy3-CAG7-OMePS accumulation in time was significantly different from Cy3-CAG3-ENA behavior, we used a two-way ANOVA. In all these cases, values were considered significantly different when: *p < 0.05, **p < 0.01, or ***p < 0.001.
Results
Gymnosis in proliferating myoblasts
Experiments in this study were performed in the immortal DM500 cell model [15], derived from a transgenic mouse model for DM1 [30]. DM500 myoblasts show nuclear accumulation of transcripts from a human DMPK transgene bearing an expanded (CTG)500 repeat. The cells show normal growth and myogenic differentiation capacity. Previous studies revealed that AON transfection characteristics did not overtly differ between DM500 and normal primary mouse or human myoblasts [7,15]. Here, we mainly used a population of DM500 myoblasts that stably expressed low levels of EGFP-MBNL1, a green fusion protein with predominant nuclear location [36], for easy microscopy visualization of localization of fluorescent AONs in the nucleus.
To study gymnosis and intracellular AON distribution, we followed the behavior of Cy3-conjugated CAG7-OMePS, a 21 nt repeat AON containing a fully modified 2′-O-methyl phosphorothioate backbone (Table 1) [15]. In proliferating myoblasts, CAG7-OMePS-positive vesicular structures in the cytoplasm were present in essentially all cells after 4 h of incubation with 200 nM naked oligo in the medium (Fig. 1A). The vesicular accumulation appeared more prominent after 24 h, and quantification of staining revealed that it became even more intense over the subsequent 2 days (Fig. 1B). Interestingly, we also noticed local accumulation of AON signal in spots in the nucleus in many, but not all, cells at >24 h of gymnosis. In addition, a diffuse nuclear staining was observed in ∼1% of cells at 72 h and ∼2% after 6 days (Fig. 1C). The fraction of Cy3-positive nuclei did not further increase after longer incubation (up to 13 days) or on use of a higher AON concentration (5 μM; data not shown).
Gymnotic uptake and distribution characteristics were entirely different from those seen with polyethyleneimine-(PEI)-mediated transfection, a commonly used procedure to deliver AONs in cultured cells [37]. The PEI transfection of DM500 myoblasts resulted in a predominant nuclear staining of Cy3-CAG7-OMePS in around 50% of all cells (Supplementary Fig. S2).
The typical uptake and distribution behavior of naked AONs was not influenced by the type of fluorophore conjugated to the AON (Fig. 1C). Neither did the conjugation position of the fluorophore, to the 5′ or the 3′ end of the AON, have any obvious effect on AON localization (Supplementary Fig. S3).
To examine the potential effects on gymnosis by the nucleotide composition of the AON, we also followed the fate of Cy3-DMD23-OMePS, an AON that induces exon 23 skipping during splicing of mouse Dmd pre-mRNA [27]. When used under the same experimental conditions, a similar localization was observed with this AON (Fig. 2A), including the predominantly vesicular and low intranuclear accumulation pattern that was found for Cy3-CAG7-OMePS (Supplementary Fig. S4). Notably, these distribution patterns were also observed after gymnosis in mouse C2C12 myoblasts and human myoblasts derived from a patient with DM1 (Supplementary Fig. S5).
FIG. 2.
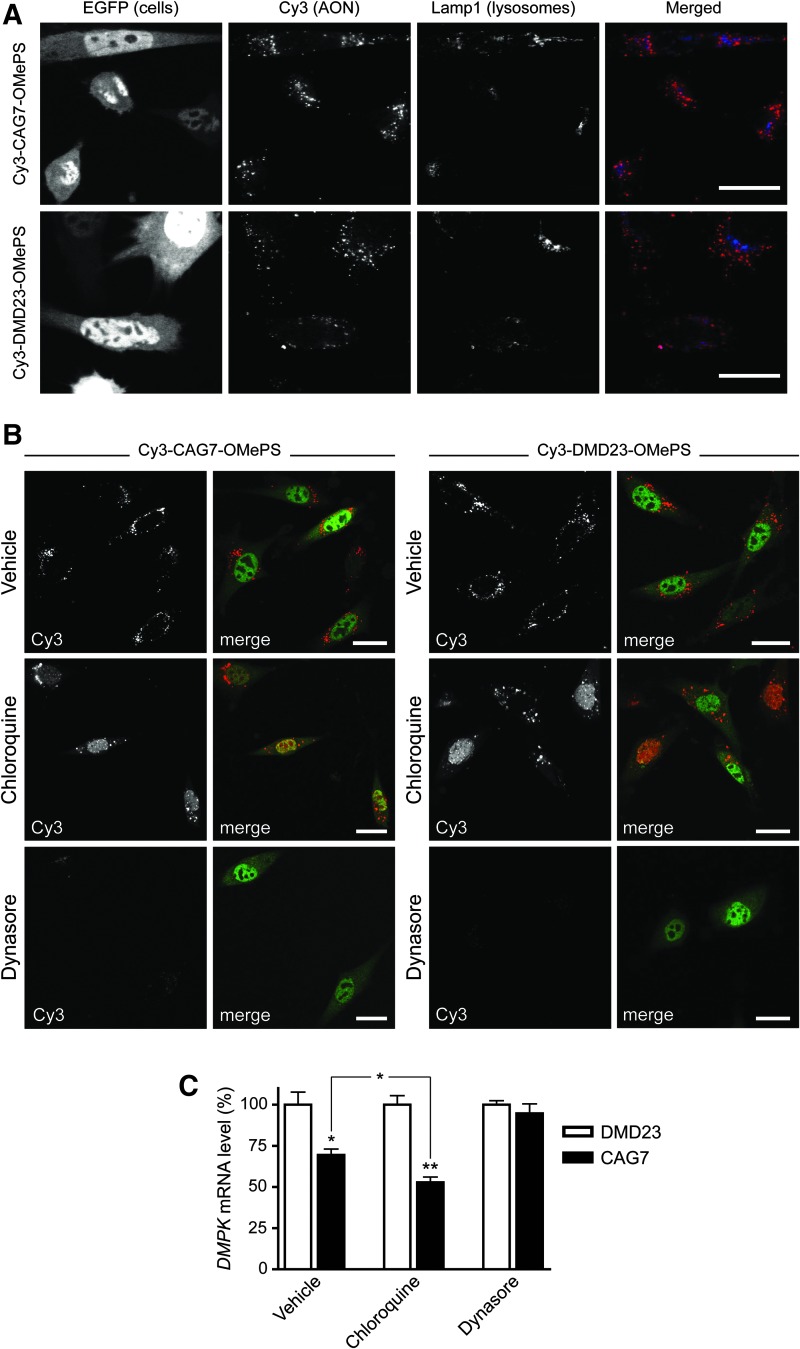
Intracellular trafficking of AONs during gymnosis. (A) Confocal images of DM500 EGFP cells after 24 h of growth in the presence of Cy3-CAG7-OMePS or Cy3-DMD23-OMePS. Lamp-1, a lysosomal marker protein, was visualized by immunochemistry and co-localized with some of the AON-positive vesicles. Scale bars indicate 25 μm. (B) Uptake of Cy3-DMD23-OMePS and Cy3-CAG7-OMePS (500 nM) in the presence of chloroquine or dynasore after 24 h of gymnosis. Merged images are a composite of Cy3 and EGFP channels. Scale bars indicate 25 μm. (C) Quantification of expanded DMPK RNA expression after 24 h of gymnosis incubation with Cy3-CAG7-OMePS in combination with chloroquine or dynasore. DMPK mRNA levels after Cy3-CAG7-OMePS and control AON treatments were compared by unpaired t-tests. CAG7-OMePS activity in the presence of chloroquine compared with vehicle was analyzed by one-way ANOVA followed by a Bonferroni's Multiple-Comparison Test (*p < 0.05, **p < 0.01).
Interfering with intracellular trafficking using endocytosis modulators
A significant fraction of the vesicles that incorporated Cy3-DMD23-OMePS or Cy3-CAG7-OMePS stained for the late endosomal/lysosomal marker Lamp1 (Fig. 2A). This suggested that at least a proportion of the vesicles that contained gymnosis-delivered OMePS AONs followed the endocytic route up to lysosomes, confirming earlier observations [22]. To study this pathway more specifically for myoblasts, we examined whether endocytic modulators such as chloroquine and dynasore would affect intracellular distribution of DM1 and DMD AONs. Chloroquine is a lysosomotropic agent that leads to endosomal disruption in vitro [38], and it can release AONs from endocytic vesicles [39]. Dynasore inhibits dynamin, a protein that is essential for clathrin-dependent endocytosis [40].
Dynasore was added 1 h before starting the incubation with AONs and kept in culture during the rest of the gymnosis period to block clathrin-dependent endocytosis during the entire process. Dynasore treatment completely blocked AON uptake and distribution, supporting the hypothesis that 2′-OMe PS AONs are internalized by clathrin-mediated endocytosis (Fig. 2B; reviewed in Ref. [22]). In contrast, chloroquine treatment resulted in a strong increase in nuclear accumulation in many cells. Chloroquine was present during the final 4 h of gymnosis, to allow AONs to reach all endosomal compartments first. The combination of these observations suggests that on gymnotic uptake of AONs, endosomal escape is required for subsequent transfer to the cell nucleus.
Gymnosis in differentiated myotubes
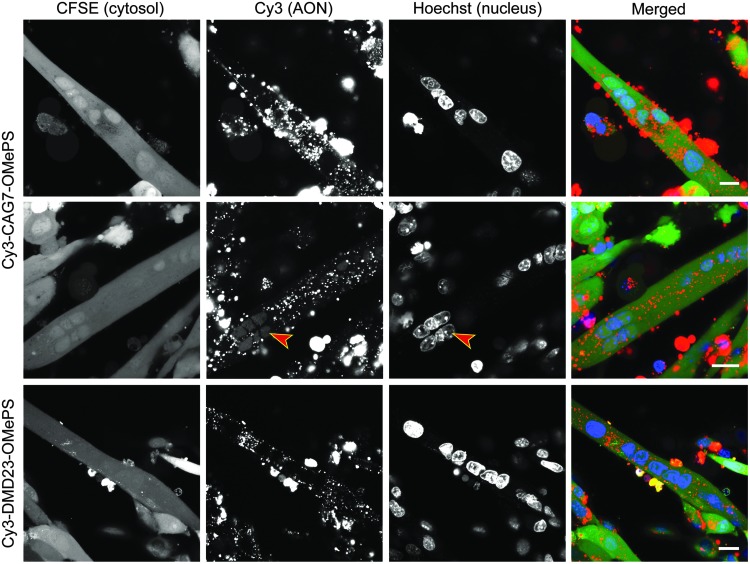
Gymnosis in non-proliferating cells is reported to be challenging [25]. We, therefore, studied independently unassisted AON uptake during the differentiation of myoblasts to myotubes in vitro. We added Cy3-CAG7-OMePS and Cy3-DMD23-OMePS to differentiating myoblast cultures for as many as 7 days. As in myoblasts, AONs accumulated in myotubes in cytoplasmic vesicles and multiple myonuclei were AON positive (Fig. 3). We questioned whether this resulted from intrinsic uptake mechanisms in these differentiated cells or whether they were donated by AON-loaded myoblasts during fusion. To evaluate this point, we performed gymnosis experiments with cell cultures that had first undergone differentiation for 3 days and were then treated with cytosine β-d-arabinofuranoside (Ara-C) for 2 days (Fig. 4A). Ara-C, a nucleoside analog that interferes with DNA replication, causes apoptosis of proliferating cells [41] and has been used to remove myoblasts during differentiation to achieve a pure myotube culture [42]. Non-fused myoblasts were largely removed from the adhered myotube layer after Ara-C treatment, and this correlated with a significant change in the gene expression of several markers reported to be up- or downregulated during myogenesis [43,44] (Supplementary Fig. S6). Gymnosis initiated after Ara-C treatment resulted in intracellular AON distribution patterns in myotubes that were indistinguishable from those observed before or in myoblasts (Fig. 3; data not shown). These experiments, thus, indicate that differentiated myotubes, such as proliferating myoblasts, are able to internalize 2′-OMe PS AONs by endocytosis.
FIG. 3.
Gymnosis during myogenic differentiation. DM500 myoblasts were grown to confluency and transferred to differentiation medium in the presence of Cy3-CAG7-OMePS or Cy3-DMD23-OMePS (500 nM). These culture conditions were continued for 7 days. After this period, a vesicular cytoplasmic localization was observed in fully differentiated myotubes (live imaging). Clear nuclear signal (arrowhead) could be observed in some myotubes. Compare this faint diffusive staining with the total absence of staining in nuclei in other myotubes. For visualization purposes, images displayed were adjusted to the maximum exposure level within the range of the negative background signal. Scale bars indicate 20 μm.
FIG. 4.
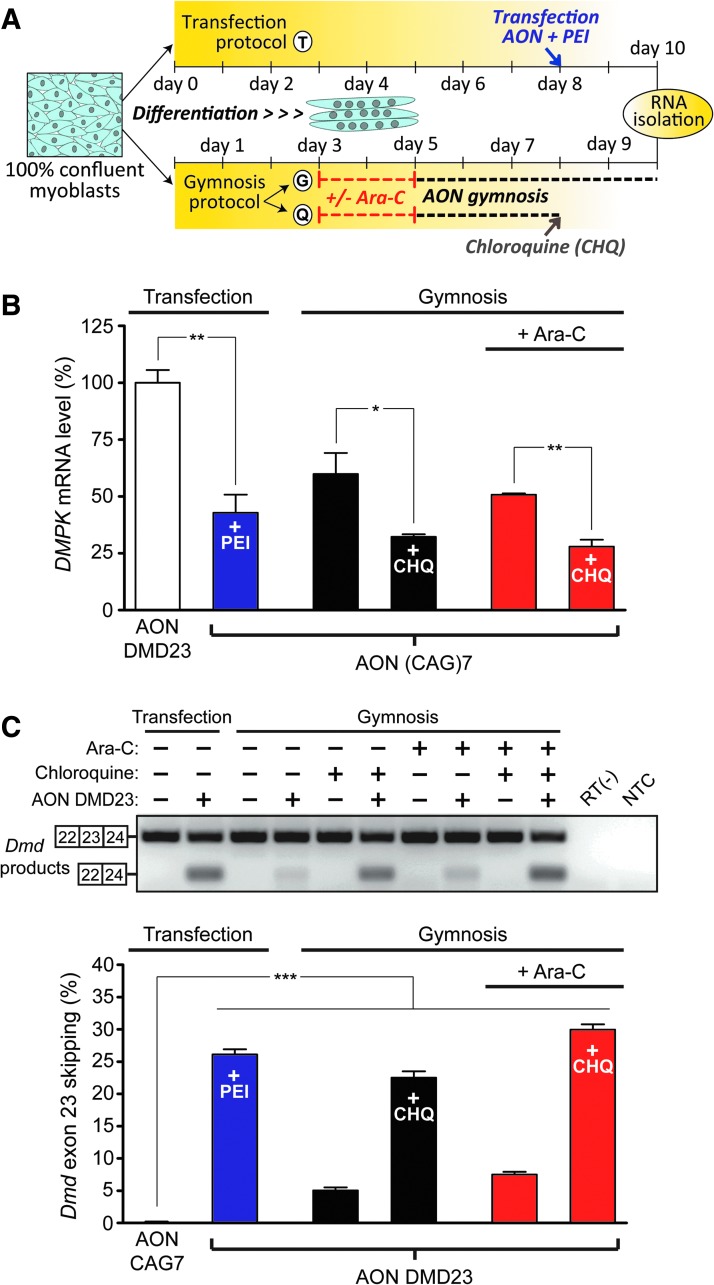
Gymnosis in differentiated DM500 myotubes. (A) Experimental protocol used for the treatment of myotubes with CAG7-OMePS and DMD23-OMePS. Differentiation of DM500 myoblasts was initiated at 100% confluency at day 0. In protocol T, as a positive control, at day 8 of differentiation, myotubes were transfected with AON (500 nM) by using PEI. Alternatively, cells were allowed to fuse for 3 days and were then incubated with Ara-C for 2 days or left untreated (protocols G and Q). At day 5, gymnosis was started and myotubes were incubated with AON (500 nM) for 5 days (protocol G) or for only 3 days, after which they were treated with CHQ followed by the addition of fresh differentiation medium without AON (protocol Q). In all protocols, RNA was isolated at day 10. (B) Analysis of DMPK mRNA expression (n = 3). All groups treated with CAG7-OMePS showed significantly lower levels compared with the matched controls that received DMD23-OMePS (unpaired t-tests; only shown for the transfection pair; **p < 0.01). We also tested whether the effect of CHQ was significant in each experimental group by t-test analysis (*p < 0.05, **p < 0.01). (C) Analysis of Dmd exon 23 skipping (n = 3). The gel image shows results of one of the triplicates, whereas the graph depicts the quantification of skipping percentage (n = 3). A t-test analysis against a zero theoretical mean was performed in each case to determine whether the average signal of Dmd skipped products was significantly different from the background (***p < 0.001). CAG7 = CAG7-OMePS; CHQ, chloroquine; DMD23, DMD23-OMePS; NTC = no template control; PEI, polyethyleneimine; RT(−) = no reverse transcriptase control.
AONs gymnotically delivered during proliferation and differentiation are active in the nucleus
To analyze whether gymnotically delivered AONs in myoblasts were released from endosomes to become active toward nuclear targets, we first examined the ability of CAG7-OMePS to silence DMPK (CUG)500 transcripts. The silencing of expanded DMPK mRNA in myoblasts could be measured and was further increased in the presence of chloroquine, whereas breakdown was prohibited in the presence of dynasore (Fig. 2C). In line with earlier observations [7,15], we did not see an effect of the Cy3 modification on nuclear activity, be it conjugated to the 5′ or 3′ end (Supplementary Fig. S3B). Not surprisingly, the bioactive effect of Cy3-CAG7-OMePS on DMPK mRNA levels in differentiating myotubes was also sensitive to chloroquine treatment and very comparable between cultures that received Ara-C treatment and those that did not (Fig. 4B).
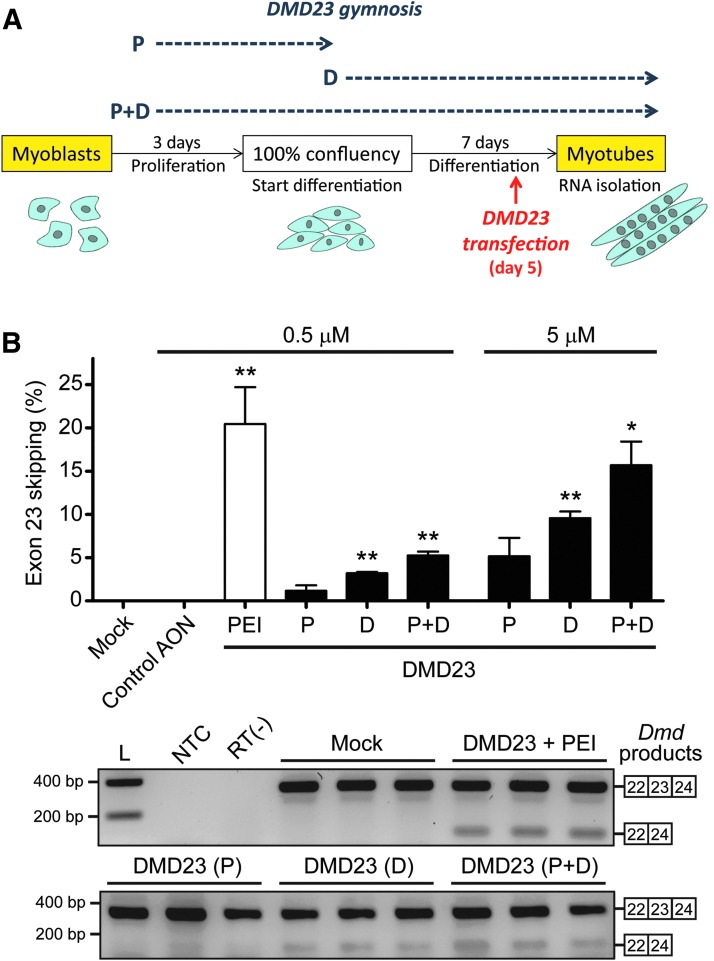
We next examined the ability of gymnotically delivered DMD23-OMePS to skip exon 23 during Dmd pre-mRNA splicing. Since DMD expression, as opposed to that of DMPK, is rather low in myoblasts but strongly induced during myogenic differentiation (data not shown; [45]), we only investigated DMD23 effects in differentiated myotubes. We found that the skipping effect of this AON was the same in Ara-C-treated and -untreated myotube cultures (Fig. 4C). Chloroquine treatment enhanced the activity of DMD23-OMePS to levels that were comparable to those achieved by PEI-mediated transfection. This suggests that intracellular trafficking of AONs in differentiated myotubes is a process that is amenable for modulation, and thus for the improvement of its translational value. Finally, we examined exon 23 skipping levels in myotubes after dedicated gymnosis protocols during either proliferation or differentiation or during both periods (Fig. 5A). DMD23-OMePS-mediated exon skipping was AON concentration dependent and most effective when applied during the entire period of proliferation and differentiation (Fig. 5B).
FIG. 5.
AON concentration- and time-dependent effects of gymnosis on Dmd exon 23 skipping. (A) Protocol used for DMD23-OMePS gymnosis in DM500 myoblasts. Gymnosis was performed during the last 2 days of proliferation (P), during 7 days of differentiation (D), or during both periods (P+D). PEI transfection was carried out as a control on day 5 of differentiation. In all cases, RNA was isolated at day 7 of differentiation. (B) Analysis of Dmd exon 23 skipping. A t-test analysis against a zero theoretical mean was performed in each case to determine whether the average signals of Dmd skipped products were significantly different from the background (*p < 0.05, **p < 0.01). Gel image compares results from gymnosis at 5 μM and transfection with 500 nM DMD23-OMePS. L, DNA size ladder.
We confirmed that gymnotically delivered AONs were also active in nuclei of cells other than the DM500 model. DMD23-OMePS efficiently mediated Dmd exon 23 skipping in differentiated C2C12 myotubes after 9 days of gymnosis (Supplementary Fig. S5). Likewise, CAG7-OMePS reduced DMPK RNA expression in human DM1 myotubes (note that these cells contain a 1:1 mix of expanded and normal-sized DMPK transcripts, the latter of which are not sensitive to CAG7-OMePS treatment [7,15]).
Effects of AON chemistry on gymnosis delivery and nuclear activity
In the course of pilot studies, wherein gymnosis was tested for a range of chemically modified CAG AONs, we observed different trafficking results. For example, Cy3-CAG3-ENA gave an almost undetectable endosomal accumulation [7] (Supplementary Fig. S7). This finding was consistent with previous observations in other cells [26], where AON uptake behavior was strongly influenced by chemical modification. As this aspect is relevant for future therapeutic applications, we tested effects of a selected set of oligo chemistries on gymnosis in our DM1 myogenic cell model. Four modifications for both AON sequences, CAG7 and DMD23, were analyzed: 2′-O-methyl phosphorothioate (OMePS), 2′-O-methyl phosphate (OMe), DNA phosphorothioate (PS), and OMePS, including a 5′ conjugation with a muscle-homing peptide (P4), which has been shown to enhance activity of DMD23-OMePS and CAG7-OMePS in vivo ([29]; Mulders et al., unpublished). Note that AON DMD23-PS was included to complete the set, but is, in fact, not useful for therapeutic purposes, since this AON may induce RNAse-H-dependent breakdown of Dmd pre-mRNAs instead of exon skipping.
By using immunofluorescent image acquisition, we were able to quantify Cy3-signals separately for the nucleus and cytoplasm in myoblasts (Fig. 6A; note that this protocol could not be applied to myotube cultures). The activity of DM1 and DMD AONs was measured in parallel in 7-day-old myotubes (Fig. 6B). Forty-eight hours of gymnosis led to a predominant cytoplasmic location for all AONs, irrespective of the chemical modification and sequence (Fig. 6C). However, a significantly lower uptake was observed for AONs with a phosphate instead of a phosphorothioate backbone, which corresponded to a lack of activity toward their nuclear target (Fig. 6D). Conjugation of peptide P4 to OMePS AONs had an inconclusive effect on gymnosis, as it increased the uptake of CAG7-OMePS, but it had essentially no effect on the uptake and cytosolic accumulation of DMD23-OMePS. Assisted delivery of the AONs by PEI transfection led mainly to nuclear localization (Fig. 6E), but with a lower efficiency for AONs with only OMe or PS backbones. The tendency of nuclear accumulation is obviously promoted by the well-known facilitating effect of PEI on endosomal escape [46,47].
Nuclear activity clearly correlated with gymnotic uptake efficiency, as assessed by cytoplasmic accumulation. The CAG7-OMePS AONs had the most, CAG7-OMe had the least, and CAG7-PS had intermediate silencing potential (Fig. 6D). Interestingly, PEI-assisted transfection yielded similar efficiency differences between the OMePS, OMe, and PS AONs and the effects on breakdown and exon skipping (with the exception of RNase H recruiting AON DMD23-PS) were moderately improved by PEI transfection (Fig. 6F). Our findings, therefore, challenge the suggested idea that a strong AON accumulation in the nucleus is required for a robust effect on nuclear targets [48,49].
Discussion
A number of studies have been reported in the past few years on effective treatment strategies using naked AONs in patients and mouse models for DMD and DM1 [6,50,51]. Since the AONs used were rather large, hydrophilic molecules that could not easily diffuse through muscle cell membranes [52], alternative cell internalization mechanisms must have been responsible for the observed biological effect in muscles. One distinct feature with regard to the cellular uptake of AONs in vivo that is specific for DMD is that due to the dystrophin deficiency some muscle fibers are more permeable to large compounds because of membrane alterations [53], whereas other fibers retain good membrane integrity [54], thereby directing treatment to the most affected ones [55]. In contrast, in muscles from DM1 patients and mouse models, all myofiber membranes appear impermeable to large molecules [56].
Supportive insight regarding normal AON delivery and intracellular trafficking mechanisms may be provided by cell culture studies, since it was shown that cells are able to internalize AONs freely from the culture medium, a process now known as gymnosis [19–21,25]. Clearly, our in vitro culture conditions were different from the conditions in vivo, for example, with respect to protein concentration (ie, 5%–20% serum in culture medium versus 100% serum in the circulation). One should, therefore, be cautious to extrapolate results obtained in cell culture directly to animal or human studies. For detailed information on the complexity of AON delivery in vivo, we refer the reader to a recent review on this topic [57].
We confirm here that gymnosis is a general characteristic of the DM500 myogenic cell culture model and was also operational in mouse C2C12 and human DM1 patient myoblasts. After several hours of incubation in the presence of AONs, vesicular localization patterns were observed in proliferating myoblasts and in differentiated myotubes. Our findings, thus, demonstrate that myotubes retain AON uptake potential after differentiation and strengthen the hypothesis that similar uptake mechanisms may be at play in muscles in vivo (knowing that differentiated myotubes in vitro exhibit several characteristics of mature muscle myofibers in vivo, such as excitation contraction coupling and expression of muscle differentiation biomarkers [58,59]).
Lehto et al. reported a difference in the uptake of a peptide-coupled morpholino (Pip6a-PMO) between myoblasts and myotubes [60]. We have not focused on the mechanistic differences of AON uptake between undifferentiated and differentiated cells. Besides, a meaningful comparison with our study is complicated, since morpholinos are uncharged (in contrast to our 2′-OMe AONs). More importantly, a positively charged peptide such as Pip6a may have dominant differential effects on endosomal escape and intracellular trafficking in myoblasts and myotubes.
The first step in unassisted AON uptake must occur at the cell membrane and involves AON binding and initial engulfment by endocytosis [61]. We found that gymnosis was blocked by dynamin inhibition, which suggests that the endocytic mechanism by which AONs enter cells is clathrin mediated [22]. Although it is not known how naked AONs are initially recognized and endocytosed in myogenic cells, a phosphorothioate (PS)-modified backbone is apparently important, as AONs in our study carrying a phosphate backbone (and either 2′-OMe or ENA ribose modification) were inactive. This idea is supported by a study showing enhanced cellular binding of AONs with PS modification compared with an unmodified phosphate backbone [62]. The importance of PS backbone is further consistent with earlier reports on successful gymnosis of a variety of AON chemistries, all carrying this modification [19,26,63,64]. The endocytic process might be triggered by the surrounding proteins that bind to PS AONs in a non-specific manner [65], rather than by the oligonucleotide itself.
In the presence of polycationic transfection reagents, such as PEI, the internalization process is mediated by the recognition of positively charged PEI-AON complexes by proteoglycan membrane receptors [66]. Initial steps in unassisted AON endocytosis may be triggered by polyanion-interacting proteins that specifically bind phosphorothioate moieties in AONs [65]. Some of the candidates involved may have been identified in a recent screen for proteins that bind phosphorothioate linkages with a high affinity [67]. The normal uptake efficiency of phosphorothioate AONs may be enhanced in vivo by conjugating peptide ligands, as was observed for P4-CAG7-OMePS [17,29,68].
A second important step required to achieve AON activity inside the cell is escape from the endosomal compartment and subsequent trafficking to the target location, that is, in our study the cell nucleus, where expanded DMPK RNAs and Dmd pre-mRNAs reside. It has been postulated that AONs may follow two distinct endocytic routes [63]: a non-productive route to lysosomal accumulation and degradation, and a productive route resulting in nuclear trafficking and effective RNA regulation. Some of the CAG7-OMePS or DMD23-OMePS AON-positive vesicles, indeed, co-localized with the lysosomal marker Lamp1, suggesting that the non-productive pathway was followed by a fraction of the internalized AONs. The non-co-localizing CAG7-OMePS/DMD23-OMePS AON fraction may be associated with the postulated productive route and may be present in early endosomes from which they escaped.
The exact mechanism by which AONs escape from endosomes is not known, but it may occur in the course of intracellular trafficking by membrane bilayer instability during vesicle fusion and budding [24]. We found that endocytic release, promoted by chloroquine treatment, indeed enhanced the activity of CAG7-OMePS and DMD23-OMePS toward their corresponding targets. Chloroquine treatment correlated with a switch from a vesicular staining pattern to a predominant nuclear signal, suggesting that soon after endosomal escape free AONs will quickly diffuse through nuclear pores into the nucleus [69,70]. A diffuse nuclear staining, including spots, has been previously reported after microinjection of AONs carrying a phosphorothioate backbone in the cytosol [69–71].
Despite the weak nuclear and strong cytoplasmic (vesicular) AON staining after gymnosis, a significant activity by both AON sequences was detected toward RNA targets that are clearly located in the nucleus [72,73]. In control experiments, we found that AON activity was higher when PEI-mediated transfection was applied, but the magnitude did not correlate with the higher nuclear AON concentration (estimated at 5–25 times higher, judging from fluorescence quantification; as a result of endosomal release induced by a proton-sponge effect of PEI [46,47]). So, how then can the apparent discrepancy between nuclear AON concentration (fluorescence intensity) and nuclear activity (RNA degradation or exon skipping) after gymnosis be explained?
We propose a few possible explanations, which are not mutually exclusive: (1) PEI molecules may remain associated and form stable complexes with AONs in the nucleus [74], thus decreasing free AON concentration and impeding proper binding to the mRNA target. (2) AON concentration in the nucleus during gymnosis may not surpass the limit of detection of our microscope in the majority of cells, but the minimal AON concentration required for a biological effect in this compartment may be much lower than generally believed and is already achieved under these conditions. Nevertheless, it is important to point out that several other factors may influence bioactive efficacy, such as levels of target RNA, AON efficiency, speed of the process, and whether one AON can modulate multiple transcripts. For instance, CAG7-OMePS AON activities after gymnosis and transfection were more similar than those of DMD23-OMePS, suggesting that the DM1 AON requires a lower concentration in the nucleus. (3) Related to the previous point, cell imaging has shown that gymnosis is an intrinsic cellular process that is active in all cells, whereas PEI-mediated transfection involves only part of the cell population (depending on transfection efficiency: around 50% in our experiments). Therefore, the proposed minimal AON concentration in the nucleus for biological effect may be present in a larger portion of the cell population during gymnosis compared with transfection.
In sum, we demonstrate that gymnosis is feasible in proliferating and non-proliferating muscle cells and we confirm the relevance of AON chemistry for uptake and intracellular trafficking with this method. Our data suggest that even low levels of AONs in the nucleus may be already sufficient for bioactivity and, thus, that gymnosis may be a useful method for a comparative analysis of therapeutic AON candidates in muscle cells in vitro.
Supplementary Material
Acknowledgments
The authors thank Jack Fransen, Cindy Dieteren, Leontien van der Bent, and Antoine Khalil for their support with microscopic techniques and use of ImageJ, and other members of the Department of Cell Biology and BioMarin Nederland for discussions. They also thank Vincent Mouly (Institut de Myologie, Paris) for human DM1 myoblasts (DM11 cell line).
Author Disclosure Statement
Funding was provided by BioMarin Nederland BV, Leiden, the Netherlands, a company that develops RNA therapeutics. A.G.-B., B.A., P.C.d.V., N.A.D., S.A.M.M., and J.C.T.v.D. are employees of BioMarin Nederland BV, which includes contribution to patent applications and participation in stock-option plans of the company. B.A., P.C.d.V., J.C.T.v.D., S.A.M.M., and D.G.W. are inventors in patents for oligonucleotide-mediated exon skipping and/or treatment of myotonic dystrophy type 1 and other neuromuscular disorders. B.N., J.K., I.D.G.v.K., H.J.E.C., and B.W. have no financial conflicts of interest.
References
- 1.Bennett CF. and Swayze EE. (2010). RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol 50:259–293 [DOI] [PubMed] [Google Scholar]
- 2.Porensky PN. and Burghes AH. (2013). Antisense oligonucleotides for the treatment of spinal muscular atrophy. Hum Gene Ther 24:489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton NP, Nelson CA, Weeden T, Taylor KM, Moreland RJ, Scheule RK, Phillips L, Leger AJ, Cheng SH. and Wentworth. BM. (2014). Antisense oligonucleotide-mediated suppression of muscle glycogen synthase 1 synthesis as an approach for substrate reduction therapy of pompe disease. Mol Ther Nucleic Acids 3:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kollberg G. and Holme E. (2009). Antisense oligonucleotide therapeutics for iron-sulphur cluster deficiency myopathy. Neuromuscul Disord 19:833–836 [DOI] [PubMed] [Google Scholar]
- 5.van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, et al. (2007). Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med 357:2677–2686 [DOI] [PubMed] [Google Scholar]
- 6.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF. and Thornton CA. (2012). Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 488:111–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez-Barriga A, Mulders SAM, van de Giessen J, Hooijer JD, Bijl S, van Kessel ID, van Beers J, van Deutekom JC, Fransen JA, et al. (2013). Design and analysis of effects of triplet repeat oligonucleotides in cell models for myotonic dystrophy. Mol Ther Nucleic Acids 2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wicklund MP. (2013). The muscular dystrophies. Continuum (Minneap Minn) 19:1535–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ. and Den Dunnen JT. (2006). Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 34:135–144 [DOI] [PubMed] [Google Scholar]
- 10.Kharraz Y, Guerra J, Pessina P, Serrano AL. and Munoz-Canoves P. (2014). Understanding the process of fibrosis in Duchenne muscular dystrophy. Biomed Res Int 2014:965631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harper PS. (2001). Myotonic Dystrophy, 3rd ed. WB Saunders, London, United Kingdom [Google Scholar]
- 12.Groenen P. and Wieringa B. (1998). Expanding complexity in myotonic dystrophy. BioEssays 20:901–912 [DOI] [PubMed] [Google Scholar]
- 13.Udd B. and Krahe R. (2012). The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol 11:891–905 [DOI] [PubMed] [Google Scholar]
- 14.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT. and Thornton CA. (2009). Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science 325:336–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulders SA, van den Broek WJAA, Wheeler TM, Croes HJE, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, et al. (2009). Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci U S A 106:13915–13920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JE, Bennett CF. and Cooper TA. (2012). RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc Natl Acad Sci U S A 109:4221–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leger AJ, Mosquea LM, Clayton NP, Wu IH, Weeden T, Nelson CA, Phillips L, Roberts E, Piepenhagen PA, et al. (2013). Systemic delivery of a peptide-linked morpholino oligonucleotide neutralizes mutant RNA toxicity in a mouse model of myotonic dystrophy. Nucleic Acid Ther 23:109–117 [DOI] [PubMed] [Google Scholar]
- 18.Pandey SK, Wheeler TM, Justice SL, Kim A, Younis H, Gattis D, Jauvin D, Puymirat J, Swayze EE, et al. (2015). Identification and characterization of modified antisense oligonucleotides targeting DMPK in mice and nonhuman primates for the treatment of myotonic dystrophy type 1. J Pharmacol Exp Ther 355:329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein CA, Hansen JB, Lai J, Wu S, Voskresenskiy A, Hog A, Worm J, Hedtjarn M, Souleimanian N, et al. (2010). Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res 38:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmajuk G, Sierakowska H. and Kole R. (1999). Antisense oligonucleotides with different backbones. Modification of splicing pathways and efficacy of uptake. J Biol Chem 274:21783–21789 [DOI] [PubMed] [Google Scholar]
- 21.Sazani P, Kang SH, Maier MA, Wei C, Dillman J, Summerton J, Manoharan M. and Kole R. (2001). Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res 29:3965–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juliano RL, Ming X, Carver K. and Laing B. (2014). Cellular uptake and intracellular trafficking of oligonucleotides: implications for oligonucleotide pharmacology. Nucleic Acid Ther 24:101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juliano RL. and Carver K. (2015). Cellular uptake and intracellular trafficking of oligonucleotides. Adv Drug Deliv Rev 87:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juliano RL, Ming X. and Nakagawa O. (2012). Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug Chem 23:147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soifer HS, Koch T, Lai J, Hansen B, Hoeg A, Oerum H. and Stein CA. (2012). Silencing of gene expression by gymnotic delivery of antisense oligonucleotides. Methods Mol Biol 815:333–346 [DOI] [PubMed] [Google Scholar]
- 26.Souleimanian N, Deleavey GF, Soifer H, Wang S, Tiemann K, Damha MJ. and Stein. CA. (2012). Antisense 2′-deoxy, 2′-fluroarabino nucleic acids (2′F-ANAs) oligonucleotides: in vitro gymnotic silencers of gene expression whose potency is enhanced by fatty acids. Mol Ther Nucleic Acids 1:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhaart IE, Tanganyika-de Winter CL, Karnaoukh TG, Kolfschoten IG, de Kimpe SJ, van Deutekom JC. and Aartsma-Rus A. (2013). Dose-dependent pharmacokinetic profiles of 2′-O-methyl phosphorothioate antisense oligonucleotides in mdx mice. Nucleic Acid Ther 23:228–237 [DOI] [PubMed] [Google Scholar]
- 28.Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J, Jadoon A, Bou-Gharios G. and Partridge T. (2005). Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc Natl Acad Sci U S A 102:198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jirka SM, Heemskerk H, Tanganyika-de Winter CL, Muilwijk D, Pang KH, de Visser PC, Janson A, Karnaoukh TG, Vermue R, et al. (2014). Peptide conjugation of 2′-O-methyl phosphorothioate antisense oligonucleotides enhances cardiac uptake and exon skipping in mdx mice. Nucleic Acid Ther 24:25–36 [DOI] [PubMed] [Google Scholar]
- 30.Seznec H, Lia-Baldini AS, Duros C, Fouquet C, Lacroix C, Hofmann-Radvanyi H, Junien C. and Gourdon G. (2000). Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum Mol Genet 9:1185–1194 [DOI] [PubMed] [Google Scholar]
- 31.Zhu CH, Mouly V, Cooper RN, Mamchaoui K, Bigot A, Shay JW, Di Santo JP, Butler-Browne GS. and Wright WE. (2007). Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell 6:515–523 [DOI] [PubMed] [Google Scholar]
- 32.Van Agtmaal EL, André LM, Willemse M, Cumming SA, van Kessel IDG, van den Broek WJAA, Gourdon G, Furling D, Mouly V, et al. (2017). CRISPR/Cas9-induced (CTG·CAG)n repeat instability in the myotonic dystrophy type 1 locus: implications for therapeutic genome editing. Mol Ther 25:24–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S. and Madden. TL. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J, et al. (2008). IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res 36:W163–W169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ. and Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 36.Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS. and Brook JD. (2002). Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet 11:805–814 [DOI] [PubMed] [Google Scholar]
- 37.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B. and Behr JP. (1995). A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A 92:7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michihara A, Toda K, Kubo T, Fujiwara Y, Akasaki K. and Tsuji H. (2005). Disruptive effect of chloroquine on lysosomes in cultured rat hepatocytes. Biol Pharm Bull 28:947–951 [DOI] [PubMed] [Google Scholar]
- 39.Mae M, El Andaloussi S, Lundin P, Oskolkov N, Johansson HJ, Guterstam P. and Langel U. (2009). A stearylated CPP for delivery of splice correcting oligonucleotides using a non-covalent co-incubation strategy. J Control Release 134:221–227 [DOI] [PubMed] [Google Scholar]
- 40.Kirchhausen T, Macia E. and Pelish HE. (2008). Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol 438:77–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant S. (1998). Ara-C: cellular and molecular pharmacology. Adv Cancer Res 72:197–233 [DOI] [PubMed] [Google Scholar]
- 42.Mastroyiannopoulos NP, Nicolaou P, Anayasa M, Uney JB. and Phylactou. LA. (2012). Down-regulation of myogenin can reverse terminal muscle cell differentiation. PLoS One 7:e29896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomczak KK, Marinescu VD, Ramoni MF, Sanoudou D, Montanaro F, Han M, Kunkel LM, Kohane IS. and Beggs AH. (2004). Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J 18:403–405 [DOI] [PubMed] [Google Scholar]
- 44.Sterrenburg E, Turk R, t Hoen PA, van Deutekom JC, Boer JM, van Ommen GJ. and den Dunnen JT. (2004). Large-scale gene expression analysis of human skeletal myoblast differentiation. Neuromuscul Disord 14:507–518 [DOI] [PubMed] [Google Scholar]
- 45.Lev AA, Feener CC, Kunkel LM. and Brown RH, Jr., (1987). Expression of the Duchenne's muscular dystrophy gene in cultured muscle cells. J Biol Chem 262:15817–15820 [PubMed] [Google Scholar]
- 46.Lin C. and Engbersen JF. (2008). Effect of chemical functionalities in poly(amido amine)s for non-viral gene transfection. J Control Release 132:267–272 [DOI] [PubMed] [Google Scholar]
- 47.Akinc A, Thomas M, Klibanov AM. and Langer R. (2005). Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med 7:657–663 [DOI] [PubMed] [Google Scholar]
- 48.Bennett CF, Chiang MY, Chan H, Shoemaker JE. and Mirabelli CK. (1992). Cationic lipids enhance cellular uptake and activity of phosphorothioate antisense oligonucleotides. Mol Pharmacol 41:1023–1033 [PubMed] [Google Scholar]
- 49.Ming X, Sato K. and Juliano RL. (2011). Unconventional internalization mechanisms underlying functional delivery of antisense oligonucleotides via cationic lipoplexes and polyplexes. J Control Release 153:83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goemans NM, Tulinius M, van den Akker JT, Burm BE, Ekhart PF, Heuvelmans N, Holling T, Janson AA, Platenburg GJ, et al. (2011). Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med 364:1513–1522 [DOI] [PubMed] [Google Scholar]
- 51.Tanganyika-de Winter CL, Heemskerk H, Karnaoukh TG, van Putten M, de Kimpe SJ, van Deutekom J. and Aartsma-Rus A. (2012). Long-term exon skipping studies with 2′-O-methyl phosphorothioate antisense oligonucleotides in dystrophic mouse models. Mol Ther Nucleic Acids 1:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dias N. and Stein CA. (2002). Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther 1:347–355 [PubMed] [Google Scholar]
- 53.Allen DG. and Whitehead NP. (2011). Duchenne muscular dystrophy—what causes the increased membrane permeability in skeletal muscle? Int J Biochem Cell Biol 43:290–294 [DOI] [PubMed] [Google Scholar]
- 54.Straub V, Rafael JA, Chamberlain JS. and Campbell KP. (1997). Animal models for muscular dystrophy show different patterns of sarcolemmal disruption. J Cell Biol 139:375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heemskerk H, de Winter C, van Kuik P, Heuvelmans N, Sabatelli P, Rimessi P, Braghetta P, van Ommen GJ, de Kimpe S, et al. (2010). Preclinical PK and PD studies on 2′-O-methyl-phosphorothioate RNA antisense oligonucleotides in the mdx mouse model. Mol Ther 18:1210–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzalez-Barriga A, Kranzen J, Croes HJ, Bijl S, van den Broek WJ, van Kessel ID, van Engelen BG, van Deutekom JC, Wieringa B, et al. (2015). Cell membrane integrity in myotonic dystrophy type 1: implications for therapy. PLoS One 10:e0121556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geary RS, Norris D, Yu R. and Bennett CF. (2015). Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 87:46–51 [DOI] [PubMed] [Google Scholar]
- 58.Mohun T. (1992). Muscle differentiation. Curr Opin Cell Biol 4:923–928 [DOI] [PubMed] [Google Scholar]
- 59.Mulders SA, van Horssen R, Gerrits L, Bennink MB, Pluk H, de Boer-van Huizen RT, Croes HJ, Wijers M, van de Loo FA, et al. (2011). Abnormal actomyosin assembly in proliferating and differentiating myoblasts upon expression of a cytosolic DMPK isoform. Biochim Biophys Acta 1813:867–877 [DOI] [PubMed] [Google Scholar]
- 60.Lehto T, Castillo Alvarez A, Gauck S, Gait MJ, Coursindel T, Wood MJ, Lebleu B. and Boisguerin P. (2014). Cellular trafficking determines the exon skipping activity of Pip6a-PMO in mdx skeletal and cardiac muscle cells. Nucleic Acids Res 42:3207–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ming X. (2011). Cellular delivery of siRNA and antisense oligonucleotides via receptor-mediated endocytosis. Expert Opin Drug Deliv 8:435–449 [DOI] [PubMed] [Google Scholar]
- 62.Zhao Q, Matson S, Herrera CJ, Fisher E, Yu H. and Krieg AM. (1993). Comparison of cellular binding and uptake of antisense phosphodiester, phosphorothioate, and mixed phosphorothioate and methylphosphonate oligonucleotides. Antisense Res Dev 3:53–66 [DOI] [PubMed] [Google Scholar]
- 63.Koller E, Vincent TM, Chappell A, De S, Manoharan M. and Bennett CF. (2011). Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res 39:4795–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wojtkowiak-Szlachcic A, Taylor K, Stepniak-Konieczna E, Sznajder LJ, Mykowska A, Sroka J, Thornton CA. and Sobczak K. (2015). Short antisense-locked nucleic acids (all-LNAs) correct alternative splicing abnormalities in myotonic dystrophy. Nucleic Acids Res 43:3318–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown DA, Kang SH, Gryaznov SM, DeDionisio L, Heidenreich O, Sullivan S, Xu X. and Nerenberg MI. (1994). Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J Biol Chem 269:26801–26805 [PubMed] [Google Scholar]
- 66.Mislick KA. and Baldeschwieler JD. (1996). Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci U S A 93:12349–12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang XH, Sun H, Shen W. and Crooke ST. (2015). Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res 43:2927–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moulton HM. and Moulton JD. (2010). Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim Biophys Acta 1798:2296–2303 [DOI] [PubMed] [Google Scholar]
- 69.Leonetti JP, Mechti N, Degols G, Gagnor C. and Lebleu B. (1991). Intracellular distribution of microinjected antisense oligonucleotides. Proc Natl Acad Sci U S A 88:2702–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fisher TL, Terhorst T, Cao X. and Wagner RW. (1993). Intracellular disposition and metabolism of fluorescently-labeled unmodified and modified oligonucleotides microinjected into mammalian cells. Nucleic Acids Res 21:3857–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lorenz P, Baker BF, Bennett CF. and Spector DL. (1998). Phosphorothioate antisense oligonucleotides induce the formation of nuclear bodies. Mol Biol Cell 9:1007–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis BM, McCurrach ME, Taneja KL, Singer RH. and Housman DE. (1997). Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci U S A 94:7388–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aartsma-Rus A. (2010). Antisense-mediated modulation of splicing: therapeutic implications for Duchenne muscular dystrophy. RNA Biol 7:453–461 [DOI] [PubMed] [Google Scholar]
- 74.Godbey WT, Wu KK. and Mikos AG. (1999). Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc Natl Acad Sci U S A 96:5177–5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.