Abstract
A competitive fluorescence polarization (FP) assay is reported for determining binding affinities of probe molecules with the pseudokinase JAK2 JH2 allosteric site. The syntheses of the fluorescent 5 and 6 used in the assay are reported as well as Kd results for 10 compounds, including JNJ7706621, NVP-BSK805, and filgotinib (GLPG0634). X-ray crystal structures of JAK2 JH2 in complex with NVP-BSK805, filgotinib, and diaminopyrimidine 8 elucidate the binding poses.
Keywords: Janus Kinase 2, JAK2, JAK2 JH2, Pseudokinase domain, Fluorescence polarization, FP, JNJ7706621, NVP-BSK805, Filgotinib (GLPG0634), Protein crystallography
Janus kinases (JAKs) are nonreceptor tyrosine kinases involved in the regulation of hematopoiesis, the immune system, and cellular metabolism. In mammals, the family has four members: JAK1–3 and tyrosine kinase 2 (TYK2). Each protein contains an N-terminal FERM domain, an SH2-like domain, a pseudokinase domain (JAK homology 2 or JH2), and the C-terminal kinase domain (JH1).1
Though JH2 domains are similar in structure to kinase domains and include an ATP binding site, they lack or possess negligible catalytic activity and serve a primarily regulatory role on JH1 kinase activity.2−5 Notably, multiple diseases are associated with mutations in the JAK2 JH2 domain; in particular, V617F has been linked to polycythemia vera (PV), essential thrombocythemia (ET), and myelofibrosis (PMF).6−10 Moreover, though disruption of ATP binding in JH2 showed only minor effects on JAK2 wild type (WT) activity, it did inhibit the hyperactivity of the pathogenic V617F mutant.11 This suggests that small molecules that bind at the JAK2 JH2 ATP site have potential for therapeutic drug development.
Although potent
inhibitors of Janus kinases have been reported
in the literature,12,13 drug discovery efforts have not
been able to address diseases caused by mutated JAK2.11 The need for binders that selectively target the JAK2 JH2
domain is therefore pressing. Since the JH2 domain is not catalytic,
an accurate and rapid direct binding assay is needed for gauging potency.
To this end, we report here such an assay using fluorescence polarization
(FP). This binding assay contrasts conventional JAK assays (e.g.,
autophosphorylation14−16 and proliferation17) by
providing quantitative measurements of binding constants, Kd.18 The use of standard
microplate readers, the ability to reanalyze assay plates, and the
fact that substrates or radiolabeled reagents are not needed make
FP attractive for drug discovery. Ten compounds, including the well-known
multikinase inhibitors JNJ7706621 (1),19 NVP-BSK805 (2),20 and filgotinib (3),21 were
tested, and three X-ray crystal structures were obtained to elucidate
the binding poses of JH2 complexes.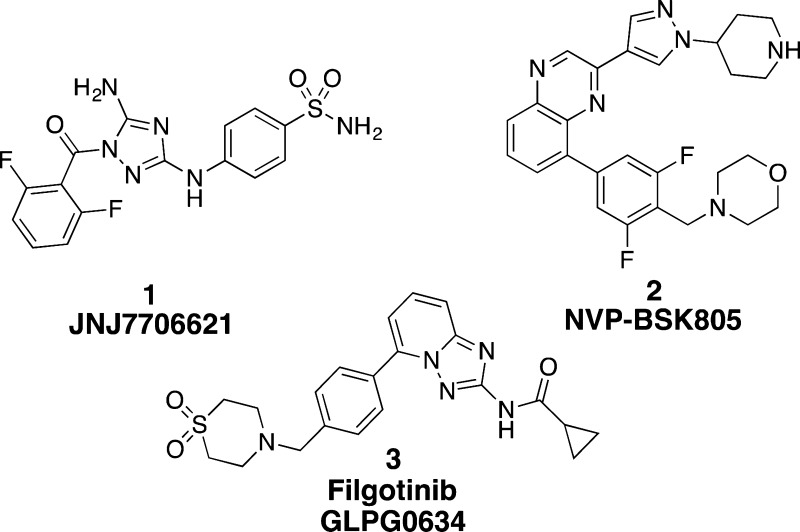
A prerequisite for FP is a fluorescent probe with high affinity for the binding site. For the JAK2 JH2 domain, the initial choice was BODIPY-ATP (4) (Invitrogen, ThermoFisher Scientific supplier), a fluorophore attached to a ribose ring.22 However, the affinity of tracer 4 toward JAK2 JH2 was weak (Kd = 7 μM) and therefore could not be used to determine accurately affinities in the nanomolar range. In addition, the concentration of protein (7 μM, Table 1) needed for the assay is not desirable for high-throughput screening.
Table 1. Concentration of Tracer and JAK2 JH2 Protein Needed When Using Tracers 4 and 5 in FP Assays.
| Tracer 4 | Tracer 5 | |
|---|---|---|
| [tracer] | 5 nM | 1.5 pM |
| [protein] | 7 μM | 200 nM |
To overcome
these problems, we designed and synthesized tracers 5 and 6 (Schemes 1 and 2) by attaching a fluorescein
isothiocyanate (FITC) to two amino analogs of 1, namely 18 and 24. A detailed description can be found
in the Supporting Information. Compound 1 was found through an in vitro screen at
the Yale Small Molecule Discovery Center and determined to have a Kd of 106 nM by isothermal titration calorimetry
(ITC) with JAK2 JH2.22 The minimum tracer
concentration that retained a satisfactory signal-to-noise ratio with 5 was found to be 1.5 pM.
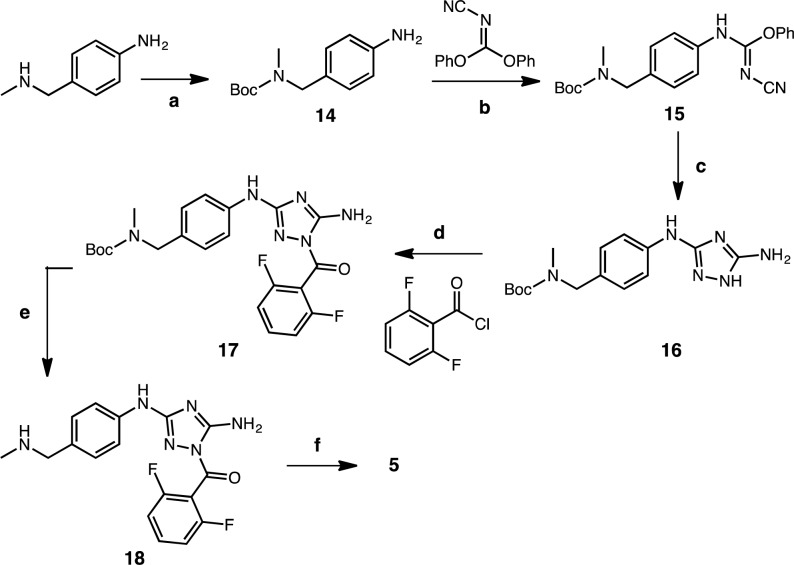
Scheme 1. Synthesis of Tracer 5.
Reagents and conditions: (a) (Boc)2O, THF, 23 °C, 20 h; (b) THF, 65 °C, 20 h; (c) Hydrazine, THF, 70 °C, 2 h; (d) Py, 18 h; (e) THF, TFA, 50 °C, 3 h; (f) Fluorescein-NCS, DIPEA, DMF, 23 °C, 1 h.
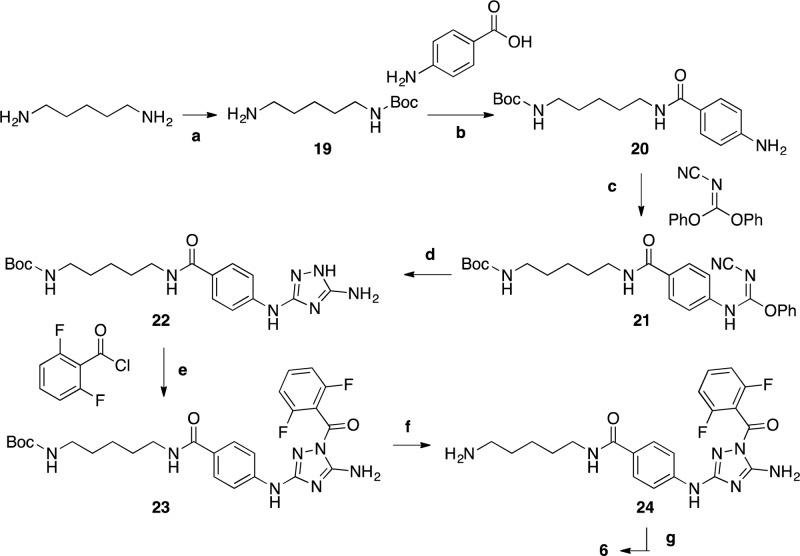
Scheme 2. Synthesis of Tracer 6.
Reagents and conditions: (a) (Boc)2O, DCM, rt, 20 h; (b) PyBOP, HOBt, Et3N, THF, rt, 20 h; (c) THF, 65 °C, 20 h; (d) hydrazine, THF, 70 °C, 2 h; (e) Py, rt, 18 h; (f) THF, TFA, 50 °C, 3 h; (g) fluorescein-NCS, DIPEA, DMF, rt, 1 h.
Saturation experiments, which involved adding incremental amounts of JAK2 JH2 (0–4.8 μM) to the tracer solution, were then carried out with measurements over 90 min and showed stable Kd values over time. The dissociation constants of tracers 5 and 6 were determined to be roughly equivalent near 0.2 μM (Figure 1B), a significant improvement over tracer 4. The lower Kd values translate to correspondently lower protein concentrations needed in the competitive assay (Table 1). Interestingly, tracer 5 showed a greater ΔFP over the range of protein concentrations than tracer 6 (2.5-fold vs 1.5-fold, Figure 1A). This suggests that the fluorescent properties of tracer 6 are affected by its binding to the protein, so 5 was selected for the subsequent competitive assays.
Figure 1.
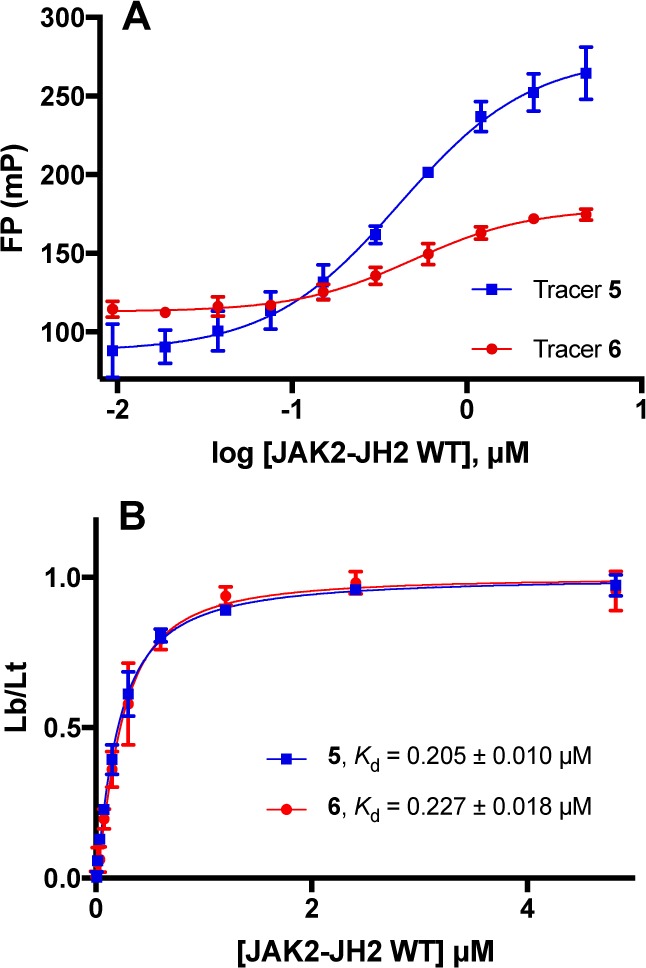
Determination of binding affinities for tracers 5 and 6 (1.5 pM) through saturation experiments. (A) Variation of FP values as a function of JAK2 JH2 WT concentration. (B) Kd determination for tracers 5 and 6. Lb/Lt = ratio of ligand bound to the total. Data from quadruplicate experiments in three independent assays. Mean ± SEM plotted for all data.
In view of the structure of 1, exploratory studies led to preparation of potential JH2 binders containing diamino-substituted heterocycles with terminal 4-cyanophenyl substituents such as 7–13 (synthetic schemes are in the Supporting Information). These compounds along with 1–3 were studied using the optimized FP assay (Table 2). The most active compound, 1, has a Kd of 0.8 μM in the FP assay. This value is 8-fold weaker than that obtained from ITC, reflecting differences in conditions, including use of 50 mM Hepes vs 20 mM Tris-Cl buffer, and less glycerol (10% vs 20%) and more protein (ca. 5 μM vs 6 nM) for ITC.22 Filgotinib (3), 7, 9, 10, and 13 showed less than ca. 10% binding in an initial screen at a concentration of 50 μM, so precise Kd values were not determined. Though the simple pyrimidine 7 was inactive, expansion at C6 did provide active compounds, with 8 showing the lowest Kd.
Table 2. Binding Affinity Values (Kd, μM) from the FP Assay.
| Compd | Kd(μM)a |
|---|---|
| 1 JNJ7706621 | 0.80 ± 0.05 |
| 2 NVP-BSK805 | 42.0 ± 3.5 |
| 3 filgotinib (GLPG0634) | 9% (50 μM) |
| 7 | 0% (50 μM) |
| 8 | 57.3 ± 2.8 |
| 9 | 3% (50 μM) |
| 10 | 11% (50 μM) |
| 11 | 122.3 ± 18.5 |
| 12 | 106.0 ± 18.8 |
| 13 | 3% (50 μM) |
Kd or % bound at indicated concentration in parentheses. Data shown from quadruplicate experiments in three independent assays. Mean ± SEM.
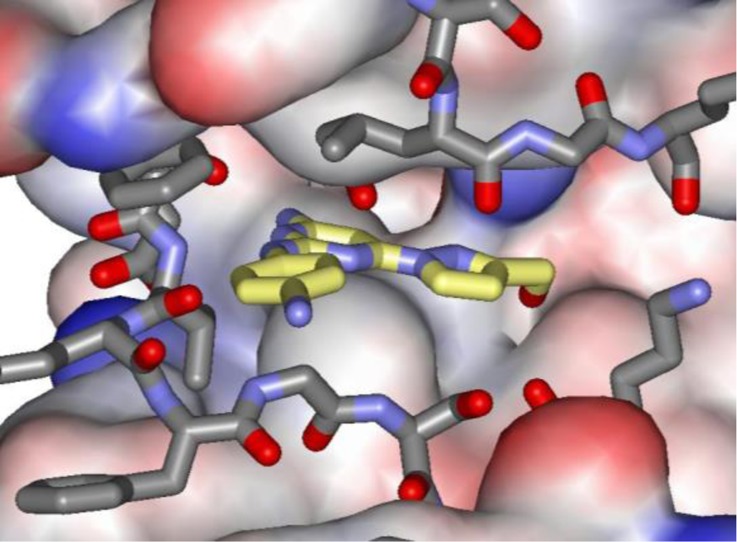
To complement these studies and provide a solid basis for structure-based design, X-ray crystal structures were pursued for complexes of the small molecules with JAK2 JH2. Success was obtained for several compounds, including 2, 3, and 8 at 2.0, 1.9, and 1.6 Å resolution, respectively. Full details are provided in the Supporting Information. The binding poses are shown in Figure 2 and confirm the expected positioning in the hinge region of the JH2 ATP site. 2 only has one hydrogen bond (yellow dashed line) with the backbone NH of Val629, while 3 adds one with the carbonyl group. However, the complex with 8 appears well-packed and has three additional hydrogen bonds with the side-chain carbonyl group of Gln626, the backbone carbonyl group of Glu627, and one between its terminal hydroxyl group and the side chain of Asn678. The lower Kd for 2 than 8 suggests that replacement of the hydroxymethylpyrazole with larger, more hydrophobic substituents could lead to enhanced binding.
Figure 2.
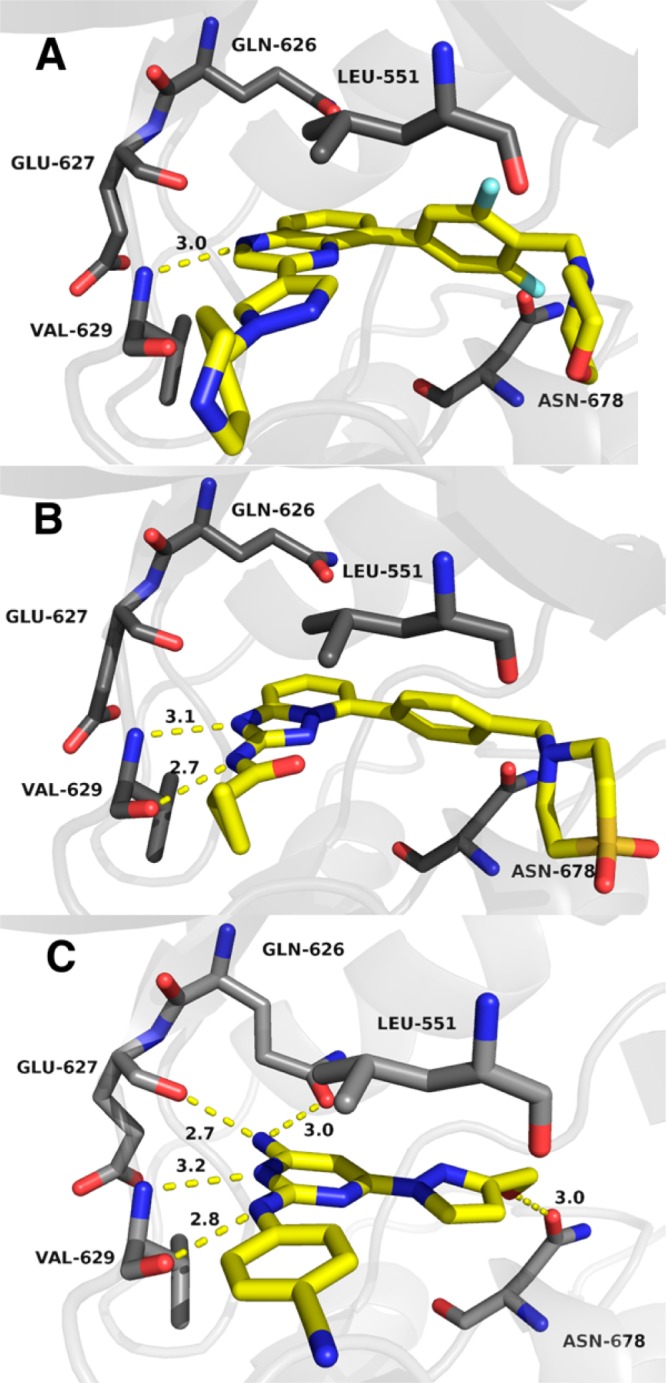
Renderings from the crystal structures of NVP-BSK805 (2) (A), filgotinib (3) (B), and 8 with wild type JAK2 JH2. Carbon atoms of 2, 3, and 8 are colored yellow. The PDB codes are 5UT4, 5UT5, and 5UT6.
In summary, the desire to discover small molecules that selectively target the JH2 domain of JAK2 led us to pursue a fluorescence polarization assay. This competitive binding assay allows fast and accurate measurements of Kd values, suitable for high-throughput screening. To this end, two fluorescein-labeled ligands (5, 6) were synthesized. The high affinity of tracer 5 (Kd = 0.2 μM) allowed for low concentrations of both tracer and protein in the FP assay. The FP assay was then applied to three well-known JAK inhibitors (1–3) and seven synthesized compounds (7–13). In conjunction with the reported crystal structures for complexes with JAK2 JH2, application of structure-based and computer-aided design to the discovery of potent JH2 binding molecules has a firm foundation.
Acknowledgments
Gratitude is expressed to the Yale-Gilead collaboration for support, to the staff of the Advanced Photon Source for assistance, and to the NIH for training support of D.E.P. (GM007324) and for support of the Yale Medical School X-ray generator and detector (Shared Instrumentation Grant 1S10OD018007).
Glossary
Abbreviations
- JAK
Janus Kinases
- JH2
pseudokinase domain
- FP
Fluorescence Polarization
- Boc
tert-butyloxycarbonyl protecting group
- DCM
dichloromethane
- THF
tetrahydrofuran
- DMF
dimethylformamide
- Py
pyridine
- TFA
trifluoroacetic acid
- DIPEA
N,N-diisopropylethylamine
- PyBop
benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate
- HOBt
hydroxybenzotriazole
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.7b00154.
Full synthetic procedures and spectral characterization data for all intermediates and final compounds 5–13; crystallographic data for complexes 2, 3, and 8 with JAK2 JH2 have been deposited in the RCSB Protein Data Bank with the PDB codes 5UT4, 5UT5, and 5UT6; experimental details of FP assays. (PDF)
Author Contributions
§ A.S.N. and L.D. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Shuai K.; Liu B. Regulation of JAK-STAT signaling in the immune system. Nat. Rev. Immunol. 2003, 3, 900–911. 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- Saharinen P.; Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of JAK2 and JAK3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J. Biol. Chem. 2002, 277, 47954–47963. 10.1074/jbc.M205156200. [DOI] [PubMed] [Google Scholar]
- Saharinen P.; Vihinen M.; Silvennoinen I. Autoinhibition of JAK2 tyrosine kinase is dependent on specific regions in its pseudokinase domain. Mol. Biol. Cell 2003, 14, 1448–1459. 10.1091/mbc.E02-06-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz A. S.; Niranjan Y.; Hammaren H.; Ungureanu D.; Ruijtenbeek R.; Touw I. P.; Silvennoinen O.; Hilhorst R. The JH2 domain and SH2-JH2 linker regulate JAK2 activity: A detailed kinetic analysis of wild type and V617F mutant kinase domains. Biochim. Biophys. Acta, Proteins Proteomics 2014, 1844, 1835–1841. 10.1016/j.bbapap.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Silvennoinen O.; Ungureanu D.; Niranjan Y.; Hammaren H.; Bandaranayake R.; Hubbard S. R. New insights into the structure and function of the pseudokinase domain in JAK2. Biochem. Soc. Trans. 2013, 41, 1002–1007. 10.1042/BST20130005. [DOI] [PubMed] [Google Scholar]
- Baxter E. J.; Scott L. M.; Campbell P. J.; East C.; Fourouclas N.; Swanton S.; Vassiliou G. S.; Bench A. J.; Boyd E. M.; Curtin N.; Scott M. A.; Erber W. N.; Green A. R. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005, 365, 1054–1061. 10.1016/S0140-6736(05)74230-6. [DOI] [PubMed] [Google Scholar]
- Levine R. L.; Wadleigh M.; Cools J.; Ebert B. L.; Wernig G.; Huntly B. J. P.; Boggon T. J.; Wlodarska L.; Clark J. J.; Moore S.; Adelsperger J.; Koo S.; Lee J. C.; Gabriel S.; Mercher T.; D’Andrea A.; Frohling S.; Dohner K.; Marynen P.; Vandenberghe P.; Mesa R. A.; Tefferi A.; Griffein J. D.; Eck M. J.; Sellers W. R.; Meyerson M.; Golub T. R.; Lee S. J.; Gilliland D. G. Cancer Cell 2005, 7, 387–397. 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- James C.; Ugo V.; Le Couedic J. P.; Staerk J.; Delhommeau F.; Lacout C.; Garcon L.; Raslova H.; Berger R.; Bennaceur-Griscelli A.; Villeval J. L.; Constantinescu S. N.; Casadevall N.; Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signaling causes polycythaemia vera. Nature 2005, 434, 1144–1148. 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Kralovics R.; Passamonti F.; Buser A. S. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 2005, 352, 1779–1790. 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- Zhao R. X.; Xing S.; Li Z.; Fu X. Q.; Li Q. S.; Krantz S. B.; Zhao Z. H. J. Identification of an acquires JAK2 mutation in Polycythemia vera. J. Biol. Chem. 2005, 280, 22788–22792. 10.1074/jbc.C500138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammaren H. M.; Ungureanu D.; Grisouard J.; Skoda R. C.; Hubbard S. R.; Silvennoinen O. ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 4642–4647. 10.1073/pnas.1423201112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan C.; Behrmann I.; Haan S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus Kinases. J. Cell. Mol. Med. 2010, 14, 504–527. 10.1111/j.1582-4934.2010.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. Companies hope for kinase inhibitor JAKpot. Nat. Rev. Drug Discovery 2011, 10, 717–718. 10.1038/nrd3571. [DOI] [PubMed] [Google Scholar]
- Changelian P. S.; Flanagan M. E.; Ball D. J.; Kent C. R.; Magnuson K. S.; Martin W. H.; Rizzuti B. J.; Sawyer P. S. Prevention of organ allograft rejection by a specific Janus Kinase 3 inhibitor. Science 2003, 302, 875–878. 10.1126/science.1087061. [DOI] [PubMed] [Google Scholar]
- Yang S. M.; Malaviya R.; Wilson L. J.; Argentieri R.; Chen X.; Yang C. M.; Wang B. B.; Cavender D.; Murray W. V. Simplified staurosporine analogs as potent JAK3 inhibitors. Bioorg. Med. Chem. Lett. 2007, 17, 326–331. 10.1016/j.bmcl.2006.10.062. [DOI] [PubMed] [Google Scholar]
- Li Z.; Xing S.; Wang S. F.; Ho W. T.; Zhao Z. Z. J. Autoinhibition of JAK2 tyrosine kinase is dependent on specific regions in its pseudokinase domain. Exp. Hematol. 2007, 35, 1624–1632. 10.1016/j.exphem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. J.; Thakur K. D.; Clark M. P.; Laughlin S. K.; George K. M.; Bookland R. G.; Davis J. R.; Cabrera E. J.; Easwaran V.; De B.; Zhang Y. G. Development of pyrimidine-based inhibitors of Janus tyrosine kinase 3. Bioorg. Med. Chem. Lett. 2006, 16, 5633–5638. 10.1016/j.bmcl.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Lea W. A.; Simeonov A. Fluorescence polarization assays in small molecule screening. Expert Opin. Drug Discovery 2011, 6, 17–32. 10.1517/17460441.2011.537322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. H.; Connolly P. J.; Huang S. L.; Wetter S. K.; Lu Y. H.; Murray W. V.; Emanuel S. L.; Gruninger R. H.; Fuentes-Pesquera A. R.; Rugg C. A.; Middleton S. A.; Jolliffe L. K. 1-acyl-1H-[1,2,4]triazole-3,5-diamine analogues as novel and potent anticancer cyclin-dependent kinase inhibitors: Synthesis and evaluation of biological activities. J. Med. Chem. 2005, 48, 4208–4211. 10.1021/jm050267e. [DOI] [PubMed] [Google Scholar]
- Baffert F.; Régnier C. H.; De Pover A.; Pissot-Soldermann C.; Tavares G. A.; Blasco F.; Brueggen J.; Chène P.; Drueckes P.; Erdmann D.; Furet P.; Gerspacher M.; Lang M.; Ledieu D.; Nolan L.; Ruetz S.; Trappe J.; Vangrevelinghe E.; Wartmann M.; Wyder L.; Hofmann F.; Radimerski T. Potent and selective inhibitor of polycythemia by the quinoxaline JAK2 inhibitor NVP-BSK805. Mol. Cancer Ther. 2010, 9, 1945–1955. 10.1158/1535-7163.MCT-10-0053. [DOI] [PubMed] [Google Scholar]
- Menet C. J.; Fletcher S. R.; Van Lommen G.; Geney R.; Blanc J.; Smits K.; Jouannigot N.; Deprez P.; van der Aar E. M.; Clement-Lacroix P.; Lepescheux L.; Galien R.; Vayssiere B.; Nelles L.; Christophe T.; Brys R.; Uhring M.; Ciesielki F.; Van Rompaey L. Triazolopyridines as selective JAK1 inhibitors: From hit identification to GLPG0634. J. Med. Chem. 2014, 57, 9323–9342. 10.1021/jm501262q. [DOI] [PubMed] [Google Scholar]
- Puleo D. E.; Kucera K.; Hammarén H.; Ungureanu D.; Newton A. S.; Silvennoinen O.; Jorgensen W. L.; Schlessinger J.. Identification and Characterization of JAK2 Pseudokinase Domain Small Molecule Binders. ACS Med. Chem. Lett. 2017, 10.1021/acsmedchemlett.7b00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.