Abstract
Some HIV-1–infected patients develop broad and potent HIV-1 neutralizing antibodies (bNAbs) that when passively transferred to mice or macaques can treat or prevent infection. However, bNAbs typically fail to neutralize coexisting autologous viruses due to antibody-mediated selection against sensitive viral strains. We describe an HIV-1 controller expressing HLA-B57*01 and HLA-B27*05 who maintained low viral loads for 30 years after infection and developed broad and potent serologic activity against HIV-1. Neutralization was attributed to three different bNAbs targeting non-overlapping sites on the HIV-1 envelope trimer (Env). One of the three, BG18, an antibody directed against the glycan-V3 portion of Env, is the most potent member of this class reported to date and, as revealed by crystallography and electron microscopy, recognizes HIV-1 Env in a manner that is distinct from other bNAbs in this class. Single-genome sequencing of HIV-1 from serum samples obtained over a period of 9 years showed a diverse group of circulating viruses, 88.5%(31 of 35) of which remained sensitive to at least one of the temporally coincident autologous bNAbs and the individual’s serum. Thus, bNAb-sensitive strains of HIV-1 coexist with potent neutralizing antibodies that target the virus and may contribute to control in this individual. When administered as a mix, the three bNAbs controlled viremia in HIV-1YU2–infected humanized mice. Our finding suggests that combinations of bNAbs may contribute to control of HIV-1 infection.
INTRODUCTION
A fraction of HIV-1–infected individuals develop broadly neutralizing antibodies (bNAbs) that show potent neutralizing activity against a range of different HIV-1 isolates (1–4). bNAbs typically develop over a period of 1 to 3 years during which time there is coevolution of circulating viral strains and antibodies (5–9). Virus and antibody coevolution starts when the infecting virus elicits early antibody responses that exhibit some levels of autologous neutralization (6, 9–11). These early antibodies put pressure on the virus and spur HIV-1 evolution. Viral strains that are sensitive to the antibodies are subjected to negative selection, resulting in the emergence of antibody-resistant HIV-1 variants that are subsequently targeted by coevolving antibody variants (5–7, 9, 12). The bNAbs that emerge have high levels of somatic mutations, suggesting that B cells that produce bNAbs develop by iterative rounds of antibody gene mutation and selection in germinal centers (2, 5, 6, 13–17). The end result of serial antibody and HIV-1 mutation is bNAbs that neutralize large numbers of heterologous viral variants but normally fail to neutralize autologous circulating viral strains (2, 6–9). Therefore, it is believed that bNAbs that develop during chronic infection are unable to control HIV-1 in the individual who develops them.
To reexamine the question of whether autologous bNAbs can coexist with bNAb-sensitive viruses and contribute to HIV-1 control, we studied an HLA (human leukocyte antigen)–B57*01HIV-1 controller (18, 19) who developed elite levels of HIV-1 neutralizing activity and maintained low levels of HIV-1 viremia for several years. Here, we describe the neutralizing antibodies that developed in this donor, the coexisting plasma viruses, and show that when combined, these bNAbs can suppress HIV-1 replication and maintain low-level viremia in vivo in HIV-1–infected humanized mice.
RESULTS
Multiple bNAbs isolated from donor EB354
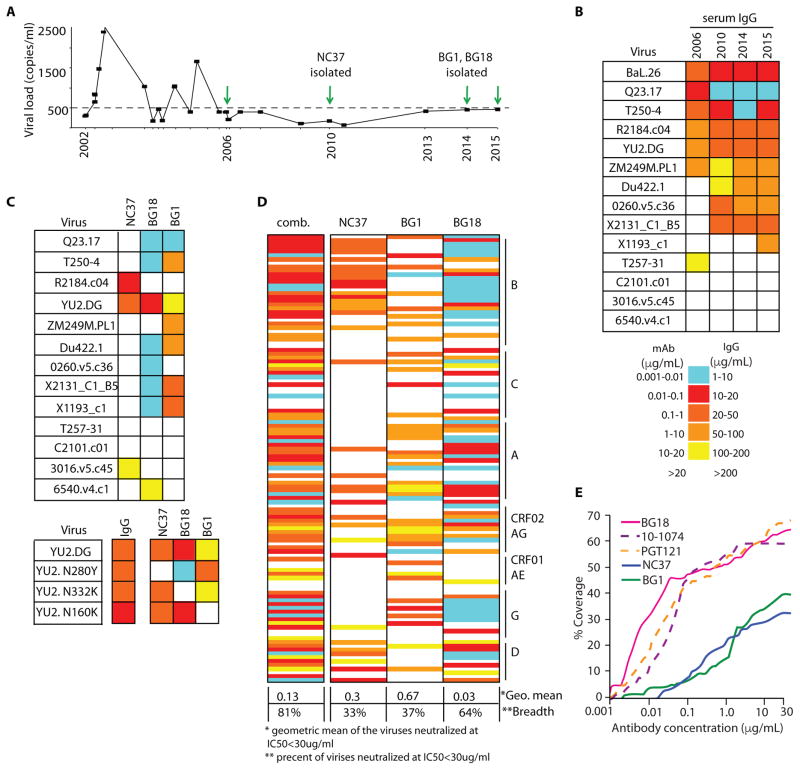
Donor EB354 was diagnosed with HIV-1 clade B in 1986. Purified immunoglobulin G (IgG) from donor EB354 was first tested for neutralization in 2006 when the viral load was <400 copies/ml (Fig. 1, A and B, and table S1). The neutralizing activity increased between 2006 and 2010, and has remained broad and potent since that time (Fig. 1B). To isolate the antibodies that account for this subject’s serologic activity, we sorted single IgG+ B cells (20–22) using four different HIV-1 baits in four separate sorts: 2CC core (23), gp140YU2 (24), a 1:1 mixture of gp14092UG37.8 (clade A) + gp140CZA79012 (clade C) (25), and BG505 SOSIP.664 (26). A total of 241 paired heavy and light chains were isolated, of which 152 antibodies formed 22 different clones (fig. S1A). Antibodies from three clones showed tier 2 neutralizing activity: BG18, BG1, and NC37 (Fig. 1C and fig. S1B). Antibody BG18 (VH4-4 and VL3-25) recapitulated most of the serologic activity, whereas antibodies NC37 (VH1-46/1-2, VK3-20) and BG1 (VH3-49 and Vk1-49) were less potent but complemented the activity of BG18 (Fig. 1C and fig. S1B).
Figure 1.
Neutralizing antibodies isolated from donor EB354.
To map the binding sites of the three neutralizing clones, we performed TZM-bl assays using HIV-1YU2 variants carrying epitope specific point mutations in Env. Whereas the polyclonal IgG from the donor showed no measurable change in sensitivity to the mutants, antibody BG18 was sensitive to YU2N332K (glycan-V3), antibody BG1 to YU2N160K (V1V2), and antibody NC37 to YU2N280Y (CD4bs) (Fig. 1C, bottom). Our epitope mapping was confirmed by competitive enzyme-linked immunosorbent assay (ELISA) (for BG18 and BG1; fig. S2) and by a 2.7 Å resolution crystal structure of NC37 Fab bound to a gp120 core (fig. S3). We conclude that BG18 recognizes the glycan patch associated with the V3 loop, that BG1 is specific for the V1V2 loops at the top of the molecule, and that NC37 is a CD4bs-specific antibody.
NC37 was isolated from a sample obtained in 2010, whereas BG1 and BG18 were isolated from peripheral blood mononuclear cells (PBMCs) collected in 2014. We examined available PBMCs from 2010 and 2013 using polymerase chain reaction (PCR) primers specific for BG1 and BG18. Whereas BG1 transcripts were detected at both time points, BG18 was first detected in 2013 and was not detected in 2010.
We conclude that BG18 emerged between 2010 and 2013, whereas BG1 and NC37 developed earlier.
BG18 neutralized 64%of viruses in a 118-virus panel (Fig. 1D) with a geometric mean IC50 of 0.03 mg/ml. Antibodies NC37 and BG1 showed geometric mean IC50 of 0.3 and 0.67 mg/ml and 33 and 37% breadth, respectively (Fig. 1, D and E, and table S2). The 1:1:1 mix of the three bNAbs neutralized 81% of the viruses in the 118-virus panel with a geometric mean IC50 of 0.130 mg/ml, indicating an additive effect (Fig. 1D, left column, and table S3).
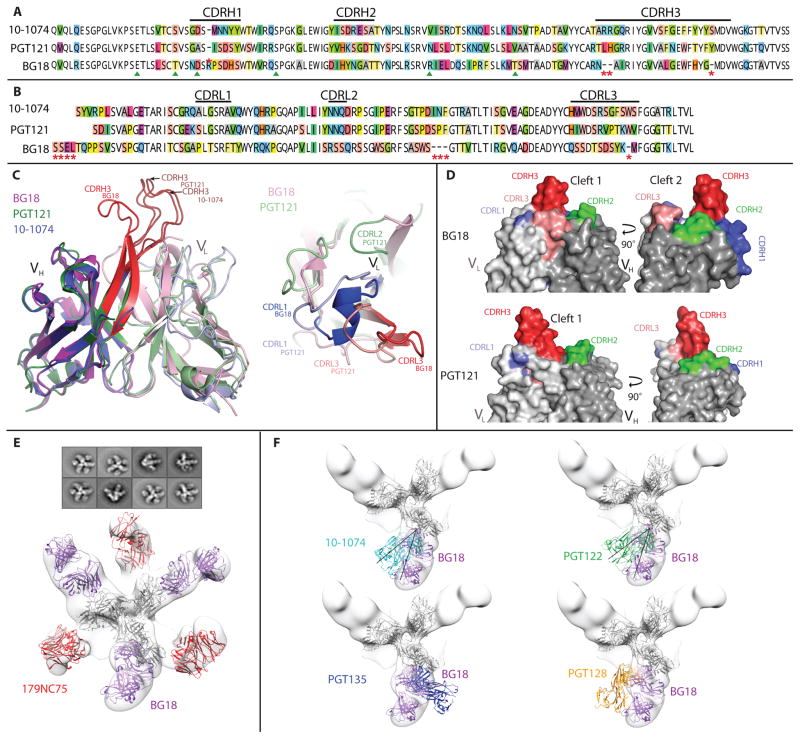
BG18 is comparable to PGT121 and 10-1074, two previously isolated potent bNAbs directed against the base of the V3 region of Env (27, 28), in terms of its breadth of coverage, but BG18 is more potent (Fig. 1E and table S2). The germline genes of BG18 and PGT121/10-1074 (VH4-4 and VH4-59, respectively) are closely related (~90% identity); however, the mature heavy chains of BG18 and PGT121/10-1074 are only about 62% identical. The light chains of BG18 and PGT121/10-1074 are more distinct (70% identity for germ lines and ~48% identity for the mature antibodies) (Fig. 2, A and B, and fig. S4). BG18 is more mutated than PGT121/10-1074; however, it does not contain any insertions or deletions, which were reported for members of PGT121/10-1074 (27). On a structural level, we found four key differences between PGT121/10-1074 and BG18. First, as revealed by the 1.3 Å resolution crystal structure of the BG18 Fab, the CDRH3 of BG18 folds on itself instead of extending fully as in PGT121/ 10-1074 (Fig. 2C, left, and table S5). This conformation of CDRH3BG18 is stabilized by hydrogen bonds. Second, as expected from the sequence alignment, the VL domain of BG18 differs in its orientation from PGT121/10-1074 (root mean square deviations of 3.1 Å were found for superimposing 93 or 92 Ca atoms in comparison to light chains in BG18-PGT121 and BG18–10-1074 superimpositions, respectively; Fig. 2C, right). Third, BG18 Fab displays a second cleft between CDRH3 and CDRL1/CDRL3, which is not found in PGT121/10-1074 (in addition to the first cleft formed between CDRH3 and CDRH2, which is found in BG18 and PGT121/10-1074) (Fig. 2D) (27, 29). Last, as revealed by a ~25 Å resolution negative-stain single-particle electron microscopy (EM) structure of BG505 SOSIP.664 trimer in complex with BG18 and the CD4bs antibody 179NC75 (30), BG18 contacts the Env trimer at a different angle, which is shifted by 34° to 41° relative to PGT122 and 10-1074, respectively (Fig. 2, E and F).
Figure 2.
Sequence and structural analysis of BG18 bNAb.
BG18 bound preferentially to syntheticV3 glycopeptides containing oligomannose N-glycans at position Asn332 gp120 but not to complex type N-glycans attached to this position or to other potential N-linked glycosylation sites within gp120 (fig. S5). Thus, BG18 appears to be more closely related to 10-1074 than to PGT121 because 10-1074 binds almost exclusively on Asn332 gp120 and not on other glycans (27, 28, 31). In a series of neutralization assays against a panel of HIV-1 pseudoviruses with deletions of specific N-linked glycosylation sites, we found that none of the glycans at the base of the V3 loop, with the exception of the Asn332 gp120, had an impact on neutralization activity (table S4). Together, these results indicate that BG18 is a new member of the Asn332 gp120-dependent class of bNAbs that is more potent and structurally different from previously isolated PGT121 and 10-1074 (27, 28, 31). When compared to the available biochemical and structural data, our findings indicate that BG18 displays recognition properties that are more similar to 10-1074 than to PGT121 (27, 28, 31).
Autologous viruses circulating between 2006 and 2015
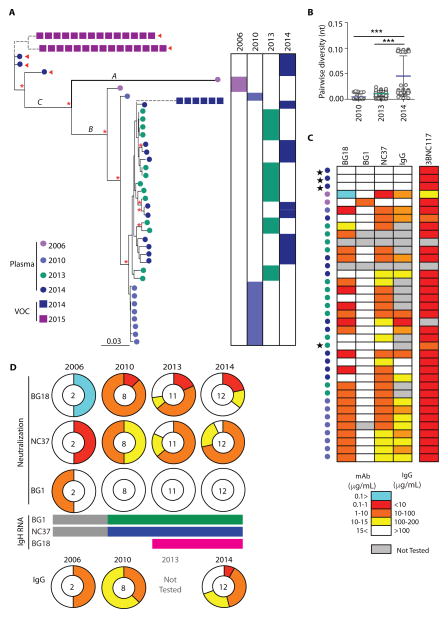
Circulating plasma viruses were characterized by single-genome sequencing (SGS) (32) at five different time points during the years 2006 to 2015 during which viremia remained <400 copies/ml. env genes were about 15%divergent from the clade B consensus sequence, as would be predicted given the length of the infection of this subject (>20 years). The low level of viremia limited sequence analysis to 37 functional in-frame transcripts, in addition to which, we also found nonfunctional transcripts at every time point (1, 33, 34). The latter were either truncated or contained stop codons in the middle of the sequence (fig. S6A). Moreover, all sequences recovered from 2015, when the viral load was <400 RNA copies/ml, were nonfunctional (fig. S6A).
The 37 functional sequences formed three clusters based on their sequence. Cluster A contained one sequence from2006 that was ~13% different from the rest of the sequences. This sequence was the closest to the clade B consensus sequence, as would be expected from the fact that this sequence was isolated from the earliest time point. The second and the biggest cluster, cluster B, contained sequences from all four time points: 1 sequence from 2006, 9 sequences from 2010, 13 sequences from2013, and 10 sequences from2014. The diversity within this cluster was low (~5%). The third cluster, cluster C, contained only three sequences, all from 2014 (Fig. 3A). In agreement with the new cluster of sequences that emerged in 2014, we saw an increase in viral diversity between 2013 and 2014 (Fig. 3B).
Figure 3.
Autologous plasma viruses in donor EB354.
We analyzed the env sequences for mutations that can be associated with resistance to the three autologous bNAbs that emerged in this individual. Cluster A and B sequences did not contain any apparent mutations that were correlated with resistance to the three autologous bNAb specificities. However, all three sequences in cluster C contained mutations that deleted the N-linked glycosylation site at Asn332 gp120 and are associated with resistance to the PGT121 class of antibodies (Fig. 3A, red arrows). In addition, they also contained a mutation associated with resistance to CD4bs antibodies (35). The sequence analysis suggests that cluster C is resistant to NC37, BG1, and BG18, whereas viruses from clusters A and B are sensitive.
Autologous virus neutralization
To determine the sensitivity of the autologous viruses to the three autologous bNAbs, we produced pseudoviruses from the cloned env genes and tested them in TZM-bl assays (36). Of the 37 env genes, 35 were produced (2 env genes did not give rise to infectious particles). Of the 35 pseudoviruses produced, only 4 were resistant to all three bNAbs (that is, were not neutralized at an antibody concentration of <15 mg/ml). These were the three viruses from cluster C and one from cluster B isolated in 2013 (Fig. 3C, black stars, and fig. S6B). The remaining 31 pseudoviruses (88.5% of all pseudoviruses) were sensitive to at least one of the three bNAbs. In addition, the pseudoviruses from 2006, 2010, and 2014 were tested for their sensitivity to the concurrent serum IgG isolated from the same time point as the viral env genes (plasma IgGfrom2013was not available for this assay). We found that 19 of the 22 viruses tested were neutralized by the contemporaneous IgG (Fig. 3C and fig. S6B).
BG18- and NC37-sensitive viruses, as well as viruses sensitive to the concurrent IgG, were found at all time points analyzed, including the times when BG18 and NC37 were isolated (Fig. 3D). In contrast, most strains, with the exception of one virus from 2006, were resistant to BG1. We conclude that most of the autologous viruses in donor EB354 escaped from BG1 but failed to escape from BG18 and NC37. Although we were unable to obtain viral sequences from plasma samples from 2015, we were able to grow virus from two of five CD4+ T cell outgrowth cultures from this time point (37). For control, we used a culture from 2014, a time point where we obtained sequence from circulating viruses. Two cultures from 2015 and one from 2014 grew out after a prolonged period (5 weeks versus typical of 2 weeks). The viruses emerging from the 2015 cultures resembled cluster C sequences in that they lacked the N-linked glycosylation site atAsn332 gp120 (Fig. 3A, violet squares) and were resistant to all three bNAbs. The viruses emerging from the 2014 cultures were closely related to one of the 2014 plasma viruses (2014_C10; fig. S6). However, whereas the virus obtained from plasma showed some sensitivity to NC37 (IC50, 4.56 mg/ml), the 2014 virus that grew out in tissue culture was resistant to all three bNAbs. Despite their resistance to the bNAbs, the tissue culture outgrowth viruses did not emerge as infectious circulating viruses in the patient and were either unfit in vivo or latent.
In vivo efficacy of BG18, NC37, and BG1
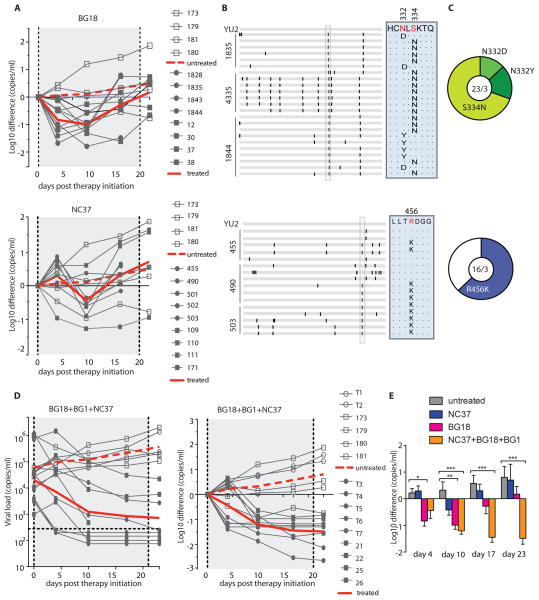
To determine whether BG18 or NC37 can exert selective pressure on HIV-1 in vivo, we infected humanized mice with HIV-1YU2 and treated with the antibodies 10 to 14 days later (38). Similar to 10-1074 (38), administration of BG18 or BG8 (a less potent clonal variant) was associated with a rapid decrease (average one log10) in viral load, followed by rebound viremia [Fig. 4A (top) and figs. S7 (top) and S8A]. As expected, all rebounding viral sequences contained a mutation that altered the N-linked glycosylation site at Asn332 gp120 (Fig. 4, B and C, top) (38).
Figure 4.
bNAb therapy in HIV YU2-infected humanized mice.
Monotherapy with NC37 also transiently suppressed viremia, but the magnitude of the decrease in viral load was less profound than that with BG18, with an average of 0.4 log10 [Fig. 4A (bottom) and fig. S7 (bottom)]. Most of the NC37-rebound viral Env sequences showed an R456K mutation that is associated with resistance toCD4bs antibodies (Fig. 4, B and C, bottom) (14, 39, 40). We conclude that BG18 and NC37 can select for HIV-1YU2 antibody-resistant variants in vivo in humanized mice.
Combinations of antibodies can control HIV-1YU2 infection in humanized mice (35, 38). To determine whether the combination of BG18, NC37, and BG1 can control viremia, we treated nine HIV- 1YU2 –infected humanized mice with a 1:1:1 combination of the three bNAbs in two independent experiments. In contrast to monotherapy with BG18 or NC37, the combination of BG18, NC37, and BG1 produced a prolonged and sustained drop in viremia (Fig. 4, D and E). Viral loads were reduced by an average of 1.48 log10 shortly after administration of the three bNAbs, and six of the nine mice showed undetectable viral loads after 3 weeks (Fig. 4D). We conclude that when applied in combination, the three bNAbs that naturally developed in donor EB354 mediate potent and durable suppression of viremia in humanized mice infected with HIV-1YU2.
DISCUSSION
Here, we describe a singular HIV-1–infected elite controller, EB354, who was followed prospectively for 9 years. During that time, EB354 donated samples for analysis at five different time points, enabling the isolation of three new bNAbs that account for serologic neutralizing activity and circulating viruses. We find that in this individual, viruses sensitive to the antibodies coexisted with the bNAbs for long periods of time.
BG18, the most potent of the three antibodies isolated from donor EB354, is directed against the Asn332 gp120-centered glycan patch at the base of the V3 loop. This is a heavily glycosylated region that is a frequent target for anti–HIV-1 bNAbs, which includes carbohydrates at positionsAsn332 gp120, Asn301 gp120, Asn386 gp120, Asn392 gp120, Asn137 gp120, and Asn156 gp120 (1, 4, 27, 31, 41, 42). A number of monoclonal bNAbs that bind to both protein and carbohydrate components at this site have been isolated, including PGT121-124, PGT128-135 (28), 10-1074 and variants (27), and the recently isolated PCDN antibodies (9). BG18 and its clonal variants resemble 10-1074 in that they rely exclusively on Asn332 gp120 (27, 31), but BG18 is more potent than published anti-V3 bNAbs and is the first to be isolated from a clade B donor.
The BG18-Env EM structure, although not of sufficient resolution to identify detailed interactions, nevertheless demonstrates that BG18 approaches Env from a different angle, which is shifted by up to 40° toward the gp120 promoter relative to the approach angles of 10-1074 and PGT122 (a closely related variant of PGT121). In addition, the BG18 Fab differs from previously characterized members of this class by a distinctive orientation and a shorter length of its CDRH3. The lack of insertions or deletions in the BG18 CDRH3 suggests that BG18-like antibodies may be easier to elicit by vaccination.
The other two neutralizing specificities, in addition to BG18, are represented by BG1 and NC37. BG1 binds to the V1V2 region of Env, another frequent target of bNAbs arising during natural infection (20), and is the first antibody in this class to be isolated from a clade B–infected donor. Its potency is similar to members of the VRC26 antibody family (43) but is less potent than PG9/16 and PGDM1400 bNAbs. Like other V1V2 bNAbs isolated to date, BG1 has a long, tyrosine-rich CDRH3. The third neutralizing specificity is represented by NC37. NC37 and its clonal variants display sequence characteristics of both VH1-2– and VH1-46–derived bNAbs; however, structural analyses suggest that NC37 recognizes Env in a VH1-46–type manner (21). As a consequence, NC37 uses its light chain to the contact loop D residue Asn280 gp120, whereas VH1-2–derived antibodies such as NIH45- 46 use their heavy chains to contact Asn280 gp120 (40, 44, 45). Superposing theNC37 Fab–gp120 complex onto an SOSIP.664 trimer structure suggests that the long NC37CDRH3makes contacts with the adjacent protomer within the Env trimer, thus NC37 recognizes a quaternary trimer epitope, the core of which overlaps with the CD4bs.
Most individuals rapidly develop strain-specific antibodies shortly after infection (6, 10, 15, 46–48). This response is associated with selection for resistant viral variants that, in some cases, elicit bNAbs (5, 12, 31, 49–51). However, the individuals studied in detail differ from EB354 in that the development of bNAbs is associated with rapid selection of autologous plasma viruses, which are resistant to coexisting bNAbs (6–9, 52, 53). Donor EB354 is significant because sensitive and resistant viral strains coexist with bNAbs, and the resistant strains fail to produce high levels of viremia because the viral strains are either in some way partially effective or kept in check by CD8+ T cells. Thus, the bNAb-sensitive plasma viruses were unable to escape immune pressure in this individual, resulting in bNAb:virus equilibrium, where the virus persists but is incapable of producing high levels of viremia.
EB354 is unusual in being both an elite controller and elite neutralizer. HIV-1 elite controllers are infected individuals who maintain low viral loads for many years (54–56). These individuals are less likely to transmit the virus (57), and they maintain long-term AIDS-free survival (58). HLA alleles B57*01 and/or B27*05 are found in 85% of HIV-1 elite controllers (19, 59), and these alleles are associated with enhancedCD8 T cell cytotoxic activity (18). Compared to viremic progressors (that is, patients with high viral loads), elite controllers are less likely to develop bNAbs (60–66). Irrespective of robust CD8+ T cell responses that may have partially controlled infection, there was sufficient HIV- 1 replication in EB354 to support bNAb development and affinity maturation.
Like several other individuals who develop serologic breadth and potency, the bNAbs that account for serologic neutralizing activity in donor EB354 recognize several nonoverlapping epitopes (5, 20, 21, 67–69). In some well-documented cases, the emergence of one bNAb lineage facilitates the development of a second one by selecting for escape variants that expose areas on Env that support further development of additional bNAbs (52). Although samples are not available to test this idea, BG1 was the earliest bNAb lineage to emerge in EB354 and may also have been a helper lineage for NC37 and/or BG18.
When combined, BG1, BG18, and NC37 durably suppressed viremia in HIV-1YU2–infected humanized mice. Although there are reports of autologous serum neutralization of circulating viruses (70), the role of monoclonal antibodies in HIV-1 control remains uncertain (51, 71–74). Our data indicate that monoclonal bNAbs and very low levels of neutralization-sensitive viruses coexist in EB354, suggesting that antibodies contribute to elite control in this individual.
HIV-1 escapes from monotherapy with bNAbs in humanized mice and humans infused with bNAbs (Fig. 4A) (35, 38, 75–78). In contrast, combination antibody therapy in humanized mice durably suppresses viremia (38, 79). In addition, infected humans treated with antibodies show enhanced humoral immunity (80) and accelerated clearance of infected cells (81), indicating that there may be immunological benefits to this form of therapy. Whether combination antibody therapy will also suppress viremia in humans remains to be determined by clinical studies. However, durable suppression of viremia in EB354 and in humanized mice treated with BG1, BG18, and NC37 suggests that combinations of bNAbs may in fact be able to contain HIV-1 infection in humans.
MATERIALS AND METHODS
Study design
The objective of this study was to investigate whether bNAbs can exhibit autologous neutralization in HIV-1 patients with suppressed viremia. Donor EB354 described in this study [also denoted as donor 622800 from the HIV Controller Consortium (82)] was selected on the basis of exceptional serum neutralization from a cohort of 394 long-term nonprogressors followed at the Ragon Institute, Boston. Staining with four different HIV-1 antigens and single-cell sorting of antigen-positive memory B cells allowed isolation of three new clones of bNAbs, two of which were analyzed structurally for their binding to HIV-1 Env protein. Autologous viruses were recovered from EB354 plasma, sequenced, and produced as cytomegalovirus (CMV)–Env pseudoviruses to test for sensitivity against the autologous bNAbs and the polyclonal IgG from the same time points. Last, the three new bNAbs were used to treat humanized HIV-1YU2–infected mice separately and in combination. Donor EB354 provided a written informed consent before participating in this study, and the Rockefeller University and the Massachusetts General Hospital Institutional Review Boards approved all studies involving patient enrollment, sample collection, and clinical followup (protocol numbers 2003P001678 and 2003P001894). For the mouse studies, this study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the Rockefeller University and in accordance with established guidelines and policies at the Rockefeller University (protocol number 13618-H).
B cell sorting and antibody isolation
Single-cell sorting of bait+CD19+IgG+ B cells from donor EB354 PBMCs was conducted as previously described (20). Memory B cells were pre-enriched with anti-CD19 magnetic beads (magnetic cell sorter) and stained using four different baits: the gp120 2CC core protein, an engineered HIV-1 gp120 displaying the CD4bs in a stabilized conformation (23); gp140YU2, an uncleaved Env trimer from HIV-1 clade B (24); a 1:1 mixture of gp14092UG37.8 (clade A) + gp140CZA79012 (clade C) (25); and the native-like, fully glycosylated and cleaved clade A HIV-1 Env, BG505 SOSIP.664 (26). Together, the three baits cover all known epitopes. Rescue primers were used to amplify both heavy chains (21) and Igl genes (83), and regular primers were used for Igk chain (84). All PCR products were sequenced and analyzed for Ig gene usage, CDR3, and the number of VH/VL somatic hypermutations [(IgBLAST (www.ncbi.nlm.nih.gov/igblast) and IMGT (www.imgt.org)]. Purified, digested PCR products were cloned into human Igg1, Igk, or Igl expression vectors as previously described (84) and produced by transient transfection of IgH, Igk, and Igl expression plasmids into exponentially growing human embryonic kidney 293-6E cells as previously described (85).
Neutralization studies
HIV-1 neutralization was evaluated using the luciferase-based TZM-bl cell assay (86). Briefly, env pseudoviruses were incubated with fivefold serial dilutions of single antibodies and applied to TZM-bl cells that carry a luciferase reporter gene. After 48 hours, cells were lysed, and luminescence was measured. IC50 and IC80 reflect single antibody concentrations that caused a reduction in relative luminescence units by 50 and 80%, respectively. Coverage curves were built using the Antibody Database computational tool (87).
Structural studies
X-ray diffraction data were collected for crystals of BG18N26Q Fab and NC102 Fab, and an NC37 Fab–93TH057 gp120 complex at the Stanford Synchrotron Radiation Light source beamline 12-2 outfitted with a Pilatus 6M pixel detector (Dectris). XDS software was used to index, integrate, and scale the data (88). The structures of BG18N26Q and NC102 Fabs were solved by molecular replacement using VHVL domains from related Fabs with CDR loops removed and CH1CL as search models and refined with Phenix (43) and manual model building in Coot (89). The NC37 Fab–93TH057 gp120 complex structure was solved by molecular replacement using the NC102 VHVL and CH1CL domains and a truncated gp120 core (from PDB 3U7Y) as search models. Data collection and refinement statistics are presented in table S4.
The structure of a BG505 SOSIP.664–BG18–179NC75 complex was solved by cryoelectron tomography/subtomogram (averaging to ~40 Å resolution) and used as a reference structure to solve a single-particle EM structure from negatively stained samples. A total of 25,639 BG505 SOSIP.664–BG18–179NC75 complex particles were picked and contrast transfer function–corrected using EMAN2.1 (90). A total of 9827 particles corresponding to good class averages were selected, and the particles were further sorted using 3D classification in RELION (91), after which, 7925 particles were selected for refinement. The resolution of the final single-particle reconstruction was ~25 Å, calculated using RELION and a gold-standard Fourier shell coefficient with a 0.143 cutoff (91). Coordinates from crystal structures were fit into the single-particle EM structure using UCSF Chimera. We first fit a BG505 SOSIP.664 structure (PDB 4TVP) into the density and then fit coordinates for BG18 and CH103 Fabs (PDB 4JAM; as a model for 179NC75 Fab) into corresponding densities individually.
Autologous virus
For SGS, HIV-1 RNA was extracted from patient plasma using the Qiagen MinElute Virus Spin Kit according to the manufacturer’s instructions. Extracted RNA was subjected to Env-specific complementary DNA synthesis using the SuperScript III Reverse Transcriptase and HIV-1–specific primer envB3out (see the Supplementary Materials for complete primers sequences). First-round PCR was performed in a 20-ml volume containing 1Å~ High-Fidelity buffer, 2 mM MgSO4, 0.2 mM deoxynucleotide triphosphates, and 0.5 U of High-Fidelity Platinum Taq using 0.2 mM each of primers envB5out and envB3out. Second-round PCR was performed using 1 ml of PCR-1 and 0.2 mM primers of envB5in and envB3in. PCR conditions were the same as PCR-1 except for 45 cycles and an increased annealing temperature of 58°C. PCR-2 products were checked using 1% 96-well E-Gel (Invitrogen). Bands from PCRs with amplification efficiencies lower than 30% were subjected to library preparation and sequenced using the Illumina Nextera DNA Sample Preparation Kit. CMV-Env expression cassettes were generated according to an established protocol (36). CMV-Env (500 ng) was cotransfected with pSG3Denv in six-well plates into 293T cells, and the supernatant was harvested after 48 hours. All plasmids were sequence-validated before expression. Supernatants were subjected to neutralization testing by TZM-bl assay as described above.
In vivo mouse model
All experiments were performed under protocols approved by the university’s IACUC. Humanized nonobese iabetic Rag1−/−Il2rgnull (NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ) mice (the Jackson Laboratory) were subcutaneously treated with 1 mg of each antibody twice a week for a period of 3 weeks (38), receiving a total of six antibody injections. Control humanized mice were reconstituted with human cells from the same donor and infected with HIV-1YU2 but not treated with antibodies. Plasma viral loads were measured weekly. The gp120 sequences from mice with rebounding virus were obtained as previously described (38).
Statistical analyses
Statistical differences between IgG neutralizing activity against HIV-1YU2 and various mutants were analyzed by Mann-Whitney test. Significant changes in viral load between mice treated with either BG18, NC37, or the combination of BG18, NC37, and BG1 or untreated controls were determined by using repeated-measures ANOVA with a Bonferroni post hoc test considering *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank the donor for his participation in this study. We thank Z. Jankovic for the Rockefeller University laboratory support; T. Eisenreich for help with mouse colony management; K. Velinson and N. Thomas for single-cell fluorescence-activated cell sorting; T. Schoofs for help with mice experiments and statistical analyses; G. Jensen and A. McDowall for use of EMs at Caltech; I. Nangiana, J. Vielmetter, and the Caltech Protein Expression Center for expression of proteins used for structural studies; J. P. Moore and A. Cupo for providing and developing the BG505.664 bait; and all the great members of the Nussenzweig and Bjorkman laboratories for helpful discussion and advice.
Funding:
This work was supported by the Robertson Foundation, the Rockefeller University, the Bill and Melinda Gates Foundation grant OPP1033115, the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery Scripps grant UM1 AI 100663, the NIH HIV Vaccine Research and Design grant 1 P01 AI100148 (to M.C.N. and P.J.B.), the NIH grant 2 P50 GM082545-06 (to P.J.B.), the Molecular Observatory and EM funding at Caltech supported by the Gordon and Betty Moore Foundation, the Melinda and Bill Gates Foundation Collaboration for AIDS Vaccine Discovery grant OPP1032144 (to M.S.S.), and the California HIV/AIDS Research Program (CHRP grant F12- CT-214 to S.A.S.). Author contributions: N.T.F. planned and performed experiments, analyzed the data, and wrote the manuscript. H.W. solved the EM structure. L.S. and S.A.S. solved and analyzed crystal structures and contributed to the manuscript preparation. L.N. propagated viral cultures and helped with mouse experiments. J.A.H. propagated viral cultures, performed SGS, and contributed to the manuscript preparation. A.H.-S and Y.B.-O. helped with humanized mice experiments. J.G. and A.G. produced antibodies. J.C.C.L. performed SGS and phylogenetic and diversity analyses and helped in analyzing the data. A.P.-T. and I.T. provided the human samples and coordinated with the donor. H.B.G. refined structural structures. M.J.v.G. and R.W.S. produced and provided reagents and critically read the manuscript preparation. A.P.W. analyzed antibody sequence data. H.C. and L.-X.W. performed the glycopeptide synthesis and binding experiments. D.S. and M.S.S. performed and analyzed the neutralization assays. D.R.B. critically read and contributed to the manuscript preparation. B.D.W. provided human samples and contributed to the manuscript preparation. A.P.W. performed sequence alignment and analyses. P.J.B. planned and analyzed structural experiments and helped write the manuscript. M.C.N. planned and supervised the experiments, analyzed data, and wrote the manuscript. Competing interests: M.C.N. and B.D.W. are Howard Hughes Medical Investigators. The other authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West AP, Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landais E, Huang X, Havenar-Daughton C, Murrell B, Price MA, Wickramasinghe L, Ramos A, Bian CB, Simek M, Allen S, Karita E, Kilembe W, Lakhi S, Inambao M, Kamali A, Sanders EJ, Anzala O, Edward V, Bekker L-G, Tang J, Gilmour J, Kosakovsky-Pond SL, Phung P, Wrin T, Crotty S, Godzik A, Poignard P. Broadly neutralizing antibody responses in a large longitudinal sub-saharan HIV primary infection cohort. PLOS Pathog. 2016;12:e1005369. doi: 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, Moore PL. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLOS Pathog. 2013;9:e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao H-X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia S-M, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BTM, Kwong PD, Mascola JR, Haynes BF. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O’Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR NISC Comparative Sequencing Program. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang G-Y, Schramm CA, Wiehe K, Alam SM, Bradley T, Gladden MA, Hwang K-K, Iyengar S, Kumar A, Lu X, Luo K, Mangiapani MC, Parks RJ, Song H, Acharya P, Bailer RT, Cao A, Druz A, Georgiev IS, Kwon YD, Louder MK, Zhang B, Zheng A, Hill BJ, Kong R, Soto C, Mullikin JC, Douek DC, Montefiori DC, Moody MA, Shaw GM, Hahn BH, Kelsoe G, Hraber PT, Korber BT, Boyd SD, Fire AZ, Kepler TB, Shapiro L, Ward AB, Mascola JR, Liao H-X, Kwong PD, Haynes BF NSCI Comparative Sequencing Program. Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell. 2016;165:449–463. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacLeod DT, Choi NM, Briney B, Garces F, Ver LS, Landais E, Murrell B, Wrin T, Kilembe W, Liang C-H, Ramos A, Bian CB, Wickramasinghe L, Kong L, Eren K, Wu C-Y, Wong C-H, Kosakovsky Pond SL, Wilson IA, Burton DR, Poignard P The IAVI Protocol C Investigators; The IAVI African HIV Research Network. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity. 2016;44:1215–1226. doi: 10.1016/j.immuni.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bar KJ, Tsao C-y, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang K-K, Montefiori DC, Liao H-X, Hraber P, Fischer W, Li H, Wang S, Sterrett BF, Keele S, Ganusov VV, Perelson AS, Korber BT, Georgiev I, McLellan JS, Pavlicek JW, Gao F, Haynes BF, Hahn BH, Kwong PD, Shaw GM. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLOS Pathog. 2012;8:e1002721. doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sather DN, Carbonetti S, Malherbe D, Pissani F, Stuart AB, Hessell AJ, Gray MD, Mikell I, Kalams SA, Haigwood NL, Stamatatos L. Emergence of broadly neutralizing antibodies and viral co-evolution in two subjects during the early stages of infection with the human immunodeficiency virus type 1. J Virol. 2014 doi: 10.1128/JVI.01816-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhiman JN, Anthony C, Doria-Rose NA, Karimanzira O, Schramm CA, Khoza D, Kitchin T, Botha G, Gorman J, Garrett NJ, Abdool Karim SS, Shapiro L, Williamson C, Kwong PD, Mascola JR, Morris L, Moore PL. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat Med. 2015;21:1332–1336. doi: 10.1038/nm.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonsignori M, Alam SM, Liao H-X, Verkoczy L, Tomaras GD, Haynes BF, Moody MA. HIV-1 antibodies from infection and vaccination: Insights for guiding vaccine design. Trends Microbiol. 2012;20:532–539. doi: 10.1016/j.tim.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu R-B, Fu BZ, Gnanapragasam PNP, Oliveira TY, Seaman MS, Kwong PD, Bjorkman PJ, Nussenzweig MC. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams M-R, Lambson BE, Ranchobe N, Ping L, Ngandu N, Abdool Karim Q, Abdool Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. Evolution of an HIV glycan–dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, Walker BD, Ho DD, Wilson PC, Seaman MS, Eisen HN, Chakraborty AK, Hope TJ, Ravetch JV, Wardemann H, Nussenzweig MC. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 18.Mendoza D, Royce C, Ruff LE, Ambrozak DR, Quigley MF, Dang T, Venturi V, Price DA, Douek DC, Migueles SA, Connors M. HLA B*5701-positive long-term nonprogressors/elite controllers are not distinguished from progressors by the clonal composition of HIV-specific CD8+ T cells. J Virol. 2012;86:4014–4018. doi: 10.1128/JVI.06982-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura T, Brockman MA, Schneidewind A, Lobritz M, Pereyra F, Rathod A, Block BL, Brumme ZL, Brumme CJ, Baker B, Rothchild AC, Li B, Trocha A, Cutrell E, Frahm N, Brander C, Toth I, Arts EJ, Allen TM, Walker BD. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J Virol. 2009;83:2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, Seaman MS, Mascola JR, Wyatt RT, Wardemann H, Nussenzweig MC. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sok D, van Gils MJ, Pauthner M, Julien J-P, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, Hua Y, Seaman MS, Moore JP, Ward AB, Wilson IA, Sanders RW, Burton DR. Recombinant HIV envelope trimer selects for quaternary dependent antibodies targeting the trimer apex. Proc Natl Acad Sci USA. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLOS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Farzan M, Wyatt R, Sodroski J. Characterization of stable soluble trimmers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2000;74:5716–5725. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacs JM, Nkolola JP, Peng H, Cheung A, Perry J, Miller CA, Seaman MS, Barouch DH, Chen B. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc Natl Acad Sci USA. 2012;109:12111–12116. doi: 10.1073/pnas.1204533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLOS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PNP, Spencer DIR, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci USA. 2012;109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong C-H, Phogat S, Wrin T, Simek; MD, Koff WC, Wilson IA, Burton DR, Poignard P Protocol G Principal Investigators. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang G-Y, Ofek G, Stewart-Jones GBE, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freund NT, Horwitz JA, Nogueira L, Sievers SA, Scharf L, Scheid JF, Gazumyan A, Liu C, Velinzon K, Goldenthal A, Sanders RW, Moore JP, Bjorkman PJ, Seaman MS, Walker BD, Klein F, Nussenzweig MC. A new glycan-dependent CD4-binding site neutralizing antibody exerts pressure on HIV-1 in vivo. PLOS Pathog. 2015;11:e1005238. doi: 10.1371/journal.ppat.1005238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garces F, Sok D, Kong L, McBride R, Kim HJ, Saye-Francisco KF, Julien J-P, Hua Y, Cupo A, Moore JP, Paulson JC, Ward AB, Burton DR, Wilson IA. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BTM, Sharp PM, Shaw GM, Hahn BH. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander L, Weiskopf E, Greenough TC, Gaddis NC, Auerbach MR, Malim MH, O’Brien SJ, Walker BD, Sullivan JL, Desrosiers RC. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J Virol. 2000;74:4361–4376. doi: 10.1128/jvi.74.9.4361-4376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type. J Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horwitz JA, Halper-Stromberg A, Mouquet H, Gitlin AD, Tretiakova A, Eisenreich TR, Malbec M, Gravemann S, Billerbeck E, Dorner M, Büning H, Schwartz O, Knops E, Kaiser R, Seaman MS, Wilson JM, Rice CM, Ploss A, Bjorkman PJ, Klein F, Nussenzweig MC. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc Natl Acad Sci USA. 2013;110:16538–16543. doi: 10.1073/pnas.1315295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirchherr JL, Lu X, Kasongo W, Chalwe V, Mwananyanda L, Musonda RM, Xia S-M, Scearce RM, Liao H-X, Montefiori DC, Haynes BF, Gao F. High throughput functional analysis of HIV-1 env genes without cloning. J Virol Methods. 2007;143:104–111. doi: 10.1016/j.jviromet.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van ’t Wout AB, Schuitemaker H, Kootstra NA. Isolation and propagation of HIV-1 on peripheral blood mononuclear cells. Nat Protoc. 2008;3:363–370. doi: 10.1038/nprot.2008.3. [DOI] [PubMed] [Google Scholar]
- 38.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu R-B, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou T, Georgiev I, Wu X, Yang Z-Y, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci USA. 2012;109:E2083–E2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gristick HB, von Boehmer L, West AP, Jr, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, Bjorkman PJ. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol. 2016;23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward AB, Wilson IA. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci. 2015;40:101–107. doi: 10.1016/j.tibs.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PNP, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O’Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng H-P, Yang A-S, Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP, Jr, Bjorkman PJ, Scheid JF, Nussenzweig MC, Shapiro L, Mascola JR, Kwong PD NISC Comparative Sequencing Program. Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell. 2015;16:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ariyoshi K, Harwood E, Chiengsong-Popov R, Weber J. Is clearance of HIV-1 viraemia at seroconversion mediated by neutralising antibodies? Lancet. 1992;340:1257–1258. doi: 10.1016/0140-6736(92)92953-d. [DOI] [PubMed] [Google Scholar]
- 47.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 49.Moore PL, Sheward D, Nonyane M, Ranchobe N, Hermanus T, Gray ES, Abdool Karim SS, Williamson C, Morris L. Multiple pathways of escape from HIV broadly cross-neutralizing V2-dependent antibodies. J Virol. 2013;87:4882–4894. doi: 10.1128/JVI.03424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, Kulp DW, Macauley MS, Sok D, Pauthner M, Menis S, Cottrell CA, Torres JL, Hsueh J, Schief WR, Wilson IA, Ward AB, Sanders RW, Burton DR. Holes in the glycan shield of the native HIV envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep. 2016;16:2327–2338. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore PL, Gray ES, Morris L. Specificity of the autologous neutralizing antibody response. Curr Opin HIV AIDS. 2009;4:358–363. doi: 10.1097/COH.0b013e32832ea7e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao F, Bonsignori M, Liao H-X, Kumar A, Xia S-M, Lu X, Cai F, Hwang K-K, Song H, Zhou T, Lynch RM, Alam SM, Moody MA, Ferrari G, Berrong M, Kelsoe G, Shaw GM, Hahn BH, Montefiori DC, Kamanga G, Cohen MS, Hraber P, Kwong PD, Korber BT, Mascola JR, Kepler TB, Haynes BF. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong L, Ju B, Chen Y, He L, Ren L, Liu J, Hong K, Su B, Wang Z, Ozorowski G, Ji X, Hua Y, Chen Y, Deller MC, Hao Y, Feng Y, Garces F, Wilson R, Dai K, O’Dell S, McKee K, Mascola JR, Ward AB, Wyatt RT, Li Y, Wilson IA, Zhu J, Shao Y. Key gp120 glycans pose roadblocks to the rapid development of VRC01-class antibodies in an HIV-1-infected chinese donor. Immunity. 2016;44:939–950. doi: 10.1016/j.immuni.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Learmont J, Tindall B, Evans L, Cunningham A, Cunningham P, Wells J, Penny R, Kaldor J, Cooper DA. Long-term symptomless HIV-1 infection in recipients of blood products from a single donor. Lancet. 1992;340:863–867. doi: 10.1016/0140-6736(92)93281-q. [DOI] [PubMed] [Google Scholar]
- 55.Keet IP, Krol A, Klein MR, Veugelers P, de Wit J, Roos M, Koot M, Goudsmit J, Miedema F, Coutinho RA. Characteristics of long-term asymptomatic infection with human immunodeficiency virus type 1 in men with normal and low CD4+ cell counts. J Infect Dis. 1994;169:1236–1243. doi: 10.1093/infdis/169.6.1236. [DOI] [PubMed] [Google Scholar]
- 56.Cao Y, Qin L, Zhang L, Safrit J, Ho DD. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 57.Gray RH, Wawer MJ, Brookmeyer R, Sewankambo NK, Serwadda D, Wabwire-Mangen F, Lutalo T, Li X, vanCott T, Quinn TC Rakai Project Team. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet. 2001;357:1149–1153. doi: 10.1016/S0140-6736(00)04331-2. [DOI] [PubMed] [Google Scholar]
- 58.Giorgi JV, Lyles RH, Matud JL, Yamashita TE, Mellors JW, Hultin LE, Jamieson BD, Margolick JB, Rinaldo CR, Jr, Phair JP, Detels R Multicenter AIDS Cohort Study. Predictive value of immunologic and virologic markers after long or short duration of HIV-1 infection. J Acquir Immune Defic Syndr. 2002;29:346–355. doi: 10.1097/00126334-200204010-00004. [DOI] [PubMed] [Google Scholar]
- 59.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, Baker B, Trocha A, Rosenberg R, Mackey E, Ueda P, Lu Z, Cohen D, Wrin T, Petropoulos CJ, Rosenberg ES, Walker BD. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 61.Pereyra F, Palmer S, Miura T, Block BL, Wiegand A, Rothchild AC, Baker B, Rosenberg R, Cutrell E, Seaman MS, Coffin JM, Walker BD. Persistent low-level viremia in HIV-1 elite controllers and relationship to immunologic parameters. J Infect Dis. 2009;200:984–990. doi: 10.1086/605446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey JR, Lassen KG, Yang H-C, Quinn TC, Ray SC, Blankson JN, Siliciano RF. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J Virol. 2006;80:4758–4770. doi: 10.1128/JVI.80.10.4758-4770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doria-Rose NA, Klein RM, Daniels MG, O’Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, Mascola JR, Connors M. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: Clustering analysis and association with clinical variables. J Virol. 2010;84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Euler Z, van Gils MJ, Bunnik EM, Phung P, Schweighardt B, Wrin T, Schuitemaker H. Cross-reactive neutralizing humoral immunity does not protect from HIV type 1 disease progression. J Infect Dis. 2010;201:1045–1053. doi: 10.1086/651144. [DOI] [PubMed] [Google Scholar]
- 65.van Gils MJ, Euler Z, Schweighardt B, Wrin T, Schuitemaker H. Prevalence of cross-reactive HIV-1-neutralizing activity in HIV-1-infected patients with rapid or slow disease progression. AIDS. 2009;23:2405–2414. doi: 10.1097/QAD.0b013e32833243e7. [DOI] [PubMed] [Google Scholar]
- 66.Rusert P, Kouyos RD, Kadelka C, Ebner H, Schanz M, Huber M, Braun DL, Hozé N, Scherrer A, Magnus C, Weber J, Uhr T, Cippa V, Thorball CW, Kuster H, Cavassini M, Bernasconi E, Hoffmann M, Calmy A, Battegay M, Rauch A, Yerly S, Aubert V, Klimkait T, Böni J, Fellay J, Regoes RR, Günthard HF, Trkola A Swiss HIV Cohort Study. Determinants of HIV-1 broadly neutralizing antibody induction. Nat Med. 2016;22:1260–1267. doi: 10.1038/nm.4187. [DOI] [PubMed] [Google Scholar]
- 67.Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, Scheid JF, Kraft Z, Liu Y, Pietzsch J, Hurley A, Poignard P, Feizi T, Morris L, Walker BD, Fätkenheuer G, Seaman MS, Stamatatos L, Nussenzweig MC. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J Exp Med. 2012;209:1469–1479. doi: 10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLOS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang G-Y, Louder MK, Schmidt SD, Altae-Tran HR, Bailer RT, McKee K, Nason M, O’Dell S, Ofek G, Pancera M, Srivatsan S, Shapiro L, Connors M, Migueles SA, Morris L, Nishimura Y, Martin MA, Mascola JR, Kwong PD. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340:751–756. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- 70.Weiss RA, Clapham PR, Weber JN, Dalgleish AG, Lasky LA, Berman PW. Variable and conserved neutralization antigens of human immunodeficiency virus. Nature. 1986;324:572–575. doi: 10.1038/324572a0. [DOI] [PubMed] [Google Scholar]
- 71.Derdeyn CA, Moore PL, Morris L. Development of broadly neutralizing antibodies from autologous neutralizing antibody responses in HIV infection. Curr Opin HIV AIDS. 2014;9:210–216. doi: 10.1097/COH.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mahalanabis M, Jayaraman P, Miura T, Pereyra F, Chester EM, Richardson B, Walker B, Haigwood NL. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naïve human immunodeficiency virus controllers. J Virol. 2009;83:662–672. doi: 10.1128/JVI.01328-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moody MA, Gao F, Gurley TC, Amos JD, Kumar A, Hora B, Marshall DJ, Whitesides JF, Xia S-M, Parks R, Lloyd KE, Hwang K-K, Lu X, Bonsignori M, Finzi A, Vandergrift NA, Alam SM, Ferrari G, Shen X, Tomaras GD, Kamanga G, Cohen MS, Sam NE, Kapiga S, Gray ES, Tumba NL, Morris L, Zolla-Pazner S, Gorny MK, Mascola JR, Hahn BH, Shaw GM, Sodroski JG, Liao H-X, Montefiori DC, Hraber PT, Korber BT, Haynes BF. Strain-specific V3 and CD4 binding site autologous HIV-1 neutralizing antibodies select neutralization-resistant viruses. Cell Host Microbe. 2015;18:354–362. doi: 10.1016/j.chom.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moog C, Fleury HJ, Pellegrin I, Kirn A, Aubertin AM. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J Virol. 1997;71:3734–3741. doi: 10.1128/jvi.71.5.3734-3741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, Salantes DB, Seamon CA, Scheinfeld B, Kwan RW, Learn GH, Proschan MA, Kreider EF, Blazkova J, Bardsley M, Refsland EW, Messer M, Clarridge NB, Tustin KE, Madden PJ, Oden K, O’Dell SJ, Jarocki B, Shiakolas AR, Tressler RL, Doria-Rose NA, Bailer RT, Ledgerwood JE, Capparelli EV, Lynch RM, Graham BS, Moir S, Koup RA, Mascola JR, Hoxie JA, Fauci AS, Tebas P, Chun T-W. Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N Engl J Med. 2016;375:2037–2050. doi: 10.1056/NEJMoa1608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP, Jr, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fätkenheuer G, Schlesinger SJ, Nussenzweig MC. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, Gordon I, Casazza J, Conan-Cibotti M, Migueles SA, Tressler R, Bailer RT, McDermott A, Narpala S, O’Dell S, Wolf G, Lifson JD, Freemire BA, Gorelick RJ, Pandey JP, Mohan S, Chomont N, Fromentin R, Chun T-W, Fauci AS, Schwartz RM, Koup RA, Douek DC, Hu Z, Capparelli E, Graham BS, Mascola JR, Ledgerwood JE VRC 601 Study Team. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med. 2015;7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PubMed] [Google Scholar]
- 78.Scheid JF, Horwitz JA, Bar-On Y, Kreider EF, Lu C-L, Lorenzi JCC, Feldmann A, Braunschweig M, Nogueira L, Oliveira T, Shimeliovich I, Patel R, Burke L, Cohen YZ, Hadrigan S, Settler A, Witmer-Pack M, West AP, Jr, Juelg B, Keler T, Hawthorne T, Zingman B, Gulick RM, Pfeifer N, Learn GH, Seaman MS, Bjorkman PJ, Klein F, Schlesinger SJ, Walker BD, Hahn BH, Nussenzweig MC. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature. 2016;535:556–560. doi: 10.1038/nature18929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Halper-Stromberg A, Lu C-L, Klein F, Horwitz JA, Bournazos S, Nogueira L, Eisenreich TR, Liu C, Gazumyan A, Schaefer U, Furze RC, Seaman MS, Prinjha R, Tarakhovsky A, Ravetch JV, Nussenzweig MC. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. 2014;158:989–999. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schoofs T, Klein F, Braunschweig M, Kreider EF, Feldmann A, Nogueira L, Oliveira T, Lorenzi JCC, Parrish EH, Learn GH, West AP, Jr, Bjorkman PJ, Schlesinger SJ, Seaman MS, Czartoski J, McElrath MJ, Pfeifer N, Hahn BH, Caskey M, Nussenzweig MC. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science. 2016;352:997–1001. doi: 10.1126/science.aaf0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu C-L, Murakowski DK, Bournazos S, Schoofs T, Sarkar D, Halper-Stromberg A, Horwitz JA, Nogueira L, Golijanin J, Gazumyan A, Ravetch JV, Caskey M, Chakraborty AK, Nussenzweig MC. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science. 2016;352:1001–1004. doi: 10.1126/science.aaf1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Freund NT, Scheid JF, Mouquet H, Nussenzweig MC. Amplification of highly mutated human Ig lambda light chains from an HIV-1 infected patient. J Immunol Methods. 2015;418:61–65. doi: 10.1016/j.jim.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 84.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klein F, Nogueira L, Nishimura Y, Phad G, West AP, Jr, Halper-Stromberg A, Horwitz JA, Gazumyan A, Liu C, Eisenreich TR, Lehmann C, Fätkenheuer G, Williams C, Shingai M, Martin MA, Bjorkman PJ, Seaman MS, Zolla-Pazner S, Karlsson Hedestam GB, Nussenzweig MC. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J Exp Med. 2014;211:2361–2372. doi: 10.1084/jem.20141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.West AP, Jr, Scharf L, Horwitz J, Klein F, Nussenzweig MC, Bjorkman PJ. Computational analysis of anti–HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc Natl Acad Sci USA. 2013;110:10598–10603. doi: 10.1073/pnas.1309215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kabsch W. XDS. Acta Crystallogr D Biol Crsytallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crsytallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 90.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 91.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Diskin R, Marcovecchio PM, Bjorkman PJ. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat Struct Mol Biol. 2010;17:608–613. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scharf L, Wang H, Gao H, Chen S, McDowall AW, Bjorkman PJ. Broadly neutralizing antibody 8ANC195 recognizes closed and open states of HIV-1 Env. Cell. 2015;162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 96.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 97.Stanfield RL, Zemla A, Wilson IA, Rupp B. Antibody elbow angles are influenced by their light chain class. J Mol Biol. 2006;357:1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 98.Amin MN, McLellan JS, Huang W, Orwenyo J, Burton DR, Koff WC, Kwong PD, Wang L-X. Synthetic glycopeptides reveal the glycan specificity of HIV-neutralizing antibodies. Nat Chem Biol. 2013;9:521–526. doi: 10.1038/nchembio.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 100.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 103.Gilbert PB, Rossini AJ, Shankarappa R. Two-sample tests for comparing intra-individual genetic sequence diversity between populations. Biometrics. 2005;61:106–117. doi: 10.1111/j.0006-341X.2005.020719.x. [DOI] [PubMed] [Google Scholar]
- 104.Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, Kosakovsky Pond SL, Mullins JI NISC Comparative Sequencing Program. DIVEIN: A web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques. 2010;48:405–408. doi: 10.2144/000113370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.