Abstract
Anaerobic ammonium oxidation (anammox) is a recently discovered microbial pathway and a cost-effective way to remove ammonium from wastewater. Anammox bacteria have been described as obligate chemolithoautotrophs. However, many chemolithoautotrophs (i.e., nitrifiers) can use organic compounds as a supplementary carbon source. In this study, the effect of organic compounds on anammox bacteria was investigated. It was shown that alcohols inhibited anammox bacteria, while organic acids were converted by them. Methanol was the most potent inhibitor, leading to complete and irreversible loss of activity at concentrations as low as 0.5 mM. Of the organic acids acetate and propionate, propionate was consumed at a higher rate (0.8 nmol min−1 mg of protein−1) by Percoll-purified anammox cells. Glucose, formate, and alanine had no effect on the anammox process. It was shown that propionate was oxidized mainly to CO2, with nitrate and/or nitrite as the electron acceptor. The anammox bacteria carried out propionate oxidation simultaneously with anaerobic ammonium oxidation. In an anammox enrichment culture fed with propionate for 150 days, the relative amounts of anammox cells and denitrifiers did not change significantly over time, indicating that anammox bacteria could compete successfully with heterotrophic denitrifiers for propionate. In conclusion, this study shows that anammox bacteria have a more versatile metabolism than previously assumed.
Anaerobic ammonium oxidation (anammox) is a recently discovered microbial pathway in the biological nitrogen cycle (9, 27) and a new cost-effective way to remove ammonia from wastewater (3, 6, 15, 16, 21, 22 34, 35). Anammox is carried out by the planctomycetes Candidatus “Brocadia anammoxidans” and Candidatus “Kuenenia stuttgartiensis” and several species of the genus Candidatus “Scalindua” (2, 7, 8, 19, 20). Nitrite is the electron acceptor for the anaerobic oxidation of ammonia to dinitrogen gas, and hydrazine is an important intermediate (32). Anammox bacteria have been described as strictly autotrophic, fixing CO2 with nitrite as the electron donor, leading to the anaerobic production of nitrate (25, 33). The overall nitrogen balance showed a ratio of 1:1.32:0.26 for the conversion of ammonium and nitrite and the production of nitrate (26). The overall anammox reaction is presented in equation 1.
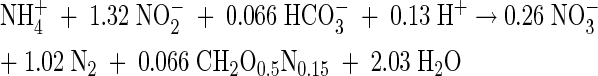 |
(1) |
Many other chemolithoautotrophs (i.e., nitrifiers) can grow mixotrophically; they can use organic compounds as a supplementory carbon source. This property is advantageous because mixotrophic growth can increase the growth rate and/or yield. In the case of anammox, this is especially advantageous, because both the growth rate (doubling time of 10 to 20 days) and yield (0.066 CO2 fixed per mol of NH4+) are very low. Previously, it was found that some organic compounds inhibit anammox (32). In this study, the effects of organic compounds on the anammox bacteria and the potential for mixotrophic growth were investigated in detail. In experiments with anammox enrichment cultures and Percoll-purified anammox cells, we show here that the anammox bacteria can use some organic compounds as substrates and are inhibited by others.
MATERIALS AND METHODS
Origin of biomass and growth conditions.
The anammox enrichment cultures were grown anaerobically at 30°C and pH 7.0 as described previously (26). The 15-liter reactor was gassed with 0.2 liter of an N2-CO2 (80/20%) gas mixture min−1 to maintain anoxic conditions and to supply CO2 for carbon fixation. The influx of fresh medium into the 15-liter reactor was adjusted to 85 ml h−1, and the same volume was removed from the reactor. The mineral medium used was described previously (32).
Continuous cultivation experiments with organic compounds.
The continuous culture experiments were performed in a 10-liter continuously stirred tank reactor with biofilm aggregates (consisting of ∼70 to 80% anammox cells [see below]) at a hydraulic retention time of 9 days (continuous exchange of medium, 46 ml h−1). The biomass aggregates were stirred with a two-blade mechanical stirrer (100 rpm). The biomass in the effluent was not recirculated. The reactor was operated at a fixed temperature of 30°C with a thermostated jacket. The pH was maintained at ∼7.0. The reactor (0.2 liter min−1) and the influent vessel (0.1 liter min−1) were gassed with an N2-CO2 (80/20%) gas mixture to maintain anaerobic conditions. In order to supply the system with sufficient electron acceptor for nitrate reduction, up to 5 mM sodium nitrate was added to the mineral medium (32). Methanol, sodium acetate, and sodium propionate were added to the reactor system from a 20 mM stock solution by a separate pump yielding final influent concentrations of 1, 2, and 4 mM, respectively. In a separate run, propionate was added from a 200 mM stock solution and gradually increased to a final concentration of 20 mM.
Batch experiments with organic compounds.
Anammox biomass from the 15-liter reactor grown before the addition of any organic compound was harvested by centrifugation (15 min at 4,000 × g) and washed twice with mineral medium (see above) without nitrogen compounds. For every batch experiment, 40 ml was taken from the 15-liter reactor. The biomass was mixed with 15 ml of mineral medium without ammonium and nitrite (the final protein concentration was 1 ± 0.2 mg ml−1), transferred to 50-ml serum bottles sealed with butyl rubber stoppers, and gassed for 15 min with the N2-CO2 (80/20%) gas mixture to remove the oxygen. To start the experiments, the substrates were added from an anoxic stock solution with a syringe. During the experiments, the cell suspensions were stirred or shaken (200 rpm) to keep the biomass aggregates in suspension. The bottles were incubated at 34°C. Gas and medium samples for further analysis were taken at appropriate time intervals.
15N-labeling studies.
Experiments with 15NH4+, 15NO2−, and 15NO3− were conducted in 10-ml serum bottles containing 5 ml of biomass aggregates (harvested as described above) and sealed with butyl rubber stoppers. After the bottles were gassed for 15 min with the N2-CO2 (80/20%) gas mixture, the experiments were started by adding the 15N substrates (final concentrations, 5 mM) from anoxic stock solutions with a syringe. During the experiments, the bottles were stirred at 150 rpm.
Percoll purification of anammox cells and microbatch experiments with purified anammox cells.
The anammox cells were purified by density gradient centrifugation as explained previously (25). Cell suspensions thus produced consisted of >99.5% target anammox cells. The purity of the cell suspensions was determined by fluorescence in situ hybridization (FISH) analysis with the appropriate oligonucleotide probes (see below).
For batch experiments, the purified cells were washed in HEPES (75 mM) supplemented with 5 mM bicarbonate buffer (pH 7.8). The cells were concentrated in the same buffer to a final concentration of 4 mg of protein ml−1. This cell suspension was transferred to a 0.6-ml Eppendorf cup, propionate and nitrate were added to a final concentration of 1.4 mM, and the open cup was placed in a 10-ml serum bottle with a butyl rubber stopper. The serum bottle was purged with a gas mixture of N2-CO2 (80/20%) to remove oxygen from the gas and liquid phases. The cell suspension was incubated for 150 min. The cells were separated from the liquid fraction by centrifugation, and the supernatant was analyzed for ammonium, nitrite, nitrate, and propionate.
Analytical procedures.
Medium samples were taken from the experiments and centrifuged to sediment the biomass, and the supernatant was stored at −20°C to await further analysis. Ammonium, nitrite, and nitrate were measured colorimetrically (29). The 15N analysis was performed by gas chromatography followed by N2 detection on an isotope ratio mass spectrometer (Delta plus; Thermo Finnigan, Breda, The Netherlands). The ratios of 28N2, 29N2, and 30N2 isotopes were measured. Production of [15N]ammonium was analyzed after conversion to N2 with hypobromite (17). The detection limit for all 15N compounds was ∼10 nM.
To test for incorporation of propionate into the biomass of anammox cells, [14C1]propionate (50 μCi/mmol) was added to a 50-ml anammox minireactor for 72 h in the presence of ammonium and nitrite. After the incubation, the biomass was harvested and washed five times to remove the radiolabel. An aliquot of the biomass was used for analysis by combined FISH and microautoradiography (10). [14C]bicarbonate was used as a positive control in these experiments. Propionate was measured on an HP 5890 gas chromatograph (230°C; innowax capillary column) equipped with a flame ionization detector. Carbon dioxide was measured on an HP6890 gas chromatograph (80°C; poropack Q in combination with a 5-Å molsieve) equipped with a thermal conductivity detector. The bacterial population was monitored by FISH and immunofluorescence analysis as described previously (13, 14, 20). The oligonucleotide probes applied were as follows: AMX368 [S-G-Amx-0368-a-A-22] covering all anammox organisms, PLA46 [S-P-Planc-0046-a-A-18] covering all Planctomycetes, EUB338 mix [S-D-Bact-0338-a-A-18] covering almost all Bacteria, ALFA968 [S-P-Alph-968-a-A-18] covering most Alphaproteobacteria, BET42a [L-P-Beta-1027-a-A-17] covering most Betaproteobacteria, and GAM42a [L-P-Gamm-1027-a-A-17] covering most Gammaproteobacteria (11). Several probes specific for certain denitrifiers (Zoogloea ramigera, Sphaerotilus natans, Paracoccus denitrificans, Alcaligenes faecalis, and the genus Azospirillum) are described in the Probe database (11). The antibodies against periplasmic nitrate reductase (NapA) of Escherichia coli were a generous gift of S. B. Mohan and J. A. Cole (Birmingham, United Kingdom). The antibody against membrane-bound nitrate reductase (NarGH) of Paracoccus pantotrophus was a generous gift of D. Richardson (Norwich, United Kingdom). The Cy3-labeled secondary antibodies were a sheep anti-rabbit antibody for NapA and a Cy3-labeled mouse anti-sheep antibody for NarGH.
RESULTS
Inhibition of anammox enrichment cultures by methanol and ethanol.
Anammox enrichment cultures were exposed to methanol and ethanol in batch (Table 1) and continuous experiments. Addition of methanol (final concentration, 0.5 mM) resulted in the immediate and complete inactivation of anammox activity. Ethanol inhibited the anammox reaction by 30% at 2 mM. When methanol (1 mM) was added to the anammox enrichment in continuous culture, the anammox reaction was immediately and irreversibly inhibited. Prolonged (1-week) flushing of the culture with medium without methanol did not restore anammox activity.
TABLE 1.
Effects of carbon sources on nitrite-reducing activity of anammox cells in batch experimentsa
| C compound | Relative nitrite-reducing activity of anammox enrichment culture (%) at carbon compound concn (mM) of:
|
|||
|---|---|---|---|---|
| 0.5 | 1 | 2 | 3 | |
| Methanol | 0 | 0 | 0 | NDc |
| Acetateb | 100 | 102 | 116 | 95 |
| Propionateb | 122 | 124 | ND | 108 |
| Ethanol | ND | 74 | 70 | ND |
| Glucose | ND | 98 | 90 | 86 |
| Formate | 98 | 89 | 72 | ND |
| Alanine | ND | 91 | 82 | ND |
The activity in the control experiment without carbon compounds (800 μmol NO2− g of protein−1 h−1) was set at 100%. An inhibitory effect of a carbon compound is indicated by values below 100%, and an enhancing effect is indicated by values above 100%. The results are mean values of eight replicate experiments. The standard deviation was <10%.
Propionate and acetate were always completely consumed.
ND, not determined.
Consumption of acetate and propionate by anammox enrichment cultures.
The anammox enrichment cultures consumed propionate and acetate in batch (Table 1) and continuous culture experiments (Fig. 1). For both, the consumption rates were 50 to 70% of the ammonium consumption rates.
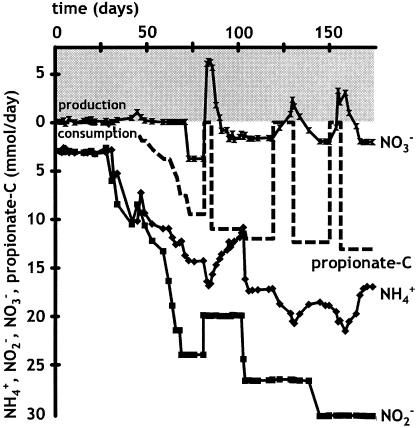
FIG. 1.
Effect of propionate on a continuous anammox enrichment culture. The consumption and/or production of ammonium (diamonds), nitrite (squares), nitrate (double triangles), and propionate (dashed line) are shown. The shaded area (above 0) represents production; the clear area (below 0) represents consumption.
Table 1 shows that acetate did not have a significant effect on the anammox stoichiometry and that propionate led to an increase in nitrite consumption relative to ammonium and to a decrease in nitrate production (Table 1 and Fig. 1). Gas chromatography analysis of the CO2 concentration in the headspace of the batch experiments showed that propionate oxidation led to CO2 production. CO2 production accounted for 50 to 70% of the consumed propionate. These results indicated that propionate was oxidized mainly to CO2, with nitrate and/or nitrite as the electron acceptor. This was confirmed in the continuous culture experiments. In these experiments, propionate addition (0.1 mM day−1, relative to 40 mM day−1 for NH4+) was started on day 19 (Fig. 1). The biomass consumed propionate at a rate of 0.2 nmol min−1 mg of protein−1 without a lag phase.
The propionate concentration in the influent of the reactor was gradually increased to 4 mM. At this still-limiting concentration, the rate of propionate consumption was ∼1 nmol min−1 mg of protein−1. No propionate was ever detected in the effluent of the reactor. During three periods, propionate addition was stopped (days 80 to 83, 118 to 128, and 149 to 154). In the first 19 days before addition and in the periods without propionate, the stoichiometry of ammonium and nitrite consumption and nitrate production was close to the predicted ratio for anammox (1:1.3:0.3). When propionate was present in the influent, the net nitrate production changed to net nitrate consumption (Fig. 1), and the stoichiometry of nitrite and ammonium consumption increased from 1.3 to 1.6. Because the culture was nitrite limited, the addition of propionate eventually led to a lower ammonium consumption rate, indicating that ammonium and propionate were competing electron donors for the electron acceptor nitrite.
Addition of glucose, formate, or alanine did not have pronounced effects on the anammox activity in batch tests (Table 1).
Identification of the bacteria responsible for propionate consumption.
Since the experiments described above were carried out with enrichment cultures (70 to 80% anammox cells, as determined by FISH with probes AMX368 and EUB338), it was still unclear what bacteria were responsible for propionate consumption. To investigate whether heterotrophic denitrifiers were involved, 0.5 g of penicillin G per liter was added directly to the reactor (days 70, 82, 110, 138, and 166). Penicillin G does not inhibit the activity of the presumably peptidoglycan-lacking anammox bacteria (32). Penicillin G had no effect on propionate consumption in the reactor, indicating that the anammox bacteria themselves might be responsible.
This was confirmed by monitoring population shifts of the enrichment culture during the 200-day duration of the continuous experiment. The population composition was determined by FISH analysis (probes Pla46 and AMX368) (Fig. 2 shows a typical result) once every 7 to 14 days. FISH analysis showed that the degree of anammox enrichment did not change significantly over time. Anammox bacteria dominated the community at ∼70 to 80% throughout the experiment. FISH analysis with probes targeting Alpha-, Beta-, and Gammaproteobacteria showed that there was no significant increase in these populations. In addition, several probes specific for certain denitrifiers (Z. ramigera, S. natans, P. denitrificans, A. faecalis, and the genus Azospirillum) did not give detectable hybridization signals (results not shown). Furthermore, immunofluorescence with antibodies raised against the periplasmic nitrate reductase NapA and the membrane-bound nitrate reductase NarGH showed that the numbers of NapA- and NarGH-positive cells (1 to 3% of the cells) did not change over time. Only when the propionate supply was increased to 20 mmol of C/day (reactor data not shown) was a gradual increase in NapA-positive cells observed (up to 20 to 30% of the cells). These cells also hybridized with the Betaproteobacteria probe. Apparently, only when the propionate supply (20 mmol of C/day) was higher than the ammonium supply (15 mmol/day) could heterotrophic denitrifiers outcompete anammox bacteria for propionate.
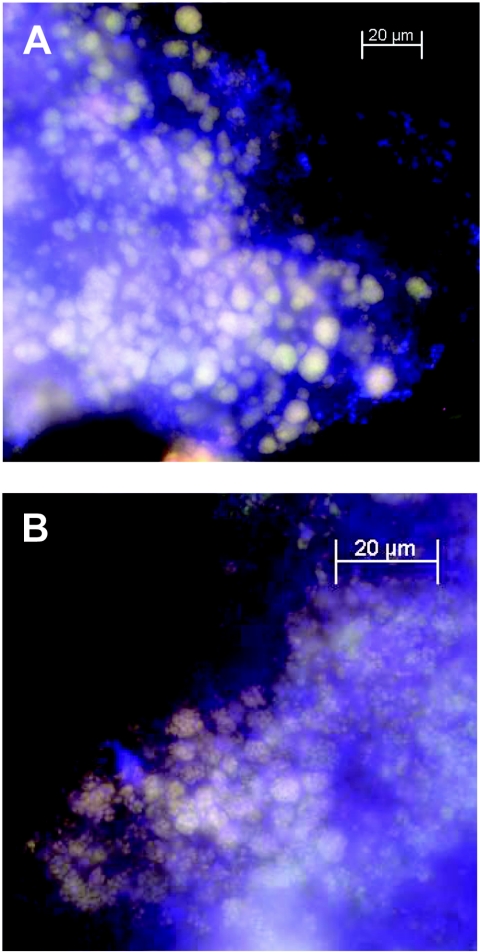
FIG. 2.
FISH analysis of anammox biomass from the chemostat reactor before and after the addition of propionate. (A) Biomass before the addition of propionate using EUB338 mix (blue), Pla46 (green), and AMX368 (red). (B) Biomass grown in the presence of 4 mM propionate for 35 days using EUB338 mix (blue), Pla46 (green), and AMX368 (red).
Finally, the capability of anammox bacteria to convert propionate was demonstrated directly by purifying the anammox cells from the enrichment culture by Percoll density gradient centrifugation. It appeared that a 99.5% pure cell suspension of anammox bacteria consumed propionate, with nitrate as the electron acceptor, at a rate of 0.8 nmol min−1 mg of protein−1, sufficient to explain propionate conversion in the continuous enrichment culture experiment.
Mechanism of propionate oxidation and nitrate reduction.
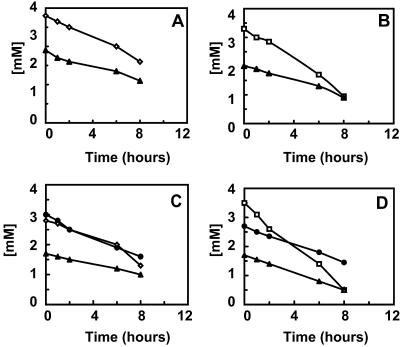
The mechanism of propionate oxidation was investigated in batch experiments with anammox enrichment cultures (Fig. 3). Propionate oxidation occurred in the presence of nitrate or nitrite (Fig. 3A and B). In the absence of nitrite and nitrate, no propionate was consumed (results not shown). The presence of ammonium was not required for propionate oxidation, indicating that anaerobic ammonium oxidation and propionate oxidation are two independent processes. When nitrate was present as the only electron acceptor, a transient accumulation of nitrite was sometimes observed. This indicated that nitrite is a free intermediate of nitrate reduction and that the reduction of nitrite was the rate-limiting step. As mentioned above, propionate was oxidized mainly to CO2. The ratio of nitrate reduction to propionate oxidation (1.6 ± 0.2) was in good agreement with the partial (±50%) oxidation of propionate to CO2. Also the observed stoichiometry of nitrite reduction and propionate oxidation (2.4 ± 0.4) agreed well with partial (±50%) oxidation of propionate. The highest rate of propionate oxidation measured in the batch tests was 2 nmol min−1 mg of protein−1.
FIG. 3.
Propionate conversion in batch tests by anammox biomass. (A) Conversion of propionate (▴) in the presence of nitrate (⋄). (B) Conversion of propionate (▴) in the presence of nitrite (□). (C) Conversion of propionate (▴) in the presence of nitrate (⋄) and ammonia (•). (D) Conversion of propionate (▴) in the presence of nitrite (□) and ammonia (•).
Incorporation of (part of) the propionate into cell material was investigated in biomass incubations with [14C]propionate. Less than 10% of the label was recovered in the biomass (compared to >50% for the [14C]bicarbonate control experiment), as determined by scintillation counting. The biomass was used for FISH-microautoradiography to determine which microorganism had incorporated the propionate. Unfortunately, the amount of label incorporation was too small to be detected via combined FISH-microautoradiography, again consistent with CO2 being the main product of propionate oxidation.
The end product of nitrite reduction was investigated by supplying [15N]nitrate and [15N]nitrite and measuring the 15N-labeling pattern of N2. When [15N]nitrate was added together with [14N]ammonium and propionate, most of the label was recovered as 14,15N2, indicating that the intermediate nitrite combined one-to-one with the ammonium present in the test. When a large amount of [14N]nitrite was present in those tests, hardly any labeled N2 was formed. This indicated that the [15N]nitrite produced from the supplied [15N]nitrate was diluted in the unlabeled nitrite pool and subsequently combined with N2. In the experiments where [15N]nitrate or [15N]nitrite was incubated with only propionate, both 14,15N2 and 15,15N2 could be detected, although the ratio varied from test to test. Thus, the experiment clearly showed that N2 was the end product of nitrate reduction.
DISCUSSION
The present study describes the effects of organic compounds on anaerobic ammonium-oxidizing bacteria. The topic is relevant to the microbiological investigation of these bacteria and their application in wastewater treatment. Anammox bacteria have a low growth rate and yield and are difficult to cultivate, and their isolation in pure culture has been unsuccessful. Knowledge of the potential for mixotrophic growth might facilitate cultivation and microbiological study in the future (29). In wastewater treatment, it is important to know the responses of anammox bacteria to the presence of organic compounds because the target wastewaters sometimes contain organic acids and alcohols resulting from incomplete or suboptimal fermentation (1, 3, 15, 16, 23, 34). The present study has documented that organic compounds can both inhibit and be converted by anammox bacteria.
The results clearly show that the exposure of anammox bacteria to alcohols, methanol in particular, should be prevented under all circumstances. Even low concentrations of methanol lead to the immediate, complete, and irreversible inhibition of the anammox process. In practice, this would mean that an anammox reactor would have to be restarted from scratch, which takes at least 100 to 200 days if no anammox seed is available (1, 15, 16, 34). This observation is highly relevant, because methanol is often used to remove nitrate in postdenitrification or to compensate for pH effects in partial nitrification (4, 5). A possible explanation for methanol inhibition is the potential conversion of methanol to formaldehyde by the anammox enzyme hydroxylamine oxidoreductase (18). Formaldehyde destroys enzyme activity by irreversibly cross-linking the peptide chains (12).
Propionate and potentially acetate were shown to be substrates for anammox bacteria. Anammox bacteria oxidized propionate to CO2 with nitrate and/or nitrite as the electron acceptor. In future studies, addition of [13C]acetate or [14C]acetate to prolonged reactor experiments must be considered in order to document the fate of acetate (24). The present study focused on propionate.
Propionate was consumed in continuous and batch incubations of anammox enrichment cultures. Theoretically, propionate must have been consumed by the anammox bacteria themselves or by another community member. To address this issue, the population composition of the enrichment culture fed with propionate, ammonium, nitrite, and nitrate was monitored for 150 days. Initially, the population consisted of 80% anammox bacteria. Based on the high biomass yield of propionate-oxidizing bacteria (1 C-mol of biomass/mol of propionate) (31) and the low biomass yield of anammox bactera (0.066 C-mol of biomass/mol of ammonium), we would have expected the degree of anammox enrichment to drop to 20% if another community member had consumed the propionate. Since the degree of anammox enrichment remained constant at 80% and denitrifiers (detected with the antibody NarA) made up only 2% of the population, it seemed that the anammox bacteria themselves were responsible for propionate consumption. As mentioned in Results, a population shift was observed only when the propionate concentration in the influent was increased from 12 to 20 mmol of propionate-C. In that case, denitrifying betaproteobacteria gradually became important community members (20 to 30% of the total population). This observation showed that propionate-consuming denitrifiers were present in the culture, but they apparently could not compete successfully for propionate with anammox bacteria at lower propionate concentrations in the feed. It is likely that the ratio of ammonium to propionate in the feed determined the competitiveness of anammox bacteria for propionate. When this ratio was above a certain threshold (propionate-C/ammonium ratio, 0.75 to 1.25) the anammox bacteria could no longer compete. The capacity of anammox bacteria for propionate consumption was demonstrated directly by the propionate consumption of 99.5% enriched suspensions of anammox cells at a rate sufficient to explain all propionate consumption in the continuous culture experiment (0.8 nmol/mg of protein/min).
The combination of CO2 measurements, 15N-labeling studies, stoichiometries of propionate and nitrite-nitrate consumption, and FISH-microautoradiography showed that CO2 and N2 were the main end products of propionate oxidation coupled to nitrite and/or nitrate reduction by anammox bacteria. At least 50% of the propionate consumed was recovered as CO2, and <10% of the propionate carbon was incorporated into cell biomass. Since not all carbon could be accounted for, it is possible that still more propionate was incorporated. In view of the autotrophic lifestyle of anammox bacteria, assimilation into the biomass (mixotrophic growth) and not oxidation to CO2 was expected.
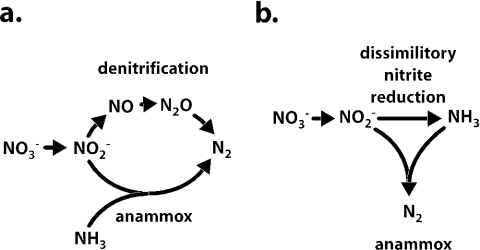
The labeling experiments clearly showed that nitrite was the intermediate of nitrate reduction. This is the first time that anammox bacteria have been shown to be capable of the reduction of nitrate to nitrite. In the absence of propionate, anammox bacteria produce nitrate from nitrite (equation 1), so apparently the reaction is reversible. The biochemical mechanism of nitrite reduction remains unclear. In principle, anammox bacteria could either make use of the denitrification pathway to produce dinitrogen gas directly or first reduce nitrite to ammonium and subsequently oxidize ammonium in the anammox reaction (Fig. 4). Future studies will concentrate on the mechanism of nitrite reduction (14) and propionate oxidation via biochemical assays, as has been described for other bacteria (24, 30).
FIG. 4.
Schematic model describing the combination of anammox, denitrification, and dissimilatory nitrate reduction to ammonia by anammox cells based on equation 1.
The direct use of nitrate as an electron acceptor is interesting for wastewater treatment and for the ecology of anammox bacteria. Nitrate is generally more abundant than nitrite, and until now, anammox bacteria have been believed to be dependent on other bacteria to reduce nitrate to nitrite. The present study showed that if a suitable electron donor (such as propionate) is available, anammox bacteria may also use nitrate directly.
Future work will focus on the question of whether feeding propionate as a supplement to anammox enrichment cultures would increase the growth rate or yield or be of use in the isolation of anammox bacteria in pure culture.
Acknowledgments
Research on anaerobic ammonium oxidation is supported by the Foundation for Applied Sciences (STW grant NPC 5987), the European Union (grant EESD EVK1-CT-2000-00054), the Foundation of Applied Water Research (STOWA), The Netherlands Foundation for Earth and Life Sciences (NWO-ALW, Biogeosphere grants 853.00.012 and 853.00.031), and Paques BV. A fellowship from the Scientific and Technical Research Council of Turkey was granted to Didem Güven, and a Spanish exchange grant supported the visit of Ana Dapena.
REFERENCES
- 1.Dapena-Mora, A., J. L. Campos, A. Mosquera-Corral, M. S. M. Jetten, and R. Méndez. 2004. Stability of the ANAMMOX process in a gas-lift reactor and a SBR. J. Biotechnol. 110:159-170. [DOI] [PubMed] [Google Scholar]
- 2.Egli, K., U. Franger, P. J. J. Alvarez, H. R. Siegrist, J. R. VanderMeer, and A. J. B. Zehnder. 2001. Enrichment and characterization of an anammox bacterium from a rotating biological contractor treating ammonium-rich leachate. Arch. Microbiol. 175:198-207. [DOI] [PubMed] [Google Scholar]
- 3.Fux, C., M. Boehler, P. Huber, I. Brunner, and H. R. Siegrist. 2002. Biological treatment of ammonium-rich wastewater by partial nitritation and subsequent anaerobic ammonium oxidation (anammox) in a pilot plant. J. Biotechnol. 99:295-306. [DOI] [PubMed] [Google Scholar]
- 4.Hao, X. D., and M. C. M. van Loosdrecht. 2004. Model-based evaluation of the influence of COD on a partial nitrification-anammox biofilm process. Water Sci. Technol. 49:83-90. [PubMed] [Google Scholar]
- 5.Hellinga, C., A. A. J. C. Schellen, J. W. Mulder, M. C. M. van Loosdrecht, and J. J. Heijnen. 1998. The SHARON process: an innovative method for nitrogen removal from ammonium-rich waste water. Water Sci. Techol. 37:135-142. [Google Scholar]
- 6.Jetten, M. S. M., M. C. Schmid, I. Schmidt, et al. 2002. Improved nitrogen removal by application of new nitrogen-cycle bacteria. Rev. Environ. Sci. Biotechnol. 1:51-63. [Google Scholar]
- 7.Jetten, M. S. M., A. O. Sliekers, M. M. M. Kuypers, et al. 2003. Anaerobic ammonium oxidation by marine and freshwater planctomycete-like bacteria. Appl. Microbiol. Biotechnol. 63:107-114. [DOI] [PubMed] [Google Scholar]
- 8.Kuenen, J. G., and M. S. M. Jetten. 2001. Extraordinary anaerobic ammonium oxidising bacteria. ASM News 67:456-463. [Google Scholar]
- 9.Kuypers, M. M. M., A. O. Sliekers, G. Lavik, M. Schmid, B. B. Jørgensen, J. G. Kuenen, J. S. Sinninghe Damste, M. Strous, and M. S. M. Jetten. 2003. Anaerobic ammonium oxidation by Anammox bacteria in the Black Sea. Nature 422:608-611. [DOI] [PubMed] [Google Scholar]
- 10.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loy, L. A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metz, B., G. F. A. Kersten, P. Hoogerhout, H. F. Brugghe, H. A. M. Timmermans, A. de Jong, H. Meiring, J. ten Hove, W. E. Hennink, D. J. A. Crommelin, and W. Jiskoot. 2004. Identification of formaldehyde-induced modifications of proteins. J. Biol. Chem. 279:6235-6243. [DOI] [PubMed] [Google Scholar]
- 13.Metz, S., W. Beisker, A. Hartmann, and M. Schloter. 2003. Detection methods for the expression of the dissimilatory copper-containing nitrite reductase gene (DnirK) in environmental samples. J. Microbiol. Methods 55:41-50. [DOI] [PubMed] [Google Scholar]
- 14.Mohan, S. B., M. C. Schmid, M. S. M. Jetten, and J. A. Cole. 2004.. Detection and widespread distribution of the nrfA gene encoding nitrite reduction to ammonia, a short circuit in the Biological Nitrogen Cycle that competes with denitrification. FEMS Microbiol. Ecol. 49:433-443. [DOI] [PubMed]
- 15.Pynaert, K., B. F. Smets, S. Wyffels, D. Beheydt, S. D. Siciliano, and W. Verstraete. 2003. Characterization of an autotrophic nitrogen-removing biofilm from a highly loaded lab-scale rotating biological contactor. Appl. Environ. Microbiol. 69:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pynaert, K., B. F. Smets, D. Beheydt, and W. Verstraete. 2004. Start-up of autotrophic nitrogen removal reactors via sequential biocatalyst addition. Environ. Sci. Technol. 38:1228-1235. [DOI] [PubMed] [Google Scholar]
- 17.Risgaard-Petersen, N., S. Rysgaard, and N. P. Revsbech. 1995. A combined microdiffusion-hypobromite oxidation method for determination of 15N isotope in NH4+. Soil Sci. Soc. Am. J. 59:1077-1080. [Google Scholar]
- 18.Schalk, J., S. DeVries, J. G. Kuenen, and M. S. M. Jetten. 2000. Involvement of a novel hydroxylamine oxidoreductase in anaerobic ammonium oxidation. Biochemistry 39:5405-5412. [DOI] [PubMed] [Google Scholar]
- 19.Schmid, M. C., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. S. M. Jetten, J. Metzger, K.-H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 20.Schmid, M., K. Walsh, R. Webb, W. I. C. Rijpstra, K.T. van de Pas-Schoonen, M. J. Verbruggen, T. Hill, B. Moffett, J. Fuerst, S. Schouten, J. S. Sinninge Damste, J. Harris, P. Shaw, M. S. M. Jetten, and M. Strous. 2003. Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst Appl. Microbiol. 26:529-538. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt, I., C. Hermelink, K. T. Van de Pas-Schoonen, M. Strous, H. J. M. Op den Camp, J. G. Kuenen, and M. S. M. Jetten. 2002. Anaerobic ammonia oxidation in the presence of nitrogen oxides (NOx) by two different lithotrophs. Appl. Environ. Microbiol. 68:5351-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt, I., A. O. Sliekers, M. Schmid, E. Bock, J. Fuerst, J. G. Kuenen, M. S. M. Jetten, and M. Strous. 2003. New concepts of microbial treatment processes for the nitrogen removal in wastewater. FEMS Microbiol. Rev. 27:481-492. [DOI] [PubMed] [Google Scholar]
- 23.Siegrist, H., D. Vogt, J. L. Garcia-Heras, and W. Gujer. 2002. Mathematical model for meso- and thermophilic anaerobic sewage sludge digestion. Environ. Sci. Technol. 36:1113-1123. [DOI] [PubMed] [Google Scholar]
- 24.Stams, A. J. M., C. Dijkema, C. M. Plugge, and P. Lens. 1998. Contribution of C-13 NMR spectroscopy to the elucidation of pathways of propionate formation and degradation in methanogenic environments. Biodegradation 9:463-473. [Google Scholar]
- 25.Strous, M., J. Fuerst, E. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. I. Webb, J. G. Kuenen, and M. S. M. Jetten. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 26.Strous, M., J. J. Heijnen, J. G. Kuenen, and M. S. M. Jetten. 1998. The sequencing batch reactor as a powerful tool to study very slowly growing micro-organisms. Appl. Microbiol. Biotechnol. 50:589-596. [Google Scholar]
- 27.Strous, M., and M. S. M. Jetten. 2004. The anaerobic oxidation of methane and ammonium. Annu. Rev. Microbiol. 58:99-117. [DOI] [PubMed] [Google Scholar]
- 28.Strous, M., J. G. Kuenen, J. A. Fuerst, M. Wagner, and M. S. M. Jetten. 2002. The anammox case—a new experimental manifesto for microbiological eco-physiology. Antonie Leeuwenhoek Int. J. Microbiol. 81:693-702. [DOI] [PubMed] [Google Scholar]
- 29.Strous, M., J. G. Kuenen, and M. S. M. Jetten. 1999. Key physiology of anaerobic ammonium oxidation. Appl. Environ. Microbiol. 65:3248-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Textor, S., V. F. Wendisch, A. A. De Graaf, U. Muller, M. I. Linder, D. Linder, and W. Buckel. 1997. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 168:428-436. [DOI] [PubMed] [Google Scholar]
- 31.Tijhuis, L., M. C. M. van Loosdrecht, and J. J. Heijnen. 1993. A thermodynamically based correlation for maintenance Gibbs energy requirements in aerobic and anaerobic chemotrophic growth. Biotechnol. Bioeng. 42:509-519. [DOI] [PubMed] [Google Scholar]
- 32.Van de Graaf, A. A., P. De Bruijn, L. A. Robertson, M. S. M. Jetten, and J. G. Kuenen. 1996. Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiology 142:2187-2196. [Google Scholar]
- 33.Van de Graaf, A. A., P. De Bruijn, L. A. Robertson, M. S. M. Jetten, and J. G. Kuenen. 1997. Metabolic pathway of anaerobic ammonium oxidation on the basis of N-15 studies in a fluidized bed reactor. Microbiology 143:2415-2421. [DOI] [PubMed] [Google Scholar]
- 34.Van Dongen, U., M. S. M. Jetten, and M. C. M. van Loosdrecht. 2001. The Sharon-anammox process for the treatment of ammonium rich wastewater. Water Sci. Technol. 44:153-160. [PubMed] [Google Scholar]
- 35.Van Loosdrecht, M. C. M., X. D. Hao, M. S. M. Jetten, and W. Abma. 2004. Use of Anammox in urban wastewater treatment. Water Sci. Technol. 4:87-94. [Google Scholar]