Abstract
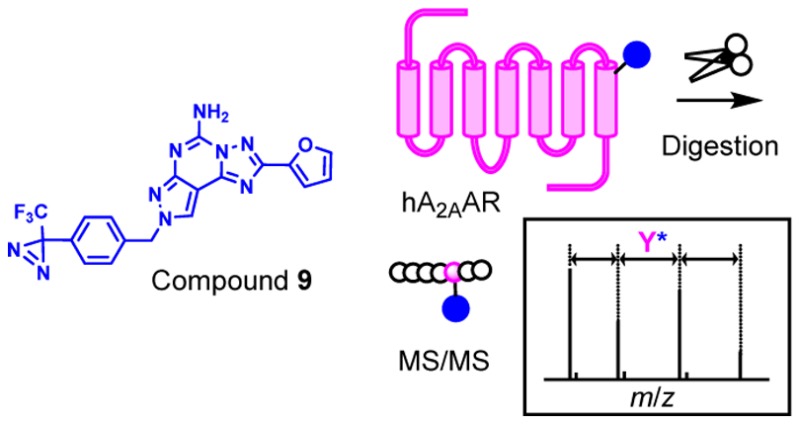
Photoaffinity labeling (PAL) is widely used for the identification of ligand-binding proteins and elucidation of ligand-binding sites. PAL has also been employed for the characterization of G protein-coupled receptors (GPCRs); however, a limited number of reports has successfully identified their cross-linked amino acids. This report is the first of its kind to determine the cross-link position of the human A2A adenosine receptor by PAL with the novel diazirine-based photoaffinity probe 9.
Keywords: Photoaffinity labeling, Mass spectrometry, A2A adenosine receptor, GPCR
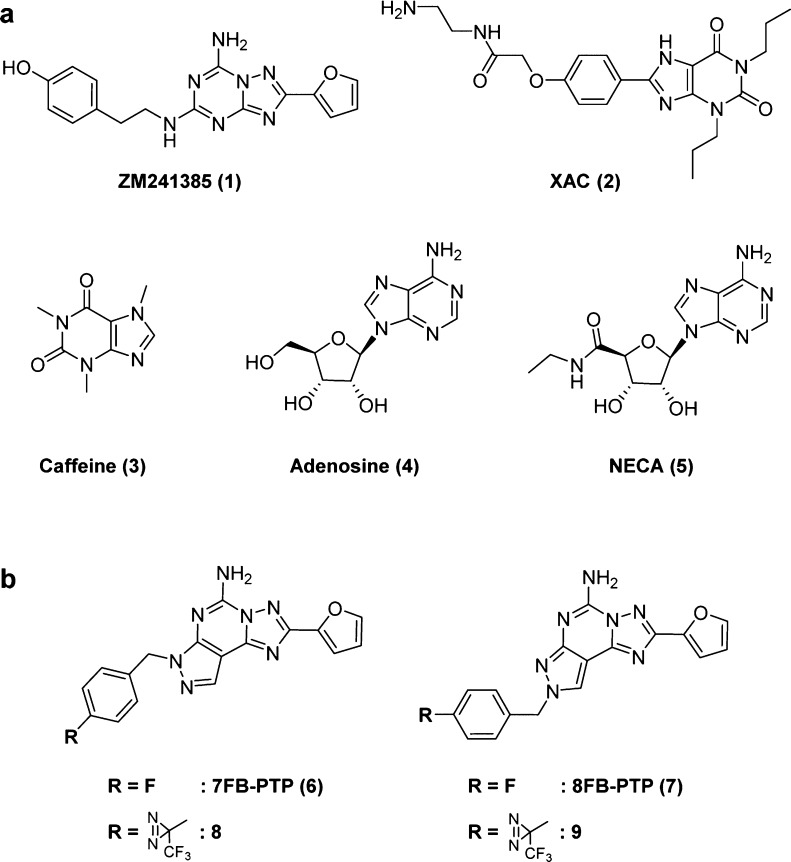
The G protein-coupled receptor (GPCR) superfamily comprises the largest groups of membrane-bound receptors. It has been estimated that GPCRs make up more than 30% of marketed drug targets.1 GPCRs share a common seven-transmembrane (TM) topology and are involved in cellular responses to a variety of extracellular signals, ranging from light, ions, odor, taste, neurotransmitters, lipids, peptides, and hormones.2 GPCRs are classified into five main classes: rhodopsin (class A), secretin (class B), glutamate (class C), adhesion, and frizzled (class F) receptors,3 in which class A contains the largest number of receptors (approximately 200 nonsensory receptors with known ligands).4 The human adenosine receptors are a member of the class A GPCRs and four different subtypes (A1, A2A, A2B, and A3) exist,5 among which A2A adenosine receptor (A2AAR) antagonism has been suggested to modulate CNS stimulatory effects by high expression in the striatum. Various A2AAR antagonists have been developed for the treatment of Parkinson’s disease and other neurodegenerative conditions.6 Additional promising applications are currently being explored, including those for Alzheimer’s disease, attention deficit hyperactivity disease, and more recently cancer immunotherapy.7,8 Thus, ARs in particular constitute attractive targets in recent drug discovery.9
The first X-ray structure of A2AAR with the high-affinity antagonist ZM241385 (1) was published by Jaakola et al.10 in 2008 to reveal the inactive states of the receptor. Since then, over 10 crystal structures of A2AAR not only with antagonists such as XAC (2) and caffeine (3), but also with agonists including adenosine (4) and NECA (5) have been published by different groups (Chart 1a).11 Owing to recent technological improvements in membrane protein crystallography, nearly 30 distinct structures of GPCRs have been characterized to date from each of the major classes A, B, C, and F, which have significantly contributed to the understanding of drug action and novel compound design in drug discovery.12−14 Despite the number of GPCR structures growing quite rapidly, it still represents a small proportion of the known GPCRs (more than 800 including taste and olfactory receptors).3 An alternative approach to structural characterization is photoaffinity labeling (PAL), which forms an irreversible covalent bond between photoreactive ligands and neighboring amino acids under the irradiation of light. PAL has proven to be a powerful tool for the identification of target proteins and the investigation of protein–protein interactions and ligand-binding sites.15,16 PAL has also been applied in the characterization of membrane-bound proteins, including GPCRs.17 Numerous reports have successfully utilized PAL with radiolabeled photoaffinity analogues to confirm the localization of GPCRs by autoradiography, and identify the cross-link sites by Edman degradation in some case. In the earlier work on PAL, Piersen et al.18 performed PAL of the canine A2AAR overexpressed in COS M6 cells with 125I-azido-PAPA-APEC and tracked the cross-link transmembrane span, but did not reveal individual amino acids. More recently, Moss et al.19 reported the site-specific chemical modification of the human A2AAR with chemically reactive agonists to covalently modify the receptor, although they did not perform PAL.
Chart 1. (a) Examples of A2AAR Ligands and (b) Photoaffinity Probe Design.
The combined approach of PAL and mass spectrometry is increasingly becoming popular for analyzing photolabeled proteins and identification of cross-link amino acids.20 However, these applications have been mostly for soluble proteins and the studies identifying cross-linked amino acids on GPCRs by mass spectrometry are scarce. This is possibly due to the low expression level of receptors, the difficulty in devising active membrane protein preparations, and the requirement of further analysis steps, such as protease digestion, following MS/MS sequencing of relatively hydrophobic peptides. Indeed, Rosa et al.21 reported in 2015 that only six GPCRs had their primary protein sequences completely determined (i.e., over 80% sequence coverage) using proteomic analyses.
For the structural study, a high level of recombinant GPCR production was achieved in a range of expression systems that included bacteria, yeast, insect, and mammalian cells.22 Fraser23 described the expression and functional purification of a glycosylation-deficient version of hA2AAR in Pichia pastoris having native-like pharmacological properties and demonstrated that the receptor retained its function during the solubilization and purification process. Thus, provided that they maintain native-like characteristics, treatment of receptors in a solution state facilitates the monitoring of the photo-cross-link reaction as well as the identification of the cross-link sites by mass spectrometry. This has prompted us to conduct PAL using solubilized hA2AAR as a model GPCR for the identification of cross-link amino acids. If successful, this strategy may be applicable for elucidating the ligand–receptor interaction of other GPCRs even if they are allosteric. In this letter, we present the findings of a PAL study on hA2AAR using a novel diazirine-based photoaffinity probe and cross-link position analysis by mass spectrometry.
Various types of photoreactive groups that show high reactivity under UV light irradiation have been reported, including arylazides, benzophenones, and diazirines. Among them, diazirines are the smallest photophor that generate a reactive carbene upon irradiation with light.24 Diazirines are also the preferred photophor for minimizing structural changes from parent molecules. In our initial hA2AAR labeling trials, we adopted the trifluoromethylphenyl diazirine (TPD) group as a photophor and the pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidine (PTP) nucleus as a template.25 7FB-PTP (6) and 8FB-PTP (7) are one of the first reported compounds having a PTP nucleus (Chart 1b). 8FB-PTP (7) is known as a highly potent antagonist (Ki rA2AAR = 1.2 nM), whereas 7FB-PTP (6) showed a decreased affinity (Ki rA2AAR = 12 nM). We designed two novel photoaffinity probes, 8 and 9, by substituting the fluorine atom on the phenyl ring at the para position of 7FB-PTP (6) and 8FB-PTP (7) for a TPD group (Chart 1b). The synthetic route is depicted in Scheme 1. The reaction of commercially available benzyl bromide 10 with 5-amino-2-(2-furyl)pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidine (11)26 in the presence of anhydrous potassium carbonate produced a mixture of N7 isomers, 8, and N8 isomers, 9, that were separable by column chromatography.
Scheme 1. Photoaffinity Probe Synthesis.
Reagents and conditions: (a) K2CO3, DMF, r.t.; 39.6% for compound 8, 19.4% for compound 9.
The hA2AAR was expressed in a Pichia pastoris system with a 10× His-tag at the C-terminal end. The sedimented membrane fractions were solubilized by overnight incubation at 4 °C in the buffer containing 16 mM Tris-HCl, pH 7.4, 120 mM NaCl, 2.0% n-dodecyl-β-d-maltoside (DDM), and 0.4% cholesteryl hemisuccinate (CHS). The receptor was purified by a cobalt-based affinity resin (TALON resin, Clontech, Palo Alto, CA, USA) and subsequently adopted in LC–MS analyses and PAL studies.
The affinity of compounds 8 and 9 for the solubilized hA2AAR was determined by fluorescence polarization (FP). Saturation binding experiments were performed using the fluorescent tracer MRS5346 (12) reported by Kecskés et al.27,28 with some modification (Figure 1a). FP signals were measured with an Infinite M1000 plate reader (TECAN, Morrisville, NC, USA) at an excitation wavelength of 470 nm and emission wavelength of 520 nm. Millipolarization (mP) values using four different concentrations of tracer 12 (8, 4, 2, and 1 nM) in the assay buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.05% DDM, 0.01% CHS) were recorded. Data analysis was performed to determine the receptor concentrations at which 50% of the tracer 12 was bound (Figure 1b).29 Binding specificity was confirmed in the presence of 10 μM of the antagonist SCH442416 (13).30 Apparent Kd values were determined by nonlinear curve fitting using PRISM (version 4.03, GraphPad Software, La Jolla, CA, USA). Based on these, we estimated that less than 2 nM (Kd = 30.8 ± 0.5 nM) of the tracer 12 approached the true Kd value for the solubilized receptor, which was similar to findings reported using membrane preparations from HEK293 cells (Kd = 16.5 ± 4.7 nM).27 Taking the S/B ratio into account, 2 nM of the tracer 12 and 25 nM of hA2AAR were used in ensuing competitive binding assays. FP competitive binding experiments were performed under the optimized conditions to determine the affinity of compounds 8 and 9 together with several adenosine ligands, including both antagonists 1, 2, and an agonist 5. The concentration of compounds required to displace 50% of the tracer 12 was elucidated (IC50). As illustrated in Figure 1c, the rank order of potency was as follows: ZM241385 (1) > compound 9 ⩾ XAC (2) > NECA (5) > compound 8. The Ki values were calculated using the Cheng–Prusoff equation, and the known ligands showed similar Ki values to those of the binding experiments with [3H]-ZM241385 using membrane preparations of hA2AAR expressed in Pichia pastoris reported by Fraser (see Supporting Information 1). The N8 isomers 9 had a high affinity (Ki = 39.7 ± 0.03 nM) comparable to that of XAC (2) (Ki = 50.1 ± 0.03 nM). The N7 isomers 8, meanwhile, showed a more than 30-fold lower affinity (Ki = 1520 ± 0.06 nM) relative to N8 isomers 9. The observed SAR was the same as that of the parent molecules; that is, 8FB-PTP (7) was a more potent antagonist than 7FB-PTP (6). As a result of the FP competitive binding experiments, we selected compound 9 for further photolabeling experiments.
Figure 1.
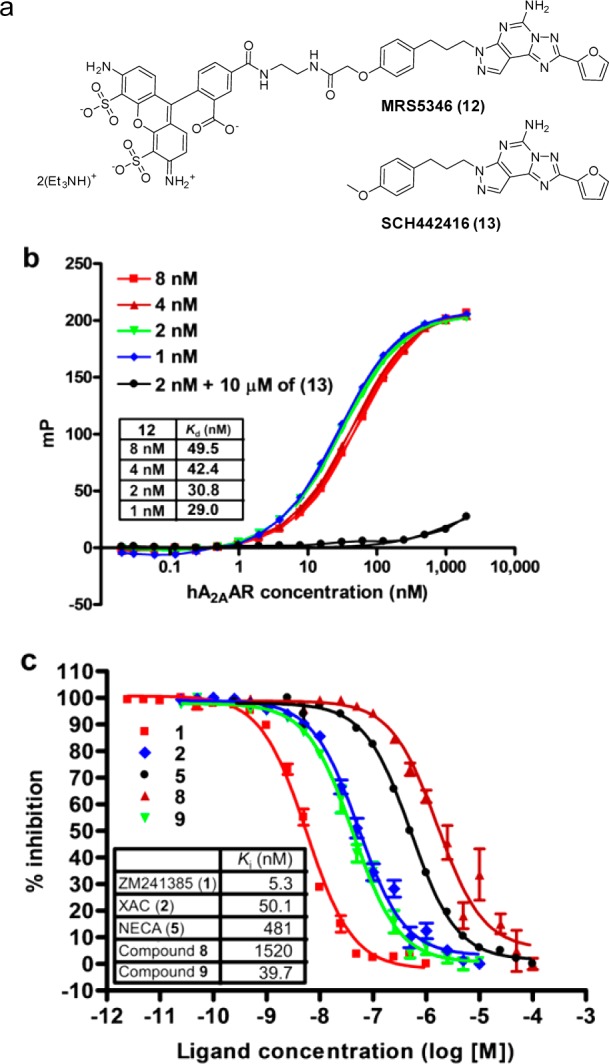
(a) Chemical structures of the fluorescent tracer MRS5346 (12) and the antagonist SCH442416 (13). (b) FP saturation binding experiments on solubilized hA2AAR with the tracer 12 at different concentrations: red squares, 8 nM; brown triangles, 4 nM; green inverted triangles 2 nM; blue diamonds, 1 nM; black circles, 2 nM in the presence of 10 μM of SCH442416 (13). (c) Normalized FP competitive binding experiments with 2 nM of the tracer 12 and 25 nM of hA2AAR: red squares, ZM241385 (1); blue diamonds, XAC (2); black circles, NECA (5); brown triangles, compound 8; green inverted triangles, compound 9. Data in parts b and c are shown as means ± SEM; N = 3.
Prior to photolabeling experiments, three reaction mixtures were prepared at 10 μM hA2AAR in the same buffer as for FP experiments: a DMSO control, a 2-fold molar excess of compound 9 relative to hA2AAR, and the same mixture containing a 5-fold molar excess of ZM241385 (1) relative to compound 9 as a competitor. After incubation at room temperature for 30 min, the mixtures were kept on ice and irradiated with a model B-100A UV light (UVP, Upland, CA, USA) for 30 min from a distance of 5 cm.
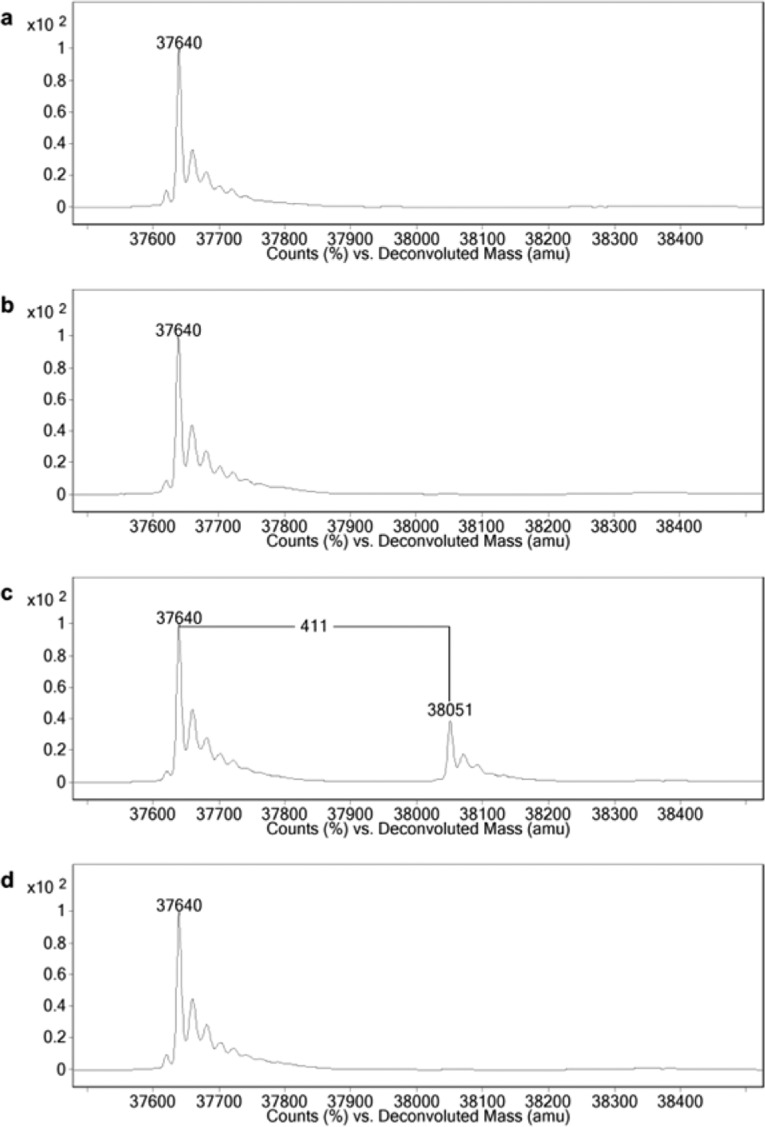
The progress of the photo-cross-link reaction was monitored by mass spectrometry using a 6520 Accurate-Mass Q-TOF instrument with a 1200 series HPLC (Agilent Technologies, Santa Clara, CA, USA). Chromatographic separation was carried out in a polystyrene divinylbenzene column (PLRP/S, Agilent Technologies)31 with a two-step short-gradient elution. The mobile phase in the first gradient step consisted of 0.1% formic acid water solution and 0.1% formic acid acetonitrile. In the second gradient step, the organic phase was changed to 0.1% formic acid isopropyl alcohol. Excess compound 9 and its phototransformation products together with the detergents and a small proportion of hA2AAR were washed out in the first gradient step, while the relatively hydrophobic hA2AAR retained in the column was eluted in the second gradient step. The multiply charged ions produced by ESI were deconvoluted to zero-charge molecular mass values using MassHunter Workstation Software Qualitative Analysis with BioConfirm Software (version B.06.00, Agilent Technologies) by means of the Maximum Entropy deconvolution algorithm. The deconvoluted mass spectrum findings for each mixture are shown in Figure 2.
Figure 2.
Deconvoluted mass spectrum of the hA2AAR. (a) Purified hA2AAR. The 37 640 Da cluster represents the mass of the hA2AAR. (b) DMSO control after UV irradiation. (c) The photolabeled hA2AAR with compound 9. The 38 051 Da cluster corresponds to compound 9 singly cross-linked to the hA2AAR with a loss of N2. (d) Competition experiment in the presence of a 5-fold molar excess of ZM241385 (1) relative to compound 9.
As shown in Figure 2a, the peak cluster was observed at 37 640 Da, which was in close agreement with the calculated mass of a 10× His-tagged hA2AAR containing four disulfide bonds (calcd for 37 639 Da), indicating there was no detectable postmodification during protein expression. In the DMSO control, no obvious peak shift was observed after UV irradiation, which suggested that receptor oxidation was almost completely suppressed during UV irradiation and highlighted one of the advantages of the diazirine photophore (Figure 2b). In the presence of compound 9, a new cluster of peaks was clearly observed at 38 051 Da after UV irradiation (Figure 2c). The mass difference of 411 Da between the two major peaks represented the cross-link of a single compound 9 molecule minus N2 with a negligible difference (calcd for 412 Da). Moreover, the absence of double-labeling peak clusters indicated that specific binding of compound 9 had occurred. In the presence of a 5-fold molar excess of ZM241385 (1), no peaks were detected around 38 051 Da, implying that compound 9 bound to the same site as did ZM241385 (1) (Figure 2d).
Since the cross-link of compound 9 could be ascertained at the protein level, the mixture was ensuingly subjected to protease digestion for identification of precise cross-link positions. The photoreacted mixture was precipitated with trichloroacetic acid, and the protein fraction was dissolved in digestion buffer containing a nonionic surfactant, 0.05% 5-cyclohexyl-1-pentyl-β-d-maltoside (CYMAL-5, Anatrace, Maumee, OH, USA)32 as well as an ionic surfactant 0.1% RapiGest (Waters, Milford, MA, USA). Disulfides were reduced by tris(2-carboxyethyl)phosphine and alkylated with iodoacetamide. Overnight proteolysis was performed with sequencing-grade chymotrypsin or trypsin (Promega, Madison, WI, USA). The reaction was terminated by the addition of trifluoroacetic acid (pH < 2) followed by incubation to decompose the ionic surfactant. After centrifugation, the supernatant was subjected to LC–MS/MS analysis, wherein multiply charged ions were automatically selected for further MS/MS sequencing analysis. Several additional ions were manually selected for the second analysis to increase peptide sequence coverage. As the results of peptide mapping by PEAKS Studio (version 7.5, Bioinformatics Solutions Inc., Waterloo, Canada), 89% coverage was obtained to include most of the residues in the extracellular and intracellular loops as well as six of seven TM domains (see Supporting Information 2). To search for cross-link peptides, we compiled a list of all theoretical digested peptide formulas, with modifications for cysteine carbamidomethylation and one molecule of compound 9–carbene addition, using in-house software. The Agilent MassHunter’s Find-by-Formula (FBF) algorithm was employed using the list to search for all possible combinations of the peptide formulas with multiply charged ions at a 10 ppm mass tolerance. The isotopic distribution pattern and the presence of other multiply charged ions for the FBF hit peptides were visually inspected. As a result of the search, quadruply charged peptide ions with m/z = 913.9158 (calcd for m/z = 913.9226, z = 4) together with triply charged peptide ions with m/z = 1218.2253 (calcd for m/z = 1218.2278, z = 3) that corresponded to a single peptide (Cys2596.61–Phe2867.51) were detected.
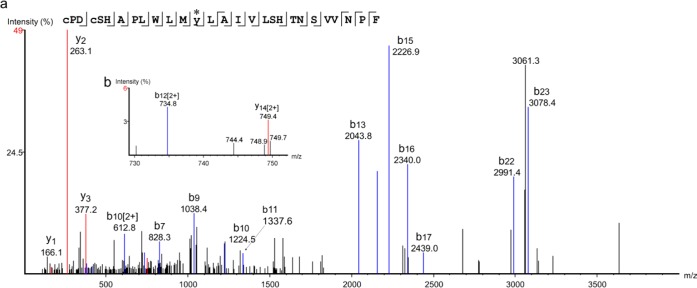
An additional LC–MS/MS analysis targeted to the quadruply charged cross-link peptide ion (m/z = 913.9226, z = 4) was carried out to determine the cross-link position. Several MS/MS product ion spectra at different collision energies (31, 34, and 37 V) were acquired, and the merged fragment ion spectra were analyzed by PEAKS Studio, which assigned sufficient b-series ions to enable sequence identification. The coexistence of b11 (without compound 9 modification) and b13 (with compound 9 modification) ions indicated the cross-linked amino acid as either Met2707.35 or Tyr2717.36 (Figure 3a). Further to the discovery of a weak doubly charged ion corresponding to b12 (with compound 9 modification), we concluded that the most plausible cross-link site was Tyr2717.36 in TM7 (Figure 3b).
Figure 3.
(a) MS/MS spectrum for the Cys2596.61–Phe2867.51 peptide labeled with compound 9 analyzed by PEAKS Studio. Carbamidomethylcysteines are designated in lower case. The asterisk indicates the cross-link position. The mass difference between C-terminal fragment ions b11 (without compound 9) and b13 (with compound 9) identified Met2707.35 or Tyr2717.36 of the hA2AAR as cross-linked amino acids. (b) An enlarged MS/MS spectrum. The weak doubly charged b12 ion indicated the most plausible cross-link site to be Tyr2717.36. The numbering of amino acids is based on the human A2A adenosine receptor.
We last investigated the precise location of Tyr2717.36 in the crystal structure of the hA2AAR. Compound 9 was docked into the ligand-binding site of the hA2AAR (PDB code: 3EML) by FRED (version 3.0.1, OpenEye Scientific Software, Santa Fe, NM, USA). One of the docking models showed the TPD moiety of compound 9 to be in close proximity to the Tyr2717.36 in TM7 (Figure 4). The tricyclic PTP part of compound 9, including the 2-furyl ring, overlapped closely with the bicyclic triazolo-triazine component of ZM241385 (1). Several mutations of Tyr2717.36 in hA2AAR have been reported. While one Tyr2717.36Ala mutant lost its binding capabilities,33,34 mutations to other aromatic residues (Phe and His) or Arg did not affect agonists and antagonists binding according to unshown data.34 A recent crystal structure of A1AR with a covalently bound xanthine-based antagonist showed that this residue was the precise point for covalent attachment of the antagonist.35 Overall, Tyr2717.36 in TM7 appeared to be the most appropriate cross-link position.
Figure 4.
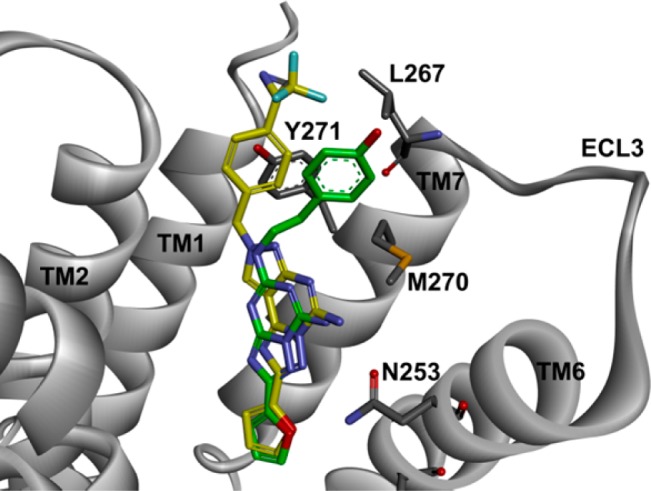
Docking model of compound 9 (yellow sticks) binding to the hA2AAR crystal structure (PDB code: 3EML) superimposed onto ZM241385 (green sticks). Key residues are shown as sticks. Tyr2717.36 in TM7 was a photolabeled amino acid. The numbering of amino acids is based on the human A2A adenosine receptor.
In conclusion, we successfully conducted PAL of detergent-solubilized hA2AAR using the novel photoaffinity probe 9 having a TPD group and identified the most likely cross-linked amino acid (Tyr2717.36) in TM7. To our knowledge, this is the first application of PAL on the hA2AAR to elucidate the cross-link position at an amino acid resolution by mass spectrometry. Further use of PAL for characterization of GPCRs is currently being planned.
Acknowledgments
The authors are grateful to Ichio Shimada (Tokyo University, Japan) for providing the cDNA of the hA2AAR and his technical advice on protein production. We thank Yasumaru Hatanaka (Toyama University, Japan) for his advice on PAL experiments. We warmly thank Toshiki Honma for FP data analysis, Shigeru Yonekubo for HPLC analyses, and Yoshinori Nonaka for NMR measurements. We also thank Kosuke Okazaki (AMED, Japan) for his insights.
Glossary
Abbreviations
- GPCRs
G protein-coupled receptors
- PAL
photoaffinity labeling
- AR
adenosine receptor
- TM
transmembrane
- TPD
trifluoromethylphenyl diazirine
- PTP
pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidine
- FP
fluorescent polarization
- mP
millipolarization
- DDM
n-dodecyl-β-d-maltoside
- CHS
cholesteryl hemisuccinate
- CYMAL-5
5-cyclohexyl-1-pentyl-β-d-maltoside
- FBF
Find-by-Formula
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.7b00138.
Experimental procedures and spectral characterization data; sequence coverage and MS/MS spectra of proteolytic digests of hA2AAR (PDF)
Author Contributions
H.M. carried out the experiments and wrote the manuscript. T.M. supervised the protein expression and purification. C.H. performed the molecular modeling studies. T.O. supervised the study. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Rask-Andersen M.; Almén M. S.; Schiöth H. B. Trends in the Exploitation of Novel Drug Targets. Nat. Rev. Drug Discovery 2011, 10, 579–590. 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- Gether U. Uncovering Molecular Mechanisms Involved in Activation of G Protein-Coupled Receptors. Endocr. Rev. 2000, 21, 90–113. 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- Fredriksson R.; Lagerström M. C.; Lundin L. G.; Schiöth H. B. The G-Protein-Coupled Receptors in the Human Genome Form Five Main Families. Phylogenetic Analysis, Paralogon Groups, and Fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Pierce K. L.; Premont R. T.; Lefkowitz R. J. Seven-Transmembrane Receptors. Nat. Rev. Mol. Cell Biol. 2002, 3, 639–650. 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Fredholm B. B.; IJzerman A. P.; Jacobson K. A.; Linden J.; Muller C. E. International Union of Basic and Clinical Pharmacology. LXXXI. Nomenclature and Classification of Adenosine Receptors-an Update. Pharmacol. Rev. 2011, 63, 1–34. 10.1124/pr.110.003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook B. C.; Jackson P. F. Adenosine A2A Receptor Antagonists and Parkinson’s Disease. ACS Chem. Neurosci. 2011, 2, 555–567. 10.1021/cn2000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C. E.Adenosine A2A Receptor Antagonists in Drug Development. In The Adenosinergic System, Current Topics in Neurotoxicity 10: A Non-Dopaminergic Target in Parkinson’s Disease; Morelli M., Simola N., Wardas J., Eds.; Springer International Publishing AG, 2015; Vol. 10, pp 39–56. [Google Scholar]
- Muller-Haegele S.; Muller L.; Whiteside T. L. Immunoregulatory Activity of Adenosine and Its Role in Human Cancer Progression. Expert Rev. Clin. Immunol. 2014, 10, 897–914. 10.1586/1744666X.2014.915739. [DOI] [PubMed] [Google Scholar]
- de Lera Ruiz M.; Lim Y. H.; Zheng J. Adenosine A2A Receptor as a Drug Discovery Target. J. Med. Chem. 2014, 57, 3623–3650. 10.1021/jm4011669. [DOI] [PubMed] [Google Scholar]
- Jaakola V. P.; Griffith M. T.; Hanson M. A.; Cherezov V.; Chien E. Y. T.; Lane J. R.; IJzerman A. P.; Stevens R. C. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science 2008, 322, 1211–1217. 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G.; Gedeon N. G.; Jankins T. C.; Jones G. B. Novel Approaches for Targeting the Adenosine A2A Receptor. Expert Opin. Drug Discovery 2015, 10, 63–80. 10.1517/17460441.2015.971006. [DOI] [PubMed] [Google Scholar]
- Shonberg J.; Kling R. C.; Gmeiner P.; Löber S. GPCR Crystal Structures: Medicinal Chemistry in the Pocket. Bioorg. Med. Chem. 2015, 23, 3880–3906. 10.1016/j.bmc.2014.12.034. [DOI] [PubMed] [Google Scholar]
- Lee S. M.; Booe J. M.; Pioszak A. A. Structural Insights into Ligand Recognition and Selectivity for Classes A, B, and C GPCRs. Eur. J. Pharmacol. 2015, 763, 196–205. 10.1016/j.ejphar.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A.; Dias J. M.; Marshall F. H. From G Protein-Coupled Receptor Structure Resolution to Rational Drug Design. J. Biol. Chem. 2015, 290, 19489–19495. 10.1074/jbc.R115.668251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka Y.; Sadakane Y. Photoaffinity Labeling in Drug Discovery and Developments: Chemical Gateway for Entering Proteomic Frontier. Curr. Top. Med. Chem. 2002, 2, 271–288. 10.2174/1568026023394182. [DOI] [PubMed] [Google Scholar]
- Hashimoto M.; Hatanaka Y. Recent Progress in Diazirine-Based Photoaffinity Labeling. Eur. J. Org. Chem. 2008, 15, 2513–2523. 10.1002/ejoc.200701069. [DOI] [Google Scholar]
- Grunbeck A.; Sakmar T. P. Probing G Protein-Coupled Receptor—Ligand Interactions with Targeted Photoactivatable Cross-Linkers. Biochemistry 2013, 52, 8625–8632. 10.1021/bi401300y. [DOI] [PubMed] [Google Scholar]
- Piersen C. E.; True C. D.; Wells J. N. 125I-2-[4-[2-[2-[(4-Azidophenyl)methylcarbonylamino]ethylaminocarbonyl]ethyl]phenyl]ethylamino-5′-N-ethylcarboxamidoadenosine Labels Transmembrane Span V of the A2A Adenosine Receptor. Mol. Pharmacol. 1994, 45, 871–877. [PubMed] [Google Scholar]
- Moss S. M.; Jayasekara P. S.; Paoletta S.; Gao Z. G.; Jacobson K. A. Structure-Based Design of Reactive Nucleosides for Site-Specific Modification of the A2A Adenosine Receptor. ACS Med. Chem. Lett. 2014, 5, 1043–1048. 10.1021/ml5002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette D.; Neamati N.; Tomer K. B.; Borchers C. H. Photoaffinity Labeling Combined with Mass Spectrometric Approaches as a Tool for Structural Proteomics. Expert Rev. Proteomics 2006, 3, 399–408. 10.1586/14789450.3.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M.; Bech-Serra J. J.; Canals F.; Zajac J. M.; Talmont F.; Arsequell G.; Valencia G. Optimized Proteomic Mass Spectrometry Characterization of Recombinant Human μ-Opioid Receptor Functionally Expressed in Pichia pastoris Cell Lines. J. Proteome Res. 2015, 14, 3162–3173. 10.1021/acs.jproteome.5b00104. [DOI] [PubMed] [Google Scholar]
- Chiu M. L.; Tsang C.; Grihalde N.; MacWilliams M. P. Over-Expression, Solubilization, and Purification of G Protein-Coupled Receptors for Structural Biology. Comb. Chem. High Throughput Screening 2008, 11, 439–462. 10.2174/138620708784911456. [DOI] [PubMed] [Google Scholar]
- Fraser N. J. Expression and Functional Purification of a Glycosylation Deficient Version of the Human Adenosine 2a Receptor for Structural Studies. Protein Expression Purif. 2006, 49, 129–137. 10.1016/j.pep.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Brunner J. New Photolabeling and Crosslinking Methods. Annu. Rev. Biochem. 1993, 62, 483–514. 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- Redenti S.; Ciancetta A.; Pastorin G.; Cacciari B.; Moro S.; Spalluto G.; Federico S. Pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidines and Structurally Simplified Analogs. Chemistry and SAR Profile as Adenosine Receptor Antagonists. Curr. Top. Med. Chem. 2016, 16, 3224–3257. 10.2174/1568026616666160506145831. [DOI] [PubMed] [Google Scholar]
- Baraldi P. G.; Cacciari B.; Spalluto G.; Pineda de las Infantas y Villatoro M. J.; Zocchi C.; Dionisotti S.; Ongini E. Pyrazolo[4,3-e]-1,2,4-triazolo[1,5-c]pyrimidine Derivatives: Potent and Selective A2A Adenosine Antagonists. J. Med. Chem. 1996, 39, 1164–1171. 10.1021/jm950746l. [DOI] [PubMed] [Google Scholar]
- Kecskés M.; Kumar T. S.; Yoo L.; Gao Z. G.; Jacobson K. A. Novel Alexa Fluor-488 Labeled Antagonist of the A2A Adenosine Receptor: Application to a Fluorescence Polarization-Based Receptor Binding Assay. Biochem. Pharmacol. 2010, 80, 506–511. 10.1016/j.bcp.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar T. S.; Mishra S.; Deflorian F.; Yoo L. S.; Phan K.; Kecskés M.; Szabo A.; Shinkre B.; Gao Z. G.; Trenkle W.; Jacobson K. A. Molecular Probes for the A2A Adenosine Receptor Based on a Pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-amine Scaffold. Bioorg. Med. Chem. Lett. 2011, 21, 2740–2745. 10.1016/j.bmcl.2010.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolovska-Coleska Z.; Wang R.; Fang X.; Pan H.; Tomita Y.; Li P.; Roller P. P.; Krajewski K.; Saito N. G.; Stuckey J. A.; Wang S. Development and Optimization of a Binding Assay for the XIAP BIR3 Domain Using Fluorescence Polarization. Anal. Biochem. 2004, 332, 261–273. 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Todde S.; Moresco R. M.; Simonelli P.; Baraldi P. G.; Cacciari B.; Spalluto G.; Varani K.; Monopoli A.; Matarrese M.; Carpinelli A.; Magni F.; Kienle M. G.; Fazio F. Design, Radiosynthesis, and Biodistribution of a New Potent and Selective Ligand for in Vivo Imaging of the Adenosine A2A Receptor System Using Positron Emission Tomography. J. Med. Chem. 2000, 43, 4359–4362. 10.1021/jm0009843. [DOI] [PubMed] [Google Scholar]
- Whitelegge J. P.HPLC and Mass Spectrometry of Integral Membrane Proteins. In The Protein Protocols Handbook; Walker J. M., Ed.; Humana Press: Totowa, NJ, 2009; pp 1149–1166. [Google Scholar]
- Zvonok N.; Yaddanapudi S.; Williams J.; Dai S.; Dong K.; Rejtar T.; Karger B. L.; Makriyannis A. Comprehensive Proteomic Mass Spectrometric Characterization of Human Cannabinoid CB2 Receptor. J. Proteome Res. 2007, 6, 2068–2079. 10.1021/pr060671h. [DOI] [PubMed] [Google Scholar]
- Guo D.; Pan A. C.; Dror R. O.; Mocking T.; Liu R.; Heitman L. H.; Shaw D. E.; IJzerman A. P. Molecular Basis of Ligand Dissociation from the Adenosine A2A Receptor. Mol. Pharmacol. 2016, 89, 485–491. 10.1124/mol.115.102657. [DOI] [PubMed] [Google Scholar]
- Kim J.; Wess J.; Michiel van Rhee A.; Schöneberg T.; Jacobson K. A. Site-Directed Mutagenesis Identifies Residues Involved in Ligand Recognition in the Human A2a Adenosine Receptor. J. Biol. Chem. 1995, 270, 13987–13997. 10.1074/jbc.270.23.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glukhova A.; Thal D. M.; Nguyen A. T.; Vecchio E. A.; Jörg M.; Scammells P. J.; May L. T.; Sexton P. M.; Christopoulos A. Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity. Cell 2017, 168, 867–877. 10.1016/j.cell.2017.01.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.